Abstract
Background
In metastatic colorectal cancer (mCRC), KRAS mutations are often associated with poorer survival; however, the prognostic impact of specific point mutations is unclear. In the phase III SUNLIGHT trial, trifluridine/tipiracil (FTD/TPI) plus bevacizumab significantly improved overall survival (OS) versus FTD/TPI alone. We assessed the impact of KRASG12 mutational status on OS in SUNLIGHT.
Patients and methods
In the global, open-label, randomized, phase III SUNLIGHT trial, adults with mCRC who had received no more than two prior chemotherapy regimens were randomized 1 : 1 to receive FTD/TPI alone or FTD/TPI plus bevacizumab. In this post hoc analysis, OS was assessed according to the presence or absence of a KRASG12 mutation in the overall population and in patients with RAS-mutated tumors.
Results
Overall, 450 patients were analyzed, including 302 patients in the RAS mutation subgroup (214 with a KRASG12 mutation and 88 with a non-KRASG12RAS mutation). In the overall population, similar OS outcomes were observed in patients with and without a KRASG12 mutation [median 8.3 and 9.2 months, respectively; hazard ratio (HR) 1.09, 95% confidence interval (CI) 0.87-1.4]. Similar OS outcomes were also observed in the subgroup analysis of patients with a KRASG12 mutation versus those with a non-KRASG12RAS mutation (HR 1.03, 95% CI 0.76-1.4). FTD/TPI plus bevacizumab improved OS compared with FTD/TPI alone irrespective of KRASG12 mutational status. Among patients with a KRASG12 mutation, the median OS was 9.4 months with FTD/TPI plus bevacizumab versus 7.2 months with FTD/TPI alone (HR 0.67, 95% CI 0.48-0.93), and in patients without a KRASG12 mutation, the median OS was 11.3 versus 7.1 months, respectively (HR 0.59, 95% CI 0.43-0.81).
Conclusions
The presence of a KRASG12 mutation had no detrimental effect on OS among patients treated in SUNLIGHT. The benefit of FTD/TPI plus bevacizumab over FTD/TPI alone was confirmed independently of KRASG12 status.
Key words: bevacizumab, KRAS mutation, metastatic colorectal cancer, overall survival, phase III, trifluridine/tipiracil
Highlights
-
•
Patients with mCRC benefit from third-line treatment with FTD/TPI irrespective of KRASG12 mutational status.
-
•
OS is improved in patients receiving FTD/TPI plus bevacizumab versus FTD/TPI, independently of KRASG12 mutational status.
-
•
KRAS mutations are not predictive of clinical outcomes with FTD/TPI as monotherapy or in combination with bevacizumab.
Introduction
Activating mutations in members of the RAS gene family, comprising the KRAS, NRAS, and HRAS viral oncogene homologs, are common in human cancers, including colorectal cancer (CRC).1 KRAS proteins translated from mutated KRAS genes are thought to be defective in their interactions with guanosine triphosphatase–activating proteins, leading to constitutive activation of KRAS and disruption of multiple downstream cellular pathways, including those involved in cell survival and proliferation.2 Mutations in KRAS are present in ∼40% of CRC cases and most commonly involve a point mutation in exon 2 at glycine residues 12 (KRASG12; ∼80%) or 13 (KRASG13; ∼20%).3,4 There are 15 different KRASG12 mutations, of which the most common in CRC is G12D (glycine to aspartic acid), followed by G12V (glycine to valine).3, 4, 5
Patients with metastatic CRC (mCRC) harboring KRAS mutations have worse survival outcomes than individuals with RAS and BRAF wild-type tumors.6, 7, 8 Although in vitro data have shown different oncogenic activities for different KRAS codon mutations,9 there are sparse clinical data to definitively indicate whether one KRAS mutation is more prognostic than another. Results from a pooled analysis of five randomized controlled trials (RCTs) in mCRC reported an association between KRAS G12C (glycine to cysteine) and G13D tumors and poor survival.6 KRAS and NRAS mutations, however, are predictive of worse treatment outcomes with the epidermal growth factor receptor (EGFR) inhibitors cetuximab and panitumumab.7,10, 11, 12 In patients carrying a wild-type KRAS gene copy, the binding of anti-EGFR antibodies to the external part of the receptor induces conformational changes that directly inhibit tyrosine kinase activity and downstream signaling. KRAS mutations, however, induce constitutive activation of the intracellular domain of the KRAS protein, thus preventing the inhibition induced by anti-EGFR antibodies. Accordingly, national and international guidelines recommend that all patients with mCRC undergo testing for RAS (KRAS and NRAS) mutations in certified laboratories before initiating treatment.13, 14, 15, 16, 17, 18
