Abstract
The crosstalk between gibberellin (GA) and abscisic acid (ABA) signaling is crucial for balancing plant growth and adaption to environmental stress. Nevertheless, the molecular mechanism of their mutual antagonism still remains to be fully clarified. In this study, we found that knockout of the rice NAC (NAM, ATAF1/2, CUC2) transcription factor gene OsNAC120 inhibits plant growth but enhances drought tolerance, whereas OsNAC120 overexpression produces the opposite results. Exogenous GA can rescue the semi-dwarf phenotype of osnac120 mutants, and further study showed that OsNAC120 promotes GA biosynthesis by transcriptionally activating the GA biosynthetic genes OsGA20ox1 and OsGA20ox3. The DELLA protein SLENDER RICE1 (SLR1) interacts with OsNAC120 and impedes its transactivation ability, and GA treatment can remove the inhibition of transactivation activity caused by SLR1. On the other hand, OsNAC120 negatively regulates rice drought tolerance by repressing ABA-induced stomatal closure. Mechanistic investigation revealed that OsNAC120 inhibits ABA biosynthesis via transcriptional repression of the ABA biosynthetic genes OsNCED3 and OsNCED4. Rice OSMOTIC STRESS/ABA-ACTIVATED PROTEIN KINASE 9 (OsSAPK9) physically interacts with OsNAC120 and mediates its phosphorylation, which results in OsNAC120 degradation. ABA treatment accelerates OsNAC120 degradation and reduces its transactivation activity. Together, our findings provide evidence that OsNAC120 plays critical roles in balancing GA-mediated growth and ABA-induced drought tolerance in rice. This research will help us to understand the mechanisms underlying the trade-off between plant growth and stress tolerance and to engineer stress-resistant, high-yielding crops.
Key words: abscisic acid, ABA, drought stress, gibberellin, GA, OsNAC120, OsSAPK9, rice, Oryza sativa L., SLR1
This study provides evidence demonstrating that the rice transcription factor OsNAC120 acts as a central regulator that balances GA-mediated plant growth and ABA-regulated drought tolerance by promoting GA biosynthesis and inhibiting ABA biosynthesis. OsNAC120 transcriptionally activates GA biosynthetic genes and represses ABA biosynthetic genes, and ABA treatment accelerates OsNAC120 degradation via the protein kinase OsSAPK9.
Introduction
As the major food crop with the largest water requirement, rice experiences particularly severe effects of drought stress on growth and yield (Gowda et al., 2011; Oladosu et al., 2019). To survive, rice has developed multifaceted strategies at different levels to adapt to major adverse environmental conditions (Gong et al., 2020; Zhang et al., 2020; Chen et al., 2021; Yang et al., 2022). The stress-triggered hormone abscisic acid (ABA) plays a key role in a broad array of plant developmental processes and adaptive stress responses to environmental stimuli (Cutler et al., 2010; Hubbard et al., 2010; Weiner et al., 2010). In particular, ABA is closely associated with the cellular dehydration process (Yamaguchi-Shinozaki and Shinozaki, 2006; Takahashi et al., 2018; Yao et al., 2018). The proposed core ABA signaling pathway in the model plant Arabidopsis is the PYR/PYL/RCAR–PP2C–SnRK2 cascade (Fujii et al., 2007; Hu et al., 2012; Fuchs et al., 2014). Generally, in the absence of ABA, the type 2C protein phosphatase (PP2C) co-receptors interact with the SNF1-related kinase 2 (SnRK2) positive effectors, resulting in inhibition of SnRK2 kinase activity via dephosphorylation. In the presence of ABA, the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) receptors and PP2Cs form a complex to prevent PP2C-mediated dephosphorylation of SnRK2s, thus activating SnRK2s and downstream ABA-responsive element binding protein/ABRE-binding factor (AREB/ABF) transcription factors by mediating their phosphorylation (Weiner et al., 2010; Li et al., 2015). All members of the SnRK2 family in rice, designated OSMOTIC STRESS/ABA-ACTIVATED PROTEIN KINASE (OsSAPK1–10), are activated via phosphorylation in response to hyperosmotic stress, and the subgroup that includes OsSAPK8, OsSAPK9, and OsSAPK10 is also activated by ABA (Kobayashi et al., 2004). Recent studies have shown that OsSAPK8, OsSAPK9, and OsSAPK10 are involved in regulation of abiotic stress response through phosphorylation of downstream transcription factors, a process that is largely dependent on ABA (Li et al., 2021; Baoxiang et al., 2022; Qin et al., 2022; Wu et al., 2022). Gibberellin (GA) and ABA are known to regulate plant growth and stress tolerance in opposite ways (Hu et al., 2019; Shohat et al., 2020). As a growth-promoting hormone, GA plays an important regulatory role in physiological processes such as seed germination, leaf expansion, stem elongation, flower formation, and fruit and seed development (Sasaki et al., 2002; Sakamoto et al., 2004; Hauvermale et al., 2012; Daviere and Achard, 2013; Fukazawa et al., 2021; Xing et al., 2023). With the help of the GA receptor GIBBERELLIN INSENSITIVE DWARF1 (GID1) and the F-box protein GID2, GA signaling is mediated by removing the inhibition of DELLA protein, a GA signaling inhibitor (Hirano et al., 2010). In general, GA signaling is controlled by GA accumulation, which is mediated by expression of genes encoding GA biosynthetic enzymes such as GA 20-oxidase (GA20ox) and GA3-oxidase (GA3ox) and the GA-deactivating enzyme GA 2-oxidase (GA2ox) in response to environmental or developmental stimuli (Hedden and Phillips, 2000; Yamaguchi, 2008; Hauvermale et al., 2012; Reinecke et al., 2013; Chen et al., 2014b). In Arabidopsis, the dioxygenases GA20ox, GA3ox, and GA2ox are the main regulatory targets of the GA signaling pathway for establishment of homeostasis (Mitchum et al., 2006; Otani et al., 2010; Barker et al., 2021). Plants balance their growth and survival by coordinately regulating growth-promoting hormone signaling and stress response signaling under adverse conditions (Verma et al., 2016; Ku et al., 2018). ABA has been proposed to inhibit GA function by reducing GA biosynthesis, resulting in stabilization of the GA repressor DELLA protein (Zentella et al., 2007; Vanstraelen and Benkova, 2012). Conversely, GA counteracts ABA signaling by enhancing ABA receptor degradation (Lin et al., 2015; Kawa, 2020). Despite recent advances, the regulatory mechanism by which ABA and GA maintain the balance between growth and stress tolerance still remains to be clarified in rice.
As plant-specific transcription factors, the NAC (NAM, ATAF1/2, CUC2) proteins play essential roles in various plant developmental processes and responses to abiotic stress (Puranik et al., 2012; Huang et al., 2015; Hong et al., 2016; Lee et al., 2017). NAC proteins function as transcriptional activators or repressors by upregulating or downregulating the expression of downstream target genes during abiotic stress, resulting in stress-tolerant or stress-sensitive phenotypes. In rice, OsNAP, SNAC3, and SNAC1 confer drought or salt tolerance by regulating stress- or ABA-responsive genes (Chen et al., 2014a; Fang et al., 2015; Li et al., 2019), whereas ONAC095 and OMTNs act as negative regulators of abiotic stress tolerance (Fang et al., 2014; Huang et al., 2016). NAC transcription factors participate in plant growth and stress response by coordinately regulating various aspects of hormone signaling under adverse conditions. Arabidopsis JUNGBRUNNEN1 (JUB1) reduces growth by inhibiting GA and BR biosynthesis through transcriptional repression of the GA biosynthetic gene GA3ox1 and the BR biosynthetic gene DWARF4 and by affecting GA signaling via upregulation of the two DELLA genes GAI and RGL1 (Shahnejat-Bushehri et al., 2016). On the other hand, JUB1 enhances abiotic stress tolerance through a gene-regulatory network that involves DREB2A (Wu et al., 2012). Our recent study showed that OsNAC016 balances plant growth and drought tolerance by coordinately regulating BR and ABA signaling in rice (Wu et al., 2022). However, to our knowledge, there has been limited research on the role of rice NAC transcription factors in integration of GA and ABA signaling to balance developmental processes and stress tolerance under environmental stress.
In this study, we found that knockout of OsNAC120 led to semi-dwarfism but conferred drought tolerance in rice. Further study showed that OsNAC120 promotes plant growth by positively regulating GA biosynthesis but impairs drought tolerance by repressing ABA biosynthesis. OsNAC120 physically interacts with both SLR1 and OsSAPK9, indicating that it acts as a central regulator in the crosstalk between the GA and ABA signaling pathways. These findings shed light on the mechanism by which OsNAC120 modulates the balance between GA-mediated plant growth and ABA-regulated drought tolerance.
