Summary
CRISPR-Cas9 short guide RNA (sgRNA) library screening is a powerful approach to understand the molecular mechanisms of biological phenomena. However, its in vivo application is currently limited. Here, we developed our previously established in vitro revival screening method into an in vivo one to identify factors involved in spermatogenesis integrity by utilizing sperm capacitation as an indicator. By introducing an sgRNA library into testicular cells, we successfully pinpointed the retinal degeneration 3 (Rd3) gene as a significant factor in spermatogenesis. Single-cell RNA sequencing (scRNA-seq) analysis highlighted the high expression of Rd3 in round spermatids, and proteomics analysis indicated that Rd3 interacts with mitochondria. To search for cell-type-specific signaling pathways based on scRNA-seq and proteomics analyses, we developed a computational tool, Hub-Explorer. Through this, we discovered that Rd3 modulates oxidative stress by regulating mitochondrial distribution upon ciliogenesis induction. Collectively, our screening system provides a valuable in vivo approach to decipher molecular mechanisms in biological processes.
Keywords: CRISPR screening, in vivo genome-wide screening, spermatogenesis, multi-omics, Hub-Explorer
Graphical abstract

Highlights
-
•
Application of CRISPR-Cas9 sgRNA library in testes
-
•
Enrichment of key sgRNAs through repeated library reconstruction
-
•
Identification of Rd3 as a factor controlling spermatogenesis
-
•
Deduction of Rd3 signaling pathway in round spermatids with Hub-Explorer
Noguchi et al. developed an in vivo genome-wide screening system to identify factors determining spermatogenesis integrity and identified RD3 as an essential factor for spermatogenesis. They also discovered that Rd3 functions as an oxidative stress modifier in round spermatids using their originally developed computational tool, Hub-Explorer.
Introduction
A biological phenotype is the consequence of the sophisticated interplay between gene expression and protein function in each cell.1,2,3 Unveiling the molecular machinery that governs biological phenomena is crucial to understanding organisms’ physiology and pathology. Over the past few decades, numerous approaches have been developed in mammalian systems to elucidate gene functions in vivo. One promising approach, in particular, is reverse genetics.4,5,6,7 While single-gene knockout mice contributed significantly to understanding gene functions at an individual level,8,9 conventional reverse genetics is unable to perturb multiple genes simultaneously within a particular tissue of each individual. In contrast, in vivo genome-wide screening is a powerful approach to comprehensively dissect molecular machinery underlying biological events.10 Several studies have used CRISPR/short guide RNA (sgRNA) libraries to demonstrate in vivo screening of different cell types, such as cardiomyocytes,11 neural cells,12,13 and hepatocytes.14,15 Most of these screening approaches used cell proliferation or locomotion as an output to enrich target cells. However, a major challenge of in vivo screening is to conduct biochemical-activity-based screening. Most biochemical activities are not directly related to growth, and most cells do not proliferate in vivo; therefore, developing a strategy to enrich target cells without cell growth is a significant issue for in vivo screening to overcome.
To achieve biochemical-activity-based screening without cell growth, we had established an in vitro screening system using a CRISPR sgRNA library, which we called “revival screening.”16 In this system, after lentiviral sgRNA introduction, target cells showing different biochemical activities are labeled with a fluorescent probe, sorted by flow cytometry based on their fluorescence intensity, and their genomic DNA (gDNA) is purified. Finally, the inserted sgRNA region is amplified from the purified gDNA by PCR to reconstitute the enriched sgRNA library. By applying this system, we successfully identified factors regulating lipid dynamics on the plasma membrane despite using dying cells, which did not exhibit growth.
Herein, we applied this system to establish an in vivo genome-wide screening system using mouse testes as a model tissue. To understand the factors involved in spermatogenesis, we focused on sperm capacitation, an important event for fertilization17,18,19 as a phenotypic output for functional sperms. We introduced a CRISPR sgRNA library directly into mice testes, and capacitation-defective sperms were collected by flow cytometry based on their Ca2+ influx, a well-known biochemical activity of capacitation.20,21,22,23 Through in vivo revival screening, we identified retinal degeneration 3 (Rd3), whose defect causes Leber congenital amaurosis (LCA) type 1224,25,26 as a factor involved in spermatogenesis. Through these achievements, we demonstrated that our in vivo genome-wide screening approach can be widely applicable to biochemical-activity-based screening in the future.
Design
We designed a testes-targeted in vivo CRISPR screening system, marked by four key achievements: (1) introduction of an sgRNA library into male germ cells via Sendai virus fusion (SVF) protein-coated lentivirus,27,28 (2) evaluation of spermatogenesis integrity by measuring levels of sperm Ca2+ influx at a high signal-to-noise ratio,20,21,22,23 (3) efficient readout of sgRNA sequences from a small number of sperms using primase-based whole-genome amplification (pWGA),29 and (4) enrichment of critical sgRNAs through repeated library reconstitution as part of our advanced revival screening method.16
Hub-Explorer is a computational analysis tool designed to unravel key regulatory pathways in particular cell types. This tool adeptly combines two fundamental components: a gene co-expression network (GCN) and Gene Ontology (GO) terms. To perform this analysis, Hub-Explorer necessitates three essential inputs: (1) a gene expression matrix, (2) a list of genes of interest, and (3) annotation files that establish connections between gene symbols and UniProt IDs. In our study, we utilized single-cell RNA sequencing (scRNA-seq) data from testicular cells as the gene expression matrix and proteome-derived data of Rd3 interactors as the list of genes of interest. For generating the GCN, Spearman’s correlation coefficient (ρ > 0.8) was utilized. Subsequently, a GO analysis was conducted for each gene within the GCN. We classified genes within the GCN by assessing the Jaccard similarity index of their GO term annotations. The final step involved pinpointing shared GO terms among these genes, enabling us to delineate the core regulatory pathways.
Results
Establishment of in vivo genome-wide screening
To develop a method for in vivo genome-wide screening using mice testes, we first established the technical basis of the screening system, by focusing on four distinct components, as illustrated in Figure 1A. Initially, efficient viral injection methods were determined by introducing lentiviruses into testes. First, lentiviruses encoding EGFP with the mitochondrial signal sequence (9 × 107 infectious titer unit [IFU]/mL) were introduced into the testes of 11 postnatal day (PND) mice according to previously reported methods30,31 such as seminiferous tubular injection and interstitial injection. Successful infection was confirmed with seminiferous tubular injection but not with interstitial injection (Figure S1A). Next, to determine the effective virus titer, two different titers of lentiviruses (2.5 × 106 and 9 × 107 IFU/mL) were introduced into mice testes by seminiferous tubular injection. Lentivirus infectivity was found to increase in a titer-dependent manner (Figure S1B). However, most of the infected cells were thought to be Sertoli cells because the EGFP signal showed a broad spreading pattern from the basal to the apical side32 (Figure S1B), and this signal co-localized with the anti-vimentin antibody signal (Figure S1C). These results are consistent with previous studies30,31,32 and suggested that not only viral titer but also virus tropism are critical factors for successful infection. Recently, a highly selective lentivirus for male germ cells was developed using SVF proteins.27,28 Utilizing this system, we endeavored to deliver lentiviruses into male germ cells. Initially, we transfected plasmids carrying SVF protein and viral components into HEK293T packaging cells. After transfection, sodium butyrate was added to enhance gene expression for generating high-titer lentiviruses. SDS-PAGE of the purified lentiviruses, followed by Coomassie brilliant blue staining, was able to detect the SVF protein consistent with its reported size33 (Figure S1D). Based on these improvements, we successfully generated a high-titer SVF lentivirus (3 × 107–3 × 108 IFU/mL). Microscope analysis showed effective viral introduction into Gcna1+ germ cells34 1 week after infection (Figures 1B and S1E). Flow cytometry analysis also confirmed that 54.2% ± 2.67% of Cdh1+ type A spermatogonia (SPGs)35 were infected by TagBFP-encoded lentivirus (Figure S1F), and TagBFP expression was sustained for 8 weeks (Figure S1G). We next examined sgRNA on-target efficiency to evaluate whether Cas9 was functional in the germ cells of CRISPR-Cas9 knockin mice. Lentivirus encoding sgRNA against Rosa26 was injected into testes via seminiferous tubule, and insertions or deletions (indels) at the Rosa26 locus in Basigin (BSG)+ germ cells36 were confirmed by Sanger sequencing (Figure 1C). Knockout effects of sgRNA against Gcna1 were also shown at the protein level in Plzf+/Sall4+ type A SPGs37,38 and primary spermatocytes with 46 double-structured chromosome39 using flow cytometry (Figure 1D). These results demonstrated successful targeting by lentiviral sgRNAs in germ cells.
Figure 1.
Establishment of in vivo genome-wide screening
(A) Experimental scheme of in vivo genome-wide screening toward spermatogenesis.
(B) Evaluation of short-term infection efficiency. Lentivirus encoding TagBFP was injected into 11 PND mice testes. One week later, cryo-sectioned testes were stained with anti-TagBFP (cyan) and anti-Gcna1 (magenta) antibodies and DAPI (white), followed by confocal microscopy analysis. Top: virus uninfected; bottom: virus infected. Left: macroscale; right: microscale. Scale bar: 200 μm.
(C) Sanger sequencing analysis against Rosa26 region in Basigin (Bsg)+ germ cells. Lentivirus encoding sgRosa26 and TagRFP was introduced into 11 PND mice testes. Ten days later, male germ cells were stained with anti-Bsg antibody. TagRFP+ and Bsg+ germ cells were sorted for gDNA extraction. Indel start position was detected by CRISP-ID, shown as an orchid-colored line. Experiment was repeated twice.
(D) Gcna1 knockout (KO) efficiency evaluation. Lentivirus encoding sgGcna1 and TagBFP was introduced into 11 PND mice testes. One week later, dissociated testes were stained with anti-Plzf, anti-Sall4, and anti-Gcna1 antibodies and DAPI. Analyzed cells are shown on the left. Gcna1 signal intensity was evaluated by median fluorescent intensity (MFI) as mean ± SD. Middle: type A spermatogonia (Plzf+/Sall4+); right: primary spermatocyte (4 chromosomes). Light orchid: uninfected cells; dark orchid: infected cells. Infected samples in type A spermatogonia were analyzed twice (n = 2), and the others were analyzed three times (n = 3).
(E) Evaluation of sperm Ca2+ influx. Adult mice sperms were capacitated (top; see the STAR Methods) and stained with Fluo-4 AM and PI. Fluo-4 AM signal in PI-negative region was analyzed. Bottom left: non-capacitated sperms; bottom right: capacitated sperms. Experiments were repeated three times (n = 3), and Fluo-4 AM-positive populations were shown as mean ± SD.
Since we successfully introduced lentiviruses into testicular cells and achieved genetic perturbation for in vivo genome-wide screening, we aimed to determine a screening target to elucidate spermatogenesis quality, which could be analyzed by flow cytometry. Among various indicators to evaluate spermatogenesis, we decided to focus on the capacitation of mature sperms, which is a crucial process for fertilization17,18,19 and reflects spermatogenesis quality.40,41,42 Among the several processes associated with capacitation, Ca2+ influx is widely known as a driving force for capacitation.20,21,22,23 When Ca2+ influx was examined by flow cytometry using the Ca2+ indicator Fluo-4 AM, a drastic intracellular Ca2+ increase was observed under capacitation conditions (Figure 1E). Scanning electron microscopy (SEM) analysis of sperm morphology was also performed to validate capacitation capability. As expected, we observed an acrosome-reaction-derived morphological change in the sperm head, which appeared after the end of capacitation43 (Figure S1H). In summary, these fine-tuned experimental conditions would enable us to perform genome-wide screening using testes.
Enrichment of essential sgRNAs associated with spermatogenesis
As we had successfully established a method to introduce a lentiviral sgRNA library into germ cells, we next validated genome-wide coverage in type A SPGs by administering the GeCKO-v2 mouse sgRNA pool B library, consisting of 62,804 unique guide sequences, into the testes of Cas9 knockin mice.44 Three days after lentiviral library injection, Plzf+ type A SPGs were sorted, and sgRNAs integrated into the gDNA of these cells were sequenced by next-generation sequencing (NGS). To statistically evaluate our screening result, we considered the following two aspects: (1) the distribution of sgRNA counts and targeted genes and (2) the profile of sgRNA counts per gene (detailed in the STAR Methods). From the result, we confirmed that there was no experimental bias in our screening (Figures S2A–S2D).
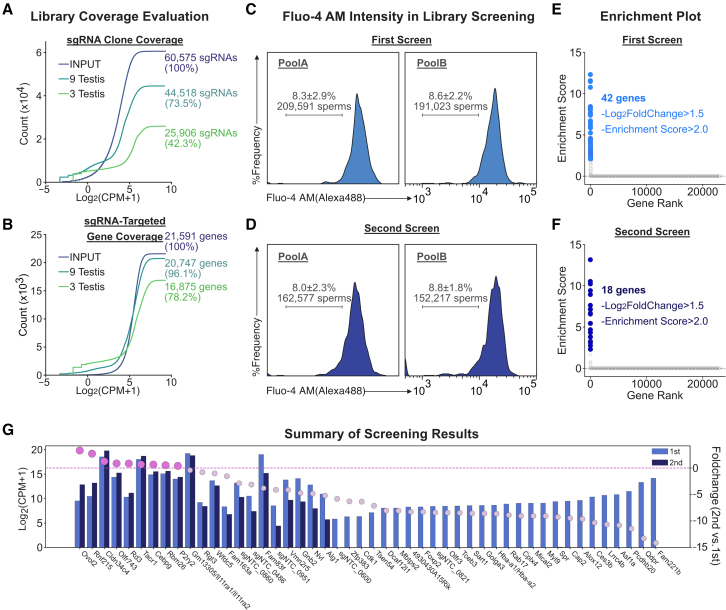
As for the evaluation of library coverage, sgRNA clone coverage analysis showed a 42.3% coverage ratio in three testes (Figure 2A). An additional six testes (total: nine testes) were further examined, and the sgRNA clone coverage was 73.5% (Figure 2A). The sgRNA-targeted gene coverage was also analyzed in three testes and nine testes. The sgRNA-targeted gene coverage in three testes was calculated to be 78.2% (Figure 2B). In nine testes, the sgRNA-targeted gene coverage was 96.1% (Figure 2B). In addition, this tendency was not altered even 1 week after infection (sgRNA clone coverage: 67.8% [Figure S2E]/sgRNA-targeted gene coverage: 94.2% [Figure S2F]), suggesting that the genome-wide coverage of sgRNAs in the testes was sufficient for screening.
Figure 2.
Enrichment of essential sgRNAs associated with spermatogenesis
(A and B) Library coverage evaluation. Pool B GeCKO-v2 sgRNA library containing 62,804 sgRNAs was introduced into 11 PND mice testes. Three days later, dissociated testes were stained with anti-Plzf and DAPI to sort Plzf+ type A spermatogonia for gDNA extraction. The amplified sgRNA regions were sequenced with the next-generation sequencing (NGS). The sgRNA clone coverage is shown in (A) and the sgRNA-targeted gene coverage is shown in (B). sgRNAs from are from input (blue), 9 testes (dark green), and 3 testes (light green). x axis: the sgRNA count (=log2(CPM + 1)); y axis: the cumulative sgRNA count.
(C and D) Sorting diagram of Fluo-4 AM-negative sperms. Left column: pool A; right columns: pool B. (C) First screen. (D) Second screen.
(E and F) Significant gene enrichment. Significant genes were extracted above log2 fold change = 1.5 and enrichment score = 2.0. Candidates in the first library are shown as light blue dots (E) and those in the second library are shown as dark blue dots (F). Enrichment score was calculated as described in the STAR Methods.
(G) Enriched candidate genes. Light blue bar: first screening; dark blue bar: second screening. Count shown as log2(CPM + 1). Fold change (count of second screening/first screening) is shown as colored dots (orchid: enriched genes; light orchid: non-enriched genes).
