Abstract
Ubiquitination often generates lysine 48-linked polyubiquitin chains that signal proteolytic destruction of the protein target. A significant subset of ubiquitination proceeds by a priming/extending mechanism, in which a substrate is first monoubiquitinated with a priming E2-conjugating enzyme or a set of E3 ARIH/E2 enzymes specific for priming. This is then followed by ubiquitin (Ub) chain extension catalyzed by an E2 enzyme capable of elongation. This report provides further insights into the priming/extending mechanism. We employed reconstituted ubiquitination systems of substrates CK1α (casein kinase 1α) and β-catenin by Cullin–RING E3 Ub ligases (CRLs) CRL4CRBN and CRL1βTrCP, respectively, in the presence of priming E2 UbcH5c and elongating E2 Cdc34b (cell division cycle 34b). We have established a new “apyrase chase” strategy that uncouples priming from chain elongation, which allows accurate measurement of the decay rates of the ubiquitinated substrate with a defined chain length. Our work has revealed highly robust turnover of monoubiquitinated β-catenin that empowers efficient polyubiquitination. The results of competition experiments suggest that the interactions between the ubiquitinated β-catenin and CRL1βTrCP are highly dynamic. Moreover, ubiquitination of the Ub-modified β-catenin appeared more resistant to inhibition by competitors than the unmodified substrate, suggesting tighter binding with CRL1βTrCP. These findings support a role for conjugated Ub in enhancing interactions with E3.
Keywords: ubiquitination, Cullin–RING E3 ubiquitin ligase, apyrase chase, priming, ubiquitin chain elongation
Ubiquitination is one of the major cellular post-translational protein modification events that covalently attach the protein substrate with ubiquitin (Ub) in either monomeric forms or polymeric forms (1, 2). Such modifications are of fundamental biological importance because they alter target protein abundance, subcellular location, or function of the target protein. Consequently, ubiquitination impacts numerous biological processes and also directly contributes to a myriad of cellular defects and disease states (1, 2). Ubiquitination requires three enzymes: the E1-activating enzyme, E2-conjugating enzyme, and E3 ligase, the last of which recognizes a substrate and hence, governs specificity of the reaction (1, 3). Cullin–RING E3 Ub ligases (CRLs) are characterized by a signature Cullin–RING heterodimeric complex (4, 5). There are six canonical cullin (CUL) proteins, CUL1, CUL2, CUL3, CUL4A, CUL4B, and CUL5, that all adopt an elongated structure to organize an E3 CRL. Typically, a CUL’s N-terminal domain interacts interchangeably with CUL-specific substrate receptors that are capable of binding a substrate. On the other hand, a CUL’s C-terminal domain binds a RING finger protein, ROC1/RBX1 for CUL1–4 or ROC2 for CUL5, to form a core ligase complex. This modular nature allows for the assembly of ∼300 structurally similar but functionally distinct CRLs in humans (4, 5).
The ubiquitination reaction directed by E3 CRLs proceeds in a sequential fashion beginning with a preinitiation stage characterized by the E3 binding to a substrate. A subsequent initiation phase enables the alignment of the substrate’s receptor lysine residue to the donor Ub’s G76, thereby promoting a nucleophilic attack that yields an isopeptide bond linkage. In the elongation stage, the substrate-linked Ub (or Ub in the distal end of an Ub chain) serves as a receptor that provides a lysine residue to attack the incoming E2–donor Ub thiol ester complex, thereby forming an Ub–Ub chain. Using the E3 anaphase-promoting complex (APC) reaction model, the Morgan group first recognized that two separate E2 enzymes are involved in initiation, typically resulting in substrate monoubiquitination, and elongation that leads to polyubiquitination (6). Wu et al. (7) and Kovacev et al. (8) subsequently described a priming/extending handoff mechanism for the ubiquitination of IκBα (nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha) and β-catenin by E3 CRL1βTrCP, in which the substrate is initially modified by single Ub moieties with E2 UbcH5c (UBE2D3) that then prime for the subsequent Ub chain elongation catalyzed by E2 Cdc34 (cell division cycle 34b; UBE2R). Note that CRL1 was originally designated SCF (Skp1–CUL1–F box protein), in which Skp1 is an adaptor protein linking CUL1 to an F-box protein that binds to a substrate (4, 5). We thus use CRL1 and SCF interchangeably in this report. The priming ubiquitination model of IκBα has been visualized in an elegant cryo-EM study by the Schulman group (9). In addition, the Schulman laboratory has elucidated a new priming mechanism (10, 11) that requires two types of E3s. It is initiated by Nedd8 (neural precursor cell expressed, developmentally downregulated 8), which is an Ub-like protein found to covalently modify a conserved lysine residue on the C terminus of a cullin protein (12). The cullin-linked Nedd8 acts to recruit E3 ARIH1, whose catalytic cysteine residue receives the Ub moiety that is transferred by the ARIH1-bound E2 UBE2L3. The ARIH1-linked Ub is then transferred to the E3 CRL–bound substrate, hence completing the priming reaction. It should be noted that the priming/extending mechanism with two different E2 enzymes extends to the ubiquitination reactions directed by non-CRL E3s (13, 14).
Poly-Ub chains on a protein substrate built by E3 CRL and collaborating E2s are critical signals for proteolytic destruction. While the overall framework of polyubiquitination has been revealed, the specific molecular details are far from completely understood. In this work, we have developed reconstituted ubiquitination systems using substrates CK1α (casein kinase 1α) and β-catenin by E3s CRL4CRBN and SCFβTrCP, respectively, in the presence of priming E2 UbcH5c and elongating E2 Cdc34b. We have established a new “apyrase chase” strategy that uncouples priming from chain elongation, which allows the accurate quantification of the decay rates of the ubiquitinated substrate with its defined chain length. The results of this study provide significant insights into E3 CRL–mediated ubiquitination.