Results
OsNAC120 affects plant growth and development by regulating GA biosynthesis in rice
We identified two independent T-DNA insertion lines, osnac120-1 and osnac120-2, which contained a single T-DNA fragment in the promoter region and the third exon of OsNAC120, respectively (Figure 1A; supplemental Figure 1A and 1B). Loss of OsNAC120 function markedly inhibited plant growth, and osnac120 mutants exhibited semi-dwarf phenotypes caused by a reduction in internode length relative to corresponding wild-type plants (Figure 1B and 1C; supplemental Figure 1C and 1D). To gain insight into the cellular and developmental basis of dwarfism in osnac120 mutants, we examined cell length in longitudinal internode sections. Compared with that of wild-type plants, cell length was significantly reduced in the first internode of osnac120 mutants (supplemental Figure 1E and 1F). These observations indicate that loss of OsNAC120 function results in a repression of cell elongation and thus leads to dwarfism in the osnac120 mutants. To further confirm the role of OsNAC120 in plant growth, we generated knockout mutants (osnac120-cr1 and -cr2) using the CRISPR/Cas9 system (Figure 1D). Similar to the T-DNA insertion mutants, the OsNAC120 knockout mutants exhibited reduced plant height (Figure 1E and 1F). We also constructed rice OsNAC120-overexpressing (OsNAC120-OE) transgenic plants (OE11 and OE20) (supplemental Figure 2A). As expected, the OsNAC120-OE plants exhibited greater plant height, indicated by increased internode length (Figure 1G and 1H; supplemental Figure 2B and 2C). These results demonstrate that OsNAC120 positively regulates rice plant height. Loss of OsNAC120 function also had adverse effects on agronomic traits (e.g., reduced panicle length and seed-setting rate), but overexpression of OsNAC120 had little effect on these traits (supplemental Figure 3). Reductions in panicle length and seed setting in the osnac120 mutants suggested that OsNAC120 is also essential for reproductive development. Consistent with these results, OsNAC120 was highly expressed in the inflorescence and seed (supplemental Figure 4). Together, these observations demonstrate that OsNAC120 is required for developmental processes and yield potential in rice. GA promotes plant height by regulating internode cell elongation, and lack of GA or defects in GA signaling cause dwarf or semi-dwarf phenotypes (Oikawa et al., 2004; Ariizumi and Steber, 2007; Ueguchi-Tanaka et al., 2008). The semi-dwarfism and shortened internodes of the osnac120 mutants (Figure 1B and 1C; supplemental Figure 1C and 1D) resembled those of GA-deficient or GA-insensitive mutants (Oikawa et al., 2004; Ariizumi and Steber, 2007; Ueguchi-Tanaka et al., 2008), and this prompted us to investigate whether GA biosynthesis or signaling was impaired in the osnac120 mutants. We first investigated OsNAC120 transcription in response to exogenous GA and found that GA treatment significantly induced OsNAC120 expression (supplemental Figure 5). Application of exogenous GA also rescued the semi-dwarf phenotype of the osnac120 mutants (Figure 1I and 1J), suggesting that GA biosynthesis might be partially inhibited in the mutants. Consistent with this result, bioactive GA levels were higher in OsNAC120-OE plants but lower in osnac120-2 mutants than in wild-type plants (Figure 1K). To further explore the role of OsNAC120 in regulation of GA biosynthesis, we examined the transcript levels of GA biosynthetic genes such as OsGA20ox family members (Kaneko et al., 2003; Abe et al., 2012) in the different genotypes. As shown in Figure 1L, expression of OsGA20ox1 and OsGA20ox3 was markedly reduced in osnac120 mutants compared with wild-type plants. These observations indicate that OsNAC120 promotes GA biosynthesis, possibly by activating the expression of GA biosynthetic genes.
Figure 1.
OsNAC120 regulates plant growth by promoting GA biosynthesis
(A) Schematic diagram showing the T-DNA insertion sites in genomic regions of osnac120 mutants. Black boxes represent exons, and lines between black boxes represent introns. The arrow indicates the transcription orientation.
(B and C) Growth of 3-month-old T-DNA insertion mutants (osnac120-1 and osnac120-2). Scale bars correspond to 20 cm (B). (C) Values are means ± SD (n = 10).
(D) Schematic diagram of OsNAC120 gene structure and single-guide RNA (sgRNA)-targeted sites. Target site position is indicated on the gene structure.
(E and F) Growth of 3-month-old CRISPR/Cas9 mutants (osnac120-cr1 and osnac120-cr2). Scale bars correspond to 20 cm (E). (F) Values are means ± SD (n = 10).
(G and H) Growth of 3-month-old OsNAC120-overexpressing lines (OE11 and OE20). Scale bars correspond to 25 cm (G). (H) Values are means ± SD (n = 10).
(I and J) Exogenous GA rescued the growth of osnac120 mutants. Scale bars correspond to 2 cm (I). (J) Three independent experiments were performed with similar results, and values are means ± SD (n = 10) from one experiment.
(K) Bioactive GA3 content in plants of different OsNAC120 genotypes. Three independent experiments were performed with similar results, and values are means ± SD (n = 4) from one experiment.
(L) Expression of the GA biosynthetic genes OsGA20ox1, OsGA20ox2, and OsGA20ox3 in leaves of 2-week-old wild-type and osnac120-2 mutant plants. OsActin and OsEF1a were used as the internal references to calculate the relative expression of target genes. Values are means ± SD (n = 3). (C and L) Significant differences were determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01; ns, not significant). (F, H, and K) Different lowercase letters indicate significant differences (p < 0.05, one-way ANOVA with Bonferroni post hoc test). (J) Significant differences were determined by two-way ANOVA with post hoc Tukey’s HSD test (p < 0.05).
OsNAC120 transcriptionally activates the GA biosynthetic genes OsGA20ox1 and OsGA20ox3
NAC transcription factors have been reported to regulate gene transcription by binding to elements containing the CACG-box in the promoter regions of target genes (Olsen et al., 2005). We found multiple CACG-box motifs in the OsGA20ox1 and OsGA20ox3 promoter regions (Figure 2A), suggesting that OsGA20ox1 and OsGA20ox3 might be targets of OsNAC120. To verify this hypothesis, we first performed chromatin immunoprecipitation–quantitative PCR (ChIP–qPCR) assays in OsNAC120-GFP plants, and we observed substantial enrichment of OsGA20ox1 and OsGA20ox3 promoter fragments that contained the CACG-box motif (Figure 2B). We next examined whether OsNAC120 binds to the CACG-box motif in vitro using an electrophoretic mobility shift assay (EMSA). When His-OsNAC120 recombinant protein was incubated with DNA probes containing CACG-box motifs of the OsGA20ox1 and OsGA20ox3 promoters, there was clear retardation of the OsNAC120 band, demonstrating that OsNAC120 could bind to the OsGA20ox1 and OsGA20ox3 promoters. The shifted binding signals became weaker and eventually disappeared when excess unlabeled probe (competitor) was added to the reaction (Figure 2C), demonstrating the specificity of OsNAC120 binding. To further verify the regulation of OsGA20ox1 and OsGA20ox3 by OsNAC120, we performed a transient transactivation assay in Nicotiana benthamiana leaves using the dual-luciferase reporter system. As shown in Figure 2D and 2E, the LUC/REN ratio and luminescence intensity were markedly higher in N. benthamiana leaves that contained OsNAC120 together with OsGA20ox1pro:LUC or OsGA20ox3pro:LUC, confirming that OsNAC120 could bind to the promoters of OsGA20ox1 and OsGA20ox3 and activate LUC expression. This result was also confirmed by transient transactivation of GUS expression in N. benthamiana (supplemental Figure 6). Our data thus convincingly demonstrate that OsNAC120 is an upstream transcriptional activator of the GA biosynthetic genes OsGA20ox1 and OsGA20ox3 and regulates their expression in vivo.
Figure 2.
OsNAC120 transcriptionally activates GA biosynthetic genes, and SLR1 impedes OsNAC120 transactivation activity via physical interaction
(A) Schematic diagram of OsGA20ox1 and OsGA20ox3 promoter regions showing the positions of key motifs and fragments amplified by ChIP–qPCR analysis. TSS, transcription start site.
(B) ChIP–qPCR assays for OsGA20ox1 and OsGA20ox3 promoter fragments enriched by anti-GFP antibody in OsNAC120-GFP plants. Values are means ± SD (n = 3 independent experiments). P1–P5 and P1–P3 represent the regions shown in (A).
(C) Electrophoretic mobility shift assays (EMSAs) demonstrating that OsNAC120 binds specifically to CACG-box motifs in the OsGA20ox1 and OsGA20ox3 promoters. OsGA20ox1pro-probe and OsGA20ox3pro-probe are oligonucleotide probes that include CACG-box motifs from the OsGA20ox1 promoter (–616 to –656 bp) and the OsGA20ox3 promoter (–220 to –260 bp).
(D and E) Transactivation activity of OsNAC120 as indicated by a dual-luciferase reporter assay (D) and luminescence intensity (E). (D) Values are means ± SD (n = 3 independent experiments). (E) Scale bars correspond to 2 cm.
(F) Subcellular localization of OsNAC120 in leaf epidermal cells of Nicotiana benthamiana. Scale bars correspond to 40 μm.
(G) Transcriptional activity of full-length or partial OsNAC120 protein in yeast.
(H) Yeast two-hybrid assays showing the full-length or partial OsNAC120 protein interacting with SLR1. DDO, SD/-Leu-Trp; QDO, SD/-Ade-Leu-His-Trp.
(I) Bimolecular fluorescence complementation (BiFC) assays for the OsNAC120–SLR1 interaction in leaf epidermal cells of N. benthamiana. Scale bars correspond to 40 μm.
(J) Influence of SLR1 on the transactivation activity of OsNAC120. Dual-luciferase reporter assays were performed in N. benthamiana leaves harboring the reporter and effector constructs, with and without 50 μM exogenous GA3 treatment for 6 h. Values are means ± SD (n = 3 independent experiments). (B and D) Significant differences were determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01; ns, not significant). (J) Different lowercase letters indicate significant differences (p < 0.05, two-way ANOVA with post hoc Tukey’s HSD test).
SLR1 physically interacts with OsNAC120 and impairs its transactivation ability
As a transcription factor, OsNAC120 was preferentially localized in the nucleus (Figure 2F). The full-length OsNAC120 protein exhibited self-activation activity in yeast, and the activation domain was located at the C terminus (OsNAC120ΔN, 161–325 amino acids), as indicated by the stronger activity of OsNAC120ΔN than of full-length OsNAC120. The N terminus (OsNAC120ΔC, 1–160 amino acids, containing the NAC conserved A–E domains) inhibited the self-activation activity (Figure 2G). DELLA protein acts as a repressor of GA signaling to inhibit downstream responses through interaction with transcription factors (Daviere and Achard, 2016; Van De Velde et al., 2017; Ito et al., 2018; Shohat et al., 2020). The transcriptional regulation of GA biosynthetic genes by OsNAC120 prompted us to ask whether it interacts with the DELLA protein SLR1. We first performed a yeast two-hybrid assay, which revealed that OsNAC120 interacts with SLR1 (Figure 2H). Deletion of the C terminus abolished the interaction of OsNAC120 with SLR1, whereas deletion of the N terminus enhanced the OsNAC120–SLR1 interaction (Figure 2H). The C terminus of OsNAC120 is therefore required for interaction with SLR1, whereas the N terminus prevents this interaction. The interaction was further confirmed by a bimolecular fluorescence complementation (BiFC) assay. As shown in Figure 2I, yellow fluorescence signals were observed only in N. benthamiana leaves that carried both the OsNAC120-nYFP and SLR1-cYFP constructs, indicating an interaction between OsNAC120 and SLR1 in vivo. No fluorescence was detected upon co-transformation with OsNAC120-nYFP and the cYFP empty vector or SLR1-cYFP and the nYFP empty vector. These observations confirm that OsNAC120 physically interacts with SLR1. To reveal the biological significance of the OsNAC120–SLR1 interaction, we next examined the effect of SLR1 on the transactivation activity of OsNAC120. Co-expression of SLR1 with OsNAC120 reduced the LUC activity driven by the OsGA20ox1 promoter compared with expression of OsNAC120 alone (Figure 2J), indicating that SLR1 blocks the transactivation activity of OsNAC120 through physical interaction. GA treatment removed the inhibition of OsNAC120 transactivation activity caused by SLR1 (Figure 2J), suggesting that GA-induced SLR1 degradation releases OsNAC120 to transcriptionally activate downstream genes.
