Abstract
Reprogramming scar fibroblasts into cardiomyocytes has been proposed to reverse the damage associated with myocardial infarction. However, the limited improvement in cardiac function calls for enhanced strategies. We reported enhanced efficacy of our miR reprogramming cocktail miR combo (miR-1, miR-133a, miR-208a, and miR-499) via RNA-sensing receptor stimulation. We hypothesized that we could combine RNA-sensing receptor activation with fibroblast reprogramming by chemically modifying miR combo. To test the hypothesis, miR combo was modified to enhance interaction with the RNA-sensing receptor Rig1 via the addition of a 5′-triphosphate (5′ppp) group. Importantly, when compared with unmodified miR combo, 5′ppp-modified miR combo markedly improved reprogramming efficacy in vitro. Enhanced reprogramming efficacy correlated with a type-I interferon immune response with strong and selective secretion of interferon β (IFNβ). Antibody blocking studies and media replacement experiments indicated that 5′ppp-miR combo utilized IFNβ to enhance fibroblast reprogramming efficacy. In conclusion, miRs can acquire powerful additional roles through chemical modification that potentially increases their clinical applications.
Keywords: MT: Non-coding RNAs, 5′-triphosphorylation, RNA modification, fibroblasts, cardiomyocytes, reprogramming, IFNβ, innate immune signaling, miRs
Graphical abstract
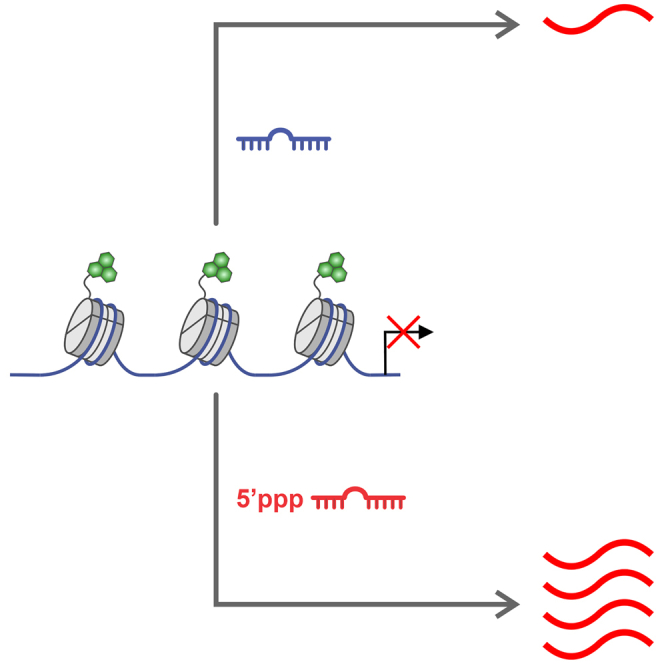
Hodgkinson and colleagues demonstrate that chemically modifying RNA to affect a biological gain-of-function improves the efficacy of RNA-based cellular reprogramming.
Introduction
Myocardial infarction remains a significant global health concern, contributing to high rates of morbidity and mortality worldwide.1,2,3 The regenerative potential of adult human hearts is limited and following cardiac injury there is an irreversible loss of functional cardiomyocytes.4 The limited efficacy of current treatment modalities in restoring myocardial function necessitates the development of innovative approaches. One such innovative approach is the direct reprogramming of scar fibroblasts into cardiomyocytes.5 Fibroblast to cardiomyocyte reprogramming has been achieved through several different methods.6,7,8 Irrespective of the method employed, functional benefits are significant with a 10%–20% increase in fractional shortening and ejection fraction.6 However, the improvements in cardiac function are too modest for clinical applications.9
We pioneered a fibroblast to cardiomyocyte reprogramming approach based on miRs. Through a screening and reductive approach, we identified miR combo (miR combo: miR-1, miR-133, miR-208, and miR-499).9 Subsequent work in our laboratory found that miR combo efficacy was significantly improved by activation of the RNA-sensing receptors Retinoic acid-inducible gene I (Rig1) and Toll-like receptor 3 (TLR3).10,11,12 In these studies, RNA-sensing receptors were activated by standard pharmacological agents such as polyinosinic-polycytidylic acid (PolyI:C), 5′ppp-dsRNA, and 3p-hpRNA.10,11,12 While useful for proof of principle experiments, there are potential issues with their use in the clinic. Clinical trials have reported toxicities including coagulation abnormalities, renal failure, and systemic cardiovascular failure.13,14 Consequently, we hypothesized that we could chemically modify the miRs of our miR combo to not only preserve their inherent miR characteristics (mRNA degradation/translation inhibition), but also allow them to function as potent RNA-sensing receptor agonists.
RNA modifications, such as nucleotide substitutions and chemical modifications, are often used to enhance stability.15 Modifications could also be employed to make an RNA recognizable by RNA-sensing receptors as these receptors recognize specific motifs. For Rig1 agonists, a 5′triphosphate (5′ppp) group must be present.16,17,18 The 5′ppp modification is emerging as a notable tool, especially with respect to RNA-based vaccines.19,20
In this study, we found that 5′ppp-modified miR combo (5′ppp-miR combo) significantly improved the efficacy of fibroblast to cardiomyocyte reprogramming. Improved efficacy was associated with Rig1 activation and the release of IFNβ.
Results
Optimizing 5′ppp-modified miR combo
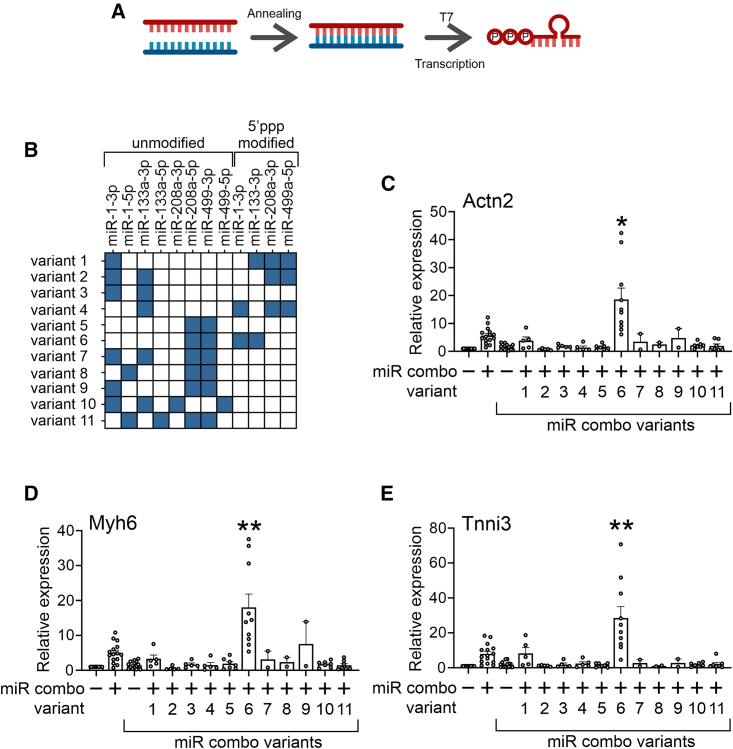
To identify the optimal combination of 5′ppp-modified miRs in miR combo, we undertook a screening experiment. Various 5′ppp-miR combo variants and unmodified controls were generated via T7 kinase reaction (Figure 1A). The variants tested in this study are shown in (Figure 1B). These 5′ppp-miR combo variants and unmodified variant controls were transfected into cardiac fibroblasts and reprogramming assessed by measuring the expression of the cardiomyocyte markers Actn2, Myh6, and Tnni3 (Figures 1C–1E). The screening process identified variant 6 as the most potent. In variant 6, miR-1 and miR-133 are unmodified while miR-208 and miR-499 are 5′ppp modified. Variant 6 was used for all subsequent experiments.