Results
In vitro ubiquitination of CK1α by E3 CRL4CRBN and lenalidomide
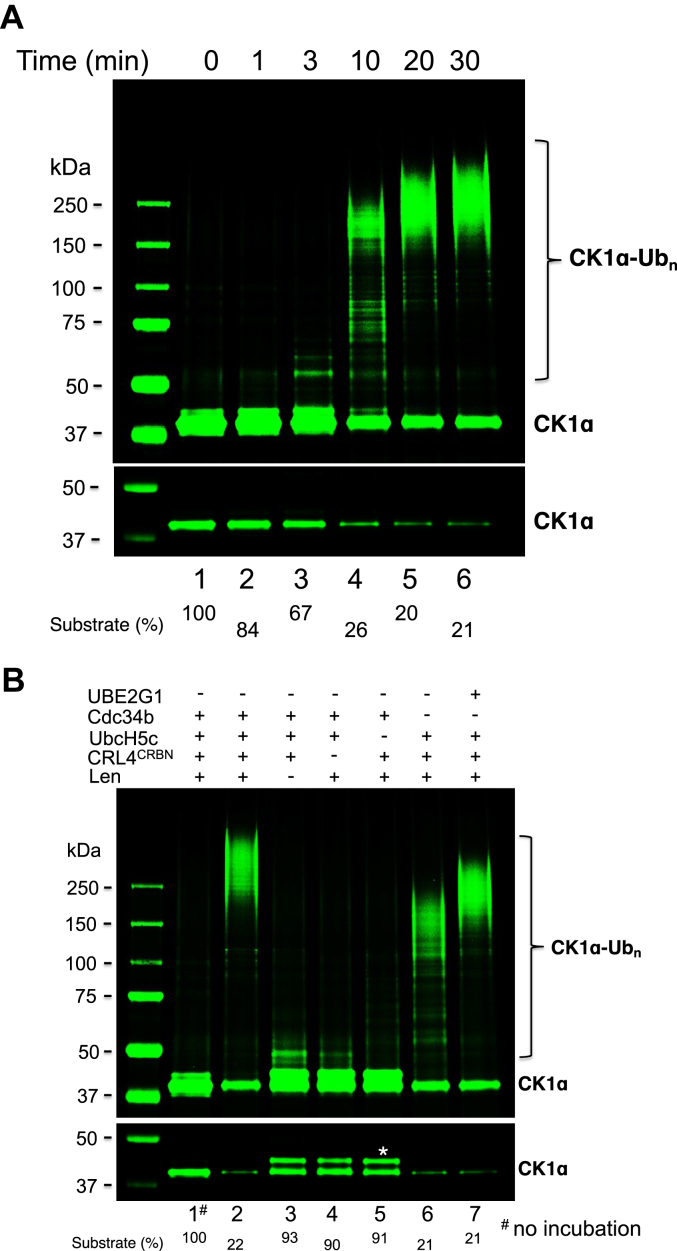
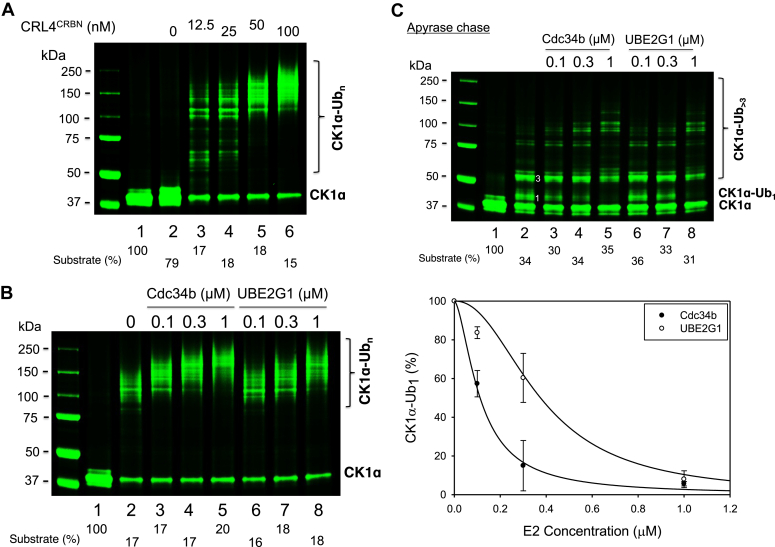
Reconstitution of ubiquitination with pure recombinant enzymes/proteins provides invaluable tools for mechanistic studies. Figure 1 shows the ubiquitination of CK1α by CRL4CRBN. This assay was developed based on previous studies that have established lenalidomide-dependent ubiquitination of CK1α by CRL4CRBN in vivo and in vitro (15, 16), which was confirmed in a recent study using an immunoprecipitation-based approach (17). In this report, we have further improved the CK1α ubiquitination assay by employing a Sortag strategy to fluorescently label CK1α, which enables better quantification. Figure 1A shows time-dependent Ub modification of fluorescent CK1α, converting the substrate to Ub-conjugated species in higher molecular weight forms. Dropout experiment was carried out to determine the requirement of CK1α ubiquitination, and the result is shown in Figure 1B. Removal of lenolidomide (lane 3), E3 CRL4CRBN (lane 4), or the priming E2-conjugating enzyme UbcH5c (lane 5) abolished the ubiquitination, demonstrating that these components are essential for the reaction. Omission of Cdc34b resulted in shorter Ub chains (compare lanes 2 and 6). Replacement of Cdc34b by UBE2G1 yielded Ub chains longer than those seen with UbcH5c alone (compare lanes 6 and 7). These findings suggest that both Cdc34b and UBE2G1 can function as elongating E2-conjugating enzyme. Note that CK1α has multiple lysine residues and, thus, the observed high molecular weight Ub-conjugated CK1α species (lane 6) was likely a result of multiple monoubiquitination as well as limited chain extension.
Figure 1.
In vitro ubiquitination of CK1α by E3 CRL4CRBN.A, time course. B, dropout. CK1α was subject to ubiquitination reaction containing E3 CRL4CRBN and lenalidomide (1 μM) with details described in the Experimental procedures section. Both high- and low-contrast fluorescent scans are shown. Species∗ most likely represents autophosphorylated form of CK1α. Percentage of CK1α consumed is indicated. CK1α, casein kinase 1α; CRBN, cereblon; CRL, Cullin–RING E3 Ub ligase.
Differential stability of UbcH5c–Ub and Cdc34b–Ub thioester complexes
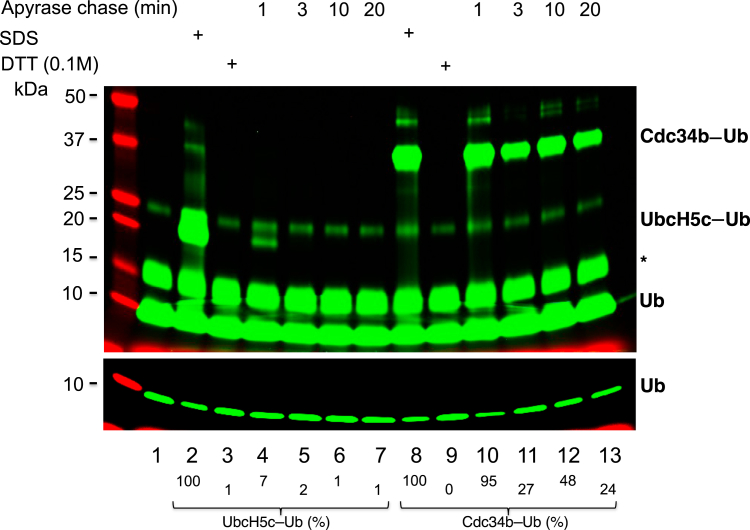
To determine and compare the stability of E2 UbcH5c–Ub and Cdc34b–Ub thioester complexes, we first formed each thioester complex with a trackable fluorescently labeled Ub and then treated the reaction mixture with apyrase, which is a potent ATP hydrolase. This treatment depleted ATP, thereby effectively preventing formation of new E2–Ub thioester complexes that require ATP hydrolysis (1). Decay/stability of the E2–fluorescent Ub complex was determined by time-course experiments. After separating reaction products by denaturing gel electrophoresis, the fluorescent Ub species were visualized by imaging on a scanner. Their results revealed that 12% and 32% of input Ub was converted to UbcH5c–Ub and Cdc34b–Ub complexes, respectively (Fig. 2, lanes 2 and 8). These species were sensitive to treatment with excess DTT (0.1 M) (Fig. 2, lanes 3 and 9), consistent with the property of thioesters. UbcH5c–Ub was highly unstable as more than 93% and 97% of the thioester complex disappeared at 37 °C within 1 and 3 min incubation after the apyrase treatment, respectively (Fig. 2, lanes 4 and 5). In contrast, Cdc34b–Ub was relatively stable as at least 24% of the thioester complex remained intact during incubation at 37 °C for 3 to 20 min after the apyrase treatment (Fig. 2, lanes 11–13). These data demonstrate a remarkable difference in the stability of E2 UbcH5c–Ub and Cdc34b–Ub thioester complexes.
Figure 2.
Stability of E2–Ub thioester complexes. The reaction is described in the Experimental procedures section. Both high- and low-contrast fluorescent scans are shown. Species∗ most likely represents dimers in the ubiquitin preparation, which takes about 1.8% of monomeric form. Percentage of E2–Ub is indicated. Ub, ubiquitin.