OsNAC120 plays a negative role in rice drought tolerance
OsNAC120 positively regulates plant growth by promoting GA biosynthesis (Figures 1 and 2A–2E), and GA has been shown to repress plant abiotic stress response (Qin et al., 2011). We therefore asked whether OsNAC120 also functions in plant abiotic stress response by examining the drought responses of wild-type, osnac120 mutant, and OsNAC120-OE plants. After drought treatment was imposed by withholding water, osnac120 plants exhibited delayed, less severe symptoms of drought stress than wild-type plants, whereas OsNAC120-OE plants showed the opposite response (Figure 3A and 3B). To further confirm the negative role of OsNAC120 in drought tolerance, we tested the drought tolerance of OsNAC120 knockout mutants generated by the CRISPR/Cas9 system. The osnac120-cr1 and -cr2 mutants displayed enhanced drought tolerance, just like that of the T-DNA insertion mutants osnac120-1 and osnac120-2 (Figure 3C). After water was withheld and then reapplied, both types of mutants had significantly higher survival rates than the wild-type plants; by contrast, the OsNAC120-OE plants exhibited markedly lower survival in these drought tolerance assays (Figure 3D–3F). Water-loss assays with detached leaves provided further evidence that osnac120 mutants were more drought tolerant than the wild-type plants, whereas OsNAC120-OE plants were more drought sensitive (Figure 3G and 3H). Together, these results show that OsNAC120 negatively regulates drought tolerance by increasing water loss in rice.
Figure 3.
OsNAC120 negatively regulates rice drought tolerance
(A–C) Phenotypes of T-DNA insertion mutants (osnac120-1 and osnac120-2), OsNAC120-overexpressing lines (OE11 and OE20), and CRISPR/Cas9 mutants (osnac120-cr1 and osnac120-cr2) in drought tolerance assays. Scale bars correspond to 10 cm. Three independent experiments were performed with similar results, and representative images are shown.
(D–F) Survival rates of plants in (A), (B), and (C) after rewatering. Values are means ± SD (n = 3 independent experiments, with 32 seedlings per independent experiment).
(G and H) Water loss rates of detached leaves from 2-month-old plants of various genetic backgrounds. Three independent experiments were performed with similar results, and values are means ± SD (n = 9) from one experiment. (D, G, and H) Significant differences between mutant or overexpression lines and wild-type plants were determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). (E and F) Different lowercase letters indicate significant differences (p < 0.05, one-way ANOVA with Bonferroni post hoc test).
OsNAC120 represses ABA biosynthesis and ABA-induced stomatal closure
Plant water loss is mainly achieved by stomatal opening in the leaves, and drought-triggered ABA production can induce stomatal closure, thereby reducing water loss (Li et al., 2017; Yao et al., 2018). We examined OsNAC120 expression in response to exogenous ABA and found that ABA induced OsNAC120 transcription (supplemental Figure 7). The reduced water loss and increased drought tolerance of osnac120 mutants (Figure 3) prompted us to explore whether OsNAC120 regulates ABA biosynthesis and ABA-induced stomatal closure. First, we examined leaf stomatal aperture of different genotypes in response to exogenous ABA treatment using scanning electron microscopy (Figure 4A). Under normal conditions, the ratio of different stomate types was comparable between wild-type and OsNAC120-OE plants, but the proportion of closed stomata was higher in osnac120 plants. Although exogenous ABA treatment increased the proportion of closed stomata in all genotypes, the proportion of closed stomata was much higher in osnac120 and lower in OsNAC120-OE than in the wild type (Figure 4B). These observations indicate that OsNAC120 represses ABA-induced stomatal closure. Similar patterns of stomatal closure were observed when water was withheld for 5 d (Figure 4C). We next evaluated ABA sensitivity by measuring the ABA-induced inhibition of primary root elongation. Primary root elongation was inhibited more severely by ABA treatment in osnac120 mutants than in wild-type plants, and the opposite result was observed in OsNAC120-OE plants (Figure 4D–4H), confirming that OsNAC120 represses the ABA response in rice. Measurement of endogenous ABA content revealed that there were differences in ABA content among the genotypes under normal conditions. When water was withheld for 5 d, ABA content increased dramatically in the osnac120 mutant, but this induction of ABA production was less pronounced in OsNAC120-OE and wild-type plants (Figure 4I). Consistent with these results, expression of ABA biosynthetic genes (OsNCED3 and OsNCED4) and drought-responsive genes (OsLEA3-1 and OsLEA3-2) was upregulated by drought stress to a greater extent in the osnac120 mutant than in the wild type (Figure 4J). Together, these findings demonstrate that OsNAC120 inhibits ABA-induced stomatal closure by inhibiting ABA biosynthesis.
Figure 4.
OsNAC120 plays a negative role in ABA signaling
(A) Images of open and closed stomata in rice leaves obtained by scanning electron microscopy. Scale bars correspond to 50 μm.
(B and C) Percentages of open and closed stomata after ABA treatment (B) and water-withholding treatment (C) in the T-DNA insertion mutant osnac120-2, the wild type (DJ), and two OsNAC120-overexpressing lines (OE11 and OE20). Values are means ± SD (n ≥ 200 stomata from 10 independent plants).
(D–F) ABA response of T-DNA insertion mutants (osnac120-1 and osnac120-2) and corresponding wild-type plants (HY, DJ). (D) Scale bars correspond to 2 cm. (E and F) Values are means ± SD (n = 10). PR, primary root.
(G and H) ABA responses of OsNAC120-overexpressing lines (OE11 and OE20). (G) Scale bars correspond to 2 cm. (H) Values are means ± SD (n = 10). Uniformly germinated seeds were grown on medium with or without 1 μM ABA and cultivated for 2 d.
(I) ABA content in plants of different genotypes, with or without drought treatment. Water was withheld from 2-week-old plants for 5 d, and leaves were collected for measurement of ABA content. Three independent experiments were performed with similar results, and values are means ± SD (n = 4) from one experiment.
(J) Relative expression of ABA biosynthetic genes and drought-responsive genes in plants of different OsNAC120 genotypes. OsActin and OsEF1a were used as the internal references to calculate the relative expression of target genes. Water was withheld from 2-week-old plants for 5 d, and leaves were collected for gene expression measurements. Values are means ± SD (n = 3). (B, C, I, and J) Different lowercase letters indicate significant differences (p < 0.05, two-way ANOVA with post hoc Tukey’s HSD test). (E, F and H) Significant differences were determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant).
OsNAC120 inhibits transcription of the ABA biosynthetic genes OsNCED3 and OsNCED4
CACG-box motifs were found in the promoters of ABA biosynthetic genes (OsNCED3 and OsNCED4) and drought-responsive genes (OsLEA3-1 and OsLEA3-2) (Figure 5A–5D), suggesting that OsNAC120 might transcriptionally regulate these genes by binding to their promoters. ChIP–qPCR assays in OsNAC120-GFP transgenic plants revealed enrichment of DNA fragments from promoters of the above-mentioned genes (Figure 5E–5H), indicating that OsNAC120 could bind to these target promoters. Next, DNA probes containing CACG-box motifs of the OsNCED3 and OsNCED4 promoters were used for EMSA. As shown in Figure 5I and 5J, His-OsNAC120 bound to these DNA probes, demonstrating the direct binding of OsNAC120 to the OsNCED3 and OsNCED4 promoter regions. The intensity of the binding signal clearly decreased when excess unlabeled probe (competitor) was added to the reaction, demonstrating the specificity of OsNAC120 binding to the target promoters. Dual-luciferase reporter assays showed that OsNAC120 transcriptionally represses these ABA biosynthetic genes and drought-responsive genes (Figure 5K), and exogenous ABA treatment alleviated this transcriptional repression (Figure 5K), suggesting that ABA treatment might destabilize the OsNAC120 protein. These results indicate that OsNAC120 represses expression of several ABA biosynthetic genes and drought-responsive genes, leading to reduced ABA biosynthesis and stomatal closure in rice.
Figure 5.
OsNAC120 transcriptionally represses ABA biosynthetic genes and drought-responsive genes
(A–D) Schematic diagram of OsNCED3, OsNCED4, OsLEA3-1, and OsLEA3-2 promoter regions showing the positions of key motifs. TSS, transcription start site.
(E–H) ChIP–qPCR assays showing OsNAC120 binding to target promoters. Chromatin was immunoprecipitated with anti-GFP antibody in OsNAC120-GFP plants, and the precipitated DNA was quantified by qPCR. The DNA enrichment values were normalized to input. Each sample contained 2 g leaves. Values are means ± SD (n = 3 independent experiments).
(I and J) EMSA showing specific binding of OsNAC120 to CACG-box motifs in the OsNCED3 and OsNCED4 promoters. OsNCED3pro-probe and OsNCED4pro-probe are oligonucleotide probes that include CACG-box motifs from the OsNCED3 promoter (–971 to –1011 bp) and OsNCED4 promoter (–202 to –242 bp).
(K) Effects of ABA on transactivation activity of OsNAC120 as indicated by dual-LUC reporter assays. Values are means ± SD (n = 3 independent experiments). (E–H and K) Significant differences were determined by Student’s t-test (∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001; ns, not significant).
OsNAC120 physically interacts with OsSAPK9
The rice SnRK2 members OsSAPK8, OsSAPK9, and OsSAPK10 participate in regulation of abiotic stress response by phosphorylating downstream transcription factors in an ABA-dependent manner (Kobayashi et al., 2004; Li et al., 2021; Baoxiang et al., 2022; Qin et al., 2022; Wu et al., 2022). We therefore asked whether these SAPKs could physically interact with OsNAC120. Yeast two-hybrid assays showed that OsNAC120 interacts with OsSAPK9 but not with OsSAPK8 or OsSAPK10 (Figure 6A; supplemental Figure 8). Furthermore, the C terminus of OsNAC120 interacts with OsSAPK9, but the N terminus prevents this interaction (Figure 6A). A GST pull-down experiment further confirmed the direct interaction between OsNAC120 and OsSAPK9 in vitro. GST-OsSAPK9, but not GST alone, pulled down a significant amount of His-OsNAC120 (Figure 6B). To further verify the OsNAC120–OsSAPK9 interaction in vivo, we performed a BiFC assay in rice protoplasts and found that OsNAC120-nYFP interacted with OsSAPK9-cYFP (Figure 6C). In addition, coimmunoprecipitation (Co-IP) analysis showed that an anti-GFP antibody could pull down OsSAPK9-3×FLAG only when OsNAC120-GFP and OsSAPK9-3×FLAG were co-incubated (Figure 6D; supplemental Figure 9), confirming that OsNAC120 and OsSAPK9 exist as a complex in plants. Overall, our results demonstrate that OsNAC120 physically interacts with OsSAPK9 in vitro and in vivo.