Trifluridine/tipiracil (FTD/TPI) is an oral combination of trifluridine (FTD), a cytotoxic thymidine-based nucleoside analog, and tipiracil hydrochloride (TPI), a thymidine phosphorylase inhibitor that prevents degradation of and improves systemic exposure to FTD.19 FTD/TPI is approved as a single agent or in combination with bevacizumab for patients with mCRC previously treated with fluoropyrimidine-, oxaliplatin-, and irinotecan-based chemotherapy, an anti-vascular endothelial growth factor biologic therapy, and, if RAS wild-type, an anti-EGFR therapy. FTP/TPI was approved as monotherapy based on the results of the phase III RECOURSE trial of FTP/TPI versus placebo.20 Subsequently, FTD/TPI in combination with bevacizumab was approved based on data from the randomized phase III SUNLIGHT study, which showed statistically significant improvements in overall survival (OS; primary endpoint) and progression-free survival (PFS; secondary endpoint) with FTD/TPI plus bevacizumab versus FTD/TPI alone.21
In both RECOURSE and SUNLIGHT, the survival benefits with FTP/TPI versus placebo and FTP/TPI plus bevacizumab versus FTP/TPI alone were observed in all prespecified subgroups, including those with and without KRAS-mutated disease.20, 21, 22, 23 In line with these findings, guidelines advocate these regimens as third-line treatment options for patients with mCRC, regardless of KRAS mutation status.15,16,18 Recently, questions have been asked regarding the relevance of KRAS mutations to predicting outcomes with FTD/TPI, and they have received discrepant answers.24, 25, 26, 27, 28 The aim of this post hoc analysis was to use data from the SUNLIGHT trial to assess the potential impact of KRASG12 mutations on survival when patients are treated with FTD/TPI plus bevacizumab, compared with the absence of KRASG12 mutations or the presence of non-KRASG12 RAS mutations, and to review the findings in the context of available literature.
Patients and methods
Study design and patients
Full details of the SUNLIGHT study design and eligibility criteria have been published previously.21,29 Briefly, SUNLIGHT was a global, open-label, randomized, phase III trial that enrolled adults with histologically confirmed, unresectable adenocarcinoma of the colon or rectum and known RAS status who had received no more than two prior chemotherapy regimens and had progressed on or were intolerant to their last line of treatment. Previous treatment included a fluoropyrimidine, irinotecan, oxaliplatin, anti-vascular endothelial growth factor, and/or (in patients with RAS wild-type tumors) anti-EGFR antibody therapy. Patients were randomized 1 : 1 to receive FTD/TPI 35 mg/m2 orally twice daily on days 1-5 and 8-12 with or without bevacizumab 5 mg/kg intravenously on days 1 and 15 of each 28-day cycle. Randomization was stratified by geographic region (North America versus Europe versus the rest of the world), time since diagnosis of metastatic disease (<18 months versus ≥18 months), and RAS status (wild-type versus mutant). RAS mutational status was tested locally, and the data were reported by individual study sites. Treatment continued until disease progression, unacceptable toxicity, or withdrawal of consent. The primary endpoint was OS, defined as time from randomization to death from any cause.
The SUNLIGHT study was carried out in accordance with the principles of the Declaration of Helsinki and the International Council for Harmonisation Good Clinical Practice guidelines. The study protocol was approved by the institutional review board(s) and/or independent ethics committee(s) at each participating site. All enrolled patients provided written informed consent.
Post hoc and statistical analysis
OS in the full analysis set (FAS; all randomized patients with confirmed KRAS mutational status) was assessed both across and within treatment groups according to the presence or absence of a KRASG12 mutation (presence of a KRASG12 mutation versus RAS wild-type or a non-KRASG12 RAS mutation). Additionally, a subgroup analysis was conducted in patients with RAS-mutated tumors to assess the OS benefit with FTD/TPI plus bevacizumab versus FTD/TPI alone stratified by KRASG12 mutational status (presence of a KRASG12 mutation versus a non-KRASG12 RAS mutation). Median OS was assessed using Kaplan–Meier methodology, hazard ratios (HRs) were calculated using Cox regression models, and confidence intervals (CIs) were calculated using Brookmeyer and Crowley’s methodology.
A stratified log-rank test with a two-sided significance level of 0.05 was used to compare the distributions of OS between the RAS mutation subgroups and to derive P values. An unstratified Cox regression model with trial group as a predictor variable was fitted for each RAS mutation subgroup, and the HR and associated 95% CI were determined for the assigned treatment.