Based on the obtained coverage ratio in testes, we introduced the GeCKO-v2 mouse sgRNA library into Cas9 knockin mice testes at 11 PND: the pool A and pool B libraries were infected into twelve testes during the first screen and into eight testes during the second screen. Eight weeks after infection, mature sperms, which developed from the infected germ cells, were obtained from the cauda epididymis, incubated with capacitation buffer, stained with Fluo-4 AM and propidium iodide (PI), and finally applied to cell sorting using flow cytometry. In the first round of screening, we sorted 209,591 Ca2+ uptake-negative sperms (corresponding to 8.3% ± 2.9% of input) from testes infected with virus from the pool A library and 191,023 Ca2+ uptake-negative sperms (corresponding to 8.6% ± 2.2% of input) from testes infected with virus from the pool B library (Figure 2C). After sorting, gDNA was purified from the sorted sperms and applied to pWGA29 (see the STAR Methods). Using the amplified gDNA, the integrated sgRNA region was then amplified by PCR and inserted into lentiviral vectors. The reconstituted sgRNA library was then applied to the next round of screening so that false positive hits could be eliminated. In the second round of screening, we sorted 162,577 Ca2+ uptake-negative sperms (corresponding to 8.0% ± 2.3% of input) from the enriched pool A library testes and 152,217 of Ca2+ uptake-negative sperms (corresponding to 8.8% ± 1.8% of input) from the enriched pool B library testes (Figure 2D). After gDNA purification from the sorted sperms and pWGA using the purified gDNA, the integrated sgRNA region was amplified by PCR. The sgRNAs from the first and second rounds of screening were then analyzed by NGS and mapped to reference sequences.
For NGS analysis, we developed the enrichment score (ES) (see STAR Methods) to precisely analyze detected sgRNAs while maintaining correlation between log2 fold change and ES at a significant value range (ES > 2.0 and log2 fold change > 1.5) (Figures S2G and S2H). According to the ES, we identified eighteen sgRNA-targeted genes in the second round of screening from forty-two sgRNA-targeted genes nominated in the first round of screening (Figures 2E and 2F). When we compared these sgRNAs derived from the second round of screening to those from the first round, we observed a significant enrichment of genes involved in reproduction, such as copulation and mating (Figure S2I). Notably, we observed an increase in the read counts of nine sgRNA-targeted genes and a decrease in all non-targeting sgRNAs in the second round of screening (Figure 2G). These results suggest that our in vivo genome-wide screening approach successfully enriched sgRNAs that regulate spermatogenesis.
Rd3 is an essential spermatogenesis factor identified through small-scale functional screening
In order to further narrow down the candidates, we analyzed gene expression and the effect of gene disruption on testis weight, sperm numbers, and Ca2+ influx (Figure 3A). We first analyzed the transcriptome dataset obtained via Bgee.45 The transcriptome analysis indicated that the expression of seven genes (Ovol2, Rnf215, Cldn34c4, Rd3, Cebpg, Rbm26, and P2ry2) out of the nine candidates was observed in adult testes (Figure 3B). Further quantitative RT-PCR (RT-qPCR) using 8 week testis RNA also confirmed this result (Figure S3A). One of the nine candidates, Olfr743 was found to be expressed at 5 and 6 weeks but decreased at 8 weeks (Figure S3B). Another candidate, Tacr1, was not expressed in testicular cells, suggesting that the false positive rate was one-ninth (Figure S3C).
Figure 3.
Rd3 is an essential spermatogenesis factor identified through small-scale functional screening
(A) Experimental scheme of shRNA screening.
(B) Transcriptome analysis in testes. Each gene expression shown as log2(TPM + 1) (see the STAR Methods).
(C) Testicular weight quantification. Data are shown as violin plot according to the following replicates: n = 24 (shLacZ), 7 (shOvol2), 10 (shRnf215), 22 (shCldn34c4), 15 (shRd3), 8 (shCebpg), 7 (shRbm26), and 10 (shP2ry2). p value: unpaired t test. Significant values (p < 0.05) are colored orange.
(D) Sperm number quantification. Data are shown as the following replicates: n = 24 (shLacZ), 8 (shOvol2), 10 (shRnf215), 22 (shCldn34c4), 15 (shRd3), 8 (shCebpg), 7 (shRbm26), and 10 (shP2ry2). Statistical evaluation is as described in (C).
(E) Sperm Ca2+ influx quantification. Ca2+ influx score is as described in Figure S3F. Data are shown as the following replicates: n = 24 (shLacZ), 8 (shOvol2), 10 (shRnf215), 22 (shCldn34c4), 15 (shRd3), 8 (shCebpg), 8 (shRbm26), and 10 (shP2ry2). Statistical evaluation is as described in (C).
(F) Histograms for the representative Fluo-4 AM signal intensity.
Based on the gene expression analysis, a small-scale short hairpin (sh)RNA screening against the seven candidates, selected based on gene expression level, was performed to validate the function of each gene. Firstly, the efficiency of each shRNA-mediated knockdown was validated in cell lines using RT-qPCR, confirming a reduction in RNA expression to 34.4% ± 14.7% (Figure S3D). Subsequently, the validated lentiviral shRNAs were injected into testes at 11 PND via their seminiferous tubules. The lentiviral infection efficiency of each shRNA into testes was determined by the expression of fluorescent proteins in Bsg+ germ cells,36 which were isolated from the dissociated testes after measuring testicular weight. From this, the average infection efficiency was found to be 24.9% ± 15.3% (Figure S3E). As for the phenotypic effect, knockdown of Ovol2, Cldn34c4, Rd3, and P2ry2 led to a decrease in testicular weight (Figure 3C). Among these, shRd3 decreased testicular weight most significantly. Considering that shRd3-encoded lentivirus had the lowest infection efficiency, silencing of Rd3 may affect testicular cell survival the most. In addition, sperm numbers were decreased after Cldn34c4, Rd3, and P2ry2 knockdown (Figure 3D). Among the seven candidates, Rd3 knockdown decreased Ca2+ influx significantly (Figures 3E, 3F, and S3F–S3H). These results strongly indicated that Rd3 was the most promising factor contributing to spermatogenesis among the identified genes.
Round spermatid is the specific cell type with high Rd3 expression in testis
RD3 has been reported as an essential factor for retinogenesis, specifically through the regulation of retinal guanylyl cyclase 1/2 in the outer segment of the retina, and loss-of-function mutations in RD3 cause LCA type 12.25,26,46 Although one report suggested that RD3 is expressed in human testes,47 the role of mouse Rd3 in spermatogenesis remains unknown. To elucidate the molecular functions of Rd3 in spermatogenesis, we first investigated Rd3 expression in testes using both transcriptome and histology approaches. We utilized the tissue-wide transcriptome dataset, also used in Figure 3B, and found that Rd3 is highly expressed in the retinal neural layer and testes (Figure 4A). Since Rd3 is highly expressed in the testes, we next utilized the testicular scRNA-seq dataset48 to identify Rd3-expressing cell types. Based on the scRNA-seq results, the cell population was grouped into seven discrete clusters (Figure 4B). When Rd3 expression was compared among these clusters, it was found to be increased during spermatogenesis and, in particular, became the highest in the latter stage of round spermatid (Figure 4C), when Acrv149 and Spaca450 were strongly expressed (Figure S4A). In addition, retinal scRNA-seq analysis also showed that rod and cone cells had the highest Rd3 expression among the twelve distinct retinal cell types51 (Figures S4B and S4C). Comparative transcript isoform analysis between the testis and retina using other transcriptome datasets (Accessions: SRR823506 and SRR34245852) revealed that the 5′ UTRs of Rd3 in these two tissues are distinct, implying that Rd3 translational regulation differs between the testis and retina (Figure S4D). We then validated Rd3 expression by RT-qPCR analysis and confirmed that Rd3 expression starts to increase 3 weeks after birth, when round spermatids increase in number,48 and reaches a maximum at 7 weeks (Figure 4D). This result was verified by RT-PCR with different amplification cycles (Figure 4E). Hybridization-chain-reaction-based fluorescent in situ hybridization and immunohistochemistry using adult mice testes showed that Rd3 mRNA and protein were detected in Acrv1+ round spermatids49 and lectin PNA+ whole spermatids53 (Figures 4F and 4G).
Figure 4.
Round spermatid is the specific cell type with high Rd3 expression in testes
(A) Tissue-wide transcriptome analysis of Rd3. Adult mice-derived Rd3 expression is shown as log2(TPM + 1) values. Testis: magenta; retinal neural layer: midnight blue.
(B) t-Distributed stochastic neighbor embedding (tSNE) plot of mouse testicular cells. Single-cell data (total: 6,693 cells) are clustered as the following cell types: spermatogonium (SG), pre-leptotene/zygotene (pL/Z), pachytene (P), diplotene (D), meiotic cell (M), round spermatid (RS), and elongating spermatid (ES).
(C) Rd3 expression profiling. x axis: cell types (total: 6,644 cells); y axis: Rd3 expression shown as Z score converted from ln(CPM + 1).
(D and E) Rd3 expression on different ages. (D) RT-qPCR. (E) RT-PCR.
(F) Hybridization-chain-reaction-based fluorescent in situ hybridization (HCR-FISH). Cryo-sectioned testes were probed with Acrv1 (magenta), Rd3 (green), DAPI (white), and lectin PNA (yellow), detected by confocal microscope. Scale bar: 200 μm.
(G) Immunohistochemistry of Rd3. Cryo-sectioned adult testes were stained with DAPI (white), lectin PNA (magenta), and Rd3 (green), detected by confocal microscope. Scale bar: 50 μm.
Identification of ciliogenesis-oriented Rd3-mitochondria axis by Hub-Explorer analysis
To uncover the function of Rd3 in round spermatids in testes, we first attempted to perform proteomics analysis to give us insights into the function of Rd3. However, conducting in vivo cell-type-specific proteomics analysis is typically challenging. Indeed, despite our efforts to identify Rd3 interactors using testicular cells, we encountered significant technical challenges in the process (i.e., low Rd3 expression level per cell, limited number of Rd3-expressing cells, and unavailability of anti-Rd3 antibody for immunoprecipitation). Therefore, we decided to use cells from the retinal cell line Y-79, which express RD3 endogenously at a high level according to the Cancer Cell Line Encyclopedia dataset,54 to perform proteomics analysis. Subsequently, gene expression of identified proteins were profiled to narrow down candidates that function in round spermatids.
To this end, using Y-79 cells, RD3 knockout (RD3−/−) cells and those expressing Spot-tagged RD3 (RD3-Spot) were generated. Next, immunoprecipitation was performed using anti-Spot nanobody-conjugated beads (Spot-Trap) on cell lysates prepared from RD3−/− cells and those with RD3-Spot, followed by mass spectrometry. As a result, 269 proteins were found to interact directly or indirectly with RD3 (Figure 5A). In order to search for proteins potentially functioning with RD3 in spermatids, gene expression of these 269 candidates was analyzed using the testicular scRNA-seq data and classified into spermatogenic cell types in Figure 4. Consequently, 49 proteins were found to be highly expressed in round and elongating spermatids (Figures 5B and 5C). GO analysis showed that most of the 49 proteins were related to mitochondria and microtubule function (Figure 5D). To validate Rd3 and mitochondrion interaction in testicular cells, we homogenized testes with Dounce homogenizer and fractionated the homogenates by sequential centrifugation and found that Rd3 was located in the mitochondria fraction (Figure S5A), suggesting that Rd3 interacts with mitochondria in testicular cells.
Figure 5.
Identification of ciliogenesis-associated Rd3-mitochondria axis by Hub-Explorer analysis
(A) Proteomics for RD3 interactors. Immunoprecipitates obtained by anti-Spot nanobody were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Significant proteins (magenta) are shown as p < 0.05 and log2 fold change (RD3-Spot-expressing cells/RD3−/− cells) ≧1.5. Experiment was repeated three times (n = 3).
(B) Gene expression profile of 269 significant proteins in testicular cells. 269 proteins gene expression levels shown as Z score from testicular gene expression matrix used in Figures 4B, 4C, and S4A. x axis: cell types; y axis: genes. Colored dots indicate the following cell types: SG, pL/Z, P, D, M, RS, and ES.
(C) Cell type classification of 269 proteins on gene expression pattern in testis. Highly expressing cell types were determined by the cell type index (CTI ≧ 0.98; see STAR Methods), shown on the y axis, and the count of proteins in each cell type is shown on the x axis. Orange-colored bars: specific group including spermatid.
(D) Gene Ontology (GO) analysis. 49 proteins analyzed by DAVID software with mouse (left column) and human (right) Ensembl IDs.
(E) Computational process of Hub-Explorer analysis.
(F) Similarity profile of 17 Rd3 interactors with hub components (Benjamini-Hochberg false discovery rate [BH-FDR] < 0.05) according to Jaccard similarity index visualized as a heatmap (both axes: 17 interactors). Each cluster is colored by 4 individual colors (C0: wine red, C1: magenta, C2: deep sky blue, and C3: blue). The annotated hierarchical cluster was used for the benchmark of K-means clustering.
(G) Profile of hub components and core signaling pathways. x axis: 17 candidates clustered and labeled as indicated in (F); y axis: hub components. Core signaling pathway: scarlet; others: Safrano pink. Numbers on the y axis indicate hub components as follows: 1, CatSper complex; 2, sperm head plasma membrane; 3, ∗centrosome; 4, membrane protein complex; 5, plasma-membrane-bounded cell projection; 6, ∗sperm midpiece; 7, ∗9 + 2 motile cilium; 8, non-membrane-bounded organelle; 9, ∗microtubule cytoskeleton; 10, intracellular non-membrane-bounded organelle; 11, protein-DNA complex; 12, ∗sperm flagellum; 13, ∗motile cilium; 14, nuclear-protein-containing complex; 15, ∗cilium; 16, nucleosome; 17, DNA packaging complex; 18, ∗cytoskeleton; 19, ribonucleoprotein complex; 20, protein-containing complex; 21, phosphatase complex; 22, ∗ciliary basal body; 23, ∗dynein complex; 24, ∗microtubule; 25, catalytic complex; 26, ∗axonemal microtubule; 27, protein serine/threonine phosphatase complex; 28, membrane; 29, ubiquitin ligase complex; 30, cytoplasmic microtubule; 31, cell projection; 32, protein phosphatase type 2A complex; 33, cellular anatomical entity; 34, intracellular-protein-containing complex; and 35, spliceosomal complex. ∗Core signaling pathway (scarlet).
(H) Intracellular localization of 17 candidate interactors with hub components in late round spermatids and early elongating spermatids.
We next asked how Rd3-mitochondria interaction regulates the function of testicular cells. To gain functional insight into these 49 genes, we developed a signaling pathway analysis tool called Hub-Explorer, which enables the identification of the cell-type-specific signaling pathway based on proteomic and transcriptomic data (Figure 5E; see the STAR Methods). While conventional proteomics studies commonly utilize GO analysis to elucidate the functions of identified proteins, GO analysis alone falls short in revealing the signaling pathways that regulate these identified proteins. To solve this issue, Hub-Explorer was designed to identify the signaling pathway using a GCN according to the following processes.
In the first step, a testicular cell-specific GCN was generated for each of the 49 genes using testicular scRNA-seq expression data. In the second step, gene components in each GCN were applied to GO analysis, and 17 out of 49 genes had the significant GO terms (=hub components: Benjamini-Hochberg false discovery rate < 0.05). In the third step, the Jaccard similarity index was calculated based on the hub components, and these 17 genes were clustered using their calculated index scores through the K-means method (k = 4). Finally, overlapping hub components were extracted as the core signaling pathway from each cluster (Figure 5F). As a result, C1 and C2 clusters, composed of 10 genes, were identified as the major cluster containing the core signaling pathway. These pathways were found to be associated with ciliogenesis, which contributes to sperm flagellar development (Figure 5G), suggesting that either the Rd3-mitochondria interaction regulates ciliogenesis or ciliogenesis regulates the Rd3-mitochondria interaction (Figures 5H and S5B). Additional analysis using Rd3-correlated genes in the testis also supported this conclusion: Rd3 is associated with ciliogenesis (Figures S5C–S5E). Although conventional GO analysis was unable to identify the ciliogenesis-related GO terms (Figure 5D), this was successfully achieved by Hub-Explorer.