Figure 1.
Optimizing 5′ppp modification of miR combo
(A) Schematic describing the methodology to generate 5′ppp-modified miRs. (B) A table listing the miR combo variants used in (C)–(E). (C–E) Cardiac fibroblasts were transfected with unmodified miR combo, miR combo variants 1–11 or the appropriate non-targeting control miRs. After 7 days, expression of the indicated cardiomyocyte-specific genes was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the unmodified non-targeting control. N = 10. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗p < 0.01, ∗p < 0.05 comparisons with unmodified miR combo.
5′ppp-miR combo enhances fibroblast to cardiomyocyte reprogramming efficacy
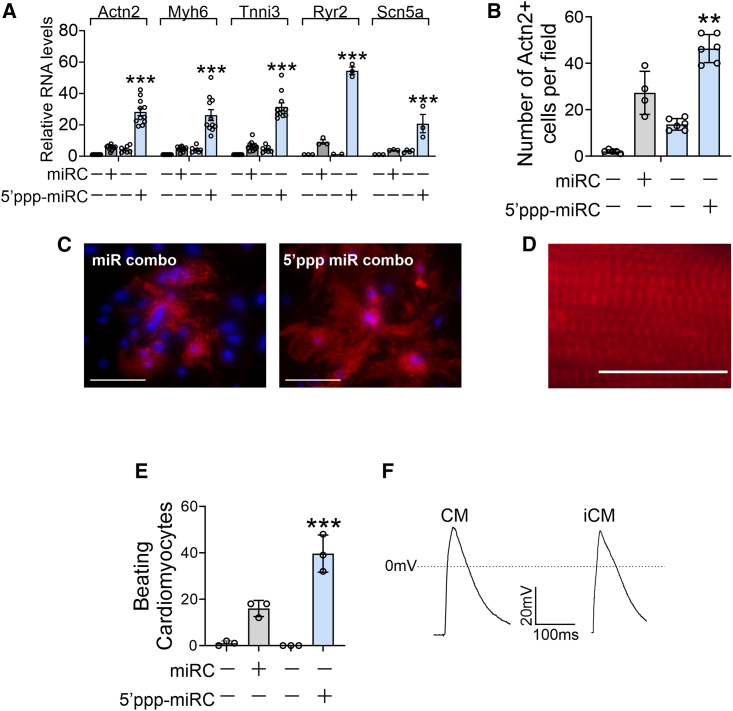
Additional experiments were performed to provide further evidence that the 5′ppp modification was enhancing reprogramming efficacy. In addition to enhancing the expression of cardiomyocyte sarcomere genes (Figure 2A), the 5′ppp modification of miR combo also increased the expression of cardiomyocyte-specific ion channels Ryr2 and Scn5a (Figure 2A). In addition to measuring changes in cardiomyocyte-specific gene expression, cells were stained with antibodies targeting the cardiomyocyte-specific protein Actn2. Actn2 immunostaining allowed for a quantification of the number of cardiomyocyte-like cells that had been generated as well as an assessment of sarcomere structure. Actn2 immunostaining indicated that 5′ppp-miR combo was more efficacious in generating cardiomyocyte-like cells (Figure 2B with sample images in Figure 2C). The cardiomyocyte-like cells also displayed distinct sarcomeres (Figure 2D). Importantly, 5′ppp-miR combo also substantially increased the number of spontaneously beating cardiomyocytes generated through reprogramming (Figure 2E, Video S1, and Video S2). Moreover, cardiomyocytes generated by the 5′ppp-miR combo displayed action potentials akin to freshly isolated neonatal cardiomyocytes (Figure 2F).
Figure 2.
5′ppp-miR combo enhances fibroblast to cardiomyocyte reprogramming efficacy
(A) Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo and appropriate non-targeting control miRs. Fourteen days after transfection days, expression of the indicated cardiomyocyte-specific genes was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the unmodified non-targeting control. N = 3–12. ANOVA (1-way) with post hoc tests were used to determine significance: ∗∗∗p < 0.001 comparisons with unmodified miR combo. (B–D) Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo and appropriate non-targeting control miRs. Fourteen days after transfection, the number of cardiomyocyte-like cells was determined by staining for the cardiomyocyte-specific protein Actn2. The number of Actn2+ cells (cardiomyocyte-like cells) per field was calculated from five independent fields (×20 magnification) from four to six independent experiments. ANOVA (1-way) with post hoc tests was used to determine significances: ∗p < 0.05 comparisons with unmodified miR combo. Representative images of unmodified and 5′ppp-modified miR combo are shown in (C) (scale bar, 20 μm). An enlarged image showing sarcomere structure in cardiomyocyte-like cells generated via 5′ppp-miR combo is shown in (D) (scale bar, 20 μm). (E) Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo or appropriate non-targeting control miRs. Fourteen days after transfection, the number of beating cells per field (×20 magnification) was counted. N = 4. ANOVA (1-way) with post hoc tests were used to determine significances: ∗p < 0.05 comparisons with unmodified miR combo. (F) Cardiac fibroblasts were transfected with 5′ppp-miR combo. Fourteen days after transfection, spontaneous calcium oscillations were measured and compared with those derived from freshly isolated neonatal cardiomyocytes. Representative traces are shown from 10 cells per group.
Fourteen days after transfection days, cells were videoed for evidence of spontaneous beating. N=3. Representative videos are shown. Unmodified miR combo.
Fourteen days after transfection days, cells were videoed for evidence of spontaneous beating. N=3. Representative videos are shown. 5’ppp-miR combo.
Taken together, these results provide further evidence that 5′ppp modification of miR combo enhances the efficacy of fibroblast to cardiomyocyte reprogramming.
Mechanism of 5′ppp-miR combo directed enhancement of reprogramming
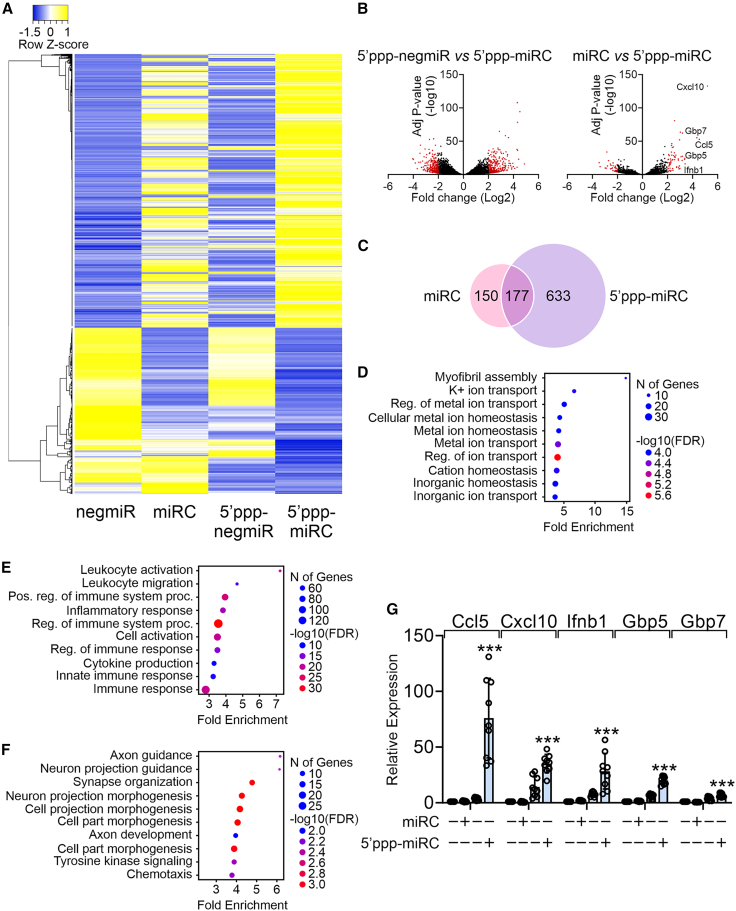
To elucidate the mechanism by which 5′ppp-miR combo enhanced the efficacy of fibroblast to cardiomyocyte reprogramming, we measured global changes in RNA expression via whole genome RNA sequencing (RNA-seq). Cardiac fibroblasts were transfected with unmodified miR combo, 5′ppp-miR combo, or their respective non-targeting controls. The 5′ppp modification was found to amplify the effect of miR combo with respect to both genes activated and genes inhibited (Figure 3A).