Results
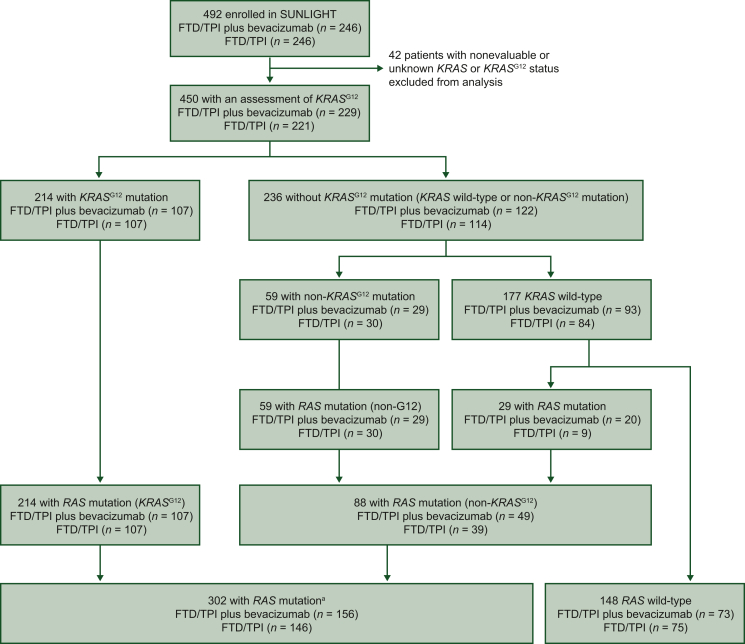
A total of 450 patients were included in the FAS population (214 with a KRASG12 mutation and 236 with RAS wild-type tumors or with a non-KRASG12 RAS mutation), and 302 patients were included in the RAS mutation subpopulation (214 with a KRASG12 mutation and 88 with a non-KRASG12 RAS mutation) (Figure 1). Baseline patient demographics among the different subgroups are summarized in Table 1.
Figure 1.
Patient analysis flowchart.aAmong the 302 patients with RAS mutations, 2 patients assessed as having wild-type disease were conserved in the analysis as having a KRASG12 mutation. FTD/TPI, trifluridine and tipiracil.
Table 1.
Baseline patient demographics and clinical characteristics
| RAS mutation (n = 302) | KRASG12 mutation (n = 214) | KRASG12C mutation (n = 24) | Non-KRASG12RAS mutation (n = 88) | RAS wild-type (n = 148) | RAS wild-type or a non-KRASG12RAS mutation (n = 236) | |
|---|---|---|---|---|---|---|
| Median age, years | 63 | 64 | 60.5 | 62 | 62 | 62 |
| Male, n (%) | 153 (50.7) | 112 (52.3) | 12 (50.0) | 41 (46.6) | 80 (54.1) | 121 (51.3) |
| Race or ethnic group, n (%) | ||||||
| White | 266 (88.1) | 186 (86.9) | 21 (87.5) | 80 (90.9) | 133 (89.9) | 213 (90.3) |
| Black | 6 (2.0) | 6 (2.8) | 0 | 0 | 1 (0.7) | 1 (0.4) |
| Asian | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.4) |
| Other | 11 (3.6) | 8 (3.7) | 3 (12.5) | 3 (3.4) | 2 (1.4) | 5 (2.1) |
| Missing | 19 (6.3) | 14 (6.5) | 0 | 5 (5.7) | 11 (7.4) | 16 (6.8) |
| Geographic region, n (%) | ||||||
| North America | 8 (2.7) | 6 (2.8) | 1 (4.2) | 2 (2.3) | 6 (4.1) | 8 (3.4) |
| European Union | 199 (65.9) | 140 (65.4) | 17 (70.8) | 59 (67.0) | 98 (66.2) | 157 (66.5) |
| Rest of the world | 95 (31.5) | 68 (31.8) | 6 (25.0) | 27 (30.7) | 44 (29.7) | 71 (30.1) |
| Primary diagnosis, n (%) | ||||||
| Colon cancer | 220 (72.9) | 162 (75.7) | 18 (75.0) | 58 (65.9) | 108 (73.0) | 166 (70.3) |
| Rectal cancer | 82 (27.1) | 52 (24.3) | 6 (25.0) | 30 (34.1) | 40 (27.0) | 70 (29.7) |
| Location of primary tumor, n (%) | ||||||
| Right side | 91 (30.1) | 65 (30.4) | 7 (29.2) | 26 (29.6) | 35 (23.6) | 61 (25.8) |
| Left side | 211 (69.9) | 149 (69.6) | 17 (70.8) | 62 (70.4) | 113 (76.4) | 175 (74.2) |
| Number of metastatic sites, n (%) | ||||||
| 1/2 | 182 (60.3) | 132 (61.7) | 14 (58.3) | 50 (56.8) | 90 (60.8) | 140 (59.3) |
| ≥3 | 120 (39.7) | 82 (38.3) | 10 (41.7) | 38 (43.2) | 58 (39.2) | 96 (40.7) |
| ECOG performance status, n (%) | ||||||
| 0 | 129 (42.7) | 93 (43.5) | 13 (54.2) | 36 (40.9) | 74 (50.0) | 110 (46.6) |
| 1 | 173 (57.3) | 121 (56.5) | 11 (45.8) | 52 (59.1) | 73 (49.3) | 125 (53.0) |
| 2 | 0 | 0 | 0 | 0 | 1 (0.7) | 1 (0.4) |
ECOG, Eastern Cooperative Oncology Group.