Figure 6.
Physical interaction of OsNAC120 with OsSAPK9
(A) Yeast two-hybrid assays showing the interaction of OsNAC120 with OsSAPK9. DDO, SD/-Leu-Trp; QDO, SD/-Ade-His-Leu-Trp.
(B) Interaction of OsNAC120 with OsSAPK9 in GST pull-down assays. GST-OsSAPK9 was used as bait, and pull-down of His-OsNAC120 was detected with anti-His antibody. A self-stain gel was used to indicate the loading control.
(C) The OsNAC120–OsSAPK9 interaction as indicated by BiFC assays in rice protoplasts. YFP, yellow fluorescent protein. Scale bars correspond to 5 μm.
(D) The OsNAC120–OsSAPK9 interaction as demonstrated by Co-IP assays. Proteins before (input) and after IP were detected with anti-GFP and anti-FLAG antibodies.
OsSAPK9 mediates OsNAC120 phosphorylation
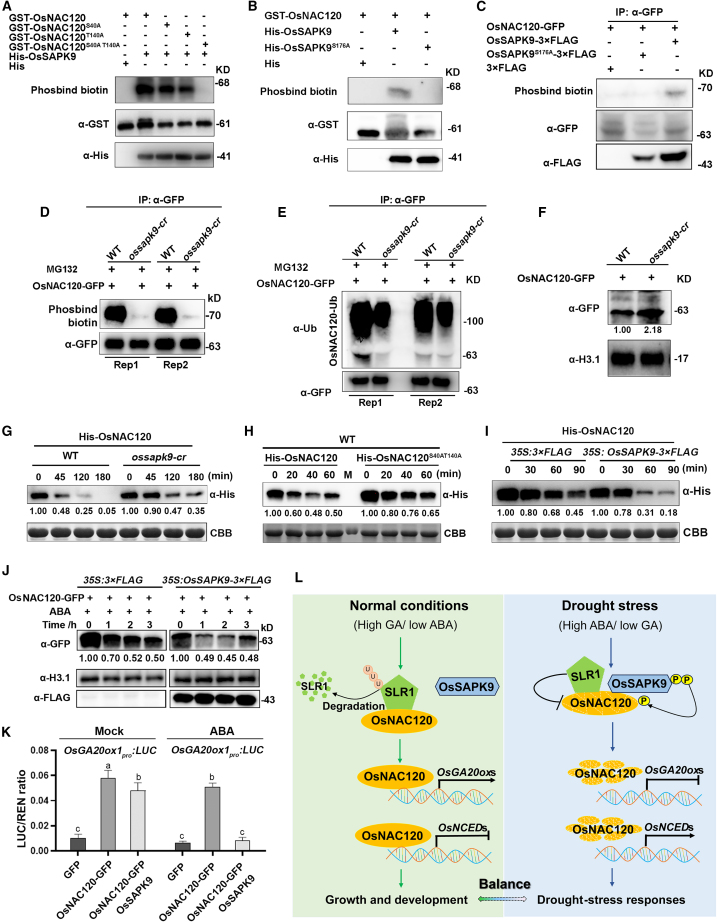
In the ABA signaling pathway, phosphorylation-activated SnRK2 proteins transmit signals to AREB/ABF transcription factors through protein phosphorylation, ultimately activating downstream ABA-responsive genes (Kagaya et al., 2002). Given the kinase properties of OsSAPK9, the interaction of OsNAC120 with OsSAPK9 implies that it may be a substrate of OsSAPK9. We first evaluated whether OsSAPK9 could phosphorylate OsNAC120 by co-expressing GST-OsNAC120 and His-OsSAPK9 in E. coli. As shown in Figure 7A, phosphorylated GST-OsNAC120 was detected by Phosbind biotin when GST-OsNAC120 was co-expressed with His-OsSAPK9, but not with His alone, indicating that His-OsSAPK9 is able to phosphorylate GST-OsNAC120. SnRK2s phosphorylate Ser/Thr residues in the RXXS/T motifs of their substrates (Fujita et al., 2013). Two potential RXXS/T phosphorylation motifs for SnRK2s were observed in OsNAC120 (supplemental Figure 10A). To identify the specific phosphorylation sites recognized by OsSAPK9, we generated dephosphorylation-mimicking forms of OsNAC120 (GST-OsNAC120S40A, GST-OsNAC120T140A, GST-OsNAC120S40A T140A) in which the conserved SnRK2 recognition sites (Ser-40 and Thr-140) were partially or completely replaced by Ala. Compared with natural GST-OsNAC120, GST-OsNAC120S40A and GST-OsNAC120T140A had much lower phosphorylation levels, and no phosphorylated band was detected in GST-OsNAC120S40AT140A (Figure 7A; supplemental Figure 10B), suggesting that Ser40 and Thr140 in OsNAC120 are the main sites recognized by OsSAPK9. Our recent study showed that OsSAPK9 displays phosphorylation activity on Ser-176 (Li et al., 2021). We therefore examined whether OsSAPK9S176A affected the phosphorylation of OsNAC120. When His-OsSAPK9S176A and GST-OsNAC120 were co-expressed in E. coli, no phosphorylated GST-OsNAC120 band was observed (Figure 7B), confirming the OsSAPK9-mediated phosphorylation of OsNAC120. To test whether OsNAC120 was phosphorylated by OsSAPK9 in vivo, OsNAC120-GFP was co-expressed with OsSAPK9-3×FLAG in N. benthamiana, and immunoprecipitated OsNAC120-GFP was detected for phosphorylation. As shown in Figure 7C, a phosphorylated band was detected when OsNAC120-GFP was co-expressed with OsSAPK9-3×FLAG but not with OsSAPK9S176A-3×FLAG, indicating that OsNAC120 can be phosphorylated by OsSAPK9 in vivo. To further examine OsSAPK9-mediated OsNAC120 phosphorylation in rice cells, we generated the ossapk9-cr knockout mutant using the CRISPR/Cas9 system (supplemental Figure 11A–11C). Drought tolerance assays revealed that the ossapk9-cr mutant was clearly less tolerant to drought stress than were wild-type plants (supplemental Figure 11D and 11E). Similarly, the T-DNA insertion mutant ossapk9 (Li et al., 2021) also exhibited reduced drought tolerance (supplemental Figure 11F and 11G). These observations indicated that OsSAPK9 knockout reduced the drought tolerance of rice, in contrast to results from osnac120 mutants. We next examined the phosphorylation of OsNAC120 in the ossapk9-cr mutant. Protein extracts from OsNAC120-GFP plants were incubated with those from wild-type (ZH11) or ossapk9-cr plants, and the immunoprecipitated OsNAC120-GFP was detected for phosphorylation. The results showed that OsNAC120-GFP phosphorylation was much lower in the ossapk9-cr mutant than in wild-type plants (Figure 7D), confirming that OsNAC120 phosphorylation is mediated by OsSAPK9 in rice cells.
Figure 7.
OsNAC120 phosphorylation mediated by OsSAPK9 destabilizes OsNAC120 protein
(A) Kinase assays for phosphorylation of OsNAC120 and its mutated versions mediated by OsSAPK9 in E. coli, detected with Phosbind biotin. An equal amount of each recombinant protein was separated on the gel and detected by immunoblotting with anti-GST and anti-His antibodies.
(B) Serine 176 in OsSAPK9 is essential for OsNAC120 phosphorylation as indicated by a kinase assay in E. coli.
(C) OsNAC120-GFP was phosphorylated by OsSAPK9-3×FLAG in Nicotiana benthamiana leaves. OsNAC120-GFP was transiently co-expressed with empty FLAG protein (3×FLAG), OsSAPK9-3×FLAG, or OsSAPK9S176A-3×FLAG in 4-week-old N. benthamiana leaves. An equal amount of each total protein was detected with Phosbind biotin, anti-GFP, and anti-FLAG antibodies, respectively.
(D and E) Phosphorylation (D) and ubiquitination (E) of OsNAC120-GFP in the wild type (ZH11) and the ossapk9-cr mutant. Protein extracts from OsNAC120-GFP plants were incubated with extracts from the wild type (ZH11) or the ossapk9-cr mutant for 6 h in the presence of 50 μM MG132, and OsNAC120-GFP was immunoprecipitated with anti-GFP antibody. An equal amount of immunoprecipitated protein was detected by Phosbind biotin, anti-ubi, and anti-GFP antibodies, respectively.
(F) Protein stability of OsNAC120-GFP in the wild type (ZH11) and the ossapk9-cr mutant. Protein extracts from OsNAC120-GFP plants were incubated with extracts from the wild type (ZH11) or the ossapk9-cr mutant for 2 h.
(G) Cell-free degradation assays for His-OsNAC120 in protein extracts from 2-week-old wild-type (ZH11) or ossapk9-cr mutant plants. His-OsNAC120 degradation was detected with anti-His antibody. An equal amount of recombinant His-OsNAC120 was incubated with equal protein extracts. The Coomassie brilliant blue (CBB)-stained ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RbcL) was used as a loading control for equal plant protein extracts.
(H) Cell-free degradation assays for His-OsNAC120 and His-OsNAC120S40A T140A (Ser-40 and Thr-140 to Ala) in protein extracts from 2-week-old wild-type (ZH11) plants.
(I) Cell-free degradation assays for His-OsNAC120 in protein extracts from rice protoplasts transiently overexpressing OsSAPK9-3×FLAG.
(J) Effects of ABA on OsNAC120 stability in rice protoplasts, with or without OsSAPK9 overexpression. OsSAPK9-3×FLAG or 3×FLAG was transiently expressed in protoplasts of OsNAC120-GFP plants and then treated with 50 μM ABA for different times. Immunoblotting analysis of histone H3.1 (H3.1) was performed as a control. Three independent experiments were performed, and representative images are shown.