Apyrase chase assay
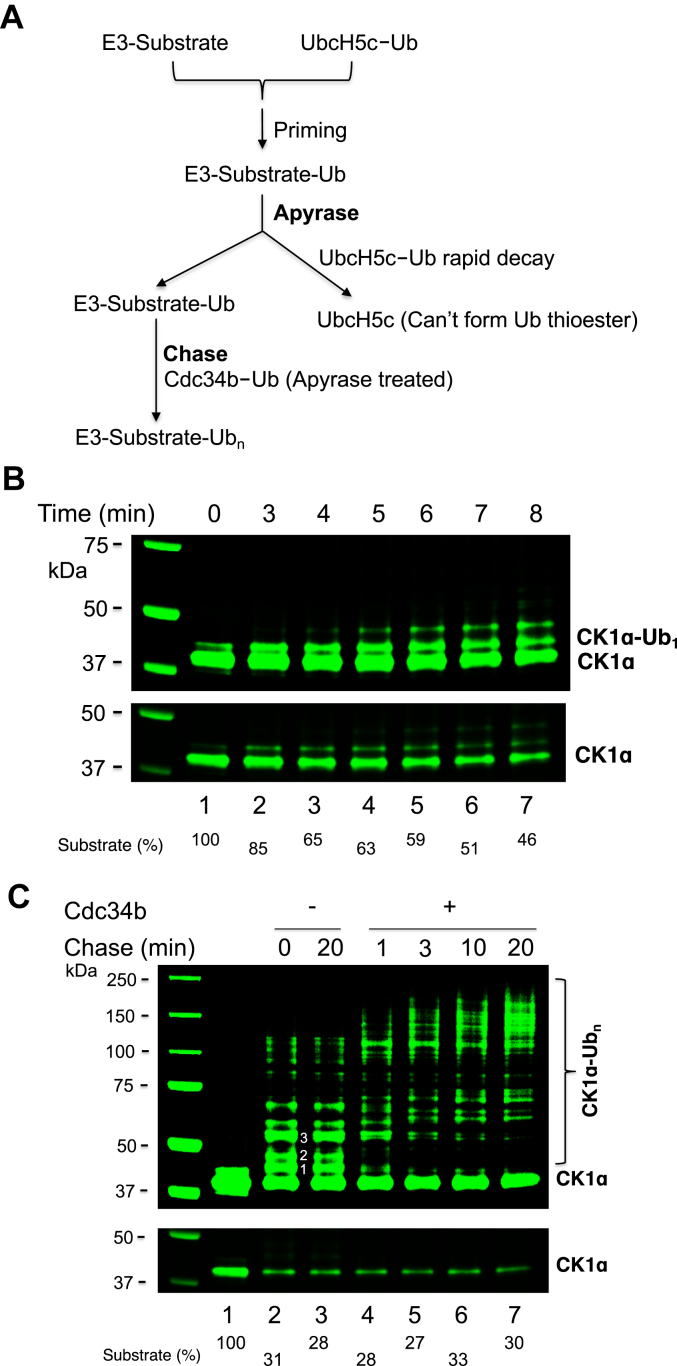
The observed differential stability of UbcH5c–Ub versus Cdc34b–Ub provides opportunity to develop an apyrase chase assay that can uncouple priming and chain elongation in ubiquitination (schemed in Fig. 3A). In this system, preassembled E3–substrate and UbcH5c–Ub are mixed, briefly incubated to form monoubiquitinated species, and then treated with apyrase to deplete ATP. As a consequence, while the substrate is monoubiquitinated, UbcH5c–Ub rapidly decays. Subsequently, preformed Cdc34b–Ub (apyrase treated) is added, and the relatively stable Cdc34b–Ub allows sufficient time for the attack of the monoubiquitinated substrate, resulting in chain elongation. This scheme thus permits accurate measurement of Cdc34-catlyzed Ub chain elongation on a substrate exclusively.
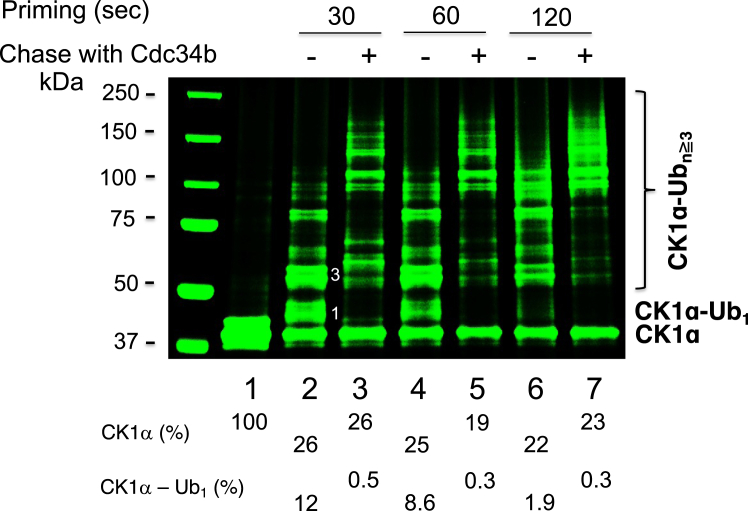
Figure 3.
“Apyrase chase.”A, scheme. B, time course of the ubiquitination of CK1α by E3 CRL4CRBN and UbcH5c. C, the “apyrase chase” reaction with CK1α, E3 CRL4CRBN, UbcH5c, and Cdc34b using the protocol described in the Experimental procedures section. Both high- and low-contrast fluorescent scans are shown. Presumed ubiquitin molecules linked to CK1α are as indicated by in-gel numbers. Percentage of CK1α consumed is indicated. CK1α, casein kinase 1α; CRL, Cullin–RING E3 Ub ligase.
We first performed a monoubiquitination time-course experiment that indicated that reaction incubation of 7 min resulted in nearly 50% substrate utilization (Fig. 3B, lane 6). Thus, a priming reaction with the combination of preassembled E3–substrate and UbcH5c–Ub was set to proceed for 7 min, apyrase treated, and then chased for varying times in the presence or the absence of Cdc34b–Ub (Fig. 3C). The results revealed several observations. First, the addition of Cdc34b–Ub did not increase the level of substrate utilization (Fig. 3C, lanes 2–7). Thus, priming with UbcH5c determines the efficiency with which a substrate is converted to Ub conjugates. Second, post the apyrase treatment, additional 20 min incubation in the absence of Cdc34b–Ub (Fig. 3C, lane 3) resulted in similar patterns of ubiquitination products (Fig. 3C, lane 2). These results are entirely consistent with the notion that the apyrase treatment causes rapid decay of UbcH5c–Ub, which inactivates UbcH5c completely. Third, the Ub conjugates built on CK1α were all extended to form longer chains (Fig. 3C, lanes 4–7). Collectively, these data validated the scheme of the apyrase chase assay as illustrated in Figure 3A.