Overall, no significant difference in survival was observed between patients with RAS wild-type (n = 148) and RAS-mutated tumors (n = 302); median OS was 9.3 months versus 8.5 months, respectively (HR 1.12, 95% CI 0.88-1.4). FTD/TPI plus bevacizumab was associated with improved OS compared with FTD/TPI alone in both subgroups. In the population with a RAS mutation, median OS (95% CI) was 10.1 months (8.9-11.3 months) with FTD/TPI plus bevacizumab versus 7.1 months (6.3-8.5 months) with FTD/TPI alone (HR 0.61, 95% CI 0.46-0.81). In the population with RAS wild-type disease, median OS (95% CI) was 11.4 months (8.6-14.9 months) with FTD/TPI plus bevacizumab versus 7.4 months (5.9-10.9 months) with FTD/TPI alone (HR 0.66, 95% CI 0.44-0.99).
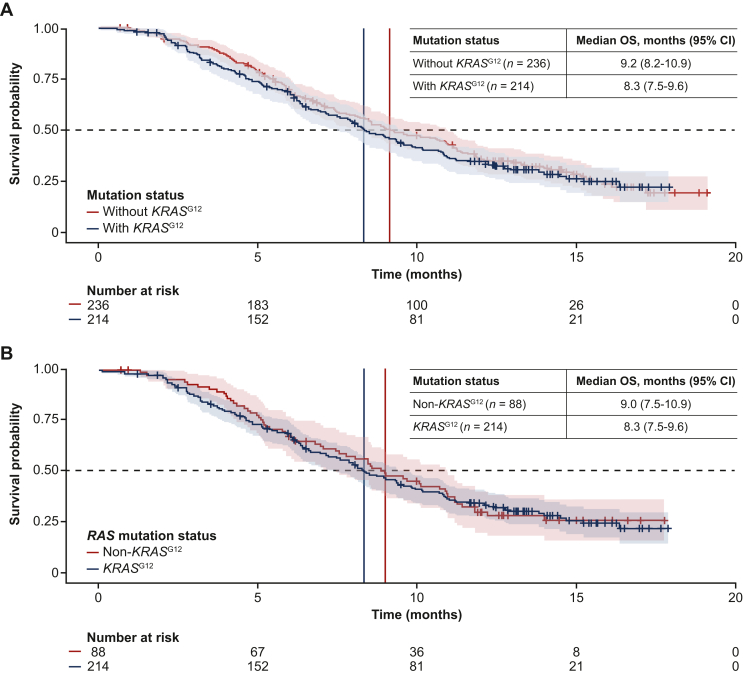
No significant difference in survival was observed between patients with and without KRASG12 mutations in the FAS (HR 1.09, 95% CI 0.87-1.4; Figure 2A), though there was a small numerical difference in the median OS (95% CI) between these two populations [8.3 months (7.5-9.6 months) versus 9.2 months (8.2-10.9 months), respectively]. Similar results were observed when the analysis was restricted to the RAS mutation subgroup (HR 1.03, 95% CI 0.76-1.4; Figure 2B). Of the 214 patients with KRASG12 mutations, 24 had confirmed KRASG12C mutations (Table 1). Median OS (95% CI) in patients with KRASG12C mutations was 8.5 months (6.0-9.6 months).
Figure 2.
OS according to KRASG12mutation status. (A) FAS population (FTD/TPI plus bevacizumab and FTD/TPI alone; n = 450). (B) RAS mutation population (n = 302). CI, confidence interval; FAS, full analysis set; FTD/TPI, trifluridine and tipiracil; OS, overall survival.
No significant differences in OS were observed according to KRASG12 mutation status within the individual treatment arms (FTD/TPI alone or FTD/TPI plus bevacizumab) in either the FAS or RAS mutation population (Figure 3).
Figure 3.
Forest plot of OS according to KRASG12mutation status in the individual treatment arms (FTD/TPI alone and FTD/TPI plus bevacizumab). (A) FAS population. (B) RAS mutation population. Interaction P values were not statistically significant [P = 0.6634 in the FAS population (versus patients without a KRASG12 mutation) and P = 0.3074 in the RAS mutation population (versus patients with a non-KRASG12RAS mutation)]. CI, confidence interval; FAS, full analysis set; FTD/TPI, trifluridine and tipiracil; HR, hazard ratio; NE, not estimable; OS, overall survival.