Figure 3.
5′ppp-miR combo promotes expression of IFNβ pathway components
Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo or appropriate non-targeting control miRs. After 4 days, RNA-seq was performed. N = 3. (A) Heatmap showing genes whose expression was significantly affected by 5′ppp-miR combo (>3-fold compared with the unmodified non-targeting control). (B) Volcano plots showing comparisons between 5′ppp-negmiR and 5′ppp-miR combo and between unmodified miR combo and 5′ppp-miR combo. Red indicates a significant increase/decrease in expression (≥3-fold). (C–F) Two gene lists were produced. One gene list comprised those genes whose expression was increased by unmodified miR combo by ≥ 3-fold when compared with the unmodified control negmiR. The second list was composed of genes whose expression was increased by 5′ppp-miR combo by ≥3-fold when compared with the 5′ppp-modified control negmiR. The Venn diagram reports the number of genes that were common and unique in both gene lists (C). Gene ontology (GO) was then performed on those common and unique genes. (D) The GO results on the common set of genes. (E) The GO results on the genes that were unique to 5′ppp-miR combo. (F) The GO results on the genes that were unique to unmodified miR combo. For (D)–(F), only the top 10 GO terms are shown. (G) Cardiac fibroblasts were transfected with unmodified or 5′ppp-modified miR combo or appropriate non-targeting control miRs. After 4 days, expression of the indicated genes was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the unmodified non-targeting control. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗∗p < 0.001 comparisons between 5′ppp-miR combo and unmodified miR combo.
To further understand the RNA-seq data, comparisons were made between 5′ppp-miR combo and either the 5′ppp-modified non-targeting control miR (5′ppp-negmiR) or unmodified miR combo. As shown in Figure 3B, 5′ppp-miR combo influenced the expression of a unique set of genes when compared with 5′ppp-negmiR and unmodified miR combo. Gene ontology (GO) analysis was used to discern the function of genes that were significantly up-regulated in the 5′ppp-miR combo group vs. the 5′ppp-negmiR and unmodified miR combo groups. Two gene lists were produced. One gene list comprised those genes whose expression was increased by unmodified miR combo by 3-fold or greater when compared with the unmodified control negmiR. The second list was composed of genes whose expression was increased by 5′ppp-miR combo by 3-fold or greater when compared with the 5′ppp-modified control negmiR. Comparing the two lists indicated that unmodified miR combo and 5′ppp-miR combo shared 177 targets, while 150 and 633 targets were unique to unmodified miR combo and 5′ppp-miR combo, respectively (Figure 3C). Analysis of the 177 shared targets identified GO terms for myofibril assembly and ion transport (Figure 3D). Both are important for cardiomyocyte differentiation and function. GO terms in the group of targets unique to 5′ppp-miR combo were all related to immunity (Figure 3E). Considering that the group of 5′ppp-miR combo genes was defined in relation to 5′ppp-negmiR, this implies that miR sequence was influencing the immune response elicited by the 5′ppp group. Interestingly, GO analysis of the targets unique to the unmodified miR combo group identified GO terms related to neuron differentiation (Figure 3F).
RNA-seq indicated that the top five genes whose expression was significantly up-regulated in the 5′ppp-miR combo group vs. the unmodified miR combo group and 5′ppp-negmiR group were Gbp5, Gbp7, Ifnb1, Ccl5, and Cxcl10 (Figure 3B). Post RNA-seq qPCR assays confirmed their significant up-regulation by 5′ppp-miR combo (Figure 3G). Again, 5′ppp-miR combo was shown to have a stronger effect than 5′ppp-negmiR.
5′ppp-miR combo improves fibroblast to cardiomyocyte reprogramming efficacy via IFNβ
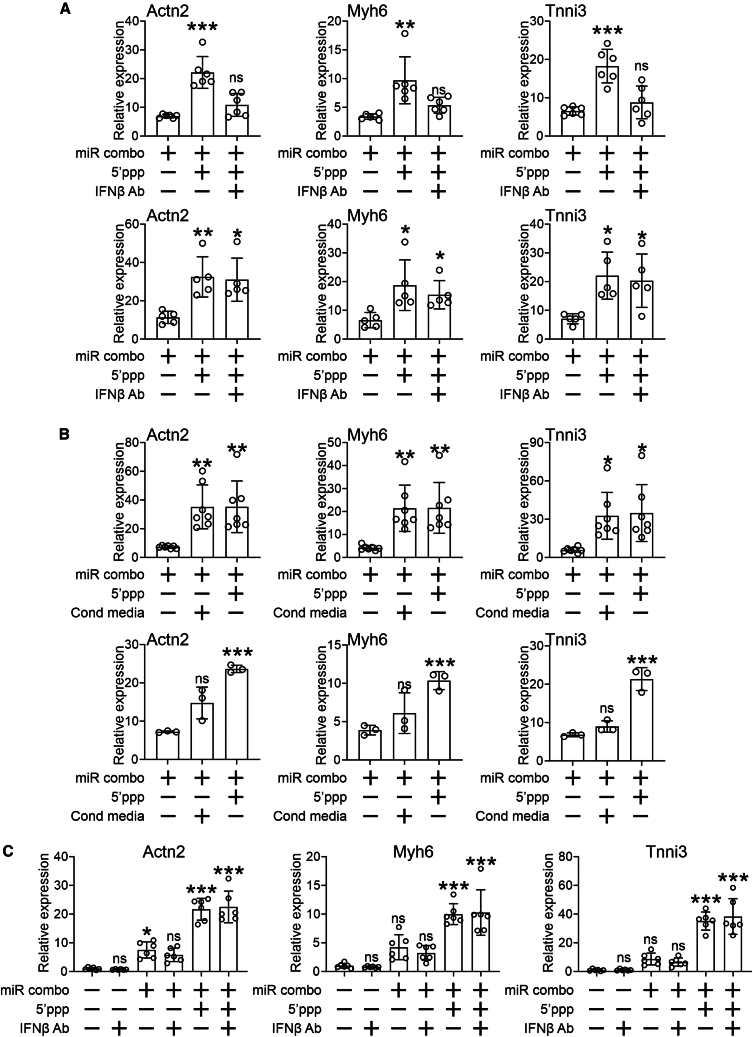
We next initiated a study to determine the protein responsible for the observed enhancement or reprogramming. Since the expression of Gbp5, Gbp7, Ccl5, and Cxcl10 is positively regulated by IFNβ, initial studies focused on IFNβ. IFNβ functions as a paracrine factor.21,22 Consequently, IFNβ blocking antibody and media replacement assays were conducted. IFNβ blocking antibodies present in the first 5 h of transfection significantly blocked the effects of 5′ppp-miR combo (Figure 4A, top panels). However, no significant effects were observed when antibodies were added 24–48 h after transfection (Figure 4A, bottom panels). These data indicated that IFNβ is needed in the early stages of reprogramming.