(K) Effect of OsSAPK9 and ABA on the transactivation activity of OsNAC120. Dual-luciferase reporter assays were performed in N. benthamiana leaves co-transfected with the reporter and effector constructs, with and without 50 μM ABA for 4 h. Values are means ± SD (n = 3 independent experiments). Different lowercase letters indicate significant differences (p < 0.05, two-way ANOVA with post hoc Tukey’s HSD test).
(L) Proposed working model showing how OsNAC120 promotes rice growth but inhibits drought tolerance. Under normal conditions of high GA/low ABA levels, GA-induced SLR1 degradation releases OsNAC120, which transcriptionally activates GA biosynthetic gene expression and represses ABA biosynthetic gene expression, thereby promoting GA-regulated plant growth. Under drought stress, ABA-activated OsSAPK9 interacts with OsNAC120, resulting in its phosphorylation and subsequent degradation; at the same time, stress-induced SLR1 impairs OsNAC120 transcriptional activity. The double restrictions on OsNAC120 function lead to repression of growth-related gene expression and alleviate the inhibitory effect of OsNAC120 on drought-responsive gene expression, thus altering the balance between growth and stress response.
OsNAC120 phosphorylation mediated by OsSAPK9 results in reduced protein stability and transactivation activity
Protein phosphorylation is closely linked to protein stability (Bigeard et al., 2014). The performance of both ossapk9-cr and ossapk9 mutants resembled that of OsNAC120-OE plants in the drought tolerance assays, suggesting that OsSAPK9 knockout might result in OsNAC120 accumulation in plants. We therefore examined OsNAC120 ubiquitination and degradation in ossapk9-cr mutants by incubating protein extracts from OsNAC120-GFP plants with those from ossapk9-cr mutants. As shown in Figure 7E, less OsNAC120-GFP was ubiquitinated in the ossapk9-cr mutant than in wild-type plants. Consistent with this result, more OsNAC120-GFP accumulated in the ossapk9-cr mutant (Figure 7F), indicating that OsNAC120 degraded more slowly in the absence of OsSAPK9. These observations indicate that OsSAPK9-mediated phosphorylation facilitates OsNAC120 ubiquitination, thus destabilizing the protein. We then performed cell-free protein degradation assays to examine the effect of OsSAPK9-mediated phosphorylation on OsNAC120 stability. Compared with protein extracts from wild-type plants, extracts from the ossapk9-cr mutant greatly delayed His-OsNAC120 degradation (Figure 7G). Next, the dephosphorylation-mimicking form of His-OsNAC120 (His-OsNAC120S40A T140A) was included in the cell-free degradation assays. As shown in Figure 7H, the stability of His-OsNAC120S40A T140A was greater than that of natural His-OsNAC120, confirming that OsSAPK9-mediated phosphorylation destabilizes OsNAC120. Consistent with this result, reduced His-OsNAC120 stability was observed in extracts from rice protoplasts transiently overexpressing OsSAPK9-3×FLAG (Figure 7I). The kinase activity of OsSAPK9 can be activated by ABA (Kobayashi et al., 2004), and we therefore investigated the effect of ABA on OsNAC120 stability in rice protoplasts, with or without OsSAPK9 overexpression. ABA treatment markedly increased the degradation rate of OsNAC120 in rice cells when OsSAPK9 was overexpressed (Figure 7J), suggesting that ABA induces OsNAC120 degradation by evoking its OsSAPK9-mediated phosphorylation. Collectively, these results demonstrate that OsSAPK9-mediated phosphorylation increases the ubiquitination of OsNAC120, leading to its destabilization.
We next used dual-luciferase reporter assays to examine the effects of ABA-activated OsSAPK9 on OsNAC120 transcriptional activity. As shown in Figure 7K, the transcriptional regulatory activity of OsNAC120 was reduced in the presence of OsSAPK9, and ABA treatment exacerbated this reduction, indicating that ABA coupled with OsSAPK9 inhibits OsNAC120 function, largely by promoting OsNAC120 degradation.
OsNAC120 is involved in maintaining the balance between GA and ABA signaling
ABA and GA antagonistically control a number of plant physiological processes, including seed germination (Zhong et al., 2015; Liu et al., 2016). Our results showed that OsNAC120 regulates both GA and ABA biosynthesis (Figures 1K and 4I) and also interacts with SLR1 and OsSAPK9 (Figures 2H and 2I and 6), suggesting a potential role for OsNAC120 in balancing GA and ABA signaling during seed germination. To assess this possibility, we examined the seed germination of different OsNAC120 genotypes in the presence of exogenous ABA or GA. In the absence of treatment, the seed germination rate was significantly lower in the osnac120-2 mutant and slightly higher in OsNAC120-OE than in the wild type (supplemental Figure 12A and 12B), suggesting that OsNAC120 positively regulates seed germination. Treatment with exogenous GA rescued the germination defects of the osnac120-2 mutant. By contrast, the inhibitory effect of ABA on seed germination was greater in the osnac120-2 mutant than in the wild-type and OsNAC120-OE lines (supplemental Figure 12C). These results demonstrate that OsNAC120 plays a crucial role in maintaining the balance between GA and ABA signaling.
Discussion
OsNAC120 regulates plant developmental processes via regulation of GA signaling in rice
This study provides evidence that OsNAC120 plays a key role in controlling plant growth by regulating GA biosynthesis in rice. First, the osnac120 mutants exhibited GA-deficient phenotypes, including reduced plant height and inhibited internode elongation (Figure 1B and 1C; supplemental Figure 1C and 1D), similar to those of the rice GA-deficient mutants d18 and sd1 (Sakamoto et al., 2004). Second, exogenous GA rescued the semi-dwarf phenotype of the osnac120 mutants (Figure 1I and 1J). Third, OsNAC120 promoted GA biosynthesis by activating expression of the GA biosynthetic genes OsGA20ox1 and OsGA20ox3 (Figures 1K and 1L, 2A-2E), and Arabidopsis GA 20-oxidase is reported to be the key synthetase for biologically active GAs (Fukazawa et al., 2017). Finally, biochemical evidence demonstrated that OsNAC120 interacts with the DELLA protein SLR1, a repressor of GA signaling (Figure 2F–2I). SLR1 physically interacts with OsNAC120 to inhibit its transcriptional regulation of GA biosynthetic genes, but this inhibition is relieved by treatment with exogenous GA (Figure 2J). These observations demonstrate that, under normal growth conditions, GA-induced SLR1 degradation releases OsNAC120 to regulate the expression of growth-related genes and thus promote plant growth. OsNAC120 therefore plays a key role in controlling plant growth by regulating GA biosynthesis in rice. In addition, the osnac120 mutants exhibited reduced panicle length and seed setting (supplemental Figure 3). Consistent with our observations, GA-deficient mutants develop abnormal floral organs, produce semifertile flowers, and are thus severely defective in flower and seed development (Sakamoto et al., 2004; Hedden and Sponsel, 2015; Xing et al., 2023). More work will be needed to gain insight into the regulation of reproductive development by OsNAC120 through modulation of GA signaling.
OsNAC120 negatively regulates rice drought tolerance by repressing ABA signaling
Drought-triggered ABA production helps plants to adapt to adverse environmental conditions, mainly by inducing stomatal closure and reducing water loss (Ding et al., 2015; Li et al., 2017; Malcheska et al., 2017; Lim et al., 2022). In our study, osnac120 mutants exhibited increased drought tolerance and reduced water loss rate (WLR) (Figure 3). Consistent with these observations, ABA and drought treatments markedly reduced stomatal aperture in the osnac120-2 mutant (Figure 4B and 4C). The increased ABA sensitivity of the osnac120 mutants (Figure 4D–4F) also demonstrated that OsNAC120 negatively regulates the ABA response. Furthermore, ABA content was significantly higher in osnac120 than in wild-type plants under drought treatment (Figure 4I). ChIP–qPCR, EMSA, and LUC/REN assays demonstrated that OsNAC120 represses expression of the ABA biosynthetic genes OsNCED3 and OsNCED4 (Figure 5). Recent research has shown that protein phosphorylation is an important mechanism for ABA signal transduction (Yang et al., 2017; Wang et al., 2020). OsSAPK9, the core kinase of ABA signaling, was found to confer drought tolerance in rice (Dey et al., 2016) (supplemental Figure 11). Here, OsNAC120 was shown to physically interact with OsSAPK9 (Figure 6), and OsSAPK9-mediated phosphorylation of OsNAC120 accelerated its ubiquitination, thus destabilizing the OsNAC120 protein (Figure 7A–7J). These results further support our speculation that the role of OsNAC120 in rice drought tolerance is dependent upon OsSAPK9-mediated phosphorylation. We also revealed the main phosphorylation sites in OsNAC120 recognized by OsSAPK9 (Figure 7A; supplemental Figure 10B) and demonstrated that their modification altered OsNAC120 stability (Figure 7H). Elucidation of the link between OsNAC120 phosphorylation and ubiquitination will help us better understand the mechanism by which OsNAC120 regulates ABA signal transduction.