Rd3 modulates mitochondrial spatial distribution under ciliogenesis-induction-derived oxidative stress
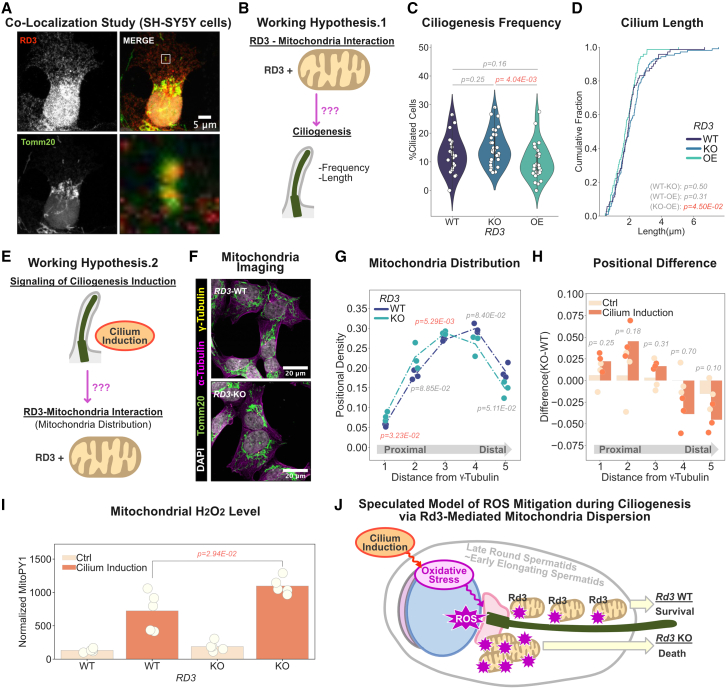
Based on the proteomics and Hub-Explorer analyses in Figures 5D and 5G, we hypothesized that the Rd3-mitochondria interaction regulates ciliogenesis or that ciliogenesis regulates the Rd3-mitochondria interaction. To examine this hypothesis, we tried to induce ciliogenesis in Y-79 cells, but induction efficiency was low. Therefore, we examined other cell lines and decided to use SH-SY5Y cells, which express RD3 endogenously54 and have ciliogenesis capability.55,56 Prior to ciliogenesis analysis, we first examined whether the RD3-mitochondria interaction occurs in SH-SY5Y cells. Using immunocytochemistry analysis, we demonstrated that RD3 co-localized with the mitochondria marker Tomm20 (Figure 6A), suggesting that RD3 also interacts with mitochondria in SH-SY5Y cells. Next, we tried to clarify the relationship between the RD3-mitochondria interaction and ciliogenesis. To this end, we induced ciliogenesis by serum starvation and investigated the frequency of cilia formation and cilium length between RD3−/− cells and those expressing exogenous RD3 (Figures 6B–6D and S6A). Despite a thorough investigation, no difference was observed, suggesting that the Rd3-mitochondria interaction does not regulate ciliogenesis (Figure 6E). Based on these results and because RD3 was also found to associate with tubulin in the proteomics study (Figure 5D), we hypothesized that ciliogenesis affects RD3-mitochondria dynamics. To examine this, we quantified mitochondrial distribution under the induction of ciliogenesis (Figure S6B). From the result, mitochondria were found to be dispersed under the induction of ciliogenesis compared to the resting state. However, in RD3−/− cells, mitochondria dispersion was not observed (Figures 6F–6H, S6C, and S6D). Previous studies have suggested that the induction of ciliogenesis is associated with accumulation of reactive oxygen species (ROS).57,58 Other studies have further shown that ROS accumulation is modulated by mitochondrial dispersion.59,60,61 Based on these findings, we hypothesized that RD3-regulated mitochondria dispersion reduces excessive ROS accumulation, promoted by ciliogenesis induction. To examine this hypothesis, we quantified the mitochondrial ROS level under ciliogenesis induction by the H2O2 probe MitoPY1 and found that ROS is more accumulated in RD3−/− cells (Figure 6I), suggesting that RD3-regulated mitochondrial dispersion decreases oxidative stress under ciliogenesis. Taken together, RD3 interacts with mitochondria, regulates mitochondria dispersion, and reduces the amount of ROS accumulation promoted by the induction of ciliogenesis (Figure 6J).
Figure 6.
RD3 modulates mitochondrial spatial distribution under ciliogenesis-induction-derived oxidative stress
(A) RD3 and mitochondria co-localization study. SPOT-tagged RD3 SH-SY5Y cells stained with anti-RD3 (red) and Alexa 488-conjugated Tomm20 (green) antibodies with DAPI (white) and imaged by confocal microscope. Scale bar: 5 μm. Cell: SH-SY5Y.
(B and E) Working hypothesis.
(C) Quantification of ciliogenesis frequency. Primary cilium frequency quantified using filtered sections (30–50 cells per section). RD3-wild type (WT): 803 cells (navy); KO: 1,269 cells (cobalt blue); and overexpression (OE): 1,141 cells (light green) are shown as a violin plot. Each frequency per section is shown as white dots. p value: an unpaired t test. p < 0.05: orange. Experiments were repeated three times (n = 3).
(D) Cilium length quantification. Primary cilium length quantified by Nikon NIS-Element software using filtered sections. RD3-WT: 71 cilia (navy); KO: 106 cilia (cobalt blue); and OE: 72 cilia (light green) are shown as a cumulative plot. Statistical analysis was conducted as described in (C).
(F) Mitochondria distribution imaging. Ciliogenesis was induced by serum starvation. Cells were stained with anti-α-tubulin (magenta), Alexa 488-conjugated Tomm20 (green), and Alexa 647-conjugated γ-tubulin (yellow) antibodies and DAPI (white), imaged by confocal microscope. Scale bar: 20 μm.
(G) Mitochondrial distribution quantification. The mitochondria distribution index calculated at 5 different spots is defined by the distance from the γ-tubulin signal. Quantification was performed by around 30 cells per replicate. Quantifications were conducted three times (n = 3). p value: an unpaired t test. p < 0.05: orange.
(H) Mitochondrial distribution differences. Distribution difference between RD3-WT and KO is shown as barplot. p value: an unpaired t test.
(I) Mitochondrial H2O2 evaluation. MitoPY1 and Hoechst33342 signals measured by plate reader. Raw MitoPY1 signal was normalized by Hoechst33342, multiplied by 1 × 104, and shown as a bar plot. p value: an unpaired t test. p < 0.05: orange. Experiments were repeated five times (n = 5).
(J) Summary of Rd3-mitochondria spatial dynamics under cilium-induction-oriented oxidative stress.
We next questioned whether these cell line results were relevant to testicular cells. Figure 3 indicates that shRd3 introduction in the testes reduced cell numbers. With the observed increase in ROS accumulation in Rd3−/− cells (Figure 6I), it is plausible to hypothesize that shRd3 might elevate ROS levels, leading to cell death. To assess the impact of shRd3 on round spermatid viability, we introduced shRd3 into testes and found that Rd3 knockdown significantly reduces the viability of Bsg+ testicular cells at 8 weeks more than at 5 weeks, suggesting a pronounced effect of Rd3 knockdown on later-stage spermatogenic cells, including round spermatids (Figure S7). To explore the relationship between this effect and ROS levels, we administered N-acetyl-cysteine (NAC), an antioxidant, and successfully mitigated the phenotype (Figure S8). These results endorse the role of Rd3 in modulating ROS accumulation during spermatogenesis, which was also corroborated by findings in Figures 3 and 6J.
Discussion
In vivo genome-wide screening approach using testicular cells
CRISPR-Cas9 sgRNA library screening is extensively utilized both in vitro and in vivo to elucidate the molecular mechanisms of biological phenomena based on cellular growth. Despite its widespread application, performing screening on biochemical activities, not directly related to growth, is challenging. Addressing this limitation, we recently established the revival screening method by leveraging repeated sorting and realized biochemical-activity-targeted screening in cell lines. Building on this achievement, this study adapted revival screening to establish a screening system targeting biochemical activity in mice testes, thereby providing proof of concept for in vivo screening.
For successful implementation of in vivo genome-wide screening, we considered that the efficiency of sgRNA introduction is a major issue to be addressed. Our research has demonstrated that the efficiency of sgRNA introduction is influenced by a variety of factors, including the method of sgRNA delivery, viral tropism, and the duration of sgRNA expression in the targeted tissues. In this study, we achieved substantial sgRNA coverage in male germ cells by employing a novel technique that combines injection through seminiferous tubules and the use of pseudo-typed lentiviruses (pLVs), especially SVF proteins. It is important to note that the choice of pLVs significantly affects sgRNA induction efficiency, underscoring the importance of selecting pLVs with an appropriate engineering history.
However, our attempts to maintain long-term sgRNA expression in male germ cells were not entirely successful. We observed a decrease in the sgRNA-derived indel population over time. This decrease occurred despite sustained infectivity of the lentivirus, as evidenced by persistent expression of a fluorescent protein marker (Figure S9). We hypothesize that the observed decrease in the sgRNA population might be attributed to activation of a DNA damage response, specifically through the ATM-γH2AX axis. This response is typically limited to the nuclei of leptotene and zygotene stages of germ cell development.62,63,64,65 The induced DNA double-strand breaks by Cas9 are likely to be a trigger for this response, leading to the reduction in sgRNA population. In the future, it may be beneficial to explore alternative techniques, such as CRISPR interference/activation (CRISPRi/a)66,67 and CRISPR-Cas13,68,69 to enhance screening efficiency in specific cell types. These alternatives could circumvent the challenges encountered with genome editing in certain cellular contexts.
Rd3 molecular function during spermatogenesis
In our study, Rd3 emerged as a critical factor influencing spermatogenesis, as demonstrated by the results presented in Figures 3 and 4. While existing research predominantly recognizes Rd3 as an inhibitor of retinal guanylyl cyclase (RetGC1/2), primarily influencing cGMP production in retinogenesis,25,26,46 its role in spermatogenesis has yet to be investigated. Hub-Explorer analysis suggests that the interaction between Rd3 and mitochondria modulates mitochondria’s localization in the context of ciliogenesis. This interaction reduces ROS accumulation by promoting mitochondrial dispersion from the tubulin organizing center. Although cilium structures are different between rod/cone cells and round spermatids, ciliogenesis is a common cell state in both cell types during development, suggesting a functional overlap of Rd3 across these processes. In vivo rescue experiments using NAC indicated a modulatory role of Rd3 in ROS management in testicular cells (Figure S8). However, the incomplete restoration observed suggests additional, unexplored mechanisms at play. Future research should focus on elucidating these interactions, in particular examining mitochondrial localization and cGMP production in round spermatids to determine if Rd3’s role is cell-type specific.
Limitations
This study has successfully established proof of concept for a testes-targeted in vivo screening system. Nonetheless, it is essential to address several limitations inherent in this system. First, a notable limitation concerns the impact of CRISPR-Cas9 editing on male germ cells. Specifically, our observations, detailed in the discussion, indicate a reduction in the Rosa26 knockout cell population despite ongoing infection. This phenomenon resulted in a constrained pool of feasible candidate sgRNAs and genes. Indeed, known Ca2+ channels were not identified in this screening. To mitigate this limitation, employing an alternative methodology, such as the use of a pooled library tailored for various cell types (including CRISPRi, CRISPRa, or CRISPR-Cas13 systems), might be beneficial.
The second major challenge involves the technical difficulty in specifically targeting type A spermatogonia for sgRNA introduction. Although SVF-pLVs effectively enhanced sgRNA delivery to type A spermatogonia, unintentional delivery to other germ cells or Sertoli cells was also noted. In our approach, we treated Cas9 mice with busulfan to deplete most germ cells except spermatogonia prior to viral introduction. However, discrepancies in infection efficiency were evident (Figure S10). Future advancements in developing lentiviral vectors specific to certain cell types are critical for more precise investigation into cell-type-specific biology and mechanisms.
In addition, our Hub-Explorer tool encounters a technical limitation due to its reliance on the type of expression data matrix utilized. Within the testicular environment, scRNA-seq data enable the capture of a spectrum of cell types in various developmental stages, allowing for the creation of an accurate GCN that mirrors expression dynamics. In contrast, the analysis of adult retinal scRNA-seq data, characterized by fully differentiated and distinct cell types, presents challenges in delineating time-scaled sequential gene expression dynamics. Therefore, the selection of appropriate datasets is vital for the effective utilization of this analytical instrument.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Plzf antibody | Santa Cruz | Catalog #: sc-28319 Clone: D-9 RRID: AB_2218941 |
| Anti-Sall4 antibody | Santa Cruz | Catalog #: sc-101147 Clone: EE-30 RRID: AB_1129262 |
| Germ cell-specific antigen antibody [TRA98] | Abcam | Catalog #: ab82527 Clone: TRA98 RRID: AB_1659152 |
| Anti-tRFP antibody | Evrogen | Catalog #: AB233 RRID: AB_2571743 |
| Anti-GFP (Green Fluorescent Protein) pAb | MBL | Catalog #: 598 RRID: AB_591819 |
| PerCP/Cyanine5.5 anti-mouse/human CD324 (E-Cadherin) Antibody | BioLegend | Catalog #: 147317 Clone: DECMA-1 RRID: AB_2750305 |
| Alexa Fluor 647 anti-mouse/human CD324 (E-Cadherin) Antibody | BioLegend | Catalog #: 147308 Clone: DECMA-1 RRID: AB_2563955 |
| PE anti-mouse CD117 (c-Kit) Antibody | BioLegend | Catalog #: 105808 Clone: RUO RRID: AB_313217 |
| PE anti-mouse CD147 Antibody | BioLegend | Catalog #: 123707 Clone: OX-114 RRID: AB_2243692 |
| Anti-RD3 antibody | Santa Cruz | Catalog #: sc-390653 Clone: A-9 RRID: N/A |
| Anti-γ-Tubulin antibody | Sigma | Catalog #: T6557 Clone: GTU-88 RRID: AB_477584 |
| Alexa Fluor 647 Anti-γ-Tubulin antibody - C-terminal | Abcam | Catalog #: ab191114 Clone: TU-30 RRID: AB_2889219 |
| ARL13B Polyclonal antibody | Proteintech | Catalog #: 17711-1-AP RRID: AB_2060867 |
| Alexa Fluor 488 Anti-Tomm20 antibody - Mitochondrial Marker | Abcam | Catalog #: ab205486 Clone: EPR15581-39 RRID: AB_2943509 |
| Anti-α-Tubulin antibody | Santa Cruz | Catalog #: sc-32293 Clone: DM1A RRID: AB_628412 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen™ | Catalog #: A-21434 RRID: AB_2535855 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen™ | Catalog #: A-21247 RRID: AB_141778 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen™ | Catalog #: A-21202 RRID: AB_141607 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen™ | Catalog #: A-21203 RRID: AB_2535789 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen™ | Catalog #: A-31571 RRID: AB_162542 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen™ | Catalog #: A21206 RRID: AB_2535792 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen™ | Catalog #: A-21207 RRID: AB_141637 |
| Biological samples | ||
| C57BL/6JJmsSlc | Japan SLC, Inc. | N/A |
| Gt(ROSA)26Sortm1.1(CAG-cas9∗,-EGFP)Fezh/J | The Jackson Laboratory (Platt et al.76) | strain: # 024858 |
| GeCKO v2 Mouse CRISPR Knockout Pooled Library | Sanjana et al.44 | Addgene: #1000000052 and #1000000053 |
| lentiGuide-Puro | Sanjana et al.44 | Addgene: #52963 |
| lentiCas9-Blast | Sanjana et al.44 | Addgene: #52962 |
| lentiGuide-mEGFP | This paper | N/A |
| lentiGuide-TagRFP | This paper | N/A |
| lentiGuide-TagBFP | This paper | N/A |
| lentiGuide-mitoEGFP | This paper | N/A |
| lentiGuide-sgGcna1-TagBFP | This paper | N/A |
| lentiGuide-sgRosa26-TagRFP | This paper | N/A |
| lentiGuide-sgRD3-Puro | This paper | N/A |
| lenti-shRNA-mEGFP | This paper | N/A |
| lenti-shOvol2-mEGFP | This paper | N/A |