In the FAS, FTD/TPI plus bevacizumab was associated with improved OS compared with FTD/TPI alone, irrespective of KRASG12 mutational status (Figure 4). In the population with a KRASG12 mutation, median OS (95% CI) was 9.4 months (8.2-10.9 months) with FTD/TPI plus bevacizumab versus 7.2 months (6.3-9.1 months) with FTD/TPI alone (HR 0.67, 95% CI 0.48-0.93). In the population without a KRASG12 mutation, median OS (95% CI) was 11.3 months (9.6-14.2 months) with FTD/TPI plus bevacizumab versus 7.1 months (5.9-8.9 months) with FTD/TPI alone (HR 0.59, 95% CI 0.43-0.81).
Figure 4.
Forest plot of OS with FTD/TPI plus bevacizumab versus FTD/TPI alone according to KRASG12mutation status in the FAS population. CI, confidence interval; FAS, full analysis set; FTD/TPI, trifluridine and tipiracil; HR, hazard ratio; OS, overall survival.
Discussion
The results of this post hoc analysis of SUNLIGHT data found no evidence to suggest that the addition of bevacizumab to FTD/TPI was less effective in improving survival outcomes in any KRAS subgroup. No significant differences in survival between patients with and without KRASG12 mutations were observed in the FAS, either across treatments (both treatment arms) or within treatment arms (FTD/TPI plus bevacizumab and FTD/TPI alone), and similar effects were observed in the RAS mutation population. The improvements in survival with FTD/TPI plus bevacizumab versus FTD/TPI alone were confirmed to be independent of KRAS mutational status.
In the SUNLIGHT trial, OS outcomes in patients who received FTD/TPI alone were comparable between those with and without a KRASG12 mutation (median 7.2 versus 7.1 months). These findings are consistent with those from a meta-analysis reported by Yoshino et al., which included updated OS data from 1375 patients enrolled in three RCTs of FTD/TPI versus placebo: RECOURSE (global), TERRA (Asia), and J003 (Japan).24 The results of the meta-analysis support the OS benefit of FTD/TPI as monotherapy in patients with KRASG12 mutations, albeit potentially with a smaller magnitude compared with that observed in patients without KRASG12 mutations. While univariate analyses suggested that the presence of a KRASG12 mutation significantly reduced the OS benefit of FTD/TPI versus placebo compared with the absence of a KRASG12 mutation (HR 0.86, 95% CI 0.70-1.05 and HR 0.62, 95% CI 0.53-0.72, respectively; interaction P = 0.0206), a multivariate analysis controlling for differences in baseline characteristics showed the OS benefit was maintained in patients with and without KRASG12 mutations (HR 0.73, 95% CI 0.59-0.89 and HR 0.63, 95% CI 0.54-0.74, respectively; interaction P = 0.2939). PFS was also significantly longer with FTD/TPI versus placebo regardless of the presence of KRASG12 mutations.24
Our findings are also in agreement with the results of a systematic review and meta-analysis reported by Huang et al., which reported that FTD/TPI monotherapy was associated with improved OS and PFS irrespective of KRAS mutational status.25 The meta-analysis included data from 2903 patients treated with FTD/TPI or placebo and/or best supportive care across the same three RCTs as in the Yoshino et al. analysis, as well as three post hoc analyses and two prospective cohort studies. Analysis of OS according to KRAS mutational status, however, was limited to results from the TERRA and J003 RCTs and a post hoc analysis of RECOURSE. FTD/TPI showed significant OS benefits versus placebo/best supportive care in both KRAS wild-type (HR 0.66, 95% CI 0.55-0.79; P < 0.00001) and KRAS mutation subgroups (HR 0.75, 95% CI 0.62-0.91; P = 0.004), along with a significant PFS improvement in both subgroups (both, HR 0.47, 95% CI 0.38-0.58; P < 0.00001). Of note, the individual studies included in the meta-analyses of Yoshino et al. and Huang et al. were not designed to assess OS benefit according to KRAS mutational status.24,25 Additionally, the latter did not include data on codon-specific KRAS mutations.25
Interestingly, the results of the current analysis differ from the findings from an observational subgroup analysis conducted by van de Haar et al.26 in a real-world cohort of 960 patients, which reported shorter OS following treatment with FTD/TPI in patients with KRASG12 mutations versus those with no KRASG12 mutation or with a KRASG13 mutation.26 The same authors also conducted an exploratory post hoc analysis using data from the phase III RECOURSE study, the results of which suggested that OS was not prolonged with FTD/TPI versus placebo in patients with a KRASG12 mutation (HR 0.97, 95% CI 0.73-1.20; P = 0.85). By contrast, patients with KRASG13-mutant tumors had significantly improved OS (HR 0.29, 95% CI 0.15-0.55; P <0.001). In this analysis, KRASG12 mutations (n = 279) were reported to be predictive of reduced OS benefit with FTD/TPI versus placebo (unadjusted interaction P = 0.0031, adjusted interaction P = 0.015); however, it should be noted that identification of a significant interaction term indicates that the degree