OsNAC120 mediates the crosstalk between ABA and GA signaling
GA and ABA play opposite physiological roles during plant growth and stress response. However, the detailed molecular mechanism underlying the mutual antagonism between these two hormones remains to be clarified. The DELLA protein has been regarded as a central connection between GA and other hormone signaling pathways (Daviere et al., 2008; Golldack et al., 2013; Claeys et al., 2014; Zheng et al., 2016; Ito et al., 2018). DELLA modulates the expression of a set of downstream genes by interacting with transcription factors involved in ABA signaling (Hu et al., 2019; Sun et al., 2020; Finkelstein and Lynch, 2022). Arabidopsis GAI (GIBBERELLIN INSENSITIVE) and RGA (REPRESSOR OF GA1-3) function in response to heat stress by interacting with the transcription factors ABSCISIC ACID-INSENSITIVE 3 (ABI3) and ABI5 (Lim et al., 2013), and RGA-LIKE2 (RGL2) inhibits seed germination by stimulating ABA synthesis and ABI5 activity (Piskurewicz et al., 2008). In our research, OsNAC120 interacted with SLR1 as well as OsSAPK9, indicating that OsNAC120 acts as a central regulator of the crosstalk between the GA and ABA signaling pathways. Our findings suggest that the OsNAC120–SLR1 interaction not only functions in control of plant growth under normal conditions but also may control plant growth under drought stress, because SLR1 expression increased significantly in response to ABA treatment (supplemental Figure 13). Our observations suggest that ABA-activated OsSAPK9 mediates OsNAC120 phosphorylation under drought stress, facilitating its degradation; this causes repression of growth-related gene expression and promotion of drought-responsive gene expression, resulting in reduced plant growth and increased drought tolerance. At the same time, ABA-induced SLR1 expression also inhibits plant growth by repressing GA signaling. It would be interesting to determine which mechanism is preferred by OsNAC120 when plants are exposed to drought stress. Together, these observations support the notion that plants sacrifice growth for survival in adverse environments in an SLR1-dependent manner. Relevant to our research, a recent study showed that salt induces SLR1 protein accumulation by increasing SLR1 expression and inhibiting GA-induced SLR1 degradation, thereby repressing rice growth under salt stress (Mo et al., 2020). In another report, the rice submergence tolerance conferred by Sub1A was mediated by SLR1 and SLRL1-inhibited GA responses (Fukao and Bailey-Serres, 2008). Consistent with the role of OsNAC120 revealed here, the Arabidopsis transcription factor INDUCER OF CBF EXPRESSION1 (ICE1) interacted with DELLA proteins and ABI5 to fine-tune GA and ABA signaling (Hu et al., 2019). Antagonistic regulation of metabolic genes is the main feature of the interaction between GA and ABA (Liu and Hou, 2018). Therefore, we can reasonably infer that the antagonistic regulation of GA and ABA metabolism by OsNAC120 occurs mainly through activation or inhibition of the corresponding biosynthetic genes (OsNCEDs/OsGA20oxs) to maintain the hormone balance during plant growth and stress response. In the future, a combination of genetic and biochemical studies is expected to produce more insights into the exact mechanism of the mutual antagonism regulated by OsNAC120.
On the basis of our results, we propose a model illustrating how OsNAC120 interacts with its partners to balance GA-mediated plant growth and ABA-regulated drought response in rice (Figure 7L). Under normal growth conditions of high GA/low ABA levels, GA-induced SLR1 degradation releases OsNAC120, which activates the expression of growth-related genes and represses ABA-responsive genes, thus promoting plant growth (Figure 7L, left). During drought stress, ABA-activated OsSAPK9 interacts with OsNAC120, resulting in its rapid phosphorylation and subsequent degradation; at the same time, stress-induced SLR1 impedes OsNAC120 transcriptional activity. The double restrictions on OsNAC120 function lead to repression of growth-related gene expression and alleviate the inhibitory effect of OsNAC120 on drought-responsive gene expression, altering the balance between growth and stress response (Figure 7L, right). OsNAC120 is therefore a central regulator that precisely controls the balance between plant growth and drought tolerance.
Methods
Plant material and growth conditions
The rice (Oryza sativa ssp. japonica) cultivars Dongjin (DJ), Hwayoung (HY), Zhonghua 11 (ZH11), and Nipponbare (NIP) were used in this study. The T-DNA insertion mutants osnac120-1 (PFG_1C-12226. L, HY background), osnac120-2 (PFG_2C-10438. L, DJ background) and ossapk9 (PFG_3A-60717. L, DJ background) were identified from the mutant database (Jeon et al., 2000; Jeong et al., 2006). The mutants osnac120-cr1 and -cr2 (NIP background) and ossapk9-cr (ZH11 background) were generated using the CRISPR/Cas9 system (Ma et al., 2015). To obtain the OsNAC120-overexpressing (OsNAC120-OE) lines, the open reading frame (ORF) of OsNAC120 was cloned into pCAMBIA1301, driven by the 35S promoter. The construct was introduced into Agrobacterium tumefaciens (GV3101) and then transformed into DJ by A. tumefaciens–mediated genetic transformation (Hiei and Komari, 2008). All transgenic lines were analyzed using stable T2–T3 progeny. Primers used are given in supplemental Table 1. Rice plants were grown in a greenhouse with a 12-h light (30°C)/12-h dark (24°C) cycle and 65% humidity in the winter or in the field under natural conditions in Chongqing, China, from May to October.
Drought tolerance and water loss assays
The drought response and WLR were investigated as described previously (Li et al., 2021). In brief, water was withheld from 4-week-old plants for drought stress treatment until the leaves of one genotype became completely wilted. The plants were then rewatered, and the number of surviving plants with green and healthy young leaves after rehydration was counted. To measure WLR, detached leaves from 2-month-old plants were placed at room temperature and their fresh weight monitored at the indicated time points. WLR was calculated from the decrease in fresh weight compared with that at time zero using the following formula: WLR (%) = (fresh weight – desiccated weight)/fresh weight × 100%. The average survival rate and WLR were calculated from three independent experiments.
Stomatal observation and hormone content determination
Two-week-old seedlings were exposed to light for at least 2 h to ensure stomatal opening. Fully expanded young leaves were then treated with 30 μM ABA in MES–KCl buffer (50 mM KCl, 10 mM MES–KOH [pH 6.15]) for 2 h. Stomatal closure was detected using a scanning electron microscope (Hitachi SU3500) with a –40°C cool stage. At least 200 stomata from each line were observed, and the proportions of open and closed stomata were calculated. Leaf samples were collected from 2-week-old seedlings for measurement of ABA and GA content using commercial kits (Ruixinbio Biological Technology, Quanzhou, China).
Subcellular localization
The ORF of OsNAC120 fused with GFP (green fluorescent protein) was cloned into the pCAMBIA1301 vector, driven by the 35S promoter (primers are listed in Supplementary Table 1), and the construct was transformed into 4-week-old N. benthamiana leaves by A. tumefaciens–mediated transient transformation (Liu et al., 2010). After infiltration, plants were incubated at 25°C for 48–72 h, and GFP fluorescence signals were detected in the leaf epidermis with a confocal laser scanning microscope (Leica SP8).
Yeast two-hybrid assays
For yeast two-hybrid assays, pGADT7 and pGBKT7 vectors (Clontech) were used to generate bait and prey constructs for co-transformation into the yeast strain Y2HGold. The assays were performed according to the manufacturer’s instructions, and interactions were identified by growth on synthetic defined medium (SD/-Ade-His-Leu-Trp). Primers used are listed in Supplementary Table 1.
RNA extraction and RT–qPCR analysis
Total RNA was extracted using the TRIzol reagent according to the manufacturer’s protocol (Invitrogen), and RT–qPCR analysis was performed with Osactin1 and OsEF1a as the internal reference genes. Relative changes in gene expression were quantified on the basis of three biological replicates (Livak and Schmittgen, 2001). Three independent experiments were performed. Primers used for RT–qPCR analysis are listed in supplemental Table 1.
BiFC assays
The BiFC vectors pFGC-nYFP and pFGC-cYFP were used (Kim et al., 2008). The ORF of OsNAC120 was cloned into pFGC-nYFP, and SLR1 or OsSAPK9 was cloned into pFGC-cYFP (primers in supplemental Table 1). The resulting cYFP and nYFP vectors were co-transformed into rice protoplasts by PEG-mediated transformation (Zhang et al., 2011; Shim et al., 2018) or into 4-week-old N. benthamiana leaves by an efficient agroinfiltration expression system (Liu et al., 2010), together with HY5-RFP as a nuclear localization marker. YFP and RFP fluorescence signals were visualized using a laser confocal microscope (Leica SP8) with the following parameters: Zoom 4.86; HyD Gain 100 for fluorescence signals; PMT Trans Gain 180 for bright signals; Frame Average 1; Line Average 1.
GST pull-down assays
For in vitro GST pull-down assays, the ORFs of OsNAC120 and OsSAPK9 were cloned into the pET-32a and pGEX-4T-1 vectors to produce His-OsNAC120 and GST-OsSAPK9 recombinant proteins in Escherichia coli DE3 (BL21). GST-OsSAPK9 was immobilized on anti-GST agarose resin, which was incubated with His-OsNAC120 at 4°C for 3 h. The resin was washed three times with incubation buffer, and the bound proteins were eluted with 3× SDS sample buffer. The pulled-down proteins were analyzed by immunoblotting with anti-His antibody (1:5000, Proteintech, 66005-1-Ig) and anti-GST antibody (1:5000, Proteintech, 66001-2-Ig). Primers used for these constructs are listed in supplemental Table 1.
Co-IP assays
Co-IP assays were performed as described previously (Liu et al., 2010), with minor modifications. In brief, FLAG-tagged OsSAPK9 and GFP-tagged OsNAC120 were transiently expressed in N. benthamiana leaves. Proteins were extracted with extraction buffer (NP40 [P0013F, Beyotime Biotechnology], 1 mM PMSF, 50 μM E64D, 50 μM MG132, 1× phosphatase inhibitors, and 1× protease inhibitor cocktail [Promega]) for 30 min, centrifuged at 4°C and 10 000 rpm for 15 min, and then incubated together. One milliliter of protein mixture was incubated with 100 μl anti-GFP antibody-coupled agarose beads (Abmart) for 3 h at 4°C. The beads were washed three times with PBS buffer, and the bound proteins were eluted with 3× SDS loading buffer. The eluted proteins were separated by SDS–PAGE and immunoblotted with anti-FLAG antibody (1:2500, Abmart, M20008) and anti-GFP antibody (1:2500, Abmart, M20004).
In vitro phosphorylation assays
In vitro phosphorylation assays were performed as described previously (Wang et al., 2020). GST-OsNAC120 was co-expressed with His-OsSAPK9 in E. coli DE3 (BL21), and recombinant proteins were purified using the corresponding antibodies. Phosphorylated bands were detected using Phosbind Biotin BTL-104 (APE × BIO) according to the manufacturer’s instructions. His-OsSAPK9 and GST-OsNAC120 were detected with anti-His antibody (1:5000, Proteintech, 66005-1-Ig) and anti-GST antibody (1:5000, Proteintech, 66001-2-Ig).
Cell-free protein degradation assays
The experiments were performed as described previously (Kong et al., 2015; Liao et al., 2017) with minor modifications. In brief, proteins were extracted from 3-week-old seedlings using extraction buffer (25 mM Tris–HCl [pH 7.5], 10 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM PMSF). The same amounts of extracts were incubated with equal amounts of His-OsNAC120 or His-OsNAC120S40A T140A at 30°C for different times in the presence of 10 mM ATP. Anti-His antibody (1:5000, Proteintech, 66005-1-Ig) was used to detect OsNAC120-His or His-OsNAC120S40A T140A protein levels by immunoblotting.