| lenti-shRnf215-mEGFP | This paper | N/A |
| lenti-shCldn34c4-TagBFP | This paper | N/A |
| lenti-shRd3-TagBFP | This paper | N/A |
| lenti-shCebpg-mEGFP | This paper | N/A |
| lenti-shRbm26-mEGFP | This paper | N/A |
| lenti-shP2ry2-TagBFP | This paper | N/A |
| lenti-Cldn34c4-HA-P2A-mEGFP | This paper | N/A |
| lenti-Ovol2-TagBFP | This paper | N/A |
| lenti-Rd3-HA-P2A-mEGFP | This paper | N/A |
| lenti-RD3-Spot-P2A-mEGFP | This paper | N/A |
| pCAG-HIVgp | Miyoshi et al.77 | RIKEN: RDB04394 |
| pCMV-VSV-G-pRSV-REV | Miyoshi et al.77 | RIKEN: RDB04393 |
| pCMV-SVF-P2A-VSV-G-pRSV-REV | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Fluo-4 AM | Dojindo | Catalog #: F311 |
| Cell Count Reagent SF | Nacalai | Catalog #: 07553-15 |
| Cellstain- PI solution | Dojindo | Catalog #: F378 |
| Cellstain- DAPI solution | Dojindo | Catalog #: D523 |
| Cellstain- Hoechst 33342 solution | Dojindo | Catalog #: H342 |
| 7-AAD Viability Staining Solution | Biolegend | Catalog #: 420404 |
| DRAQ5™ | Biostatus | Catalog #: DR50200 |
| lectin PNA From Arachis hypogaea (peanut), Alexa Fluor™ 647 Conjugate | Invitrogen™ | Catalog #: L32460 |
| lectin PNA From Arachis hypogaea (peanut), Alexa Fluor™ 488 Conjugate | Invitrogen™ | Catalog #: L21409 |
| Critical commercial assays | ||
| Pre-washed Sterilized Glass Capillary Tubing w/Internal Glass Fiber | Narishige | Catalog #: GDC-1 |
| QIAamp DNA mini kit | QIAGEN | Catalog #: 51304 |
| QuantiFluor ONE dsDNA | Promega | Catalog #: E4871 |
| MegaX DH10B T1R Electrocomp™ cells | Invitrogen™ | Catalog #: C640003 |
| MitoPY1 | Tocris | Catalog #: 4428 |
| Next Generation Micropipette Puller | SHUTTER INSTRUMENT | Catalog #: P-1000 |
| NEBuilder HiFi DNA Assembly | NEB | Catalog #: E2621 |
| Liberase™ TM | Roche | Catalog #: 5401127001 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems™ | Catalog #: 4368814 |
| TB Green Premix Ex Taq™ II (Tli RNaseH Plus) | Takara | Catalog #: RR820A |
| HCR™ RNA-FISH Bundle | Molecular Instruments | N/A |
| Taq DNA Polymerase | Ampliqon | Catalog #: A111103 |
| ChromoTek Spot-Trap Magnetic Agarose | Proteintech | Catalog #: emta-20 |
| Sera-Mag™ SpeedBeads Protein A/G | Cytiva | Catalog #: 17152104011150 |
| 4BB™ TruePrime Whole Genome Amplification (WGA) Kit | 4basebio29 | Catalog #: 380100 |
| Deposited data | ||
| Library coverage analysis | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool A (input) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool A (1st round) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool A (2nd round) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool B (input) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool B (1st round) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| sgRNA library Pool B (2nd round) | This paper | https://doi.org/10.5281/zenodo.10528508 |
| RD3 interactome data from LC-MS/MS | This paper | ProteomeXchange: PXD042933 Zenodo DOI: https://doi.org/10.5281/zenodo.10528508 |
| Tissue-wide bulk transcriptome dataset | Bgee suite45 | N/A |
| Testis RNA-seq data | https://www.refine.bio/samples/SRR823506 | Accession: SRR823506 |
| Retina RNA-seq data | Grant et al.52 | Accession: SRR342458 |
| Testis scRNA-seq data | Hermann et al.48 | GEO: GSE109033 |
| Retina scRNA-seq data | van Hove et al.51 | Accession: E-MTAB-9061 |
| Experimental models: Cell lines | ||
| HEK293T | Pear et al.70 | S. Nagata lab, Osaka Univ RRID: CVCL_0063 |
| NIH3T3 | Jainchill et al.71 | S. Nagata lab, Osaka Univ RRID: CVCL_0594 |
| Y-79 | Reid et al.72 | National Institutes of Biomedical Innovation, Health and Nutrition RRID: CVCL_1893 |
| F9 | Bernstine et al.73 | RIKEN BRC RRID: CVCL_0259 |
| HT-1080 | Rasheed et al.74 | M. Matsuda lab, Kyoto Univ RRID: CVCL_0317 |
| SH-SY5Y | Biedler et al.75 | Y. Kimura lab, Kyoto Univ RRID: CVCL_0019 |
| Software and algorithms | ||
| 10x Genomics Cell Ranger 6.0.0 | Zheng et al.78 | https://github.com/10XGenomics/cellranger |
| ApE | Davis et al.79 | https://jorgensen.biology.utah.edu/wayned/ape/ |
| CRISP-ID v1.1 | Dehairs et al.80 | http://crispid.gbiomed.kuleuven.be |
| Customized python script for statistical analysis | This paper | https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024 |
| Cutadapt v4.1 | Martin et al.81 | https://github.com/marcelm/cutadapt |
| DAVID | Sherman et al.82 | https://david.ncifcrf.gov |
| docker-cellranger | litd/docker-cellranger | https://github.com/litd/docker-cellranger |
| DSIR | Vert et al.83 | http://biodev.cea.fr/DSIR/DSIR.html |
| FastQC v0.12.1 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| FlowJo | BD Life Sciences | https://www.flowjo.com |
| goatools v1.2.4 | Klopfenstein et al.84 | https://github.com/tanghaibao/goatools |
| guide-caller v1.0.0 | This paper | https://github.com/SuzukiLab-icems/guide-caller |
| HISAT2 v2.2.1 | Perțea, M et al.85 | http://daehwankimlab.github.io/hisat2/ |
| Hub-Explorer v1.0.0 | This paper | https://github.com/SuzukiLab-icems/Hub-Explorer |
| Igv v2.16.1 | Robinson et al.86 | https://software.broadinstitute.org/software/igv/home |
| ImageJ | Schneider et al.87 | https://imagej.github.io/software/imagej/ |
| MAGeCK v0.5.9.4 | Li et al.88 | https://sourceforge.net/p/mageck/wiki/Home/ |
| Matplotlib v3.7.0 | Hunter.89 | https://github.com/matplotlib/matplotlib |
| NIS-Elements Viewer | Nikon | https://www.microscope.healthcare.nikon.com/ja_JP/products/software/nis-elements/viewer |
| Numpy v1.21.2 | van der Walt et al.90 | https://github.com/numpy/numpy |
| Pandas v1.5.3 | McKinney.91 | https://pandas.pydata.org |
| Primer Blast | Ye et al.92 | https://www.ncbi.nlm.nih.gov/tools/primer-blast/ |
| Proteome Discoverer™ software v2.5 | Thermo Scientific | https://www.thermofisher.com |
| Samtools v1.17 | Li et al.93 | http://www.htslib.org |
| Seaborn v0.12.1 | Waskom.94 | https://github.com/mwaskom/seaborn |
| Scanpy v1.9.2 | Wolf et al.95 | https://github.com/scverse/scanpy |
| Scikit-learn v0.0.post1 | Pedregosa et al.96 | https://github.com/scikit-learn/scikit-learn |
| Scipy v1.8.0 | Virtanen et al.97 | https://github.com/scipy/scipy |
| lightSra2Count | This paper | https://github.com/SuzukiLab-icems/Sra2Count/tree/main/lightSra2Count |
| SRA toolkit v3.0.5 | SRA Toolkit | https://hpc.nih.gov/apps/sratoolkit.html |
| StringTie v2.2.1 | Perțea et al.98 | https://github.com/gpertea/stringtie |
| TrimGalore v0.6.10 | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Suzuki (jsuzuki@icems.kyoto-u.ac.jp).
Materials availability
The plasmids and cell lines generated in this study are available on request to the lead contact.
Data and code availability
The MS proteomics data have been deposited in the ProteomeXchange Consortium via the jPOST partner repository with the dataset identifiers PXD042933. All fastq files and processed proteomics data are deposited to Zenodo repository (https://doi.org/10.5281/zenodo.10528508). All the analysis script including Hub-Explorer is deposited to GitHub repository (https://github.com/SuzukiLab-icems) and Zenodo repository (https://doi.org/10.5281/zenodo.10528508).
Experimental model and subject details
Animals
C57BL/6JJmsSlc mice were purchased from Japan SLC, Inc., and Gt(ROSA)26Sortm1.1(CAG-cas9∗,-EGFP)Fezh/J mice were purchased from Jackson Laboratory (JAX: 026179). Mice were housed at 5 mice per cage and maintained in a temperature- (25 ± 2°C) and humidity- (50 ± 10%) controlled conventional animal room under a 12 h light/dark cycle with unrestricted access to food and water at the Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University. All animal studies were conducted according to the Regulations on Animal Experimentation at Kyoto University based on the International Guiding Principles for Biomedical Research Involving Animals. Furthermore, the Animal Experiment Committee of Kyoto University approved all procedures employed in the present study (Permission Number: 49-4).
Cell lines
HEK293T and NIH3T3 cells were gifted from Dr. Shigekazu Nagata (Osaka University). Y-79 and F9 cells were acquired from the National Institutes of Biomedical Innovation, Health and Nutrition, and RIKEN BRC, respectively. HT-1080 and SH-SY5Y cells were gifted from Dr. Michiyuki Matsuda (Kyoto University) and Dr. Yasuhisa Kimura (Kyoto University), respectively. Cells were cultured in DMEM media (Wako) with 1% Penicillin Streptomycin (PS) solution (Nacalai) and 10% Fetal Bovine Serum (FBS) (Gibco). Y-79 cells were cultured in RPMI media (Wako) with 1% PS and 20% FBS. SH-SY5Y cells were cultured in DMEM/Ham’s F-12 media (Nacalai) with 1% PS and 10% FBS. Each cell line was maintained in a 37°C, 5% CO2 incubator. For proteomics study in Figure 5, Y-79 RD3 knockout (RD3−/−) cells and Spot-tagged RD3 expressing (RD3-Spot) cells were utilized. RD3-Spot cells were established by restoring Spot-tagged RD3 expression in RD3−/− cells. The Spot-tagged RD3 sequence was designed to delete the PAM sequence corresponding to the sgRD3 target site with the mutagenesis primer set described in Table S1 by PrimeSTAR MAX DNA polymerase (Takara).
Plasmid construction
For constructing Sendai Virus Fusion (SVF) protein-containing lentivirus packaging plasmid (pCMV-SVF-P2A-VSV-G-pRSV-REV), the SVF-full length coding sequence (GenBank: U86411.1) with the P2A sequence was synthesized and obtained from Sangon Biotech, then inserted into a pCMV-VSV-G-pRSV-REV plasmid acquired from RIKEN77 by In-Fusion HD Cloning Kit (Takara) (Figure S1D). To construct lenti-shRNA-mEGFP, the sgRNA scaffold sequence was removed from the lentiGuide-mEGFP plasmid (derived from lentiGuide-Puro44) using PrimeSTAR Max DNA Polymerase. The shRNA was designed following RIKEN’s protocol (https://dnaconda.riken.jp/Form_PDF/lntRNAien.pdf). All sequences of sgRNA/shRNA oligo and cloning primers used in this study are listed in Table S1.
Method details
Lentivirus production
Lentivirus production was performed according to Li et al.99 and RIKEN’s protocol (https://dnaconda.riken.jp/Form_PDF/lntPrepen.pdf). HEK293T cells were seeded onto sixteen 10 cm culture dishes (Nunc) at 4×106 cells/dish. After 24 h incubation, the medium was changed to 5 mL fresh medium without PS and FBS, and incubated for an extra 1 h at 37°C, 5% CO2. Transfection was conducted by the calcium phosphate precipitation method. In brief, the plasmid mixture was prepared by mixing 272 μg target plasmid, 160 μg pCAG-HIVgp, and 160 μg pCMV-SVF-P2A-VSV-G-pRSV-REV, and made up to 7,200 μL with DDW. Next, 800 μL of 1 M CaCl2 (Wako) was applied to the bottom of the tube containing this plasmid mixture and vortexed vigorously for 10 s. Then, 8,000 μL of 2×BBS (50 mM BES (Nacalai), 280 mM NaCl (Wako) and 1.5 mM Na2HPO4 (Wako)) buffer was added dropwise to the plasmid mixture while gently mixing. After incubation at RT for 30 min, 1 mL of the plasmid mixture was applied to HEK293T cells, incubated at 37°C, 5% CO2 for 6 h, and the medium was then changed to fresh medium containing 10% FBS and 9 mM sodium butylate (Wako). After incubation at 37°C, 5% CO2 for 40–44 h, the supernatants were collected and divided into two 50 mL tubes to remove cell debris by centrifugation (4°C, 900 G, 10 min). Finally, the supernatant was filtered through a 0.45 μm PES filter (Merck Millipore), divided into eight 15 mL tubes by layering onto 1 mL of 20% sucrose (Wako) solution, and centrifuged at 4°C, 15,000 rpm for 4 h with an angle rotor (Hitachi) to pellet the virus. The virus pellet was resuspended with 500 μL chilled HBSS (Nacalai), layered onto the 1 mL of 20% sucrose solution in 1.5 mL tube, and additionally centrifuged at 4°C, 16,000 rpm for 2 h (Hitachi). The centrifuged virus pellet was vigorously resuspended with 90 μL chilled HBSS, separated into 5 μL aliquots, and stored in a −80°C freezer. For confirming SVF protein expression, the virus aliquot was boiled with Sample Buffer containing 2-ME at 95°C for 5 min and applied to SDS-PAGE, followed by Coomassie Brilliant Blue (CBB) staining.
Titer evaluation for the lentivirus encoding fluorescent protein
Titer evaluation was performed, following the LV-MAX Lentiviral Production System USER GUIDE (Gibco, Cat.A35684, Publication No.MAN0017000). First, HT-1080 cells were seeded onto 24 multi-well plates (Nunc) at 3.5×104 cells/well. After 4 h incubation, serially diluted virus solutions (0, 1,000, 2,000, 4,000, 8,000, 16,000, 32,000-fold dilution) were applied to cells and incubated at 37°C, 5% CO2 for an additional 72 h. Next, cells were detached with 0.25% trypsin EDTA (Wako) and analyzed by FACS Lyric (BD) to quantify the percentage of fluorescent protein-positive population. The following formula was used to calculate the infectious titer unit (IFU/ml):
An estimated fold dilution at 20% was calculated by the approximate linearization method (by Microsoft Excel) according to infection efficiency values under 20%.
Titer evaluation for the lentivirus encoding puromycin-resistant protein
HT-1080 cells were seeded onto 96 multi-well plates (Nunc) at 7×103 cells/well in triplicate following the LV-MAX Lentiviral Production System USER GUIDE. After 4 h incubation, serially-diluted virus solutions (0, 1,000, 2,000, 4,000, 8,000, 16,000, 32,000-fold dilution) were applied to cells, and 10 μg/mL puromycin (InvivoGen) was added the next day. After 72 h incubation, 10 μL Cell Count Reagent SF (Nacalai) was added to each well, incubated at 37°C, 5% CO2 for around 1 h, and cell viability was quantified by SYNERGY H1 microplate reader (BioTeK) at 450 nm absorbance. The infectious titer unit (IFU/ml) was calculated as described above regarding %cell viability as %infection.
Determination of mice age for virus injection
The VSVg-coated lentiviruses demonstrate limited infectivity in spermatogonia. However, the application of Sendai Virus Fusion protein facilitates the infection of VSVg-coated lentiviruses into male germ cells, including spermatogonia, even in the testes of adult mice.27,28 In addition, virus infection has to be performed before the initiation of blood-testis barrier formation, at 15 PND. In light of this technical context, our research aimed to optimize the age of mice while considering the following two issues: 1. The number of spermatogonia is maximized. 2. The number of spermatocytes is not increased excessively. Based on these considerations, we determined that the most suitable age for viral injection is at 11 PND.
Capacitation medium
Non-capacitation medium (NCM) was prepared according to RIKEN’s protocol (https://mus.brc.riken.jp/ja/wp-content/uploads/manual/IVF_with_frozen_sperm_ver4.pdf) with 101.6 mM NaCl (Wako), 4.7 mM KCl (Wako), 0.4 mM KH2PO4 (Wako), 0.2 mM MgSO4 (Wako), 2.78 mM D-Glucose (Wako), 23.3 mM Na-Lactate solution (Wako), 0.34 mM Na-Pyruvate (Wako), 1% Penicillin-Streptomycin (Nacalai), 20 mM HEPES-NaOH (Nacalai) and 3 mg/mL fatty-acid free BSA (Sigma), and finally adjusted to pH 7.4. Capacitation medium (CM) was prepared by adding 4 mM CaCl2 (Wako). In the experiment, CM was mixed with 1 M NaHCO3 solution (final: 50 mM) and diluted with NCM containing sperms to be 2 mM CaCl2 and 25 mM NaHCO3.