of benefit with FTD/TPI versus placebo was different between the populations with and without KRASG12 mutations, not that the KRASG12 mutation subgroup did not benefit from treatment. The analyses reported by van de Haar et al. had some limitations, including the fact that the real-world study was not designed to identify the OS benefit in patients with or without codon-specific KRAS mutations. There were also potential confounding factors due to imbalances in baseline characteristics and prior treatment, and no sensitivity analysis was conducted to account for heterogeneity bias. Additional limitations of the real-world cohort analysis include the limited quality of documentation (e.g. a lack of detail on what inclusion and exclusion criteria were considered for data selection), and small patient numbers (n = 37) in the discovery cohort that identified KRASG12 mutations as a potential biomarker of resistance. Interestingly, OS outcomes in the real-world cohort and the control arm of the RECOURSE study appear discordant and inconsistent with historical data. Median OS in the KRASG13 population, for example, was ∼15 months in the real-world cohort treated with FTD/TPI, compared with 8.7 months for the KRASG13 mutation FTD/TPI subgroup of the RECOURSE population, and just 2.9 months in the KRASG13 mutation placebo group. By comparison, median OS in a pooled analysis of 571 patients with KRASG13-mutant, chemo-refractory mCRC treated with cetuximab-based treatment was 7.6 months.30
As per the aforementioned analyses, the fact that the SUNLIGHT trial was not designed to assess the impact of codon-specific KRAS mutations is also a limitation of the current analysis, along with the post hoc nature of the subgroup analysis. Furthermore, in the analysis restricted to the RAS mutation subgroup, patients with KRASG12-mutant tumors who were treated with FTD/TPI alone comprised <10% of the study population, thereby precluding meaningful interpretation of the comparison between populations with a KRASG12 mutation versus a non-KRASG12 RAS mutation. Moreover, data on KRASG13 or specific KRASG12 mutations were not reported because the numbers of patients with available data in the SUNLIGHT trial were too small to allow meaningful comparisons. Therefore, like the previously reported observational data and meta-analyses,24, 25, 26 it is not possible to speculate on the prognostic/predictive value of individual KRASG12 point mutations in patients receiving FTD/TPI with or without bevacizumab. Further insights in this regard would be of value, particularly with respect to the KRASG12C mutation, which has been established as a strong negative prognostic factor in patients with mCRC.31 Lastly, data on KRAS mutational status were collected upon diagnosis of metastatic disease, whereas the SUNLIGHT study was conducted in the later-line setting; therefore, it is possible that RAS mutations may have emerged throughout treatment and some patients might have been misclassified. For example, in a recent analysis of the phase III FIRE-4 study of first-line folinic acid, fluorouracil, and irinotecan plus cetuximab in patients with RAS wild-type disease per tissue biopsy, serial liquid biopsy detected a RAS mutation in 13% of patients.32 Higher-sensitivity KRAS mutation testing methods may also be needed to support detailed interpretation of the tumor genotype. Hyperselective circulating tumor DNA analysis of baseline samples from the phase III PARADIGM trial of first-line panitumumab, for example, showed that 8% of patients had a KRAS/NRAS mutation.33
Conclusions
The results of this post hoc analysis, based on results from an RCT that had a high quality of data collection and homogeneous patient population, show no evidence that KRAS mutations have an impact on OS with the combination of FTD/TPI plus bevacizumab. The combination provides clinical benefit to all patient populations, including those with or without KRASG12-mutant mCRC. Overall, based on available data, and to the best of our knowledge, KRAS mutations are not predictive of clinical outcomes with FTD/TPI either as monotherapy or in combination with bevacizumab.
Acknowledgements
We thank and acknowledge all the participants, their families, and study personnel for participating in the SUNLIGHT study. Medical writing assistance was provided by Envision Pharma Group and funded by Taiho Oncology, Inc.
Funding
This work was supported by Servier International Research Institute (France) and Taiho Oncology, Inc. (USA) (no grant number).
Role of the funder
The sponsors (Servier International Research Institute and Taiho Oncology, Inc.) were involved in the study design and were responsible for the overall study management, data management, and statistical analysis. The sponsors were involved in the writing of this report, alongside the authors, all of whom had access to the raw data. The corresponding author had full access to all the data in the study and had the final responsibility for the decision to submit for publication.