Figure 4.
5′ppp-miR combo improves fibroblast to cardiomyocyte reprogramming efficacy via IFNβ
(A) Cardiac fibroblasts were transfected with 5′ppp-miR combo, unmodified miR combo, or their respective non-targeting controls. Five (top panels) or 24 (bottom panels) hours after transfection an isotype control or IFNβ blocking antibody was added to the media. Cardiomyocyte-specific gene expression was then measured 14 days after transfection by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are shown as a fold change to the unmodified non-targeting control miR. N = 5–6 per group. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns, not significant, comparisons to unmodified miR combo + isotype control group. (B) Cardiac fibroblasts were transfected with 5′ppp-miR combo. Media was collected from 5′ppp-miR combo transfected cells 24 (top panels) or 72 (bottom panels) hours after transfection and placed onto cells 24 hours after transfection with unmodified miR combo. Non-targeting miRs were used as a control. Cardiomyocyte-specific gene expression measured by qPCR 14 days after transfection. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to non-targeting control miR. N = 4. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns, not significant, comparisons to the unmodified miR combo group. (C) Six hours prior to transfection, cardiac fibroblasts were incubated with either an isotype control or IFNβ blocking antibody. After the 6 h incubation with antibodies, the cells were transfected with 5′ppp-miR combo, unmodified miR combo, or the unmodified non-targeting control. Cardiomyocyte-specific gene expression was then measured 14 days after transfection by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are shown as a fold change to the unmodified non-targeting control miR. N = 6 per group. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗∗p < 0.001, ns, not significant, comparisons to unmodified miR combo + isotype control group.
To verify the blocking antibody experiments, conditioned media studies were conducted. Media was collected from 5′ppp-miR combo transfected cells and placed onto cells transfected with unmodified miR combo. The effects on unmodified miR combo were dependent on when the media was collected from the 5′ppp-miR combo transfected cells. Media collected in the first 24 h improved the efficacy of unmodified miR combo (Figure 4B, top panels). However, media collected 72 h post-transfection had no effect on unmodified miR combo efficacy (Figure 4B, bottom panels).
Importantly, IFNβ did not alter fibroblast plasticity with respect to reprogramming as the addition of IFNβ blocking antibodies prior to transfection did not affect the ability of 5′ppp-miR combo to induce cardiomyocyte-specific gene expression (Figure 4C).
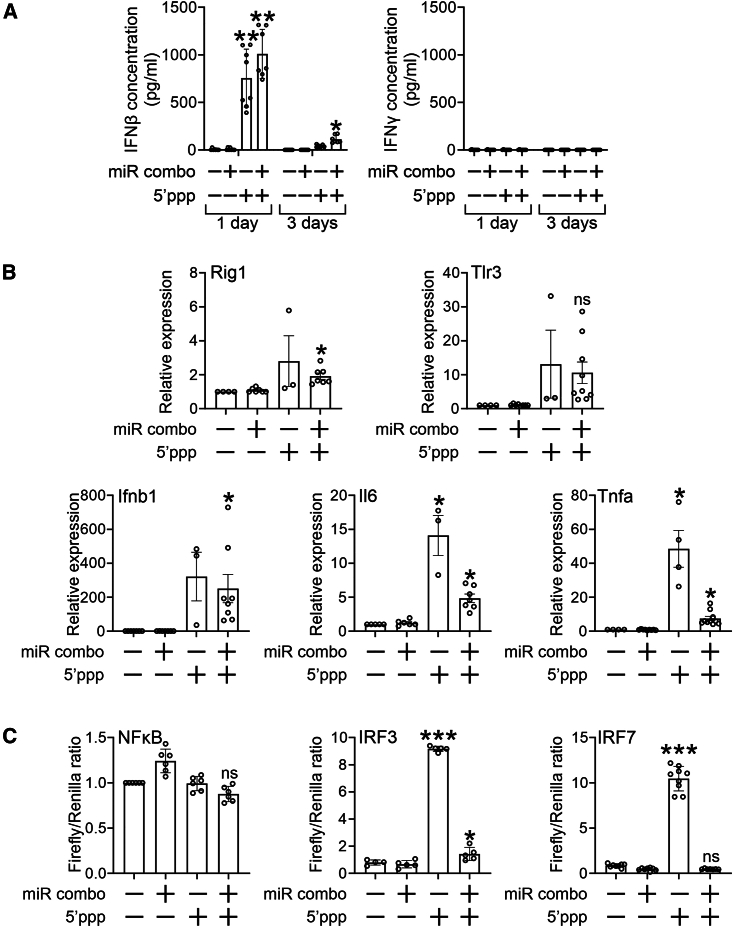
These time-dependent effects on gene transcription were dependent on when IFNβ was present in the media. ELISA indicated high levels of IFNβ present in the media for the first 24 h post 5′ppp-miR combo transfection and virtually absent by 72 h (Figure 5A).
Figure 5.
5′ppp-miR combo is selective for the IFNβ pathway
(A) Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo or the appropriate non-targeting control miRs. One and 4 days after transfection, the media was collected. IFNβ and IFNγ amounts were measured by ELISA. N = 7. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗p < 0.01, ∗p < 0.05 comparisons to the unmodified non-targeting miR group. (B) Cardiac fibroblasts were transfected with unmodified or 5′ppp-miR combo or the appropriate non-targeting control miRs. One day after transfection, expression of the indicated RNA-sensing receptor pathway components was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the unmodified non-targeting control. N = 3–7. ANOVA (1-way) with post hoc tests were used to determine significances: ∗p < 0.05, ns, not significant, comparisons to the unmodified non-targeting miR group. (C) Cardiac fibroblasts were transfected with unmodified or 5′ppp-modified miR combo and appropriate non-targeting control miRs. After 6 h, NF-κB, IRF3, and IRF-7 activity was determined by dual-luciferase assay. N = 5–6. ANOVA (1-way) with post hoc tests was used to determine significances: ∗∗∗p < 0.001; ∗∗p < 0.01, ∗p < 0.05, ns, not significant, comparisons with unmodified non-targeting miR group.
5′ppp-miR combo is a biased agonist
Having demonstrated that 5′ppp-miR combo strongly induced IFNβ, we wanted to determine the effect on other RNA-sensing receptor mediators. In contrast to IFNβ, IFNγ was virtually absent in the media (Figure 5A). Studying this apparent bias in more detail, while IFNβ transcripts were very abundant following 5′ppp-miR combo transfection, there were substantially fewer numbers of TNFα and IL6 transcripts (Figure 5B). Interestingly, the miR combo miRs were also found to down-regulate the expression of TNFα and IL6 (Figure 5B). Rig1 and TLR3 expression was only marginally affected by 5′ppp-miR combo (Figure 5B).
Subsequent experiments were conducted on RNA-sensing receptor activated transcription factors NF-κB, IRF3, and IRF7. NF-κB is predominantly involved in the expression of pro-inflammatory cytokines, while IRF3/7 regulates IFN expression.23,24 In agreement with our previous data,12 unmodified miR combo weakly activated NF-κB (Figure 5C). The 5′ppp modification of miR combo appeared to weakly inhibit NF-κB (Figure 5C). In contrast, 5′ppp-miR combo predominantly activated IRF3 and IRF7 (Figure 5C). Activation of IRF3 was affected by the miR combo miRs, as IRF3 activation by 5′ppp-miR combo was weaker than that found with the 5′ppp-non-targeting miR (Figure 5C).
5′ppp-miR combo mediates its effects on IFNβ and reprogramming via Rig1
With respect to the receptor mediating the effects of 5′ppp-miR combo, we focused on Rig1 and TLR3. We have implicated both receptors in cellular reprogramming.10,11 The 5′ppp modification of miRs was expected to enhance miR interactions with Rig1.