Sperm capacitation and flow cytometry analysis
Sperm capacitation was induced according to Xia et al.23 500 μL NCM and 400 μL CM were prewarmed at 37°C in different 1.5 mL tubes on a heat-block. Sperms were extracted from the cauda epididymis (8 weeks∼) into 500 μL NCM, mildly mixed, and incubated on a heat block for 5 min. After 5 min incubation, 400 μL supernatant was transferred to the preheated 400 μL CM containing 0.8 μL Propidium Iodide (Dojindo) and 1.6 μL Fluo-4 AM (Dojindo) and mixed mildly, followed by further incubation at 37°C on a heat block for 60 min with the lid open to facilitate sperm reaction. Finally, the capacitated sperms were analyzed by FACS Aria IIIu (BD) with a 100 μm nozzle while kept warm by setting the chamber temperature to 42°C in order to maintain sperm function.
Immunohistochemistry (IHC)
IHC was conducted by referring to Niedenberger et al.100 Testes were extracted from several mice at the ages indicated in each figure legend, fixed with 4% PFA (Nacalai) in DPBS at 4°C for 3–8 h (∼4 weeks) or overnight (8 weeks∼), embedded in Tissue-Tek O.C.T compound (Sakura Finetek) and stored at −80°C until sectioning. For immunostaining, embedded testes were sectioned by a cryostat (Leica) at 20 μm, washed three times with DPBS (Nacalai), and blocked with Blocking One Histo (Nacalai) at RT for 10 min. In the case of Figure 4G, the section was additionally fixed with 4% PFA in DPBS at RT for 10 min and washed three times before proceeding onto the blocking step. The section was washed three times and applied to antigen retrieval with HistoVT One (Nacalai) at 70°C for 20 min. After washing the section, the primary antibody (see each figure legend) in DPBS (Nacalai) with 0.1% Triton X-100 (Wako), 1% donkey serum (Sigma), 5 mg/mL of Probumin (Merck Millipore), and 0.01% of Proclin 950 (Sigma) was applied, and incubated at 4°C overnight. For a secondary antibody staining, the antibody solution was applied to the washed section and incubated at RT for 2 h. The secondary antibody solution was prepared below: Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (Invitrogen, A-21434, 1:400) for Gcna1 staining, Donkey anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Invitrogen, A-21206, 1:400) for TagBFP and EGFP staining, Donkey anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Invitrogen, A-21202, 1:400) for Rd3 staining. Finally, the section was washed twice and mounted with FluorSave (Merck Millipore), and stored at 4°C until imaging by Nikon Ti Eclipse Confocal Microscope. For Figures S1A–S1C, we aimed to evaluate the infection efficiency at single-cell resolution; hence, we localized EGFP to a specific organelle by employing a mitochondria target signal-fused EGFP.
Testis dissociation
Extracted testes were dissociated with 1.5 mL dissociation buffer (DPBS including 0.2 U/ml Liberase (Roche), 5 U/μL DNase I (Takara), 5 mM MgSO4 (Wako)) at 37°C for 30 min while shaking at 200 rpm. Dissociated testicular cells were pipetted 20 times with 10% FBS and 2 mM EDTA (Wako) and filtered with pluriStrainer 40 μm (pluriSelect Life Science UG & Co.KG). Filtered cells were washed twice with 500 μL DPBS for downstream analysis.
In vivo knockout study by indel analysis
The SVF-encapsulated lentivirus encoding sgRosa26 and TagRFP was purified as described in the ‘Lentivirus production’ section and introduced into Cas9+ mice testes at 11 PND via the seminiferous tubule. At 21 PND, the testes were extracted and dissociated, following protocols described in the ‘Testis dissociation’ section and stained with anti-Basigin (Bsg) antibody (Biolegend, 123701, 1:100) on ice for 30 min. Subsequently, Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Invitrogen, A-21247, 1:400) and DAPI (Dojindo, 1:1,000) were applied to cells, and incubated on ice for 30 min and TagRFP+/Bsg+ germ cells were sorted by FACS Aria IIIu and genomic DNA (gDNA) was extracted by the QIAamp DNA mini kit (QIAGEN). The Rosa26 region was amplified from the gDNA with PrimeSTAR GXL DNA Polymerase (Takara), sequenced by Sanger sequencing at Eurofins Genomics Japan, Inc and analyzed by CRISP-ID software80 to detect the indel position. The primer set for PCR is described in Table S1.
In vivo knockout study by Gcna1 knockout analysis
The SVF-encapsulated lentivirus encoding sgGcna1 and TagBFP was introduced into 11 PND Cas9+ mice testes. One week later, dissociated testicular cells were acquired, following protocols described in the ‘Testis dissociation’ section, fixed with 4% PFA in DPBS at RT for 10 min and permeabilized with chilled 70% EtOH on ice for 30 min as given below: fixed cells were resuspended with 300 μL chilled DPBS, and chilled 700 μL absolute EtOH was layered followed by vigorously vortexing. Cells were washed twice and stained with anti-Gcna1 (Abcam, ab82527, 1:100), Sall4 (Santa Cruz, sc-101147, 1:50), Plzf (Santa Cruz, sc-28319, 1:50), and TagBFP (Evrogen, AB223, 1:100) antibodies at 4°C overnight. After staining with Alexa Fluor 647 (for Gcna1)/Alexa Fluor 488 (for Sall4 and Plzf)/Alexa Fluor 594 (for TagBFP) conjugated-secondary antibodies (Invitrogen, 1:400) and DAPI (Dojindo, 1:1,000), the Plzf+/Sall4+ population (for Type A Spermatogonia) and the four chromosomes population (for Primary Spermatocytes) were gated. Gcna1 Median Fluorescent Intensity (MFI) was analyzed for knockout evaluation by comparing the uninfected (TagBFP−) and infected (TagBFP+) cells.
Infection efficiency investigation by flow cytometry
The SVF-encapsulated lentivirus encoding TagBFP was introduced into 11 PND Cas9+ mice testes. One week later, testes were extracted, dissociated, and stained with PerCP/Cyanine5.5 anti-mouse/human Cdh1 antibody (Biolegend, 147317, 1:20). After 30 min incubation on ice, Cdh1+ Type A Spermatogonia were gated and analyzed by FACS Aria IIIu to evaluate the percentage of TagBFP+ population.
Scanning electron microscope (SEM)
Sperms were capacitated, following protocols described in the ‘Sperm capacitation and flow cytometry analysis’ section. As a control, non-capacitated sperms were prepared by incubating sperms without HCO3−. Sperms were washed twice with chilled DPBS and fixed with freshly prepared 2% glutaraldehyde and 4% PFA in DPBS solution. After incubation on ice for 1 h, the sperms were washed twice with chilled DPBS, seeded on 0.1% poly-L-lysine coated glass (25φ) at 5×105 sperms, incubated until the glass was semi-dried, washed with 6 mL DPBS, applied to post-fixiation with 1% OsO4 for 2 h, dehydrated, dried, and finally coated with a thin layer of platinum for analysis with a JEOL JSM-7900F scanning electron Microscope (JEOL Tokyo, Japan).
Sperm in vivo genome-wide screening
library introduction
GeCKO v2 Mouse CRISPR Knockout Pooled Library44 was separately used as Pool A and B. First, lentivirus encoding each library was purified, following protocols described in the ‘Lentivirus production’ section, 4–5 μL lentivirus solutions (0.4–1.4×108 IFU/ml) were injected into testes via the seminiferous tubules, following Ogawa et al.101 In brief, a borosilicate glass injection pipette with 1 mm outer diameter, 0.6 mm inner diameter, and 90 mm length (Narishige, GDC-1) was drawn by Next Generation Micropipette Puller (SHUTTER INSTRUMENT) and snapped by a steel micro tweezers to sharpen the glass tips. Finally, the pipette was filled with the virus solutions for injection.
The expected Multiplicity Of Infection (MOI) in this screening system is around 1∼1.5 according to 30–50% infection efficiency in testicular cells. In a conventional CRISPR screening approach, in which sgRNAs are analyzed after performing a one-time screening, 0.3 MOI (single virus infection) is considered appropriate. On the other hand, our “Revival Screening” aims to cover as many sgRNAs as possible without any omissions during screening and to enrich only essential sgRNAs by repeating the screening process. The combination of repeated screening and statistical analysis reduce the effects of multiple sgRNA integration.
capacitation induction and sorting
Matured sperms were extracted from the cauda epididymis (8–10 weeks), capacitated, following protocols described in the ‘Sperm capacitation and flow cytometry analysis’ section and analyzed by FACS Aria IIIu with a 100 μm nozzle. The negative population of Fluo-4 AM was determined according to an un-capacitated sperm-derived intensity (see Figures 2C and 2D) and directly sorted into a 5 mL tube containing 280 μL chilled RLT buffer (QIAGEN) while kept warm by setting the chamber temperature to 42°C for maintenance of sperm function. After sorting, the lysates were inverted 20 times, centrifuged at 4°C, 1,000 G for 15 min and stored at −80°C until gDNA extraction. Under this condition, the number of sorted sperms is expected to be 20,000 sperms per testis, and the final volume is 350 μL due to accumulation of the FACS flow solution (70 μL/20,000 sperms).
genome DNA extraction
Frozen lysates were slowly thawed on ice and mixed with 150 mM DTT (Wako), 200 μg/mL Proteinase K (Sigma) and 200 μg/mL RNaseA (Sigma). DTT solution was prepared just prior to use. After incubation at 56°C for 2 h while shaking, samples were vortexed with the same volume of the buffer AL (QIAGEN) for 5–10 s and vigorously shaken with the same amount of absolute EtOH (final: 33%) for 5–10 s. Subsequently, according to the manufacturer’s protocol, gDNA was purified by QIAamp DNA mini kit (QIAGEN). In the elution step, gDNA was eluted twice with 50 μL of 70°C pre-warmed DDW and the first and second elusions were combined for the following analysis. Finally, the gDNA was combined in two different tubes (Pool A and Pool B) and applied to QuantiFluor ONE dsDNA system (Promega) to measure the concentration. The concentration of the obtained gDNA was around 68.2 ± 12.4 pg/μL (total average amount: 67.90 ± 10.55 ng per screening) and the estimated viral copy number was 78,492 ± 12,201 copies per screening.
primase-based Whole Genome Amplification (pWGA)
All extracted gDNA was applied to pWGA using the 4BB TruePrime WGA kit (4basebio). This kit leverages the TthPrimPol primase instead of random hexamer oligos to achieve unbiased amplification in terms of DNA copy amount.29 In this study, for scaling up the gDNA input capacity, an alternative highly-concentrated denaturation buffer (1 M KOH solution (Nacalai)) and neutralization buffer (400 mM HCl (Nacalai) and 600 mM Tris-HCl pH 7.5 (Nippon Gene)) were utilized instead of the manufacturer’s prepared buffer. Finally, input was increased from 2.5 μL to 24.7 μL and applied to phi29 polymerase-based isothermal amplification at 30°C for 6 h. The total amount of amplified gDNA product was 300 μg.
library reconstruction
Detailed methods were described by Maruoka et al.16 In brief, sgRNA regions were amplified by PCR (33 cycles) from the pWGA products with the indicated primer set.
-
(1)
Fwd: GTTTTAAAATGGACTATCATATGC
-
(2)
Rev: TATCCATCTTTGCACCCGGGC
The purified amplicon harboring sgRNA regions were assembled with a SmaⅠ(NEB)- and NdeⅠ(NEB)- digested lentiGuide-TagBFP plasmid by NEBuilder HiFi DNA Assembly (NEB) at 52°C for 1 h, electroporated into MegaX DH10B T1R Electrocomp Cells (Invitrogen), and incubated at 37°C with supplemented Recovery Medium (Invitrogen) for 2 h, followed by expansion on LB agar. After confirming library variety by counting the colonies formed (around 1×107 cells), the reconstructed library-encoding plasmid was purified with QIAGEN Plasmid Maxi Kit (QIAGEN).
next generation sequencing
After repeating steps 1–5 for two rounds to enrich sgRNAs of significance, these enriched sgRNAs were amplified by PCR for 15 cycles with the following primers containing an adaptor sequence and sample barcodes (N5-6) of Pool A and Pool B.
Fwd: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCG
ATCT(N5-6)TCTTGTGGAAAGGACGAAACACCG
Rev: CAAGCAGAAGACGGCATACGAGATTCTACTATTCTTTCCCCTGCACTGT
The sample barcodes N5-6 encode TAGCT(N5) and ATCGAC(N6). Finally, equal amounts of the purified amplicons were mixed together and sequenced with the Illumina HiSeq2500 platform at Macrogen Japan.
sgRNA coverage analysis
library introduction
The Pool B GeCKO v2 Mouse CRISPR Knockout Pooled Library (containing 62,804 unique guide sequences)44 was introduced into 11 PND Cas9+ mice testes via seminiferous tubules, following protocols described in the ‘Sperm in vivo genome-wide screening’ section.
testicular cell dissociation and sorting
Three days or one week later, testes were dissociated as described in the ‘Testis dissociation’ section to prepare the following four samples: 1. Three testes from three days treatment, 2. Nine testes from three days treatment, 3. Three testes from one week treatment 4. Nine testes from one week treatment. Dissociated testicular cells were permeabilized with 70% EtOH on ice for 30 min, washed twice with DPBS containing 5% Probumin (Merck Millipore), 0.1% Donkey Serum (Sigma), and 0.01% of Proclin 950 (Sigma), incubated with anti-Gcna1 (Abcam, ab82527, 1:200) and Plzf (Santa Cruz, sc-28319, 1:50) antibodies at 4°C overnight, washed twice, and stained with Alexa Fluor 555 (for Gcna1)- and Alexa Fluor 488 (for Plzf)- conjugated secondary antibodies (Invitrogen, 1:400) and DAPI (Dojindo, 1:1,000). The Gcna1+/Plzf+ Type A Spermatogonia were gated and sorted by FACS Aria IIIu, following the same procedure as sperm sorting for subsequent gDNA extraction.
genomic DNA extraction and next generation sequencing
Genomic DNA was extracted in the presence of 200 μg/mL Proteinase K (Sigma) and 200 μg/mL RNaseA (Sigma), following protocols described in the ‘Sperm in vivo genome-wide screening’ section. The sgRNA regions were amplified by PCR with the following primers containing an adaptor sequence and four sample barcodes (N4: CAAG, N5: TAGCT, N6: ATCGAC, N7: GGTACAG)
-
(1)
Fwd: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
(N4-7)TCTTGTGGAAAGGACGAAACACCG
-
(2)
Rev: CAAGCAGAAGACGGCATACGAGATTCTACTATTCTTTCCCCTGCACTGT
The purified amplicons were mixed with equal amount and sequenced with the Illumina HiSeq2500 platform at Macrogen Japan.
sgRNA data processing
The sgRNA sequences were readout by our original shell script, named guide-caller v1.0.0 (https://github.com/SuzukiLab-icems/guide-caller) following a general analysis pipeline, including FastQC, Cutadapt,81 and MAGeCK.88 In brief, the 20 bp sgRNA sequences were extracted by Cutadapt81 with two rounds of trimming from 51 bp with the sequences read according to the following parameters: “-u {N4:28 N5:29 N6:30 N7:31}” for the first trimming, and “-u {N4:-3 N5:-2 N6:-1 N7:0}” for the second trimming. The trimmed reads were mapped to an annotation list by MAGeCK.88 In the GeCKO v2 sgRNA library, several sgRNAs are annotated to multiple genes concurrently, for instance genes with family members (ex. Il11ra1, Il11ra2 and Gm13305). Therefore, such genes were merged into a single target, and this change was reflected in the annotation list as a modified list for our analysis. Raw count data is deposited in Table S2.