Disclosure
JTab has received fees for advisory/consultancy roles from Array BioPharma, AstraZeneca, Bayer, BeiGene, Biocartis, Boehringer Ingelheim, Chugai Pharmaceutical Co., Ltd., F. Hoffmann-La Roche, Foundation Medicine, Genentech, Genmab A/S, HalioDX SAS, Halozyme, Imugene, Inflection Biosciences, Ipsen, Kura Oncology, Lilly, Menarini, Merck Serono, Merrimack Pharmaceuticals, Merus, Molecular Partners, MSD, Novartis, Peptomyc, Pfizer, Pharmacyclics, ProteoDesign SL, Rafael Pharmaceuticals, Roche Diagnostics, Sanofi, Seagen, Servier, Symphogen, Taiho, and VCN Biosciences. JTai has received honoraria for speaker or advisory role from Amgen, Astellas, AstraZeneca, Bristol Myers Squibb, Merck Serono, MSD, Novartis, Pierre Fabre, Roche, Servier, and Takeda. MF has received fees for consultancy or participation in advisory boards from AstraZeneca, Bayer Corporation, Bristol-Myers Squibb, Eisai Oncology, Entos Pharmaceuticals, Janssen, Merck, Mirati Therapeutics, Nouscom, Pfizer, Roche/Genentech, Taiho Oncology, and Xenthera; fees for an editorial board role from Mirati Therapeutics; and research grant support from Agenus, Bristol-Myers Squibb, Genentech/imCORE, and Verastem Oncology. GWP has received fees for advisory/consultancy roles from Amgen, AstraZeneca, Bayer, Bristol-Myers Squibb, CECOG, Daiichi Sankyo, Incyte, Lilly, Merck Serono, MSD, Novartis, Pierre Fabre, Roche, Sanofi, Servier, and Takeda. EVC has received grants or contracts from Amgen, Bayer, Boehringer Ingelheim, Bristol Myers Squibb, Eli Lilly and Company, Ipsen, Merck Sharp & Dohme, Merck KGaA, Novartis, Roche, and Servier; and consulting fees from AbbVie, ALX, Amgen, Array, Astellas, AstraZeneca, Bayer, Beigene, Boehringer Ingelheim, Bristol Myers Squibb, Daiichi Sankyo, Eli Lilly and Company, GlaxoSmithKline, Incyte, Ipsen, Merck Sharp & Dohme, Merck KGaA, Mirati, Novartis, Nordic, Pierre Fabre Oncologie, Pfizer, Roche, Seagen, Servier, Takeda, Terumo, Taiho Pharmaceutical, and Zymeworks. FC reports grants or contracts from Roche, Pfizer, and Pierre Fabre; and payment or honoraria from Merck KGaA, Bayer, Pierre Fabre, Servier, MSD, and Roche. NA and DS are employees of Servier. EC is an employee of Taiho Oncology, Inc. TY has received research grants from Amgen, Chugai Pharmaceutical Co., Ltd., Daiichi Sankyo, Eisai, FALCO Biosystems, Genomedia Inc., Molecular Health, MSD, Nippon Boehringer Ingelheim, Ono Pharmaceutical Co., Ltd., Pfizer, Roche Diagnostics, Sanofi, Sysmex, and Taiho Oncology; honoraria from Bayer Yakuhin, Chugai Pharmaceutical Co., Ltd., Merck, MSD K.K., Ono Pharmaceutical Co., Ltd., and Takeda; and consulting fees from Sumitomo Corporation. RJM has declared no conflicts of interest.
References
- 1.Hobbs G.A., Der C.J., Rossman K.L. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang L., Guo Z., Wang F., Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther. 2021;6:386. doi: 10.1038/s41392-021-00780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng M., Zhong K., Jiang T., Liu Z., Kwan H.Y., Su T. The current understanding on the impact of KRAS on colorectal cancer. Biomed Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111717. [DOI] [PubMed] [Google Scholar]
- 5.Zhu G., Pei L., Xia H., Tang Q., Bi F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol Cancer. 2021;20:143. doi: 10.1186/s12943-021-01441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Modest D.P., Ricard I., Heinemann V., et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol. 2016;27:1746–1753. doi: 10.1093/annonc/mdw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richman S.D., Seymour M.T., Chambers P., et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 8.Strickler J.H., Yoshino T., Stevinson K., et al. Prevalence of KRAS G12C mutation and co-mutations and associated clinical outcomes in patients with colorectal cancer: a systematic literature review. Oncologist. 2023;28:e981–e994. doi: 10.1093/oncolo/oyad138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stolze B., Reinhart S., Bulllinger L., Fröhling S., Scholl C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep. 2015;5:8535. doi: 10.1038/srep08535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lièvre A., Bachet J.B., Le Corre D., et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 11.Karapetis C.S., Khambata-Ford S., Jonker D.J., et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 12.Douillard J.Y., Oliner K.S., Siena S., et al. Panitumumab–FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 13.Sepulveda A.R., Hamilton S.R., Allegra C.J., et al. Molecular biomarkers for the evaluation of colorectal cancer: guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35:1453–1486. doi: 10.1200/JCO.2016.71.9807. [DOI] [PubMed] [Google Scholar]
- 14.Benson A.B., III, Venook A.P., Al-Hawary M.M., et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19:329–359. doi: 10.6004/jnccn.2021.0012. [DOI] [PubMed] [Google Scholar]
- 15.National Comprehensive Cancer Network (NCCN®) NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Colon Cancer. Version 3.2023. 2023. https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf Available at.