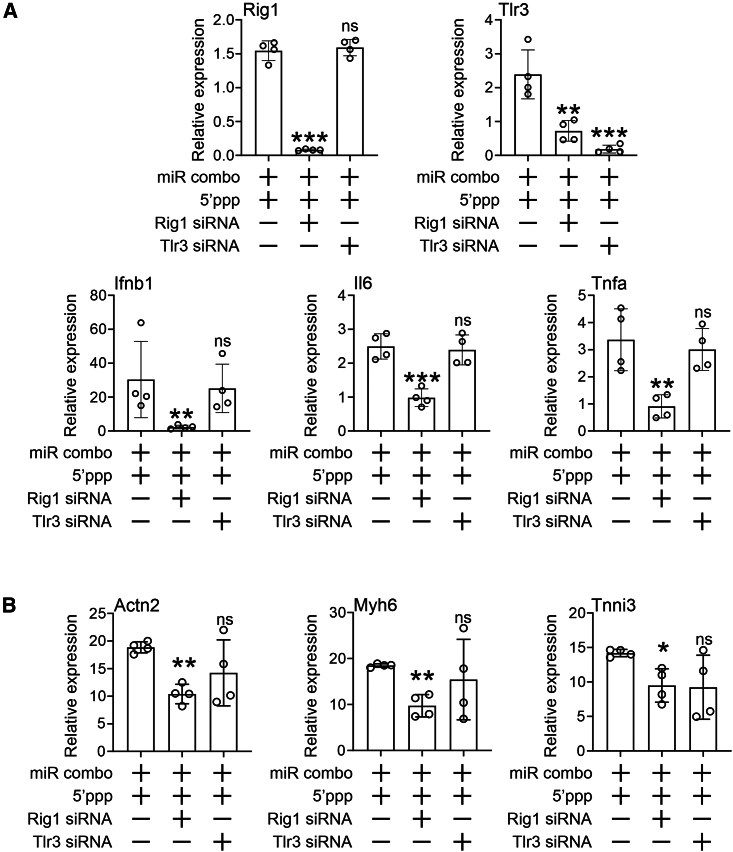
To investigate the role of Rig1 and TLR3 in mediating the effects of 5′ppp-miR combo, knockdown experiments were performed. Knockdown of both receptors was robust (Figure 6A). While TLR3 knockdown had no effect on Rig1 expression, knockdown of Rig1 reduced TLR3 expression by ∼60% (Figure 6A). TLR3 knockdown had no effect on the ability of 5′ppp-miR combo to induce expression of the RNA-sensing receptor mediators IFNβ, TNFα, and IL6 (Figure 6A). In contrast, 5′ppp-miR combo lost the ability to induce these genes in the absence of Rig1 (Figure 6A).
Figure 6.
Rig1 mediates the effects of 5′ppp-miR combo on the IFNβ pathway and reprogramming
(A) Cardiac fibroblasts were transfected with 5′ppp-miR combo and siRNAs (Rig1, TLR3 targeting, or non-targeting control siRNA). One day after transfection, expression of Rig1, Tlr3 and other RNA-sensing receptor pathway components was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the non-targeting control siRNA group. N = 4. ANOVA (1-way) with post hoc tests were used to determine significances: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; ns, not significant, comparisons to the 5′ppp-miR combo + non-targeting siRNA group. (B) Cardiac fibroblasts were transfected with 5′ppp-miR combo and siRNAs (Rig1, TLR3 targeting, or non-targeting control siRNA). Transfection with unmodified non-targeting miRs served as an additional control. After 14 days, expression of the indicated cardiomyocyte-specific genes was determined by qPCR. Expression levels were normalized to the housekeeping gene Gapdh and are represented as a fold change to the unmodified non-targeting control miR. N = 4. ANOVA (1-way) with post hoc tests was used to determine significances: ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; ns, not significant, comparisons to the 5′ppp-miR combo + non-targeting siRNA group.
Further evidence for the role of Rig1 came from an analysis of fibroblast to cardiomyocyte reprogramming. Knockdown of TLR3 was ineffective in inhibiting the ability of 5′ppp-miR combo to induce the expression of cardiomyocyte genes (Figure 6B). In contrast, induced cardiomyocyte gene expression was ablated by Rig1 knockdown (Figure 6B). Taken together, the data indicate that Rig1 is the receptor of 5′ppp-miR combo.
Discussion
In this study, we optimized fibroblast to cardiomyocyte reprogramming by modifying miR combo with a 5′ppp group. Furthermore, we showed that the increased reprogramming efficacy was due to a Rig1-IFNβ signaling pathway.
Bypassing the induced pluripotent stem cell (iPSC) stage and directly reprogramming fibroblasts to cardiomyocytes has garnered considerable interest in the field of cardiac regeneration.5,25,26 Although numerous studies in recent years have substantiated the efficacy of the approach, the improvements observed in cardiac function remain relatively modest.27,28 As a result, there has been a focus on improving efficacy.6,29,30,31,32 In this regard, we discovered a role for the innate immune response and specifically RNA-sensing receptors.10,11,12 We initially identified a role for TLR312 and later work identified a role for Rig1.10,11 These two RNA-sensing receptors also appear to be important for transcription factor-based fibroblast to cardiomyocyte reprogramming.33,34
In this study, we tested the hypothesis that the efficacy of fibroblast to cardiomyocyte reprogramming could be enhanced by modifying our miR combo to function additionally as an RNA-sensing receptor agonist. Results obtained were consistent with our hypothesis. When compared with unmodified miR combo, the inclusion of a 5′ppp group significantly increased the efficacy of fibroblast to cardiomyocyte reprogramming. This increased efficacy was evident not only in the expression of cardiomyocyte-specific factors, but also in sarcomere maturation and in the number of beating cardiomyocytes generated.
RNA-sensing receptors are believed to act independent of sequence.35,36 Rig1 binds specifically to RNA molecules with a 5′ppp group. Thus, we expected that 5′ppp modification of all four miRs of miR combo would have the greatest effect on reprogramming efficacy. Unexpectedly, this was not observed. Indeed, the greatest effect on miR combo efficacy was observed when only of the two of the constituent miRs of miR combo, miR-208 and miR-499, were modified. Moreover, we also observed that 5′ppp-miR combo differed from 5′ppp-negmiR in its ability to induce immunity-related responses such as key transcription factors, pro-inflammatory proteins, and interferons. Taken together, these data imply that sequence is an important determinant of RNA-sensing receptor binding. Despite what is often believed, RNA molecules are highly structured.37 For each RNA sequence there are a unique set of thermodynamically likely structures; those that correspond to the lowest energy state. As such, it can be envisaged that certain RNA structures enclose the 5′ppp group and essentially hide it from Rig1 recognition. The implications of sequence and RNA structure dictating ligand recognition by RNA-sensing receptors could be important in clinical applications. There are a number of clinical trials under way to manipulate RNA-sensing receptors, most notably as a cancer treatment. The focus has been on the moiety that is recognized by the receptor; for example, the 5′ppp group needed for Rig1 binding. However, our data suggest that sequence may also need to be taken into account, as this may either aid or inhibit recognition.
We identified a crucial role for IFNβ. What is currently unclear is the mechanism by which IFNβ regulates cardiomyocyte gene expression. We are currently undertaking studies in this area. They are complex studies because IFNβ does not directly influence gene expression, instead influencing biological responses through binding to the IFN receptor IFNAR. As might be expected, published IFNAR pathways are focused on immune responses such as parasite encapsulation. The links to cell fate determination are not immediately apparent. Similarly, we also noted that 5′ppp-miR combo was acting as a biased Rig1 agonist, significantly inducing IFNβ to the exclusion of other IFNs and other cytokines. Further studies are being undertaken to understand the source of bias. It is interesting, however, as biased RNA-sensing receptor agonists have not been reported. Such biased agonists could be very useful clinically. RNA-sensing receptor agonists are attracting much interest for the treatment of cancer. The problem has been their propensity to induce inflammation through induction of TNFα and IL6. Our data suggest that this could be avoided through biased agonists such as 5′ppp-miR combo.