Relative luciferase activity measurement
pGreenII cloning vectors were used for the transient transactivation assay (Hellens et al., 2005). To generate the reporter, the promoter region of the candidate target gene was cloned into pGreenII 0800-LUC to drive the LUC gene. To generate the effector, the ORF of OsNAC120 was inserted into pGreenII 62-SK. The effector and reporter constructs were co-transfected into N. benthamiana leaves. Plants were incubated for 2–3 d, and relative luciferase (LUC) activity was measured using a fluorescence detection CCD camera (Uvitec Alliance Q9) with D-luciferin as the substrate for LUC or with a dual-luciferase reporter assay kit (Promega, E1910). Primers used for these constructs are listed in supplemental Table 1.
ChIP–qPCR analysis
Chromatin extraction and immunoprecipitation were performed as described previously (Saleh et al., 2008) with minor modifications. In brief, chromatin was isolated from crosslinked leaves of 2-week-old OsNAC120-GFP transgenic plants. Isolated chromatin was sonicated to obtain DNA fragments ranging from 200 to 500 bp. DNA–protein complexes were immunoprecipitated with anti-GFP antibody (1:2500, Abmart, M20004), and the immunoprecipitated DNA fragments were detected by qPCR analysis with gene-specific primers. Primers used are listed in supplemental Table 1.
EMSA
Recombinant His-OsNAC120 protein was produced as described above for the GST pull-down assays. Oligonucleotide probes containing CACG-box motifs from the target promoters were synthesized and labeled using a Biotin 3′ End DNA Labeling Kit (Thermo), and EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo) according to the manufacturer’s instructions. Probes used are listed in supplemental Table 1.
Statistical analysis
Significant differences between the two groups were determined using Student’s t-test (∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001). Significant differences among more than two groups were determined using one-way or two-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test (p < 0.05) or Tukey’s honestly significant difference (HSD) test (p < 0.05). All statistical analyses were performed using GraphPad Prism 9.0.
Accession numbers
Sequence data from this article can be found in the rice genome annotation project databases under the following accession numbers: OsNAC120 (LOC_Os10g33760), SLR1 (LOC_Os03g49990.1), OsSAPK8 (LOC_Os03g55600.1), OsSAPK9 (LOC_Os12g39630.1), OsSAPK10 (LOC_Os03g41460.1), OsGA20ox1 (LOC_Os03g63970.1), OsGA20ox3 (LOC_Os07g07420), OsLEA3-1 (LOC_Os05g46480.1), OsLEA3-2 (LOC_Os03g20680.1), OsNCED3 (LOC_Os03g44380.1), OsNCED4 (LOC_Os07g05940.1), OsActin1 (LOC_Os03g50885), and OsEF1a (LOC_Os03g0178000).
Data and code availability
All data generated or analyzed during this study can be found within the manuscript and its supporting materials.
Funding
This work was supported by the National Natural Science Foundation of China (32071985) and the Chongqing Special Key Project for Technological Innovation and Application Development (CSTB2022TIAD-KPX0018, CSTB2022TIAD-KPX0016, CSTB2022TIAD-KPX0015).
Author contributions
J.H. designed the research. Z.X. performed the research. N.L. helped to construct the vectors. J.L., Y.S., and C.Z. helped with greenhouse work. S.T. and T.Q. helped with data analysis. Z.X. wrote the manuscript, and J.H. revised the manuscript.
Acknowledgments
We thank Prof. Zhixiang Chen (Purdue University) for providing the BiFC vectors pFGC-nYFP and pFGC-cYFP. No conflict of interest is declared.
Published: December 26, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Abe A., Takagi H., Fujibe T., Aya K., Kojima M., Sakakibara H., Uemura A., Matsuoka M., Terauchi R. OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 2012;125:647–657. doi: 10.1007/s00122-012-1857-z. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Steber C.M. Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell. 2007;19:791–804. doi: 10.1105/tpc.106.048009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baoxiang W., Bo X., Yan L., Jingfang L., Zhiguang S., Ming C., Yungao X., Bo Y., Jian L., Jinbo L., et al. A Novel mechanisms of the signaling cascade associated with the SAPK10-bZIP20-NHX1 synergistic interaction to enhance tolerance of plant to abiotic stress in rice (Oryza sativa L.) Plant Sci. 2022;323 doi: 10.1016/j.plantsci.2022.111393. [DOI] [PubMed] [Google Scholar]
- Barker R., Fernandez Garcia M.N., Powers S.J., Vaughan S., Bennett M.J., Phillips A.L., Thomas S.G., Hedden P. Mapping sites of gibberellin biosynthesis in the Arabidopsis root tip. New Phytol. 2021;229:1521–1534. doi: 10.1111/nph.16967. [DOI] [PubMed] [Google Scholar]
- Bigeard J., Rayapuram N., Pflieger D., Hirt H. Phosphorylation-dependent regulation of plant chromatin and chromatin-associated proteins. Proteomics. 2014;14:2127–2140. doi: 10.1002/pmic.201400073. [DOI] [PubMed] [Google Scholar]
- Chen X., Wang Y., Lv B., Li J., Luo L., Lu S., Zhang X., Ma H., Ming F. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 2014;55:604–619. doi: 10.1093/pcp/pct204. [DOI] [PubMed] [Google Scholar]
- Chen Y., Hou M., Liu L., Wu S., Shen Y., Ishiyama K., Kobayashi M., McCarty D.R., Tan B.C. The maize DWARF1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166:2028–2039. doi: 10.1104/pp.114.247486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shen J., Zhang L., Qi H., Yang L., Wang H., Wang J., Wang Y., Du H., Tao Z., et al. Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol. Plant. 2021;14:1297–1311. doi: 10.1016/j.molp.2021.05.001. [DOI] [PubMed] [Google Scholar]
- Claeys H., De Bodt S., Inzé D. Gibberellins and DELLAs: central nodes in growth regulatory networks. Trends Plant Sci. 2014;19:231–239. doi: 10.1016/j.tplants.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Davière J.M., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- Davière J.M., Achard P. A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant. 2016;9:10–20. doi: 10.1016/j.molp.2015.09.011. [DOI] [PubMed] [Google Scholar]
- Davière J.M., de Lucas M., Prat S. Transcriptional factor interaction: a central step in DELLA function. Curr. Opin. Genet. Dev. 2008;18:295–303. doi: 10.1016/j.gde.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Dey A., Samanta M.K., Gayen S., Maiti M.K. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016;16:158. doi: 10.1186/s12870-016-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Zhang B., Qin F. Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell. 2015;27:3228–3244. doi: 10.1105/tpc.15.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Xie K., Xiong L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014;65:2119–2135. doi: 10.1093/jxb/eru072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Liao K., Du H., Xu Y., Song H., Li X., Xiong L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015;66:6803–6817. doi: 10.1093/jxb/erv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. Overexpression of ABI5 binding proteins suppresses inhibition of germination due to overaccumulation of DELLA proteins. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23105537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Tischer S.V., Wunschel C., Christmann A., Grill E. Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. USA. 2014;111:5741–5746. doi: 10.1073/pnas.1322085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yoshida T., Yamaguchi-Shinozaki K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plantarum. 2013;147:15–27. doi: 10.1111/j.1399-3054.2012.01635.x. [DOI] [PubMed] [Google Scholar]
- Fukao T., Bailey-Serres J. Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA. 2008;105:16814–16819. doi: 10.1073/pnas.0807821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Ohashi Y., Takahashi R., Nakai K., Takahashi Y. DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell. 2021;33:2258–2272. doi: 10.1093/plcell/koab102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa J., Mori M., Watanabe S., Miyamoto C., Ito T., Takahashi Y. DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol. 2017;175:1395–1406. doi: 10.1104/pp.17.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions. Plant Cell Rep. 2013;32:1007–1016. doi: 10.1007/s00299-013-1409-2. [DOI] [PubMed] [Google Scholar]
- Gong Z., Xiong L., Shi H., Yang S., Herrera-Estrella L.R., Xu G., Chao D.Y., Li J., Wang P.Y., Qin F., et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020;63:635–674. doi: 10.1007/s11427-020-1683-x. [DOI] [PubMed] [Google Scholar]
- Gowda V.R., Henry A., Yamauchi A., Shashidhar H.E., Serraj R. Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 2011;122:1–13. [Google Scholar]
- Hauvermale A.L., Ariizumi T., Steber C.M. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol. 2012;160:83–92. doi: 10.1104/pp.112.200956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Hedden P., Sponsel V. A century of gibberellin research. J. Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008;3:824–834. doi: 10.1038/nprot.2008.46. [DOI] [PubMed] [Google Scholar]
- Hirano K., Asano K., Tsuji H., Kawamura M., Mori H., Kitano H., Ueguchi-Tanaka M., Matsuoka M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell. 2010;22:2680–2696. doi: 10.1105/tpc.110.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Zhang H., Huang L., Li D., Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 2016;7:4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Wang F.Z., Liu Z.N., Liu Y.P., Yu X.L. ABA signaling mediated by PYR/PYL/RCAR in plants. Yi Chuan. 2012;34:560–572. doi: 10.3724/sp.j.1005.2012.00560. [DOI] [PubMed] [Google Scholar]
- Hu Y., Han X., Yang M., Zhang M., Pan J., Yu D. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell. 2019;31:1520–1538. doi: 10.1105/tpc.18.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D., Wang S., Zhang B., Shang-Guan K., Shi Y., Zhang D., Liu X., Wu K., Xu Z., Fu X., Zhou Y. A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell. 2015;27:1681–1696. doi: 10.1105/tpc.15.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Hong Y., Zhang H., Li D., Song F. Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 2016;16:203. doi: 10.1186/s12870-016-0897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Okada K., Fukazawa J., Takahashi Y. DELLA-dependent and -independent gibberellin signaling. Plant Signal. Behav. 2018;13 doi: 10.1080/15592324.2018.1445933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeong D.H., An S., Park S., Kang H.G., Park G.G., Kim S.R., Sim J., Kim Y.O., Kim M.K., Kim S.R., et al. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 2006;45:123–132. doi: 10.1111/j.1365-313X.2005.02610.x. [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Hobo T., Murata M., Ban A., Hattori T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor. Plant Cell. 2002;14:3177–3189. doi: 10.1105/tpc.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Ashikari M., Matsuoka M. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 2003;35:104–115. doi: 10.1046/j.1365-313x.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- Kawa D. APC/C(TE) shapes rice architecture from top to bottom. Plant Cell. 2020;32:1786–1787. doi: 10.1105/tpc.20.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.C., Lai Z., Fan B., Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Yamamoto S., Minami H., Kagaya Y., Hattori T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell. 2004;16:1163–1177. doi: 10.1105/tpc.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Cheng J., Zhu Y., Ding Y., Meng J., Chen Z., Xie Q., Guo Y., Li J., Yang S., Gong Z. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015;6:8630. doi: 10.1038/ncomms9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku Y.S., Sintaha M., Cheung M.Y., Lam H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.K., Chung P.J., Jeong J.S., Jang G., Bang S.W., Jung H., Kim Y.S., Ha S.H., Choi Y.D., Kim J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017;15:754–764. doi: 10.1111/pbi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Shen H., Wang T., Wang X. ABA regulates subcellular redistribution of OsABI-LIKE2, a negative regulator in ABA signaling, to control root architecture and drought resistance in Oryza sativa. Plant Cell Physiol. 2015;56:2396–2408. doi: 10.1093/pcp/pcv154. [DOI] [PubMed] [Google Scholar]