Reverse transcription (RT)-PCR and quantitative RT-PCR (RT-qPCR)
Testis RNA was extracted from dissociated testicular cells using Cas9+ mice at different ages (1–8 weeks) by RNeasy Mini Kit (QIAGEN) and converted to cDNA by High-Capacity cDNA Reverse Transcription Kit (Invitrogen) from 1 μg RNA. The cDNA was diluted 20-fold with DDW in 2.5 ng/μL, amplified with the Rd3-specific primer set (Table S1) by Ampliqon polymerase (Ampliqon) for 33 and 40 cycles through RT-PCR, and the same amount of cDNA was amplified with the targeted primer sets (Table S1) by TB Green Premix Ex Taq I (Takara) for 40 cycles in RT-qPCR.
shRNA-based small-scaled screening
validation of knockdown efficiency
SVF-encapsulated lentiviruses encoding shRNAs targeting LacZ, Ovol2, Rnf215, Cldn34c4, Rd3, Cebpg, Rbm26, and P2ry2 were applied to the following cells: NIH3T3-Ovol2OE (for shOvol2), NIH3T3 (for shRnf215 and shCebpg), F9-Cldn34c4OE (for shCldn34c4), NIH3T3-Rd3OE (for shRd3), Ba/F3 (for shRbm26), and F9 (for shP2ry2). Each knockdown efficiency was evaluated by qRT-PCR.
investigation of shRNA effect on the spermatogenesis
Validated shRNA-encoded lentiviruses were introduced into 10–11 PND Cas9+ mice testes via the seminiferous tubule. After 8–10 weeks, testes and cauda epididymis-derived sperms were extracted for analysis. For testes analysis, testes were initially used for testis weight measurement, subsequently dissociated, stained with PE-conjugated anti-mouse Bsg Antibody (Biolegend, 123707, 1:100), 7-AAD (Biolegend, 1:100), and DRAQ5 (Biostatus, 1:200), and finally 7-AAD-/Bsg+ living germ cells were used to evaluate infection efficiency based on the population of fluorescent protein-positive cells. For sperm analysis, sperms were capacitated as described in the ‘Sperm capacitation and flow cytometry analysis’ section. Sperm number was counted from 10 μL sperm solution (from 800 μL total volume), and Fluo-4 AM intensity was quantified by FACS Aria IIIu after 1 h incubation. All raw data were deposited to Table S3.
scRNA-seq data processing
For adult testicular scRNA-seq data analysis, the following data matrices were utilized: GSE109033_AdultMouseBarcode_filtered_matrix.mtx, barcodes.tsv and genes.tsv.48 For adult retinal scRNA-seq data analysis, deposited fastq file (Accession: E-MTAB-906151) was obtained via ArrayExpress (https://www.ebi.ac.uk/biostudies/arrayexpress) and processed by CellRanger docker image (litd/docker-cellranger) to generate the filtered matrices. These filtered matrices were processed by Scanpy95 to cells and genes with the following criteria: 1. Cells with less than 500 (testes) or 200 (retina) detected genes 2. Genes with fewer than 3 cells. Finally, cell types were annotated according to consensus markers48,51 for each cell type (Figures S4A and S4C). Detailed parameters were described in our analysis script (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024), following the recommended procedure from the developer.
RNA-seq data processing for Rd3 5′UTR comparative study
Analysis flow was described in detail in our original shell script, named lightSra2Count (https://github.com/SuzukiLab-icems/Sra2Count/tree/main/lightSra2Count) by utilizing prefetch, fasterq-dump, trim-galore, HISAT2,85 and Samtools.93 Briefly, mouse-derived testis (Accession: SRR823506) and retina (Accession: SRR34245852) transcriptome SRA files were obtained from the SRA run selector, converted to fastq file by fasterq-dump, trimmed with an automatic adaptor identification process implemented by trim-galore and finally aligned to the mouse reference genome (mm10) by HISAT2 to generate bam files. The aligned information was visualized as a Sashimi plot by the Igv genome browser.86
Hybridization chain reaction fluorescent in situ hybridization (HCR-FISH)
The testis section was prepared according to the IHC procedure and applied to the HCR-FISH process according to the manufacturer’s protocol from Molecular Instruments. In brief, the section was washed to remove the O.C.T. compound, additionally fixed with 4% PFA in DPBS at RT for 10 min, permeabilized with 0.5% Triton X-100 (Wako) at RT for 1 h, washed three times with 5xSSC buffer (Nacalai) with 0.1% Tween 20 (Biorad), equilibrated with HCR hybridization buffer at 37°C for 1 h, hybridized by manufacturer-designed probes targeting Acrv1 and Rd3 (final concentration: 16 nM), and incubated in a humidified chamber at 37°C overnight. The hybridization probes were designed to target multiple spots (maximum: 20 spots). Finally, 10 μL fluorescent probes with identifiers (Acrv1: B1-Alexa Flour 594, Rd3: B3-Alexa Flour 647) in 600 μL amplification buffer were applied to the section for the chain reaction. After washing out excess probes, lectin PNA (Invitrogen, 1:200) and DAPI (Dojindo, 1:1,000) were applied to the section, mounted with FluorSave (Merck Millipore), and incubated at 4°C overnight. Nikon Ti Eclipse Confocal Microscope was used to image the section.
Mass spectrometry analysis
Immunoprecipitation was performed, following Nishino et al.102 In brief, RD3−/− and RD3-Spot Y-79 cells were seeded onto a 10 cm dish (Nunc) at 5.0×107 cells/10 mL medium, incubated at 37°C, 5% CO2 for 24 h, fixed by adding 27 μL of 37% formaldehyde (Wako) (final: 0.1%), incubated at RT for 10 min, quenched with 1 mL of 1 M Glycine-NaOH pH 7.5 (Nacalai) (final: 100 mM) at RT for 4 min, washed with 10 mL DPBS, divided into 1.5 mL tubes at 1.0x107 cells, and stocked at −80°C as a cell pellet. For immunoprecipitation, the cell pellet was lysed by 500 μL RIPA buffer containing 5 μL Protease Inhibitor Cocktail (Nacalai) and 1 μL Benzonase Nuclease (Sigma), incubated at 4°C for 1 h then centrifuged at 4°C, 20,000 G for 10 min to separate the supernatant. A slurry containing ChromoTek Spot-Trap Magnetic Agarose (Chromotek) was washed three times with 500 μL RIPA buffer before 10 μL was added to the supernatant and incubated at 4°C for 2 h. After the incubation, the immunoprecipitated product was washed three times with 300 μL RIPA buffer containing 3 μL of Protease Inhibitor Cocktail (Nacalai), transferred into a new 1.5 mL tube to remove non-specific binding proteins and additionally washed with 300 μL of 50 mM Ammonium Bicarbonate (ABC) buffer. The final products were resuspended with 50 μL of ABC buffer and transferred into a new 1.5 mL tube for LC-MS/MS analysis. Proteins on the beads were digested by adding 200 ng trypsin/Lys-C mix (Promega) and incubating at 37°C overnight. The resulting digests were reduced, alkylated, acidified, and desalted using GL-Tip SDB. The eluates were evaporated and dissolved in 0.1% trifluoroacetic acid and 3% acetonitrile (ACN). LC-MS/MS analysis of the resultant peptides was performed on an EASY-nLC 1200 UHPLC connected to an Orbitrap Fusion mass spectrometer through a nanoelectrospray ion source (Thermo Fisher Scientific). The peptides were separated on a C18 reversed-phase column (75 μm × 150 mm; Nikkyo Technos) with a linear 4–32% ACN gradient for 0–100 min, followed by an increase to 80% ACN for 10 min and a final hold at 80% ACN for 10 min. The mass spectrometer was operated in data-dependent acquisition mode with a maximum duty cycle of 3 s. MS1 spectra were measured with a resolution of 120,000, an AGC target of 4e5, and a mass range of 375-1,500 m/z. HCD MS/MS spectra were acquired in the linear ion trap with an AGC target of 1e4, an isolation window of 1.6 m/z, a maximum injection time of 35 msec, and a normalized collision energy of 30. Dynamic exclusion was set to 20 s. Raw data were directly analyzed against the SwissProt database restricted to Homo sapiens using Proteome Discoverer 2.5 (Thermo Fisher Scientific) with Sequest HT search engine. The search parameters were as follows: (a) trypsin as an enzyme with up to two missed cleavages; (b) precursor mass tolerance of 10 ppm; (c) fragment mass tolerance of 0.6 Da; (d) carbamidomethylation of cysteine as a fixed modification; (e) acetylation of protein N-terminus and oxidation of methionine as variable modifications. Peptides were filtered at a false discovery rate of 1% using the Percolator node. Label free quantification was performed according to the intensities of precursor ions using the Precursor Ions Quantifier node. Normalization was performed such that the total sum of abundance values for each sample over all peptides was the same.
Primary cilia imaging
Primary cilium imaging was performed according to Miyamoto et al.103 First, SH-SY5Y cells were seeded onto an 8-well culture slide (Invitrogen) at 2.5–5.0×104 cells per well, incubated at 37°C, 5% CO2 for 24 h, and ciliogenesis was induced by changing the medium to fresh serum-free medium. After 24 h incubation, cells were fixed with chilled absolute MeOH at RT for 10 min, blocked with Blocking One Histo (Nacalai) at RT for 10 min, stained with anti-ARL13B (Proteintech, 17711-1-AP, 1:400) and γ-Tubulin (Sigma, T6557, 1:500) at 4°C overnight, washed twice, incubated with the Alexa Fluor 488 (for ARL13B)- and Alexa Fluor 594 (for γ-Tubulin)- conjugated secondary antibody (Invitrogen, 1:400) with DAPI (Dojindo, 1:1,000) at RT for 2 h, washed twice, mounted with FluorSave (Merck Millipore), and stored at 4°C until imaging by Nikon Ti Eclipse Confocal Microscope.
RD3 and mitochondria co-localization imaging
SH-SY5Y cells were seeded onto an 8-well culture slide (Invitrogen) at 1×104 cells per well, incubated at 37°C, 5% CO2 for 24 h, fixed with 4% PFA in DPBS at RT for 10 min, blocked with Blocking One Histo (Nacalai) at RT for 10 min, stained with anti-RD3 antibody (Santa Cruz, sc-390653, 1:100) at 4°C overnight, washed twice, stained with anti-mouse Alexa Fluor 647-conjugated secondary antibody (Invitrogen, A-31571, 1:400) at RT for 2 h, washed three times, stained with Alexa Fluor 488-conjugated anti-Tomm20 antibody (Abcam, ab205486, 1:500) and DAPI (Dojindo, 1:1,000) at RT for 1 h, washed twice, mounted with FluorSave (Merck Millipore), incubated at 4°C overnight, and stored at 4°C until imaging by Nikon Ti Eclipse Confocal Microscope.
Mitochondria distribution imaging
Mitochondria distribution imaging was performed, following Onodera et al.104 First, SH-SY5Y cells (RD3 wildtype and knockout cells) were seeded onto an 8-well culture slide (Invitrogen) at 5×103 cells (for Control) and 1×104 cells (for Cilium Induction) per well, and incubated at 37°C, 5% CO2 for 24 h. Next, ciliogenesis was induced by changing the media for fresh serum-free media, incubated at 37°C, 5% CO2 for 24 h, fixed by adding formaldehyde (final: 2%) in DPBS at 37°C for 10 min, washed twice, permeabilized with chilled absolute MeOH on ice for 5 min, washed twice, stained with anti-α-Tubulin antibody (Santa Cruz, sc-32293, 1:250) at 4°C overnight, washed twice, stained with anti-mouse Alexa Fluor 594-conjugated secondary antibody (Invitrogen, 1:400) at RT for 2 h, washed three times, stained with Alexa Fluor 488-conjugated anti-Tomm20 antibody (Abcam, ab205486, 1:500), Alexa Fluor 647-conjugated anti-γ-Tubulin antibody (Abcam, ab191114, 1:500) and DAPI (Dojindo, 1:1,000) at RT for 1 h, washed twice, mounted with FluorSave (Merck Millipore), then stored at 4°C until imaging by Nikon Ti Eclipse Confocal Microscope.
MitoPY1 assay
MitoPY1 staining was performed according to Dickinson et al.105 Briefly, SH-SY5Y cells (RD3 wildtype and knockout cells) were seeded onto a 6-well culture plate (Nunc) at 1×105 cells per well, incubated at 37°C, 5% CO2 for 24 h, and ciliogenesis was induced. After 24 h ciliogenesis induction, cells were detached with 150 μL Accutase (Nacalai), mixed with 850 μL DPBS, centrifuged at 4°C, 400 G for 5 min, resuspended with 250 μL DPBS containing 10 μM MitoPY1 (Tocris) and 2 μg/mL Hoechst 33342 (Dojindo), incubated at 37°C for 60 min, centrifuged at 4°C, 400G for 5 min, resuspended with 200 μL DPBS, then analyzed by Synergy H1 plate reader (MitoPY1: ex.488 nm/em.527 nm; Hoechst33342: ex.350 nm/em.461 nm). The raw MitoPY1 signal was normalized by Hoechst33342 signal, and multiplied by 1×104 for quantification.
Population study of testicular cells
shLacZ or shRd3 was inserted into a lentiviral vector encoding TagBFP and introduced into 11 PND mice testes via the seminiferous tubule. After mice body and testes weight were measured at 5 or 8 weeks, testes were dissociated and 2×106 cells were used for antibody staining with PE-conjugated anti-mouse Bsg Antibody (Biolegend, 123707, 1:100), 7-AAD (Biolegend, 1:100), and DRAQ5 (Biostatus, 1:100). For evaluating the Bsg+ population, 7-AAD-/TagBFP+ cells were analyzed by FACS Aria IIIu.
Rd3 knockdown study under N-acetyl-cysteine treatment
shLacZ or shRd3 was assembled into a lentiviral vector encoding TagBFP and introduced into 11 PND mice testes via the seminiferous tubule. From 28 PND, 100 mg/kg N-acetyl-cysteine (NAC) was administrated via i.p. every single day. At 8 weeks, testes were analyzed according to the ‘Population study of testicular cells’ section.
Quantification of lentivirus infection efficiency under Busulfan treatment
Busulfan administration was performed as described in Kanatsu-Shinohara et al.106 In brief, 0, 15, 30 and 45 mg/kg Busulfan solutions were prepared in DMSO and administrated to 14 PND mice via i.p. administration. Eight days later, mice body and testes were measured. To study the testicular cell population, testes were dissociated and incubated with anti-Gcna1 (Abcam, ab82527, 1:100) and Plzf (Santa Cruz, sc-28319, 1:50) antibodies at 4°C overnight, washed twice, and stained with Alexa Fluor 647 (for Gcna1)- and Alexa Fluor 488 (for Plzf)- conjugated secondary antibodies (Invitrogen, 1:400) and DAPI (Dojindo, 1:1,000). Testicular cell populations indicated in Figures S10D and S10E were quantified by FACS Aria IIIu. For the evaluation of lentiviral infection efficiency, 7.91×107 IFU/ml SVF-coated lentiGuide-mEGFP was introduced into testes via seminiferous tubules. Seven days later, testes were dissociated and stained with PE-conjugated anti-mouse Kit Antibody (Biolegend, 105808, 1:100), Alexa Fluor 647-conjugated anti-mouse Cdh1 antibody (Biolegend, 147308, 1:100), 7-AAD (Biolegend, 1:100), and Hoechst33342 (Dojindo, 1:50), and finally analyzed by FACS Aria IIIu.
Fractionation of testicular cell lysate and western blotting
Organelle fractionation from testes was performed according to Ge et al.107 In brief, 4 testes from 35 to 42 PND mice were dissociated in Liberase, split into two 1.5 mL tubes, and washed twice with chilled 1 mL DPBS, followed by resuspension with 1.0 mL mitochondria isolation buffer (5 mM Tris-HCl (pH 7.5), 1 mM EDTA (pH 8.0), 1 mM DTT, 250 mM sucrose and protease inhibitor cocktail). Subsequently, cell homogenates were prepared by Dounce homogenization (20 times strokes) on ice, subjected to sequential differential centrifugation at 300G, 1,000G, 3,000G, 5,000G, 8,000G and 20,000G at 4°C for 5 min, and lysed with RIPA buffer containing protease inhibitor cocktail, followed by western blotting.