Further studies will be needed to determine if our findings translate into human cardiac fibroblasts. Previous work in the laboratory demonstrated that miR combo reprogrammed cardiac fibroblasts derived from mice, dogs, pigs, and humans.38 Considering that the effects of miR combo appear to be conserved, the implication is that the mechanisms by which miR combo reprograms fibroblasts are similarly conserved. Thus, it would seem likely that 5′ppp modification of miR combo would similarly improve reprogramming efficacy in human cardiac fibroblasts. The Chiono group has also successfully reprogrammed human cardiac fibroblasts with miR combo.39 Work from the group also suggests methods for delivery and testing of 5′ppp-miR combo. The 5′ppp-miR combo could be delivered systemically with modifications to the RNA to prevent nuclease degradation. The other option would be a carrier and the aforementioned group has shown that certain lipoprotein complexes improve miR combo efficacy.40 Thus, there is a possibility by combining a modified miR combo with an optimized carrier that reprogramming efficacy could be improved still further. With respect to moving 5′ppp-miR combo forward clinically, future testing could be carried out in cardiac tissue mimics,41 which represent a different approach to testing and screening drugs prior to pre-clinical models such as the pig.
In summary, this study highlights the potential of modifying miRs for clinical applications. In addition, by modulating the innate immune response, and understanding the role of subsequent signaling pathways, the study sheds light on potential targets, such as IFNβ, to further improve cardiac regeneration strategies.
Materials and methods
In vitro synthesis of 5′ppp-miR
MiRs with a 5′ triphosphate end were produced via a HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs, E2040S/E2050S). We initially created a DNA template by annealing matching oligonucleotides. The annealing solution was incubated at 90°C–95°C for 1 min and gradually cooled to room temperature over a minimum of 30 min using a Thermocycler. Each oligonucleotide template featured a T7 RNA polymerase promoter, followed by the miR sequence. The miR combo miRs miR-1, miR-133, miR-208a, and miR-408 each have two forms labeled 3p and 5p. Both 3p and 5p forms were tested in this study.
miR-1-3p Sense (S): ata 5′cat act tct tta cat tcc act ata gtg agt cgt att a3′, miR 1-3p antisense (AS): 5′taa tac gac tca cta tag tgg aat gta aag aag tat gta t3′
miR-1-5p (S): 5′tgg gca tat aaa gaa gta tgt ttc tat agt gag tcg tat ta3′, (AS):5′taa tac gac tca cta tag aaa cat act tct tta tat gcc ca3′, miR-208a-3p sense (S): 5′taa tac gac tca cta tag ata aga cga gca aaa agc ttg t3’; (AS): 5′ aca agc ttt ttg ctc gtc tta tct ata gtg agt cgt att a3′.
miR-208a-5p (S): 5′taa tac gac tca cta tag gag ctt ttg gcc cgg gtt ata c3′, (AS): 5′gta taa ccc ggg cca aaa gct cct ata gtg agt cgt att a3′.
miR-133a-3p (S): 5′taa tac gac tca cta tag ttt ggt ccc ctt caa cca gct g3′, (AS): 5′cag ctg gtt gaa ggg gac caa act ata gtg agt cgt att a3′.
miR-133a-5p (S): 5′taa tac gac tca cta tag gct ggt aaa atg gaa cca aat3′, (AS): 5′att tgg ttc cat ttt acc agc cta tag tga gtc gta tta3′.
miR-499a-3p (S): 5′taa tac gac tca cta tag gaa cat cac agc aag tct gtg ct3′, (AS): 5′agc aca gac ttg ctg tga tgt tcc tat agt gag tcg tat ta3′.
miR-499a-5p (S): 5′taa tac gac tca cta tag tta aga ctt gca gtg atg ttt3′, (AS): 5′aaa cat cac tgc aag tct taa cta tag tga gtc gta tta3′.
miR non-targeting negative control (S):5′ taa tac gac tca cta tag ggt tcg tac gta cac tgt tca3′, (AS): 5′tga aca gtg tac gta cga acc cta tag tga gtc gta3′.
Transcription was performed according to the manufacturer’s instructions using 1 μg of DNA template and 2 μL of T7 RNA polymerase (NEB). The resultant RNA was isolated using Monarch RNA Cleanup Kit (New England Biolabs, T2040L). Size and integrity of the miR was confirmed by gel electrophoresis. An outline of the procedure is shown in Figure 1A.
Cell isolation and culture
Neonatal cardiac fibroblasts were isolated from 2-day-old mice (C57BL/6) according to the protocol established in Jayawardena et al.6 After extraction, fibroblasts were cultivated in growth medium consisting of DMEM (ATCC, 30–2002) augmented with 15% v/v fetal bovine serum (Genessee), and 1% v/v penicillin/streptomycin (GIBCO, 15140-122). Fibroblasts were passaged at 70%–80% confluence using 0.05% w/v trypsin solution (GIBCO, Waltham, MA; catalog number 25300-054). All research was sanctioned by the Division of Laboratory Animals (DLAR) at Duke University, as well as the Duke Institutional Animal Care and Use Committee (IACUC). The approved protocol number is A035-22-02.
miR and siRNA transfection
Freshly isolated fibroblasts were designated as passage zero, with all subsequent experimental procedures conducted on cells at passage two. For all investigations, cells were plated at a density of 5,000 cells/cm2 in growth medium. After a 24-h interval, the cells were transfected with transfection reagent (Dharmafect-I, Horizon Discovery, T-2001-03) with 5 nmol miR (consisting of unmodified and 5′ppp-modified non-targeting control and miR combo miRs). Where appropriate, 5nmol siRNA from Horizon Discovery Bio (non-targeting, D-001810-03-05; siIFNβ L-043699-00-0005; siGbp7, L-061204-01-0005; siGbp5, L-054703-00-0005; siCcl5, L-047423-00-0005; siCxcl10, L-042605-01-0005; siRig1, L-065328-00-0005; and siTLR3, L-059850-00-0005) was transfected alongside the miRs. Transfections were performed according to the guidelines provided by the manufacturer. Transfection complexes were subsequently removed after 24 h and the cells were cultured in growth media for the remainder of the experiment.42
Treatment with neutralizing antibodies
At defined time points after transfection (5 or 24 h), 5 μg/mL IFNβ neutralizing antibody (BioLegend, 508108)43 or 5 μg/mL IgG Isotype Control (BioLegend, 400940) was added directly to the culture media.44 After a 3-day incubation period, the media was collected. Debris was removed by centrifugation (12,000 × g, 5 min, 4°C).
qPCR
Total RNA was extracted using Quick-RNA MiniPrep Kit according to the manufacturer’s instructions (Zymo Research, Irvine, CA, USA). Total RNA was converted to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Waltham, MA, USA) in a 40-μL reaction according to the manufacturer’s instructions. The resulting cDNA (4 μL) was used in a standard qPCR reaction involving FAM conjugated gene specific primers and TaqMan Gene Expression Master Mix (Applied Biosystems, Waltham, MA, USA). The following qPCR primers were acquired from Thermo Fisher (Waltham, MA, USA) Gapdh(Mm99999915_m1),Tnni3(Mm00437164_m1), Actn2 (Mm00473657_m1), Myh6 (Mm00440359_m1), Tnfα (Mm00440359_m1), Il6 (Mm00446190_m1), Ifnb1 (Mm00439552_s1), (Mm00523797_m1), Tlr3 (Mm01207404_m1), Rig1 (Mm01216853_m1). Expression values were calculated by normalizing to the housekeeping gene Gapdh.