- 16.National Comprehensive Cancer Network (NCCN®) NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) for Rectal Cancer. Version 5.2023. 2023. https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf Available at.
- 17.Cervantes A., Adam R., Roselló S., et al. Metastatic colorectal cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:10–32. doi: 10.1016/j.annonc.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Yoshino T., Cervantes A., Bando H., et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the diagnosis, treatment and follow-up of patients with metastatic colorectal cancer. ESMO Open. 2023;8 doi: 10.1016/j.esmoop.2023.101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenz H.J., Stintzing S., Loupakis F. TAS-102, a novel antitumor agent: a review of the mechanism of action. Cancer Treat Rev. 2015;41:777–783. doi: 10.1016/j.ctrv.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer R.J., Van Cutsem E., Falcone A., et al. Randomized trial of TAS-102 for refractory metastatic colorectal cancer. N Engl J Med. 2015;372:1909–1919. doi: 10.1056/NEJMoa1414325. [DOI] [PubMed] [Google Scholar]
- 21.Prager G.W., Taieb J., Fakih M., et al. Trifluridine-tipiracil and bevacizumab in refractory metastatic colorectal cancer. N Engl J Med. 2023;388:1657–1667. doi: 10.1056/NEJMoa2214963. [DOI] [PubMed] [Google Scholar]
- 22.Van Cutsem E., Mayer R.J., Laurent S., et al. The subgroups of the phase III RECOURSE trial of trifluridine/tipiracil (TAS-102) versus placebo with best supportive care in patients with metastatic colorectal cancer. Eur J Cancer. 2018;90:63–72. doi: 10.1016/j.ejca.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabernero J., Argiles G., Sobrero A.F., et al. Effect of trifluridine/tipiracil in patients treated in RECOURSE by prognostic factors at baseline: an exploratory analysis. ESMO Open. 2020;5:E000752. doi: 10.1136/esmoopen-2020-000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshino T., Van Cutsem E., Li J., et al. Effect of KRAS codon 12 or 13 mutations on survival with trifluridine/tipiracil in pretreated metastatic colorectal cancer: a meta-analysis. ESMO Open. 2022;7 doi: 10.1016/j.esmoop.2022.100511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang F., Yang H., Bao W., et al. Efficacy and safety of trifluridine/tipiracil (TAS-102) in patients with metastatic colorectal cancer: a systematic review and meta-analysis. Clin Transl Oncol. 2024;26:468–476. doi: 10.1007/s12094-023-03268-5. [DOI] [PubMed] [Google Scholar]
- 26.van de Haar J., Ma X., Ooft S.N., et al. Codon-specific KRAS mutations predict survival benefit of trifluridine/tipiracil in metastatic colorectal cancer. Nat Med. 2023;29:605–614. doi: 10.1038/s41591-023-02240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Haar J., Valeri N., Voest E.E. Third-line therapy in metastatic colorectal cancer. N Engl J Med. 2023;389:190. doi: 10.1056/NEJMc2306486. [DOI] [PubMed] [Google Scholar]
- 28.Tabernero J., Fakih M., Prager G.W. Third-line therapy in metastatic colorectal cancer. Reply. N Engl J Med. 2023;389:190–191. doi: 10.1056/NEJMc2306486. [DOI] [PubMed] [Google Scholar]
- 29.Tabernero J., Taieb J., Prager G.W., et al. Trifluridine/tipiracil plus bevacizumab for third-line management of metastatic colorectal cancer: SUNLIGHT study design. Future Oncol. 2021;17:1977–1985. doi: 10.2217/fon-2020-1238. [DOI] [PubMed] [Google Scholar]
- 30.De Roock W., Jonker D.J., Di Nicolantonio F., et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. J Am Med Assoc. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 31.Ottaiano A., Sabbatino F., Perri F., et al. KRAS p.G12C mutation in metastatic colorectal cancer: prognostic implications and advancements in targeted therapies. Cancers (Basel) 2023;15:3579. doi: 10.3390/cancers15143579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stintzing S., Heinemann V., von Weikersthal L.F., et al. Phase III FIRE-4 study (AIO KRK-0114): influence of baseline liquid biopsy results in first-line treatment efficacy of FOLFIRI/cetuximab in patients with tissue RAS-WT mCRC. J Clin Oncol. 2023;41:3507. [Google Scholar]
- 33.Shitara K., Muro K., Watanabe J., et al. Negative hyperselection of patients with RAS wild-type metastatic colorectal cancer for panitumumab: a biomarker study of the phase III PARADIGM trial. J Clin Oncol. 2023;41:11. [Google Scholar]