Western blotting was performed in accordance with Maruoka et al.16 In brief, 20 μg lysates from each fraction were boiled in Sample Buffer containing 2-ME at 95°C for 5 min, applied to SDS-PAGE at 35 mA for 30 min, transferred to a PVDF membrane at 65 mA for 60 min, blocked with PVDF Blocking Reagent for Can Get Signal (TOYOBO), and incubated with anti-RD3 (Santa Cruz, sc-390653, 1:500) and Tomm22 (Atlas Antibody, HPA003037, 1:1,000) antibodies in Can Get Signal Immunoreaction Enhancer Solution 1 (TOYOBO) at 4°C overnight. Next, PVDF membranes were washed three times, incubated with 5,000-fold-diluted HRP-conjugated goat anti-mouse or rabbit Igs (Dako) in Can Get Signal Immunoreaction Enhancer Solution 2 (TOYOBO) at 4°C for 2 h, washed three times and reacted with Amersham ECL Select Western Blotting Detection Reagent (Cytiva), followed by visualization with FUSION chemiluminescence imaging system (Vilber).
Quantification and statistical analysis
Quantification and statistical analysis for experimental data
All experiments were performed with biological replicates described in each figure legend. In the shRNA-based small-scale screening, the number of mice was indicated as “n.” Outlier removal was only conducted in mass spectrometry analysis. All animal data were used for analysis. p value was calculated according to an unpaired t test by the Python scipy.stats.ttest_ind function97 with default parameter. p < 0.05 was used as a threshold for evaluation of significance.
Tissue-wide comparative study for characterizing gene expression
Tissue-wide RNA-seq data were downloaded from the Bgee suite45 in TPM-valued format and filtered into specific mice data containing C57BL/6J mice and C57BL/6J hybrid mice with 129SvEvTac, DBA/2J, and CAST/EiJ. The total dataset was constructed from 20 studies harboring 34 tissues from 252 samples. The dataset information is described in detail in Table S4. For gene expression profiling, all datasets were combined, and the target genes were analyzed by an original Python script (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024). Finally, each TPM value was converted into Log2(TPM+1).
Rd3 expression profiling in testicular cells and retinal cells
Testicular and retinal scRNA-seq data were processed as outlined in the ‘scRNA-seq data processing’ section. The data analysis was executed in several stages: Initially, raw gene counts were transformed into ln(CPM+1) using Python scanpy.pp.normalize_total and scanpy.pp.log1p functions.95 This transformation was a precursor to the smoothing process by a moving average, which was performed using the scanpy.pl.paga_path function (n_avg = 50),95 to generate a gene expression matrix. This matrix was then utilized for Rd3 expression profiling and subsequent analysis with Hub-Explorer. In this process, we integrated every sequential group of 50 neighboring cells into a single cell representation (resulting in a reduction from 6,693 to 6,644 cells for testicular data and from 4,599 to 4,550 cells for retinal data). Accordingly, cell type information was reassigned to each identified cell cluster using Python numpy.floor function.90 Finally, gene expressions were normalized to a Z score across all cell types employing the scipy.stats.zscore function,97 and visualized in Figures 4C, S4A and S4C.
Analysis for in vivo genome-wide screening
sgRNA read counts were summarized as a raw count table (values: read counts/columns: sgRNA IDs/index: gene names) by an original Python script contained in guide-caller v1.0.0 (https://github.com/SuzukiLab-icems/guide-caller/blob/main/v1.0.0/matrix_shaper.py), and every count was converted into Log2(CPM +1) and an Enrichment Score (ES). The ES was calculated by modifying Log2FoldChange formula so that detected and undetected sgRNAs are precisely distinguished while maintaining correlation between Log2FoldChange and ES at the significant region (Log2FoldChange≧1.5). This score was calculated by comparing the test data (test) to control data (ctrl) as described below:
The correlation between ES and Log2FoldChange was validated in Figures S2G and S2H. The analysis script was deposited to GitHub repository (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024).
Analysis for library coverage investigation
The sgRNA read count table was generated, following protocols described in the ‘Analysis for in vivo genome-wide screening’ section, and every read count was converted into Log2(CPM+1) and shown as a cumulative plot by the Python seaborn.ecdfplot function.94 The analysis script was deposited to the GitHub repository (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024).
To evaluate the cumulative count of the nine testes-derived sgRNAs at day three (Figure 2A, coverage = 73.5%, total count of detected sgRNA = 44,518), 35,334 sgRNAs were detected over 29.78 counts (which satisfy Log2(CPM+1) = 2, ∗total sgRNA counts = 9,926,238) (79.4%), that we considered as the threshold for a valid count of sgRNAs. According to this criterion, we evaluated the number of genes and total sgRNA counts. As a result, we confirmed that there was no biased count of most of the sgRNAs based on the equal distribution around 2–8 Log2(CPM+1) (Figures S2A and S2B). From these data, we determined that sgRNA coverage was sufficient for screening. Further analysis showed that over half of the genes were targeted by more than two sgRNAs (each gene is targeted by three sgRNAs) in Type A Spermatogonia’s genome from nine testes, but not three testes, at day three and one week (Figures S2C and S2D).
Hub-Explorer
The analysis package was deposited to the GitHub repository with detailed analysis flow (https://github.com/SuzukiLab-icems/Hub-Explorer). In brief, this analysis proceeded as described below.
1. Generation of the cell-type specific gene co-expression (GCN) network:
According to the input expression matrix from scRNA-seq data, the Spearman correlation coefficient between targets and every gene was calculated by the Python scipy.stats.spearmanr function97 in a multiprocessing manner to extract highly-correlated pairs above Spearman’s ρ = 0.8 as the gene co-expression network (GCN) for each target.
2. Hub components identification and back-annotation to target genes:
Each GCN was applied to gene ontology (GO) analysis iteratively using goatools84 to extract significant GO terms (BH_FDR<0.05) as hub components, and these hub components were back-annotated to the targets.
3. Similarity-based K-means clustering for core signaling hub identification:
The Jaccard Similarity Index was calculated as described below for evaluating similarity among targets according to the hub components:
Finally, the Python sklearn.cluster.KMeans function96 was utilized to cluster the targets according to the Jaccard Similarity Index, and the overlapping hub components were extracted as the core signaling pathway. As a benchmark for the K-means method, a dendrogram-based clustering was conducted and labeled on the Jaccard Similarity Index-based clustermap by the Python seaborn.clustermap function.94
Mass spectrometry analysis for profiling RD3 interactors
Three replicates were analyzed in each sample from RD3−/− and RD3-Spot Y-79 cells. Raw spectra data were processed by Proteome Discoverer 2.5 (Thermo Scientific) to generate a normalized signal intensity table, and the original Python script removed outlier values according to the Z test-based approach (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024) below p value = 0.01. This script could detect high outliers from the RD3−/− cells-derived data and a low outliers from the RD3-Spot cells-derived data for enhancing detection sensitivity. Subsequently, Log2FoldChange was calculated by dividing the signal intensities of RD3-Spot cells by those of RD3−/− cells, and the p value was determined from the Python scipy.stats.ttest_ind function97 following unpaired t test. Finally, RD3 interactors were extracted according to Log2FoldChange ≧ 1.5 and p value < 0.05 (Table S5).
Testicular cell identification highly expressing Rd3 interactors
The 269 candidate proteins obtained in Figure 5A were clustered according to testicular cell expression pattern. A detailed analysis script was deposited to the GitHub repository (https://github.com/SuzukiLab-icems/utils_for_Cell_Genomics_2024). In brief, gene expression data of these 269 proteins were acquired from testis scRNA-seq data used in Figures 4 and 5 and converted into a Z score from ln(CPM+1) by the Python scipy.stats.zscore function.97 The following formula was utilized to identify the cell type highly expressing each of the 269 genes:
According to the CTI value, 269 genes were categorized into a specific group above CTI = 0.98, and the number of assigned groups was summarized and counted (Figure 5C, and Table S6).
Gene ontology (GO) analysis
The mouse- and human-derived Ensembl IDs of targeted genes/proteins were applied to DAVID software,82 analyzed in the GOTERM_DIRECT categories and filtered into significant GO terms below a p value described in each figure legend.
Mitochondrial distribution analysis
Mitochondrial distribution was quantified, following Onodera et al.104 by MetaMorph software (Molecular Devices) with the originally developed macro program as the journal function in this software. In brief, fluorescent images of nuclei (DNA), mitochondria (Tomm20), microtubule (α-Tubulin), and microtubule organizing center (MTOC: γ-Tubulin) were loaded to automatically or manually determine the thresholds for detecting Tomm20 and α-Tubulin fluorescent signal. In the manual determination, Tomm20 signal intensity was adjusted to be the most sensitive while removing background noise. This manual adjustment was repeated twice, and the same result was confirmed. The selected single cells were applied to automatic alignment with γ-Tubulin staining (MTOC), and boundaries of the intracellular regions (#1 to #5) were generated. Finally, the Tomm20 fluorescence signal intensity was measured at each region, and converted into the density score by calculating the ratio comparing signal intensities among each region (Figure S6B).
Ciliogenesis evaluation
Primary cilium was distinguished with a co-stained pattern of γ-Tubulin and ARL13B. As technical replicates, ten sections were acquired per sample, and the cilium length and number of cilia were measured by Nikon NIS-Elements software. Finally, these experiments were performed three times as biological replicates to quantify the population of ciliary cells and length of induced cilia. The cilium frequency was calculated by dividing the number of primary cilia by the total number of cells.
Acknowledgments
We thank Takuya Yamamoto, Kyoto University, for discussion of Hub-Explorer; Tomonori Nakamura and Yukiko Ishikura, Kyoto University, for advice on germ cell experimentation; So Nagaoka, Nara Medical University, for discussion of plasmid design; Kouichi Hasegawa, CUORiPS Inc., for the technical discussion of virus injection methods; Yasutaka Okabe, Osaka University, for advice on the testis dissociation protocol; and the Division of Electron Microscopic Study, Center for Anatomical Studies, Graduate School of Medicine, Kyoto University, for SEM imaging and discussion. We thank members of the Suzuki lab for discussions, especially Wen Ann Wee for English editing. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (grant no. 21H00230 to J.S.), the Japan Science and Technology Agency (JST) Fusion Oriented Research for Disruptive Science and Technology (FOREST) (grant no. JPMJFR2162 to J.S.), the Joint Usage and Joint Research Programs of the Institute of Advanced Medical Sciences of Tokushima University (to J.S.), a Grant-in-Aid for Research Activity Start-up (grant no. 22K20972 to Y.N.), JST SPRING (grant no. JPMJSP2110 to Y.N.), and The Sasakawa Scientific Research Grant from the Japan Science Society (grant no. 2022-4005 to Y.N.).
Author contributions
Y.N. and J.S. designed the overall research and interpreted experimental results. Y.N. conducted most experiments, including in vivo genome-wide screening establishment, computational analysis, and in vivo/in vitro molecular biology experiments. Y.O. performed the quantification study of mitochondrial distribution. T.M. designed the primary cilium experiment. M.M. realized the concept of the repeated sorting-based sgRNA enrichment method, called revival screening. H.K. performed the mass spectrometry analysis. Y.N. and J.S. wrote the manuscript.
Declaration of interests
J.S. and Y.N. are inventors on a patent application of the in vivo genome-wide screening method toward spermatogenesis.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work, the authors used Grammarly and ChatGPT4 for English language editing after completing the initial writing by the authors. After using these tools, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Published: March 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2024.100510.
Supplemental information
SG: Spermatogonium; pL/Z: preLeptotene/Zygotene; P: Pachytene; D: Diplotene; M: Meiotic cells; RS: Round Spermatid; ES: Elongating Spermatid. Each expression level is normalized in Z-score (see 'Rd3 expression profiling in testicular cells and retinal cells' in the STAR Methods).
References
- 1.Mani R., St.Onge R.P., Hartman J.L., Giaever G., Roth F.P. Defining genetic interaction. Proc. Natl. Acad. Sci. USA. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T., et al. A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D., et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith V., Botstein D., Brown P.O. Genetic footprinting: a genomic strategy for determining a gene’s function given its sequence. Proc. Natl. Acad. Sci. USA. 1995;92:6479–6483. doi: 10.1073/pnas.92.14.6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen G., Hazendonk E., Thijssen K.L., Plasterk R.H.A. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 1997;17:119–121. doi: 10.1038/ng0997-119. [DOI] [PubMed] [Google Scholar]
- 6.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional Characterization of the S. cerevisiae Genome by Gene Deletion and Parallel Analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 8.Tsien J.Z., Chen D.F., Gerber D., Tom C., Mercer E.H., Anderson D.J., Mayford M., Kandel E.R., Tonegawa S. Subregion- and Cell Type–Restricted Gene Knockout in Mouse Brain. Cell. 1996;87:1317–1326. doi: 10.1016/S0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 9.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T., et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn M., Santinha A.J., Platt R.J. Moving from in vitro to in vivo CRISPR screens. Gene and Genome Editing. 2021;2 doi: 10.1016/j.ggedit.2021.100008. [DOI] [Google Scholar]
- 11.VanDusen N.J., Lee J.Y., Gu W., Butler C.E., Sethi I., Zheng Y., King J.S., Zhou P., Suo S., Guo Y., et al. Massively parallel in vivo CRISPR screening identifies RNF20/40 as epigenetic regulators of cardiomyocyte maturation. Nat. Commun. 2021;12:4442. doi: 10.1038/s41467-021-24743-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruetz T.J., Kashiwagi C.M., Morton B., Yeo R.W., Leeman D.S., Morgens D.W., Tsui C.K., Li A., Bassik M.C., Brunet A. In vitro and in vivo CRISPR-Cas9 screens reveal drivers of aging in neural stem cells of the brain. bioRxiv. 2021 doi: 10.1101/2021.11.23.469762. Preprint at. [DOI] [Google Scholar]
- 13.Wertz M.H., Mitchem M.R., Pineda S.S., Hachigian L.J., Lee H., Lau V., Powers A., Kulicke R., Madan G.K., Colic M., et al. Genome-wide In Vivo CNS Screening Identifies Genes that Modify CNS Neuronal Survival and mHTT Toxicity. Neuron. 2020;106:76–89. doi: 10.1016/j.neuron.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keys H.R., Knouse K.A. Genome-scale CRISPR screening in a single mouse liver. Cell Genomics. 2022;2 doi: 10.1016/j.xgen.2022.100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia Y., Li L., Lin Y.H., Gopal P., Shen S., Zhou K., Yu X., Sharma T., Zhang Y., Siegwart D.J., et al. In vivo CRISPR screening identifies BAZ2 chromatin remodelers as druggable regulators of mammalian liver regeneration. Cell Stem Cell. 2022;29:372–385.e8. doi: 10.1016/j.stem.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maruoka M., Zhang P., Mori H., Imanishi E., Packwood D.M., Harada H., Kosako H., Suzuki J. Caspase cleavage releases a nuclear protein fragment that stimulates phospholipid scrambling at the plasma membrane. Mol. Cell. 2021;81:1397–1410.e9. doi: 10.1016/j.molcel.2021.02.025. [DOI] [PubMed] [Google Scholar]
- 17.Austin C. Observations on the Penetration of the Sperm into the Mammalian Egg. Aust. Jnl. Of Bio. Sci. 1951;4:581. doi: 10.1071/BI9510581. [DOI] [PubMed] [Google Scholar]
- 18.Chang M.C. Fertilizing Capacity of Spermatozoa deposited into the Fallopian Tubes. Nature. 1951;168:697–698. doi: 10.1038/168697b0. [DOI] [PubMed] [Google Scholar]
- 19.Austin C.R. The ‘Capacitation’ of the Mammalian Sperm. Nature. 1952;170:326. doi: 10.1038/170326a0. [DOI] [PubMed] [Google Scholar]
- 20.Loeb J. On the Nature of the Conditions Which Determine or Prevent the Entrance of the Spermatozoon Into the Egg. Am. Nat. 1915;49:257–285. doi: 10.1086/279480. [DOI] [Google Scholar]