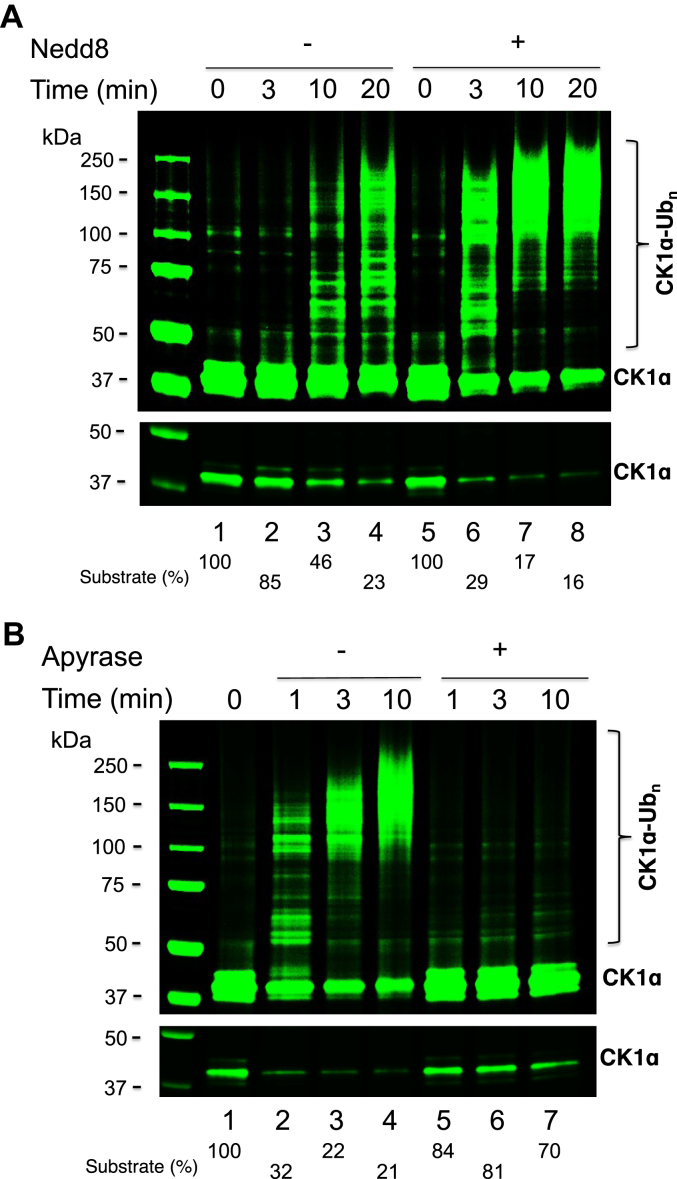
Stimulation of CK1α ubiquitination by Nedd8 modification
Consistent with previous observations (16), CK1α ubiquitination was stimulated by prior Nedd8 modification to CUL4A (Fig. 4A). However, the ubiquitination was abolished if the preassembled UbcH5c–Ub and Cdc34b–Ub were treated with apyrase before mixing with Nedd8–E3–substrate (Fig. 4B). Given that the apyrase treatment caused rapid decay of UbcH5c–Ub, but left Cdc34b–Ub intact (Fig. 2), the observed near complete inhibition of the ubiquitination (Fig. 4B) suggests that the ubiquitination completely depends on priming with UbcH5c–Ub.
Figure 4.
Stimulation of the CRL4CRBN-mediated-ubiquitination of CK1α by Nedd8.A, time course of CK1α ubiquitination by CRL4CRBN and Nedd8–CRL4CRBN. B, time course of CK1α ubiquitination by Nedd8–CRL4CRBN with E2–Ub (UbcH5c–Ub and Cdc34b–Ub) treated with or without apyrase. Reactions were carried out as described in the Experimental procedures section. Both high-and low-contrast fluorescent scans are shown. Percentage of CK1α consumed is indicated. CK1α, casein kinase 1α; CRBN, cereblon; CRL, Cullin–RING E3 Ub ligase; Nedd8, neural precursor cell expressed, developmentally downregulated 8.
Nedd8–CRL4CRBN was found to significantly enhance the priming with UbcH5c. Using the apyrase chase assay (Fig. 3A), Nedd8–CRL4CRBN was used with Cdc34b–Ub for elongation (Fig. 5). The results showed that incubation of Nedd8–E3–substrate with UbcH5c–Ub for 30 s converted >70% of CK1α to ubiquitinated species (Fig. 5B, lane 2). By contrast, similar substrate utilization efficiency required 7 min (420 s) incubation in the priming reaction with the unmodified E3 (Fig. 3, B and C). These findings suggest that Nedd8 modification stimulated priming with UbcH5c by a factor of >10-fold. The initial priming product CK1α–Ub1 formed within 30 s of incubation (Fig. 5, lane 2) and its level decreased with longer incubation (compare lanes 2, 4, and 6), most likely reflecting the ability of UbcH5c to catalyze multimonoubiquitination on different substrate lysine residues. As shown, CK1α–Ub1 disappeared as a result of chase with Cdc34b (lanes 3, 5, and 7). In addition, Cdc34b significantly enhanced production of Ub chains of larger size (lanes 3, 5, and 7). These findings are consistent with the ability of Cdc34b to catalyze Ub chain elongation.
Figure 5.
The “apyrase chase” reaction with CK1α, Nedd8–CRL4CRBN, UbcH5c, and Cdc34b. Priming reactions were carried out for indicated times followed by elongation with Cdc34 for 3 min at 37 °C. Reactions are carried out and analyzed as described for Figure 3C. CK1α, casein kinase 1α; CRBN, cereblon; CRL, Cullin–RING E3 Ub ligase; Nedd8, neural precursor cell expressed, developmentally downregulated 8.
Effects of E3 concentration and elongating E2s in CK1α ubiquitination
Results of titration of Nedd8–CRL4CRBN concentrations revealed intriguing reaction properties (Fig. 6A). First, at all E3 concentrations tested, >80% of CK1α was converted to Ub conjugates. Note that 250 nM of the substrate was used for reaction. Thus, as low as 12.5 nM concentration of E3 was able to consume 200 nM of substrate to form reaction products. Second, low E3 concentrations (12.5 and 25 nM) resulted in Ub conjugates that varied in size, ranging from low molecular weight species to those with longer chains. Third, at high E3 concentrations (50 and 100 nM), predominant reaction products were larger sized Ub conjugates. These data suggest an ability of Nedd8–CRL to rapidly prime its cognate substrate with mono-Ub and then engage with the ubiquitinated substrate for chain elongation, likely requiring multiple rounds of E3 binding.
Figure 6.
E3 and E2 titration.A, titration of Nedd8–CRL4CRBN. B, comparison of Cdc34b and UBE2G1 in the ubiquitination of CK1α. The reaction was carried out as described for Figure 1. C, comparison of Cdc34b and UBE2G1 in the “apyrase chase” reaction. Priming reaction with UbcH5c was 30 s at 37 °C. The elongation with Cdc34b was 3 min at 37 °C. Graphs integrate data in triplets with indicated error bars. CK1α, casein kinase 1α; CRBN, cereblon; CRL, Cullin–RING E3 Ub ligase; Nedd8, neural precursor cell expressed, developmentally downregulated 8.
Recent studies demonstrate that Cdc34a–UBE2R1, Cdc34b–UBE2R2, and UBE2G1–Ubc7 are all capable of working with E3 CRL for Ub chain elongation (18, 19, 20). We first compared Cdc34b and UBE2G1 in their ability to support CK1α ubiquitination. An E2 titration experiment showed that Cdc34b formed Ub chains in a size that was considerable longer than those observed with UBE2G1 (Fig. 6B). It was noted that in the absence of elongating E2, UbcH5c was able to form ubiquitinated products with considerably large size with 10 min incubation (Fig. 6B, lane 2). To more precisely compare the ability of elongating E2, we employed the apyrase chase assay (Fig. 3A). E2 titration showed that Cdc34b was markedly more effective than UBE2G1 in consuming the monoubiquitinated CK1α (Fig. 6C, graph). Together, these data demonstrate that Cdc34b is more active than UBE2G1 in supporting CK1α ubiquitination in vitro.