Immunofluorescence
Cells were fixed with 2% v/v paraformaldehyde (EMS, Durham, NC, USA) as described.45 The fixed cells were blocked with antibody buffer (5% w/v BSA and 0.1% v/v Tween 20 in PBS) for 1 h at room temperature and then incubated with an Actn2 antibody (Sigma, A7811, 1:100) in antibody buffer at 4°C overnight. Cells were then rinsed three times with PBS and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibodies (Invitrogen, goat anti-mouse 594 nm), at a 1:500 dilution in the antibody buffer. Nuclear staining was achieved by a 30-min incubation at room temperature with 4′, 6-diamidino-2-phenylindole (DAPI) at a concentration of 1 μg/mL in antibody buffer. After washing with PBS to remove any unbound complexes, the cells were visualized, and their immunofluorescence was assessed using a Zeiss Axiovert 200 inverted microscope. A blinded investigator captured six random images per well.
Luciferase reporter assay
The NF-κB luciferase reporter plasmid was procured from Promega (pGL4.32[luc2P/NF-κB-RE/Hygro], E8491). Luciferase reporter plasmids pGL4-IRF3-RE and pGL4-IRF7-RE incorporating five copies of either the IRF3 or IRF7 response element, were generated by digesting pGL4.32 plasmid with BmtI and HandIII and treating it with T4 DNA polymerase.
IRF7-RE S: 5′ acc gct agc gaa aat gaa aat gaa aat gaa aat gaa aat gaaaat gaa aat gaa aat gaa aat gaa aat aag ctt ggg 3′
IRF7-RE AS: 5′ ccc aag ctt att ttc att ttc att ttc att ttc att ttc att ttc att ttc att ttc att ttc att ttc gct agc ggt 3′
IRF3-RE S: 5′acc gct agc gaa agg gaa agg gaa agg gaa agg gaa agg gaa agg gaa agg gaa agg gaa agg gaa agg aag ctt ggg 3′
IRF3-RE AS: 5′ ccc aag ctt cct ttc cct ttc cct ttc cct ttc cct ttc cct ttc cct ttc cct ttc cct ttc cct ttc gct agc ggt 3′
In transient transfections, cells were co-transfected with the specified plasmids and the pRL-SV40 plasmid that carries the Renilla luciferase gene (5:1 ratio firefly luciferase plasmid to Renilla luciferase plasmid). Cells underwent lysis 6 h or 24 h post-transfection by application of the passive lysis buffer, a component of the dual-luciferase assay kit. (Promega, Dual-Luciferase Reporter Assay System, E1910). A luminometer was utilized to assess luciferase activity. The outcome of this evaluation is represented as the ratio of the luciferase to Renilla values.
RNA-seq
Neonatal cardiac fibroblasts were transfected with unmodified or 5′ppp-modified miRs, as described. Four days after transfection, total RNA was extracted using the Quick-RNA MiniPrep Kit (Zymo Research, 11–328). High-throughput sequencing was performed by the Duke Genomic Core. Libraries were generated with a HiSeq 4000 kit (Illumina). Libraries were pooled and run in duplicate (50 base pairs paired-end) with an Illumina HiSeq 4000. Sequencing depth was >25 × 106 individual reads per sample. Individual bioinformatics programs within the Galaxy suite were used to analyze gene expression as described.46
ELISA
IFNβ and IFNγ were measured by ELISA Kits (R&D Systems, Mouse IFN-beta DuoSet ELISA, DY8234-05; Mouse IFN-gamma DuoSet ELISA, DY485-05) according to the manufacturer’s protocol. Media was assayed and the amount of IFNβ and IFNγ in pg/mL in the culture media was determined via a standard curve.
Statistics
Independent t tests were used for experiments with two groups. For experiments with more than two groups, ANOVA was used with Tukey’s correction to determine significance between groups.
Data and materials availability
All data needed to evaluate the conclusions in the paper are present in the paper. The raw sequencing files for the RNA-seq are publically available from the NIH Single-Read Archive: PRJNA1067687.
Acknowledgments
Work in this study was funded by the Edna & Fred Mandel Jr Foundation (no agency ID).
Author contributions
X.W., data curation, investigation, formal analysis, writing – original draft, writing – review & editing. S.S.B., investigation, validation. R.E.P., writing – review & editing. V.J.D., supervision. C.P.H., formal analysis, funding acquisition, investigation, methodology, project administration, supervision, validation, writing – review & editing.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102160.
References
- 1.Garry G.A., Olson E.N. Cardiac Reprogramming: Toward a Total Eclipse of the Failing Heart. Circulation. 2023;147:239–241. doi: 10.1161/CIRCULATIONAHA.122.062691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Savarese G., Lund L.H. Global Public Health Burden of Heart Failure. Card. Fail. Rev. 2017;3:7–11. doi: 10.15420/cfr.2016:25:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Towbin J.A., Bowles N.E. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 4.Prabhu S.D., Frangogiannis N.G. The Biological Basis for Cardiac Repair After Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadahiro T., Yamanaka S., Ieda M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ. Res. 2015;116:1378–1391. doi: 10.1161/CIRCRESAHA.116.305374. [DOI] [PubMed] [Google Scholar]
- 6.Jayawardena T.M., Finch E.A., Zhang L., Zhang H., Hodgkinson C.P., Pratt R.E., Rosenberg P.B., Mirotsou M., Dzau V.J. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ. Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu J., Hodgkinson C.P., Pratt R.E., Lee J., Sullenger B.A., Dzau V.J. Enhancing cardiac reprogramming via synthetic RNA oligonucleotides. Mol. Ther. Nucleic Acids. 2021;23:55–62. doi: 10.1016/j.omtn.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baksh S.S., Hu J., Pratt R.E., Dzau V.J., Hodgkinson C.P. Rig1 receptor plays a critical role in cardiac reprogramming via YY1 signaling. Am. J. Physiol. Cell Physiol. 2023;324:C843–C855. doi: 10.1152/ajpcell.00402.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodgkinson C.P., Pratt R.E., Kirste I., Dal-Pra S., Cooke J.P., Dzau V.J. Cardiomyocyte Maturation Requires TLR3 Activated Nuclear Factor Kappa B. Stem Cell. 2018;36:1198–1209. doi: 10.1002/stem.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson R.A., DeVita V.T., Levy H.B., Baron S., Hubbard S.P., Levine A.S. A phase I-II trial of multiple-dose polyriboinosic-polyribocytidylic acid in patieonts with leukemia or solid tumors. J. Natl. Cancer Inst. 1976;57:599–602. doi: 10.1093/jnci/57.3.599. [DOI] [PubMed] [Google Scholar]
- 14.Krown S.E., Kerr D., Stewart W.E., 2nd, Field A.K., Oettgen H.F. Phase I trials of poly(I,C) complexes in advanced cancer. J. Biol. Response Mod. 1985;4:640–649. [PubMed] [Google Scholar]