- 21.Visconti P.E., Galantino-Homer H., Moore G.D., Bailey J.L., Ning X., Fornes M., Kopf G.S. The Molecular Basis of Sperm Capacitation. J. Androl. 1998;19:242–248. doi: 10.1002/j.1939-4640.1998.tb01994.x. [DOI] [PubMed] [Google Scholar]
- 22.Suarez S.S. Control of hyperactivation in sperm. Hum. Reprod. Update. 2008;14:647–657. doi: 10.1093/humupd/dmn029. [DOI] [PubMed] [Google Scholar]
- 23.Xia J., Ren D. The BSA-induced Ca(2+) influx during sperm capacitation is CATSPER channel-dependent. Reprod. Biol. Endocrinol. 2009;7:119. doi: 10.1186/1477-7827-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman J.S., Chang B., Kannabiran C., Chakarova C., Singh H.P., Jalali S., Hawes N.L., Branham K., Othman M., Filippova E., et al. Premature truncation of a novel protein, RD3, exhibiting subnuclear localization is associated with retinal degeneration. Am. J. Hum. Genet. 2006;79:1059–1070. doi: 10.1086/510021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azadi S., Molday L.L., Molday R.S. RD3, the protein associated with Leber congenital amaurosis type 12, is required for guanylate cyclase trafficking in photoreceptor cells. Proc. Natl. Acad. Sci. USA. 2010;107:21158–21163. doi: 10.1073/pnas.1010460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peshenko I.V., Olshevskaya E.V., Azadi S., Molday L.L., Molday R.S., Dizhoor A.M. Retinal Degeneration 3 (RD3) Protein Inhibits Catalytic Activity of Retinal Membrane Guanylyl Cyclase (RetGC) and Its Stimulation by Activating Proteins. Biochemistry. 2011;50:9511–9519. doi: 10.1021/bi201342b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe S., Kanatsu-Shinohara M., Shinohara T. Sendai virus-mediated transduction of mammalian spermatogonial stem cells. Biol. Reprod. 2019;100:523–534. doi: 10.1093/biolre/ioy192. [DOI] [PubMed] [Google Scholar]
- 28.Shinohara T., Kanatsu-Shinohara M. Transgenesis and Genome Editing of Mouse Spermatogonial Stem Cells by Lentivirus Pseudotyped with Sendai Virus F Protein. Stem Cell Rep. 2020;14:447–461. doi: 10.1016/j.stemcr.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picher Á.J., Budeus B., Wafzig O., Krüger C., García-Gómez S., Martínez-Jiménez M.I., Díaz-Talavera A., Weber D., Blanco L., Schneider A. TruePrime is a novel method for whole-genome amplification from single cells based on TthPrimPol. Nat. Commun. 2016;7 doi: 10.1038/ncomms13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanatsu-Shinohara M., Toyokuni S., Shinohara T. Transgenic Mice Produced by Retroviral Transduction of Male Germ Line Stem Cells In Vivo1. Biol. Reprod. 2004;71:1202–1207. doi: 10.1095/biolreprod.104.031294. [DOI] [PubMed] [Google Scholar]
- 31.Ikawa M., Tergaonkar V., Ogura A., Ogonuki N., Inoue K., Verma I.M. Restoration of spermatogenesis by lentiviral gene transfer: Offspring from infertile mice. Proc. Natl. Acad. Sci. USA. 2002;99:7524–7529. doi: 10.1073/pnas.072207299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F., Zhang Q., Cao J., Huang Q., Zhu X. The microtubule plus end-binding protein EB1 is involved in Sertoli cell plasticity in testicular seminiferous tubules. Exp. Cell Res. 2008;314:213–226. doi: 10.1016/j.yexcr.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Kinoh H., Inoue M., Washizawa K., Yamamoto T., Fujikawa S., Tokusumi Y., Iida A., Nagai Y., Hasegawa M. Generation of a recombinant Sendai virus that is selectively activated and lyses human tumor cells expressing matrix metalloproteinases. Gene Ther. 2004;11:1137–1145. doi: 10.1038/sj.gt.3302272. [DOI] [PubMed] [Google Scholar]
- 34.Enders G.C., May J.J. Developmentally Regulated Expression of a Mouse Germ Cell Nuclear Antigen Examined from Embryonic Day 11 to Adult in Male and Female Mice. Dev. Biol. 1994;163:331–340. doi: 10.1006/dbio.1994.1152. [DOI] [PubMed] [Google Scholar]
- 35.Tokuda M., Kadokawa Y., Kurahashi H., Marunouchi T. CDH1 Is a Specific Marker for Undifferentiated Spermatogonia in Mouse Testes. Biol. Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- 36.Chen H., Fok K.L., Yu S., Jiang J., Chen Z., Gui Y., Cai Z., Chan H.C. CD147 is required for matrix metalloproteinases-2 production and germ cell migration during spermatogenesis. Mol. Hum. Reprod. 2011;17:405–414. doi: 10.1093/molehr/gar013. [DOI] [PubMed] [Google Scholar]
- 37.Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- 38.Hobbs R.M., Fagoonee S., Papa A., Webster K., Altruda F., Nishinakamura R., Chai L., Pandolfi P.P. Functional Antagonism between Sall4 and Plzf Defines Germline Progenitors. Cell Stem Cell. 2012;10:284–298. doi: 10.1016/j.stem.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vara C., Paytuví-Gallart A., Cuartero Y., le Dily F., García F., Salvà-Castro J., Gómez-H L., Julià E., Moutinho C., Aiese Cigliano R., et al. Three-Dimensional Genomic Structure and Cohesin Occupancy Correlate with Transcriptional Activity during Spermatogenesis. Cell Rep. 2019;28:352–367.e9. doi: 10.1016/j.celrep.2019.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Plessis S.S., Cabler S., McAlister D.A., Sabanegh E., Agarwal A. The effect of obesity on sperm disorders and male infertility. Nat. Rev. Urol. 2010;7:153–161. doi: 10.1038/nrurol.2010.6. [DOI] [PubMed] [Google Scholar]
- 41.Palmer N.O., Bakos H.W., Fullston T., Lane M. Impact of obesity on male fertility, sperm function and molecular composition. Spermatogenesis. 2012;2:253–263. doi: 10.4161/spmg.21362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin S.K., Yang W.X. Factors and pathways involved in capacitation: how are they regulated? Oncotarget. 2016;8:3600–3627. doi: 10.18632/oncotarget.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujihara Y., Murakami M., Inoue N., Satouh Y., Kaseda K., Ikawa M., Okabe M. Sperm equatorial segment protein 1, SPESP1, is required for fully fertile sperm in mouse. J. Cell Sci. 2010;123:1531–1536. doi: 10.1242/jcs.067363. [DOI] [PubMed] [Google Scholar]
- 44.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bastian F.B., Roux J., Niknejad A., Comte A., Fonseca Costa S.S., de Farias T.M., Moretti S., Parmentier G., de Laval V.R., Rosikiewicz M., et al. The Bgee suite: integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 2021;49:D831–D847. doi: 10.1093/nar/gkaa793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molday L.L., Jefferies T., Molday R.S. Insights into the role of RD3 in guanylate cyclase trafficking, photoreceptor degeneration, and Leber congenital amaurosis. Front. Mol. Neurosci. 2014;7 doi: 10.3389/fnmol.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aravindan S., Somasundaram D.B., Kam K.L., Subramanian K., Yu Z., Herman T.S., Fung K.M., Aravindan N. Retinal Degeneration Protein 3 (RD3) in normal human tissues: Novel insights. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-13337-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernströer B., et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018;25:1650–1667. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddi P.P., Flickinger C.J., Herr J.C. Round Spermatid-Specific Transcription of the Mouse SP-10 Gene Is Mediated by a 294-Base Pair Proximal Promoter1. Biol. Reprod. 1999;61:1256–1266. doi: 10.1095/biolreprod61.5.1256. [DOI] [PubMed] [Google Scholar]
- 50.Fujihara Y., Herberg S., Blaha A., Panser K., Kobayashi K., Larasati T., Novatchkova M., Theussl H.C., Olszanska O., Ikawa M., et al. The conserved fertility factor SPACA4/Bouncer has divergent modes of action in vertebrate fertilization. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2108777118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Hove I., De Groef L., Boeckx B., Modave E., Hu T.T., Beets K., Etienne I., Van Bergen T., Lambrechts D., Moons L., et al. Single-cell transcriptome analysis of the Akimba mouse retina reveals cell-type-specific insights into the pathobiology of diabetic retinopathy. Diabetologia. 2020;63:2235–2248. doi: 10.1007/s00125-020-05218-0. [DOI] [PubMed] [Google Scholar]
- 52.Grant G.R., Farkas M.H., Pizarro A.D., Lahens N.F., Schug J., Brunk B.P., Stoeckert C.J., Hogenesch J.B., Pierce E.A. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee M.C., Damjanov I. Anatomic distribution of lectin-binding sites in mouse testis and epididymis. Differentiation. 1984;27:74–81. doi: 10.1111/j.1432-0436.1984.tb01410.x. [DOI] [PubMed] [Google Scholar]
- 54.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bae J.E., Kang G.M., Min S.H., Jo D.S., Jung Y.K., Kim K., Kim M.S., Cho D.H. Primary cilia mediate mitochondrial stress responses to promote dopamine neuron survival in a Parkinson’s disease model. Cell Death Dis. 2019;10:1–15. doi: 10.1038/s41419-019-2184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shankar S., Hsu Z.T., Ezquerra A., Li C.C., Huang T.L., Coyaud E., Viais R., Grauffel C., Raught B., Lim C., et al. Α γ-tubulin complex-dependent pathway suppresses ciliogenesis by promoting cilia disassembly. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111642. [DOI] [PubMed] [Google Scholar]
- 57.Tian W.N., Braunstein L.D., Apse K., Pang J., Rose M., Tian X., Stanton R.C. Importance of glucose-6-phosphate dehydrogenase activity in cell death. American Journal of Physiology-Cell Physiology. 1999;276:C1121–C1131. doi: 10.1152/ajpcell.1999.276.5.C1121. [DOI] [PubMed] [Google Scholar]
- 58.Pandey S., Lopez C., Jammu A. Oxidative stress and activation of proteasome protease during serum deprivation-induced apoptosis in rat hepatoma cells; inhibition of cell death by melatonin. Apoptosis. 2003;8:497–508. doi: 10.1023/a:1025542424986. [DOI] [PubMed] [Google Scholar]
- 59.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 60.Youle R.J., van der Bliek A.M. Mitochondrial Fission, Fusion, and Stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vincent A.E., Turnbull D.M., Eisner V., Hajnóczky G., Picard M. Mitochondrial Nanotunnels. Trends Cell Biol. 2017;27:787–799. doi: 10.1016/j.tcb.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24:927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 63.Mahadevaiah S.K., Turner J.M.A., Baudat F., Rogakou E.P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W.M., Burgoyne P.S. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 64.Bellani M.A., Romanienko P.J., Cairatti D.A., Camerini-Otero R.D. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm -/- spermatocytes. J. Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- 65.Hirano K., Nonami Y., Nakamura Y., Sato T., Sato T., Ishiguro K., Ogawa T., Yoshida S. Temperature sensitivity of DNA double-strand break repair underpins heat-induced meiotic failure in mouse spermatogenesis. Commun. Biol. 2022;5:1–16. doi: 10.1038/s42003-022-03449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C., et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H., et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu D., Cai Y., Tang L., Han X., Gao F., Cao H., Qi F., Kapranov P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020;10:1794. doi: 10.1038/s41598-020-58104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S., Li X., Xue W., Zhang L., Yang L.Z., Cao S.M., Lei Y.N., Liu C.X., Guo S.K., Shan L., et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods. 2021;18:51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 70.Pear W.S., Nolan G.P., Scott M.L., Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jainchill J.L., Aaronson S.A., Todaro G.J. Murine Sarcoma and Leukemia Viruses: Assay Using Clonal Lines of Contact-Inhibited Mouse Cells. J. Virol. 1969;4:549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reid T.W., Albert D.M., Rabson A.S., Russell P., Craft J., Chu E.W., Tralka T.S., Wilcox J.L. Characteristics of an Established Cell Line of Retinoblastoma2. JNCI. Journal of the National Cancer Institute. 1974;53:347–360. doi: 10.1093/jnci/53.2.347. [DOI] [PubMed] [Google Scholar]
- 73.Bernstine E.G., Hooper M.L., Grandchamp S., Ephrussi B. Alkaline Phosphatase Activity in Mouse Teratoma. Proc. Natl. Acad. Sci. USA. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasheed S., Nelson-Rees W.A., Toth E.M., Arnstein P., Gardner M.B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::AID-CNCR2820330419>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 75.Biedler J.L., Helson L., Spengler B.A. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 1973;33:2643–2652. [PubMed] [Google Scholar]
- 76.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miyoshi H., Blömer U., Takahashi M., Gage F.H., Verma I.M. Development of a Self-Inactivating Lentivirus Vector. J. Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8 doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davis M.W., Jorgensen E.M. ApE, A Plasmid Editor: A Freely Available DNA Manipulation and Visualization Program. Frontiers in Bioinformatics. 2022;2 doi: 10.3389/fbinf.2022.818619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dehairs J., Talebi A., Cherifi Y., Swinnen J.V. CRISP-ID: decoding CRISPR mediated indels by Sanger sequencing. Sci. Rep. 2016;6 doi: 10.1038/srep28973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 82.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vert J.P., Foveau N., Lajaunie C., Vandenbrouck Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinf. 2006;7:520. doi: 10.1186/1471-2105-7-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klopfenstein D.V., Zhang L., Pedersen B.S., Ramírez F., Warwick Vesztrocy A., Naldi A., Mungall C.J., Yunes J.M., Botvinnik O., Weigel M., et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hunter J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007;9:90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- 90.van der Walt S., Colbert S.C., Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011;13:22–30. doi: 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- 91.Mckinney W. pandas: a Foundational Python Library for Data Analysis and Statistics. Python High Performance Science Computer. 2011;14 [Google Scholar]
- 92.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., Durbin, R., and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed]
- 94.Waskom M.L. seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021. doi: 10.21105/joss.03021. [DOI] [Google Scholar]
- 95.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 97.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s4152-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li M., Husic N., Lin Y., Snider B.J. Production of Lentiviral Vectors for Transducing Cells from the Central Nervous System. JoVE. 2012:4031. doi: 10.3791/4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niedenberger B.A., Geyer C.B. Advanced immunostaining approaches to study early male germ cell development. Stem Cell Res. 2018;27:162–168. doi: 10.1016/j.scr.2018.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ogawa T., Aréchaga J.M., Avarbock M.R., Brinster R.L. Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- 102.Nishino K., Kosako H. Immunoprecipitation-Mass Spectrometry for Analysis of Post-Translational Modifications and Interactomes of Target Proteins. Preprint at Proteome Letters. 2022 doi: 10.14889/jpros.7.1_9. [DOI] [Google Scholar]
- 103.Miyamoto T., Hosoba K., Ochiai H., Royba E., Izumi H., Sakuma T., Yamamoto T., Dynlacht B.D., Matsuura S. The Microtubule-Depolymerizing Activity of a Mitotic Kinesin Protein KIF2A Drives Primary Cilia Disassembly Coupled with Cell Proliferation. Cell Rep. 2015;10:664–673. doi: 10.1016/j.celrep.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Onodera Y., Nam J.M., Horikawa M., Shirato H., Sabe H. Arf6-driven cell invasion is intrinsically linked to TRAK1-mediated mitochondrial anterograde trafficking to avoid oxidative catastrophe. Nat. Commun. 2018;9:2682. doi: 10.1038/s41467-018-05087-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dickinson B.C., Lin V.S., Chang C.J. Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat. Protoc. 2013;8:1249–1259. doi: 10.1038/nprot.2013.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kanatsu-Shinohara M., Toyokuni S., Morimoto T., Matsui S., Honjo T., Shinohara T. Functional Assessment of Self-Renewal Activity of Male Germline Stem Cells Following Cytotoxic Damage and Serial Transplantation. Biol. Reprod. 2003;68:1801–1807. doi: 10.1095/biolreprod.102.012575. [DOI] [PubMed] [Google Scholar]
- 107.Ge L., Melville D., Zhang M., Schekman R. The ER–Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2 doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SG: Spermatogonium; pL/Z: preLeptotene/Zygotene; P: Pachytene; D: Diplotene; M: Meiotic cells; RS: Round Spermatid; ES: Elongating Spermatid. Each expression level is normalized in Z-score (see 'Rd3 expression profiling in testicular cells and retinal cells' in the STAR Methods).
Data Availability Statement
The MS proteomics data have been deposited in the ProteomeXchange Consortium via the jPOST partner repository with the dataset identifiers PXD042933. All fastq files and processed proteomics data are deposited to Zenodo repository (https://doi.org/10.5281/zenodo.10528508). All the analysis script including Hub-Explorer is deposited to GitHub repository (https://github.com/SuzukiLab-icems) and Zenodo repository (https://doi.org/10.5281/zenodo.10528508).