In vitro ubiquitination of β-catenin by E3 CRL1–SCFβTrCP
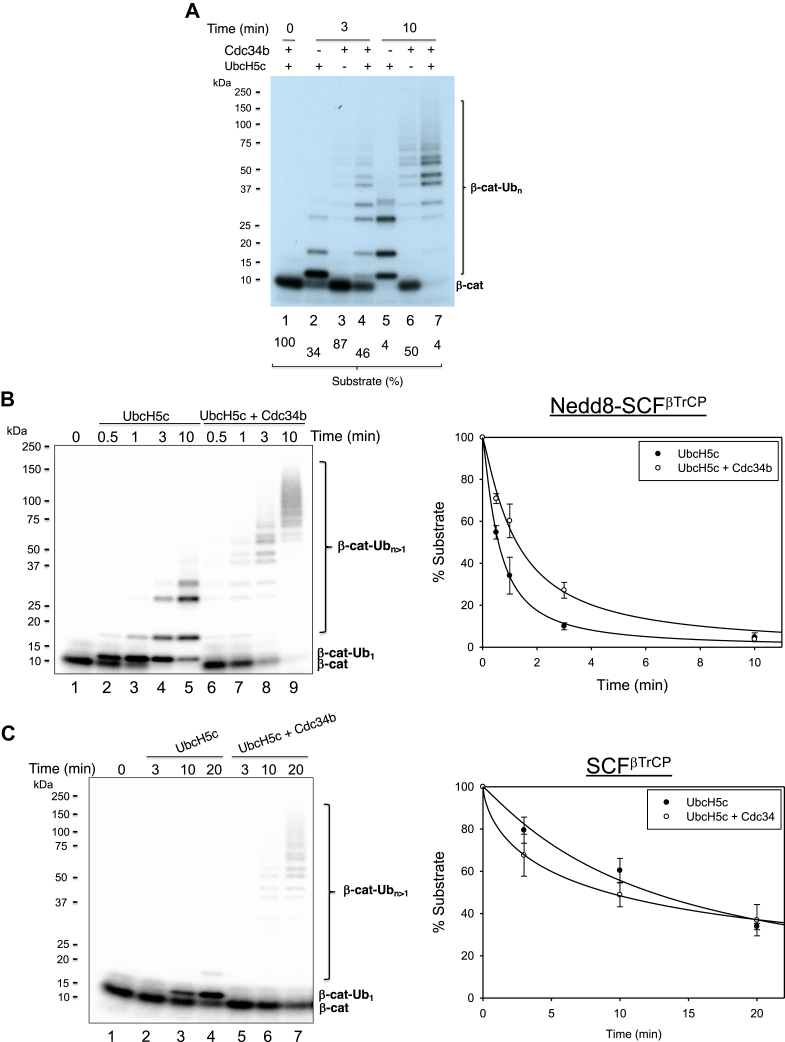
In vitro ubiquitination of β-catenin by E3 SCFβTrCP employs the substrate’s 22-amino acid phosphodegron peptide. This peptide exhibits binding to βTrCP (beta-transducin repeat–containing protein) as revealed by a previously reported cocrystallographic study (21) and supports efficient in vitro ubiquitination by SCFβTrCP (8, 22). Figure 7A shows time-dependent ubiquitination of the radioactive β-catenin degron peptide by Nedd8–SCFβTrCP with UbcH5c as a priming E2 and Cdc34b as an elongating E2.
Figure 7.
In vitro ubiquitination of β-catenin by E3 CRL1–SCFβTrCP.A, effects of UbcH5c and Cdc34b. B, time course with UbcH5c alone as well as UbcH5c and Cdc34b in the presence of Nedd8–SCFβTrCP. C, time course with UbcH5c alone as well as UbcH5c and Cdc34b in the presence of SCFβTrCP. The reaction was carried out using the protocol described in the Experimental procedures section. Graphs integrate data in triplets with indicated error bars. βTrCP, beta-transducin repeat–containing protein; CRL, Cullin–RING E3 Ub ligase; SCF, Skp1–CUL1–F box protein.
We performed time-course experiments with UbcH5c alone or UbcH5c plus Cdc34b by SCFβTrCP in unmodified (Fig. 7C) or Nedd8-conjugated forms (Fig. 7B). In the presence of Nedd8–SCFβTrCP, the addition of Cdc34b markedly stimulated production of ubiquitinated species with large size as expected (Fig. 7B). However, it appears that in short incubation periods (30 s to 3 min), the presence of Cdc34b reduced levels of substrate utilization (Fig. 7B, graph). On the other hand, in the presence of unmodified SCFβTrCP, the addition of Cdc34b slightly increased the conversion of substrate to ubiquitinated products (Fig. 7C, graph). We speculated that the differential effects of Cdc34b on substrate consumption (Fig. 7, B and C) are due to different reaction kinetics. In the presence of Nedd8–SCFβTrCP with faster kinetics, the E3 works with UbcH5c to rapidly monoubiquitinate the substrate. Cdc34b may compete with UbcH5c for binding to E3–substrate, thereby resulting in observed lower substrate consumption rate. In the presence of SCFβTrCP with slower kinetics, both UbcH5c and Cdc34b work with the E3 more slowly for Ub modification, producing an E2 additive effect.
Note that in reactions described in this work, the level of substrate is in excess to E3. In our previously published study (8), the ubiquitination of β-catenin was tested in a range of substrate and E3 SCFβ-TrCP concentrations. The results show that in all range of concentrations tested, including E3 in excess to substrate, UbcH5c is far more effective than Cdc34 in priming the substrate for ubiquitination.
Building ubiquitinated substrate intermediate models with defined chain length
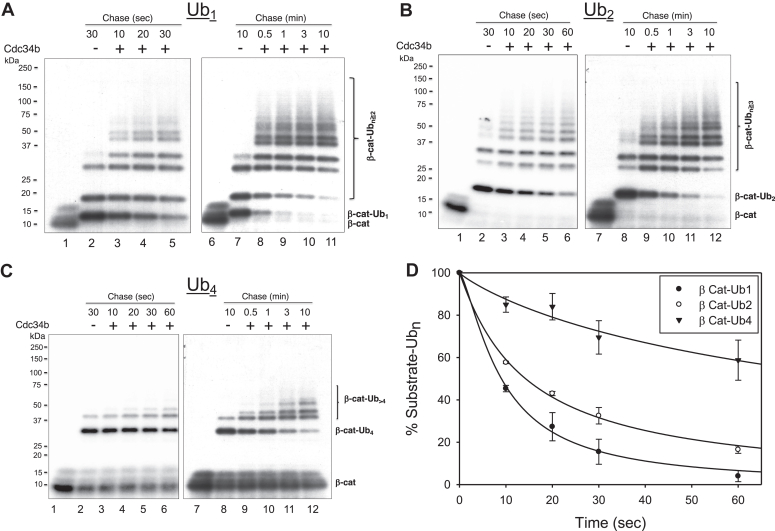
A single lysine receptor within the β-catenin peptide degron substrate provides an opportunity to build Ub conjugates with varying chain length, including mono-Ub (Fig. 8A), di-Ub (Fig. 8B), and tetra-Ub (Fig. 8C). The monoubiquitinated β-catenin was generated using the apyrase chase assay (Fig. 3A). Incubation of Nedd8–SCFβTrCP and UbcH5c–Ub at 37 °C for 3 min followed by the apyrase treatment formed β-catenin–Ub1 (Fig. 8A, lanes 2 and 7), which typically represented more than 50% of the reaction products (sum of β-catenin–Ub1, β-catenin–Ub2, β-catenin–Ub3, and β-catenin–Ub4). No new β-catenin–Ub1 could form because of ATP depletion that inactivates UbcH5c. These conditions permitted chase reaction with relatively stable Cdc34b–Ub, allowing accurate measurement of the turnover of β-catenin–Ub1. As shown by time-course experiments (Fig. 8A), β-catenin–Ub1 rapidly disappeared as a result of Cdc34b–Ub addition with more than 50% conversion to longer products within 10 s of incubation (Fig. 8D). Note that other species including β-catenin–Ub2, β-catenin–Ub3, and β-catenin–Ub4 are both reaction inputs and products. Measurement of their decay rates requires methods such as differential equations (23).
Figure 8.
Elongation of ubiquitinated β-catenin with defined chain length.A, decay of β-catenin–Ub1. B, decay of β-catenin–Ub2. C, decay of β-catenin–Ub4. D, graph compares turnover rates of β-catenin–Ub1, β-catenin–Ub2, and β-catenin–Ub4 at time scale of 10–60 s. It integrates data in triplets with indicated error bars. The reaction was carried out using the protocol described in the Experimental procedures section. Ub, ubiquitin.
β-catenin–Ub2 was built using similar procedure as described for β-catenin–Ub1, except that in the priming reaction, commercially purchased mixed Ub chains of Ub2–7 were used in lieu of Ub. Incubation of Nedd8–SCFβTrCP and UbcH5c–Ub2–7 at 37 °C for 10 min followed by the apyrase treatment formed β-catenin–Ub2 (Fig. 8C, lanes 2 and 8), which was the predominant reaction product (>80% of all products formed). Using the same “apyrase chase” strategy as described previously, the rate with which β-catenin–Ub2 disappeared was determined (Fig. 8B). In comparison to β-catenin–Ub1, the β-catenin–Ub2 turnover rate was measurably reduced (Fig. 8D).