- 15.Liu A., Wang X. The Pivotal Role of Chemical Modifications in mRNA Therapeutics. Front. Cell Dev. Biol. 2022;10:901510. doi: 10.3389/fcell.2022.901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki T., Kawai T. Toll-like receptor signaling pathways. Front. Immunol. 2014;5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abbas Y.M., Pichlmair A., Górna M.W., Superti-Furga G., Nagar B. Structural basis for viral 5'-PPP-RNA recognition by human IFIT proteins. Nature. 2013;494:60–64. doi: 10.1038/nature11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y., An C., Zhang X., Li K., Cheng F., Cui B., Song Z., Liu D., Zhang J., He Q., et al. A Novel Targeted RIG-I Receptor 5'Triphosphate Double Strain RNA-Based Adjuvant Significantly Improves the Immunogenicity of the SARS-CoV-2 Delta-Omicron Chimeric RBD-Dimer Recombinant Protein Vaccine. Viruses. 2023;15 doi: 10.3390/v15051099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni R.R., Rasheed M.A.U., Bhaumik S.K., Ranjan P., Cao W., Davis C., Marisetti K., Thomas S., Gangappa S., Sambhara S., Murali-Krishna K. Activation of the RIG-I pathway during influenza vaccination enhances the germinal center reaction, promotes T follicular helper cell induction, and provides a dose-sparing effect and protective immunity. J. Virol. 2014;88:13990–14001. doi: 10.1128/JVI.02273-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael Lavigne G., Russell H., Sherry B., Ke R. Autocrine and paracrine interferon signalling as 'ring vaccination' and 'contact tracing' strategies to suppress virus infection in a host. Proc. Biol. Sci. 2021;288:20203002. doi: 10.1098/rspb.2020.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil S., Fribourg M., Ge Y., Batish M., Tyagi S., Hayot F., Sealfon S.C. Single-cell analysis shows that paracrine signaling by first responder cells shapes the interferon-beta response to viral infection. Sci. Signal. 2015;8:ra16. doi: 10.1126/scisignal.2005728. [DOI] [PubMed] [Google Scholar]
- 23.Cao X. New DNA-sensing pathway feeds RIG-I with RNA. Nat. Immunol. 2009;10:1049–1051. doi: 10.1038/ni1009-1049. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez Pulido M., Sáiz M. Molecular Mechanisms of Foot-and-Mouth Disease Virus Targeting the Host Antiviral Response. Front. Cell. Infect. Microbiol. 2017;7:252. doi: 10.3389/fcimb.2017.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto K., Akiyama M., Tamura F., Isomi M., Yamakawa H., Sadahiro T., Muraoka N., Kojima H., Haginiwa S., Kurotsu S., et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell. 2018;22:91–103.e5. doi: 10.1016/j.stem.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Qian L., Srivastava D. Direct cardiac reprogramming: from developmental biology to cardiac regeneration. Circ. Res. 2013;113:915–921. doi: 10.1161/CIRCRESAHA.112.300625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur K., Hadas Y., Kurian A.A., Żak M.M., Yoo J., Mahmood A., Girard H., Komargodski R., Io T., Santini M.P., et al. Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol. Ther. 2021;29:3042–3058. doi: 10.1016/j.ymthe.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohamed T.M.A., Stone N.R., Berry E.C., Radzinsky E., Huang Y., Pratt K., Ang Y.S., Yu P., Wang H., Tang S., et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Y., Londono P., Cao Y., Sharpe E.J., Proenza C., O'Rourke R., Jones K.L., Jeong M.Y., Walker L.A., Buttrick P.M., et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H., Dickson M.E., Kim M.S., Bassel-Duby R., Olson E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA. 2015;112:11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamakawa H., Muraoka N., Miyamoto K., Sadahiro T., Isomi M., Haginiwa S., Kojima H., Umei T., Akiyama M., Kuishi Y., et al. Fibroblast Growth Factors and Vascular Endothelial Growth Factor Promote Cardiac Reprogramming under Defined Conditions. Stem Cell Rep. 2015;5:1128–1142. doi: 10.1016/j.stemcr.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Wang L., Vaseghi H.R., Liu Z., Lu R., Alimohamadi S., Yin C., Fu J.D., Wang G.G., Liu J., Qian L. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y., Liu Z., Welch J.D., Gao X., Wang L., Garbutt T., Keepers B., Ma H., Prins J.F., Shen W., et al. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell. 2019;25:149–164.e9. doi: 10.1016/j.stem.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y., Aryal S., Geng X., Zhou X., Fast V.G., Zhang J., Lu R., Zhou Y. TBX20 Improves Contractility and Mitochondrial Function During Direct Human Cardiac Reprogramming. Circulation. 2022;146:1518–1536. doi: 10.1161/CIRCULATIONAHA.122.059713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 36.Liu G., Gack M.U. Distinct and Orchestrated Functions of RNA Sensors in Innate Immunity. Immunity. 2020;53:26–42. doi: 10.1016/j.immuni.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicens Q., Kieft J.S. Thoughts on how to think (and talk) about RNA structure. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2112677119. e2112677119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baksh S.S., Hodgkinson C.P. Conservation of miR combo based direct cardiac reprogramming. Biochem. Biophys. Rep. 2022;31:101310. doi: 10.1016/j.bbrep.2022.101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paoletti C., Marcello E., Melis M.L., Divieto C., Nurzynska D., Chiono V. Cardiac Tissue-like 3D Microenvironment Enhances Route towards Human Fibroblast Direct Reprogramming into Induced Cardiomyocytes by microRNAs. Cells. 2022;11:800. doi: 10.3390/cells11050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicoletti L., Paoletti C., Tarricone G., Andreana I., Stella B., Arpicco S., Divieto C., Mattu C., Chiono V. Lipoplexes for effective in vitro delivery of microRNAs to adult human cardiac fibroblasts for perspective direct cardiac cell reprogramming. Nanomedicine. 2022;45:102589. doi: 10.1016/j.nano.2022.102589. [DOI] [PubMed] [Google Scholar]
- 41.Visone R., Paoletti C., Cordiale A., Nicoletti L., Divieto C., Rasponi M., Chiono V., Occhetta P. In Vitro Mechanical Stimulation to Reproduce the Pathological Hallmarks of Human Cardiac Fibrosis on a Beating Chip and Predict The Efficacy of Drugs and Advanced Therapies. Adv. Healthc. Mater. 2024;13:e2301481. doi: 10.1002/adhm.202301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jayawardena T., Mirotsou M., Dzau V.J. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol. Biol. 2014;1150:263–272. doi: 10.1007/978-1-4939-0512-6_18. [DOI] [PubMed] [Google Scholar]
- 43.Creasy B.M., McCoy K.L. Cytokines regulate cysteine cathepsins during TLR responses. Cell. Immunol. 2011;267:56–66. doi: 10.1016/j.cellimm.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He C., Jiang B., Wang M., Ren P., Murtada S.I., Caulk A.W., Li G., Qin L., Assi R., Lovoulos C.J., et al. mTOR inhibition prevents angiotensin II-induced aortic rupture and pseudoaneurysm but promotes dissection in Apoe-deficient mice. JCI Insight. 2022;7:e155815. doi: 10.1172/jci.insight.155815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X., Hodgkinson C.P., Lu K., Payne A.J., Pratt R.E., Dzau V.J. Selenium Augments microRNA Directed Reprogramming of Fibroblasts to Cardiomyocytes via Nanog. Sci. Rep. 2016;6:23017. doi: 10.1038/srep23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hodgkinson C.P., Gomez J.A., Baksh S.S., Payne A., Schmeckpeper J., Pratt R.E., Dzau V.J. Insights from molecular signature of in vivo cardiac c-Kit(+) cells following cardiac injury and beta-catenin inhibition. J. Mol. Cell. Cardiol. 2018;123:64–74. doi: 10.1016/j.yjmcc.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fourteen days after transfection days, cells were videoed for evidence of spontaneous beating. N=3. Representative videos are shown. Unmodified miR combo.
Fourteen days after transfection days, cells were videoed for evidence of spontaneous beating. N=3. Representative videos are shown. 5’ppp-miR combo.