- Li J., Li Y., Yin Z., Jiang J., Zhang M., Guo X., Ye Z., Zhao Y., Xiong H., Zhang Z., et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017;15:183–196. doi: 10.1111/pbi.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Chang Y., Ma S., Shen J., Hu H., Xiong L. Genome-wide identification of SNAC1-targeted genes involved in drought response in rice. Front. Plant Sci. 2019;10:982. doi: 10.3389/fpls.2019.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yu B., Wu Q., Min Q., Zeng R., Xie Z., Huang J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D., Cao Y., Sun X., Espinoza C., Nguyen C.T., Liang Y., Stacey G. Arabidopsis E3 ubiquitin ligase PLANT U-BOX13 (PUB13) regulates chitin receptor LYSIN MOTIF RECEPTOR KINASE5 (LYK5) protein abundance. New Phytol. 2017;214:1646–1656. doi: 10.1111/nph.14472. [DOI] [PubMed] [Google Scholar]
- Lim C., Kang K., Shim Y., Yoo S.C., Paek N.C. Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 2022;188:1900–1916. doi: 10.1093/plphys/kiab492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., Kim J., Kang H., Kim D.H., Kawakami N., Choi G. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell. 2013;25:4863–4878. doi: 10.1105/tpc.113.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wu F., Sheng P., Zhang Z., Zhang X., Guo X., Wang J., Cheng Z., Wang J., Wang H., Wan J. The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015;6:7981. doi: 10.1038/ncomms8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 2010;61:893–903. doi: 10.1111/j.1365-313X.2009.04109.x. [DOI] [PubMed] [Google Scholar]
- Liu X., Hou X. Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front. Plant Sci. 2018;9:251. doi: 10.3389/fpls.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hu P., Huang M., Tang Y., Li Y., Li L., Hou X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016;7 doi: 10.1038/ncomms12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., Wang B., Yang Z., Li H., Lin Y., et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Malcheska F., Ahmad A., Batool S., Müller H.M., Ludwig-Müller J., Kreuzwieser J., Randewig D., Hänsch R., Mendel R.R., Hell R., et al. Drought-enhanced xylem sap sulfate closes stomata by affecting ALMT12 and guard cell ABA synthesis. Plant Physiol. 2017;174:798–814. doi: 10.1104/pp.16.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Mo W., Tang W., Du Y., Jing Y., Bu Q., Lin R. PHYTOCHROME-INTERACTING FACTOR-LIKE14 and SLENDER RICE1 interaction controls seedling growth under salt stress. Plant Physiol. 2020;184:506–517. doi: 10.1104/pp.20.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Koshioka M., Kojima K., Yoshida H., Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004;55:687–700. doi: 10.1007/s11103-004-1692-y. [DOI] [PubMed] [Google Scholar]
- Oladosu Y., Rafii M.Y., Samuel C., Fatai A., Magaji U., Kareem I., Kamarudin Z.S., Muhammad I., Kolapo K. Drought resistance in rice from conventional to molecular breeding: a review. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20143519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005;169:785–797. doi: 10.1016/j.tplants.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Otani M., Yoon J.M., Park S.H., Asami T., Nakajima M. Screening and characterization of an inhibitory chemical specific to Arabidopsis gibberellin 2-oxidases. Bioorg. Med. Chem. Lett. 2010;20:4259–4262. doi: 10.1016/j.bmcl.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 2012;17:369–381. doi: 10.1016/j.tplants.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Qin C., Fang Q., Fan X., Chen M., Jiang M. Phosphorylation of DUF1639 protein by osmotic stress/ABA-activated protein kinase 10 regulates abscisic acid-induced antioxidant defense in rice. Biochem. Biophys. Res. Commun. 2022;604:30–36. doi: 10.1016/j.bbrc.2022.03.003. [DOI] [PubMed] [Google Scholar]
- Qin F., Kodaira K.S., Maruyama K., Mizoi J., Tran L.S.P., Fujita Y., Morimoto K., Shinozaki K., Yamaguchi-Shinozaki K. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011;157:1900–1913. doi: 10.1104/pp.111.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke D.M., Wickramarathna A.D., Ozga J.A., Kurepin L.V., Jin A.L., Good A.G., Pharis R.P. Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol. 2013;163:929–945. doi: 10.1104/pp.113.225987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Miura K., Itoh H., Tatsumi T., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Agrawal G.K., Takeda S., Abe K., et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134:1642–1653. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 2008;3:1018–1025. doi: 10.1038/nprot.2008.66. [DOI] [PubMed] [Google Scholar]
- Sasaki A., Ashikari M., Ueguchi-Tanaka M., Itoh H., Nishimura A., Swapan D., Ishiyama K., Saito T., Kobayashi M., Khush G.S., et al. A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Shahnejat-Bushehri S., Tarkowska D., Sakuraba Y., Balazadeh S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants. 2016;2 doi: 10.1038/nplants.2016.13. [DOI] [PubMed] [Google Scholar]
- Shim J.S., Oh N., Chung P.J., Kim Y.S., Choi Y.D., Kim J.K. Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 2018;9:310. doi: 10.3389/fpls.2018.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shohat H., Illouz-Eliaz N., Kanno Y., Seo M., Weiss D. The tomato DELLA protein PROCERA promotes abscisic acid responses in guard cells by upregulating an abscisic acid transporter. Plant Physiol. 2020;184:518–528. doi: 10.1104/pp.20.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Tuan P.A., Izydorczyk M.S., Ayele B.T. Ethylene regulates post-germination seedling growth in wheat through spatial and temporal modulation of ABA/GA balance. J. Exp. Bot. 2020;71:1985–2004. doi: 10.1093/jxb/erz566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N., Fukuda H., Yamaguchi-Shinozaki K., Shinozaki K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature. 2018;556:235–238. doi: 10.1038/s41586-018-0009-2. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Hirano K., Hasegawa Y., Kitano H., Matsuoka M. Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell. 2008;20:2437–2446. doi: 10.1105/tpc.108.061648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Velde K., Ruelens P., Geuten K., Rohde A., Van Der Straeten D. Exploiting DELLA signaling in cereals. Trends Plant Sci. 2017;22:880–893. doi: 10.1016/j.tplants.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Benková E. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 2012;28:463–487. doi: 10.1146/annurev-cellbio-101011-155741. [DOI] [PubMed] [Google Scholar]
- Verma V., Ravindran P., Kumar P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16:86. doi: 10.1186/s12870-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hou Y., Qiu J., Wang H., Wang S., Tang L., Tong X., Zhang J. Abscisic acid promotes jasmonic acid biosynthesis via a 'SAPK10-bZIP72-AOC' pathway to synergistically inhibit seed germination in rice (Oryza sativa) New Phytol. 2020;228:1336–1353. doi: 10.1111/nph.16774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J.J., Peterson F.C., Volkman B.F., Cutler S.R. Structural and functional insights into core ABA signaling. Curr. Opin. Plant Biol. 2010;13:495–502. doi: 10.1016/j.pbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Allu A.D., Garapati P., Siddiqui H., Dortay H., Zanor M.I., Asensi-Fabado M.A., Munné-Bosch S., Antonio C., Tohge T., et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24:482–506. doi: 10.1105/tpc.111.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Liu Y., Xie Z., Yu B., Sun Y., Huang J. OsNAC016 regulates plant architecture and drought tolerance by interacting with the kinases GSK2 and SAPK8. Plant Physiol. 2022;189:1296–1313. doi: 10.1093/plphys/kiac146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing M.Q., Chen S.H., Zhang X.F., Xue H.W. Rice OsGA2ox9 regulates seed GA metabolism and dormancy. Plant Biotechnol. J. 2023;21:2411–2413. doi: 10.1111/pbi.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yang L., Chen Y., Xu L., Wang J., Qi H., Guo J., Zhang L., Shen J., Wang H., Zhang F., et al. The OsFTIP6-OsHB22-OsMYBR57 module regulates drought response in rice. Mol. Plant. 2022;15:1227–1242. doi: 10.1016/j.molp.2022.06.003. [DOI] [PubMed] [Google Scholar]
- Yang W., Zhang W., Wang X. Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017;15:4–14. doi: 10.1111/pbi.12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L., Cheng X., Gu Z., Huang W., Li S., Wang L., Wang Y.F., Xu P., Ma H., Ge X. The AWPM-19 family protein OsPM1 mediates abscisic acid influx and drought response in rice. Plant Cell. 2018;30:1258–1276. doi: 10.1105/tpc.17.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R., Zhang Z.L., Park M., Thomas S.G., Endo A., Murase K., Fleet C.M., Jikumaru Y., Nambara E., Kamiya Y., Sun T.P. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell. 2007;19:3037–3057. doi: 10.1105/tpc.107.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhao Y., Zhu J.K. Thriving under stress: how plants balance growth and the stress response. Dev. Cell. 2020;55:529–543. doi: 10.1016/j.devcel.2020.10.012. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7:30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Gao Z., Zhu Z. DELLA-PIF modules: old dogs learn new tricks. Trends Plant Sci. 2016;21:813–815. doi: 10.1016/j.tplants.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Zhong C., Xu H., Ye S., Wang S., Li L., Zhang S., Wang X. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiol. 2015;169:2288–2303. doi: 10.1104/pp.15.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study can be found within the manuscript and its supporting materials.