Finally, β-catenin–Ub4 was assembled with a priming reaction with commercially purchased Ub4. Incubation of Nedd8–SCFβTrCP and UbcH5c–Ub4 at 37 °C for 10 min followed by the apyrase treatment formed β-catenin–Ub4 (Fig. 8C, lanes 2 and 8), which was the predominant reaction product (>80% of all products formed). As shown, β-catenin–Ub4 was used effectively for chain elongation with longer incubation of 3 and 10 min (Fig. 8C, lanes 11 and 12). However, at the time scale of 10 to 60 s of incubation, the turnover rate of β-catenin–Ub4 was significantly slower than β-catenin–Ub1 and β-catenin–Ub2 (Fig. 8D). Thus, increasing Ub chain length appeared to decrease the decay rate.
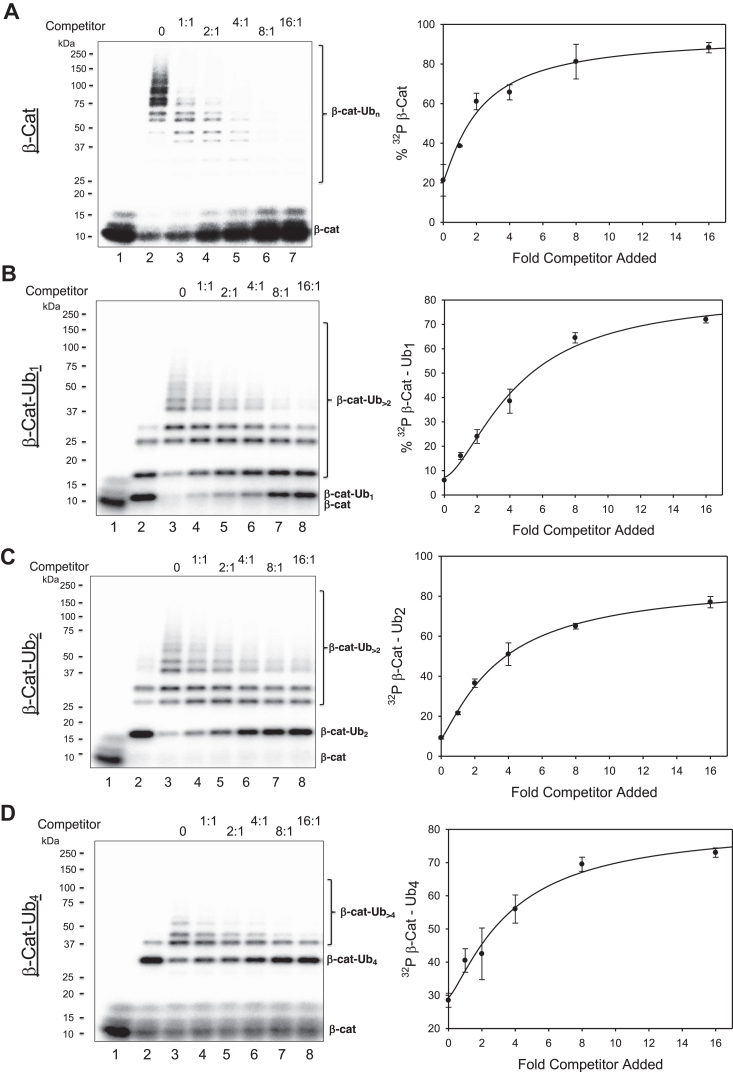
Probing E3 binding to substrate or ubiquitinated substrate using competition experiments
E3 binding to substrate and substrate–Ub conjugates is key to ubiquitination. To gain insights into this process, we sought to perform competition experiments to probe the dynamics of the E3 complexes with substrate or substrate–Ub conjugates. We first tested the ability of excess unlabeled β-catenin degron peptides to compete with the same but radioactively labeled (32P) β-catenin peptide substrate. The results showed that addition of competitors led to significant reduction of ubiquitinated products with concomitant accumulation of unmodified probe (Fig. 9A), thus validating the effectiveness of the competitor in competing with 32P–β-catenin for ubiquitination.
Figure 9.
Competition.A, effects of increasing concentrations of competitor (unlabeled β-catenin phosphor-degron peptide) on the ubiquitination of radioactive β-catenin. The molar ratio of the competitor versus the radioactive substrate is indicated. B–D, increasing concentrations of competitor were added to preformed β-catenin–Ub1 (pane B), β-catenin–Ub2 (pane C), or β-catenin–Ub4 (pane D), followed by chase with Cdc34b–Ub. The reactions were carried out using the protocol described in the Experimental procedures section. Graphs integrate data in duplicates with indicated error bars. Ub, ubiquitin.
We next assessed the ability of the competitor to compete with 32P–β-catenin–Ub1, 32P–β-catenin–Ub2, or 32P–β-catenin–Ub4. These three conjugates were assembled as described previously (Fig. 8). Increasing amounts of the competitors were added for a brief incubation. The resulting mixture was then combined with Cdc34b–Ub (apyrase treated) for chain elongation (10 min). In the absence of the competitor, >90% of 32P–β-catenin–Ub1 (Fig. 9B, lane 3), or 32P–β-catenin–Ub2 (Fig. 9C, lane 3), or >70% of 32P–β-catenin–Ub1 (Fig. 9D, lane 3) decayed to form longer Ub chain products, in keeping with previous observations (Fig. 8). The results of competition experiments suggest a few points. First, unlabeled β-catenin peptides were able to inhibit the ubiquitination of 32P–β-catenin–Ub1 (Fig. 9B), 32P–β-catenin–Ub2 (Fig. 9C), or 32P–β-catenin–Ub4 (Fig. 9D), suggesting their ability to displace the ubiquitinated substrates for binding with the E3. Thus, the interactions between the ubiquitinated substrates and the E3 are dynamic. Second, the competitors inhibited the utilization of 32P–β-catenin (Fig. 9A) more effectively than the turnover of 32P–β-catenin–Ub1 (Fig. 9B), 32P–β-catenin–Ub2 (Fig. 9C), or 32P–β-catenin–Ub4 (Fig. 9D). Third, the competitors blocked 32P–β-catenin to form Ub chains in a robust fashion (Fig. 9A, lanes 3 and 4). By comparison, however, the same competitors appeared far less effective in inhibiting Ub chain elongation with preformed 32P–β-catenin–Ub1 (Fig. 9B), 32P–β-catenin–Ub2 (Fig. 9C), or 32P–β-catenin–Ub4 (Fig. 9D). Given that the strength of competition is likely correlated with the binding affinity between substrate–Ub-modified substrate and the E3, the observed lesser inhibition by the competitors against the Ub-modified substrate may reflect tighter binding of the E3 to the ubiquitinated probe. Collectively, these findings suggest a role for conjugated Ub in enhancing interactions with E3 (see the Discussion section).
Discussion
This study has developed reconstituted ubiquitination systems with substrates CK1α (Figs. 1–6) and β-catenin (Figs. 7–9) by E3s CRL4CRBN and SCFβTrCP, respectively, in the presence of priming E2 UbcH5c and elongating E2 Cdc34b. We have established a new “apyrase chase” strategy (Fig. 3A) that is based on remarkable differences in E2–Ub stability that feature both the rapid turnover of UbcH5c–Ub in contrast to relatively stable Cdc34b–Ub (Fig. 2), and the ability of potent ATP hydrolase apyrase to prevent de novo synthesis of E2–Ub thioesters required for ubiquitination. The “apyrase chase” uncouples priming from chain elongation without the need to use affinity immobilization–based methods (7). The results of this study provide significant insights into E3 CRL–mediated ubiquitination as summarized later.
Priming
Priming determines substrate consumption
Previous studies have demonstrated a decisive role for priming E2 such as UbcH5c in ubiquitination (7, 8). This work confirms and extends the dominance of UbcH5c in committing substrates for Ub modification. With CK1α, no detectable substrate utilization was observed in the reaction lacking UbcH5c (Fig. 1B, lane 5). Similar effects were detected in reactions containing active Cdc34b–Ub but lacking UbcH5c–Ub (Fig 4B, lanes 5–7), conditions that were created by treating E2–Ub (UbcH5c–Ub and Cdc34b–Ub) with apyrase before mixing with Nedd8–CRL4CRBN–CK1α. With β-catenin, substrate utilization was largely blocked in reactions lacking UbcH5c as well (Fig. 7A, lane 3).
Role of Nedd8 in priming
Previous studies have documented a significant activating role for Nedd8 modification in E3 CRL–mediated ubiquitination (12). Nedd8 conjugation to CUL causes multiple conformational changes, and the CUL-linked Nedd8 provides new protein–protein interaction surface (24, 25, 26). Collectively, these effects promote productive interactions with Ub-carrying enzymes (26). Results of this work further support the activating role of Nedd8. Neddylation markedly activated both CK1α (Fig. 4A) and β-catenin (Fig. 7, B and C) ubiquitination. Nedd8 modification stimulated priming with UbcH5c by a factor of >10-fold (compare Figs. 3, B and C and 5).
Monoubiquitination empowers robust polyubiquitination
“Apyrase chase” allowed us to accurately measure the decay rate of the monoubiquitinated substrate β-catenin–Ub1. The turnover of β-catenin–Ub1 was highly robust as 55% of the input (∼1.1 pmol) was consumed within 10 s incubation (Fig. 8B, lane 3). Thus, monoubiquitination empowers robust Ub chain elongation. The results of recent high-resolution studies have shown that substrate, E3, E2, and Ub form multifaced productive complexes (9). Presumably, β-catenin–Ub1 forms a highly productive elongation complex containing E3 and elongating E2–Ub thioester. It is possible that modification of β-catenin by a single Ub moiety transforms the conjugate into a state that is most favorable for binding to both E3 SCFβTrCP and E2 Cdc34–Ub thioester. In this regard, β-catenin-linked mono-Ub may provide additional contact surface(s) for E3 component and elongating E2, thereby stabilizing/orientating substrate–E3–E2–Ub interactions/conformations in favor of Ub chain elongation. Future biochemical and structure work are required to precisely define the role of the substrate-conjugated Ub in forming more productive elongation complexes.
Action of E3
Near complete consumption of CK1α required Nedd8–CRL4CRBN at levels far less than those of the substrate (Fig. 6A). Nedd8–CRL4CRBN therefore exhibits an ability to turnover substrate at a high rate. Given that the substrate utilization was determined by the action of UbcH5c during priming, it suggests that priming is a reaction that utilizes the ability of E3 to rapidly produce monoubiquitinated species, which then enables robust chain elongation.
Ub chain elongation
Decreased decay rates of substrate–Ub conjugates are correlated with increasing Ub chain length
In addition to β-catenin–Ub1, we have assembled β-catenin–Ub2 (Fig. 8B, lane 2) and β-catenin–Ub4 (Fig. 8C, lane 2). β-catenin–Ub2 decayed rapidly albeit with a rate measurably slower than β-catenin–Ub1 (Fig. 8D). β-catenin–Ub4 has a much slower turnover rate (Fig. 8D). Our results are consistent with previous report by the Deshaies group (23), who used the quench flow approach of millisecond resolution to observe progressive decrease in transfer rates in the ubiquitination of β-catenin phosphodegron peptide by SCFβTrCP and Cdc34. Our “apyrase chase” assay provides an alternative strategy to accurately determine the decay rates of ubiquitinated substrates with defined chain length.
Of note, we have previously shown that K48–tetra-Ub slows down ubiquitination using preformed β-catenin–Ub4 (8). In the previous study, however, the decay rate of β-catenin–Ub4 was significantly slower than that seen in this study. The previous preparation of preformed β-catenin–Ub4 involved heat denaturation or contains low levels of active priming E2 UbcH5c, which likely contributed to the observed rate discrepancy.
Redundancy of elongating E2
Recent studies demonstrate that Cdc34a–UBE2R1, Cdc34b–UBE2R2, and UBE2G1–Ubc7 are all capable of working with E3 CRL for Ub chain elongation (18, 19, 20). However, there appears to be conflicting views concerning E2 specificity for cooperating with E3 CRL to catalyze polyubiquitination. In 2018, using genetic screens, the Rolfe (18) and Ebert (19) groups have independently reported identification of UBE2D3–UbcH5c and UBE2G1 as priming and elongating E2 enzymes, respectively, for supporting the pomaidomide/lenalidomide-dependent degradation of neomophic substrate IKZF by E3 CRL4CRBN. These studies highlighted a critical role for UBE2G1 in the ubiquitination and degradation of neomophic substrates by CRL4, raising a possibility whether CRL4 works with UBE2G1 specifically. However, the results of subsequent studies appear at odds with such an E2-specificity hypothesis. The Kleiger group has performed a synthetic lethality screen with Cdc34a/b double knockout and has identified UBE2G1 as a top synthetic lethal interactor (20). Simultaneous silencing of Cdc34a, Cdc34b, and UBE2G1 is needed to cause accumulation of SCF substrates p27 and cyclin E as well as CRL2 substrate HIF1α. Moreover, UBE2G1 was able to support the ubiquitination of SCF substrates albeit with lower enzyme efficiency in comparison with Cdc34a/b. These results support a hypothesis that Cdc34a, Cdc34b, and UBE2G1 play redundant role in supporting Ub chain elongation by E3 CRL. In this work, we have shown that Cdc34b is more effective than UBE2G1 in supporting the ubiquitination of lenalidomide-dependent ubiquitination of CK1α by CRL4CRBN (Figs. 1B and 6, B and C). Our results align with the elongating E2 redundancy hypothesis for E3 CRL–mediated ubiquitination (20).
Role of conjugated Ub in ubiquitination
Previous studies on E3 APC–cyclosome (APC/C) have revealed a positive feedback mechanism called processive affinity amplification (PAA) (27, 28). The results of a single-molecule study have shown that initial ubiquitination of a number of substrates by APC/C and E2 UBE2C has greatly increased the substrate’s binding affinity to the E3, resulting in higher on rate and lower off rate (27). Subsequent high-resolution structural experiments have further suggested a role for the substrate-linked Ub in binding to APC/C’s RING domain, leading to increased processivity (28). This PAA mechanism is thought necessary for the selection of APC/C substrates for productive ubiquitination because these substrates typically contain degrons with D and KEN boxes that are characterized by low binding affinity to the E3 (27). Our competition experiments suggest a role for β-catenin-linked Ub in enhancing interactions with E3 SCFβ-TrCP (Fig. 9) and thus bears a level of similarity to the PAA mechanism. However, our work differs from the APC/C–PAA study in significant ways. First, unlike low-affinity interactions between APC/C and substrates, β-catenin–SCFβ-TrCP and CK1α–CRL4CREN in this study are of high-affinity interactions, possessing binding Kd of 2 nM (29) and 75 nM (30), respectively. Second, the maximal effects of APC/C–PAA appear to require a large number (>10) of substrate-conjugated Ub molecules (27). By comparison, in the ubiquitination of either β-catenin by SCFβ-TrCP or CK1α by CRL4CREN, priming through monoubiquitination by UbcH5c is the rate-limiting step. Ub chain elongation rate decreases progressively as the chain grows (Fig. 8).
Experimental procedures
Protein preparations
Substrates
CK1α cloning and expression
Cloning and expression of baculovirus-driven human CK1α in infected insect cells were done by Genscript. Briefly, human CK1α sequence was cloned into the baculoviral expression vector pFASTBAC1, with Strep tag and thrombin site sequences inserted in the 5′ end, and Sortase tag (LPETGG) and FLAG tag sequences inserted in the 3′ end. The tagged protein was expressed in 2 l of infected SF9 cells. About 14 g worth of cell pellets were obtained.
CK1α purification and labeling
About 4 g of cell pellet was thawed and resuspended in 30 ml lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 2 mM MgCl2, 10% glycerol, 1% Triton X-100, 25 units/ml Benzonase [EMD Millipore], 2 mM PMSF, and 1× Halt protease inhibitor cocktail [ThermoFisher]). The resuspended cells were sonicated for 4 × 30 s pulses and then incubated at 4 °C for 30 min with agitation. The extract was then clarified by centrifugation at 17,000g for 30 min.
Extracts were incubated with 1.5 ml of FLAG beads (Sigma) for 1 h at 4 °C and then washed extensively with wash buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 2 mM CaCl2, 0.05% NP-40, and 10% glycerol). Bound protein was slowly eluted using 5 ml of elution buffer (1 mg/ml 3× FLAG peptide in wash buffer) in three fractions. A total of 280 mg protein was eluted, with most of the protein in fraction 1 (200 mg in 2 ml).
About 100 mg of purified CK1α was mixed with Alexa Fluor 647 maleimide-labeled Sortag peptide (GGGGGFVC custom labeled by ThermoFisher) in a 1:3 ratio. Sortase A5 protein (active motif) was added (20:1 substrate to enzyme ratio) and incubated for 1 h at room temperature. The labeled material was then passed through a MidiTrap G-25 column (GE Healthcare). Approximately 25 mg of labeled protein was recovered. The final product is fluorescently labeled CK1α-LPET-GGGGGFVC-647.
The β-catenin phosphor-degron peptide substrate was prepared as previously described (8, 22). This substrate contains chemically incorporated phosphoserines at residues 33 and 37 as well as an engineered cAMP phosphorylation consensus sequence. The β-catenin peptide substrate was 32P-labeled by cAMP kinase.
Other proteins
The following reagents were prepared using established protocols or purchased. β-TrCP–Skp1, Ub E1, UbcH5c, Cdc34a, and Ubc12 were prepared using methods described by Wu et al. (31). E3 CRL4CRBN, ROC1/Rbx1–CUL1, Cdc34b, UBE2G1, Nedd8, Nedd8 E1, human Ub, fluorescein Ub, and Ub chains (Ub2–7) were purchased from R&D Systems. Ub chain Ub4 was purchased from Enzo. Nedd8–ROC1/Rbx1–CUL1 was a gift of Ning Zheng of the University of Washington, Seattle.
Decay of E2–Ub thioester
E2–Ub thioester was formed by a reaction (10 μl) containing 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), fluorescein Ub (1 μM), E1 (100 nM), and UbcH5c (1 μM) or Cdc34b (1 μM). The reaction was incubated at room temperature for 5 min. Formation of E2–Ub thioester was quenched by the addition of SDS loading buffer (without DTT). To destabilize E2–Ub, DTT was added to a final concentration of 50 mM. To determine the stability of E2–Ub, apyrase was added to the preformed E2–Ub thioester complex to a concentration of 6 mU/μl, and the resulting mixture continued incubation at 37 °C for times as indicated. After separation by 4 to 20% SDS-PAGE, the reaction products were visualized and quantified by fluorescent imaging (Typhoon).
Nedd8 modification
E3 CRL4CRBN (100 nM) was incubated in a reaction (3 μl) containing 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Nedd8 (10 μM), Nedd8 E1 (7.5 nM), and Ubc12 (1.7 μM). The reaction was incubated at room temperature for 5 min.
Protocols for ubiquitination
Substrate ubiquitination
For ubiquitination experiments described in Figures 1, 3B, 4A, 6, A and B, and 7, the reaction was initiated by combining preformed mixtures that contained E2–S–Ub and E3–substrate, respectively. The E2 charging reaction was assembled typically in a mixture (2.5–5 μl) that contained 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA, Ub (20 μM), or Ub chains in concentrations as specified, E1 (100 nM), and UbcH5c (1–2 μM), or Cdc34 (1–3 μM; or in concentration as specified). The reaction was incubated for 10 min at room temperature. To assemble the E3–substrate complex, a mixture (2.5–5 μl) containing E3 (100 nM; or in concentration as specified), fluorescently or radioactively labeled substrate (CK1α, 250 nM; β-catenin, 240 nM), lenalidomide (1 μM) as specified, 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, 0.1 mg/ml BSA was incubated for 10 min at room temperature. Finally, the E2–S–Ub and E3–substrate mixtures were combined (in a final volume of 10 μl with 70 mM sodium glutamate) and incubated at 37 °C for times as indicated. After separation by 4 to 20% SDS-PAGE, the reaction products were visualized and quantified by fluorescent or phosphor imaging (Typhoon).
Note that Nedd8–SCFβTrCP was assembled by incubating Nedd8–ROC1–CUL1 (100 nM) and β-TrCP–Skp1 (100 nM) at room temperature for 10 min.
Apyrase chase
For “apyrase chase” experiments described in Figures 3C, 5, 6C and 8, the reaction is as follows. The preformed E3–substrate complex and UbcH5c–Ub or UbcH5c–Ubn, both generated as described previously, were combined for incubation at 37 °C for times as indicated. Apyrase (6 mU/μl) was added to the reaction mixture and incubated at 37 °C for 1 min. The preformed Cdc34b–Ub, generated as described previously, was treated with apyrase (6 mU/μl) at 37 °C for 1 min. These two apyrase-treated reaction mixtures were then combined for the chase reaction at 37 °C for times as indicated.
Competition
For the experiment described in Figure 9A, increasing concentrations of unlabeled β-catenin peptide were mixed with the radioactive β-catenin substrate in a reaction (2.5 μl) containing 50 mM Tris–HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 0.5 mM DTT, and 0.1 mg/ml BSA. This substrate mix was then combined with E3 to form the E3–subtrate complex, and then added with E2–Ub, as described previously. For experiments described in Figure 9, B–D, ubiquitinated substrate with defined chain length was formed by incubating E3–substrate with UbcH5c–Ubn and then treated with apyrase using the protocol as described previously. Increasing concentrations of unlabeled β-catenin peptide were added, and the reaction continued incubation for 10 min at room temperature. Finally, preformed Cdc34b–Ub was added, and the reaction continued at 37 °C for 10 min as indicated.
Data availability
All data are contained within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank I. Lu for technical assistance and N. Zheng for reagents.
Author contributions
K. W. and Z.-Q. P. methodology; K. W., R. J. D., and Z.-Q. P. formal analysis; K. W. and Z.-Q. P. investigation; Z.-Q. P. writing–original draft.
Funding and additional information
This work was supported by the National Institutes of Health grant CA251425 (to Z.-Q. P. and R. J. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
References
- 1.Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Rajalingam K., Dikic I. SnapShot: expanding the ubiquitin code. Cell. 2016;164:1074.e1. doi: 10.1016/j.cell.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 3.Morreale F.E., Walden H. Types of ubiquitin ligases. Cell. 2016;165:248–248.e1. doi: 10.1016/j.cell.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Petroski M.D., Deshaies R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Sarikas A., Hartmann T., Pan Z.Q. The cullin protein family. Genome Biol. 2011;12:220. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodrigo-Brenni M.C., Morgan D.O. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 7.Wu K., Kovacev J., Pan Z.Q. Priming and extending: an UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacev J., Wu K., Spratt D.E., Chong R.A., Lee C., Nayak J., et al. A snapshot at ubiquitin chain elongation: lysine 48-tetra-ubiquitin slows down ubiquitination. J. Biol. Chem. 2014;289:7068–7081. doi: 10.1074/jbc.M113.530576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek K., Krist D.T., Prabu J.R., Hill S., Klügel M., Neumaier L.M., et al. NEDD8 nucleates a multivalent cullin-RING-UBE2D ubiquitin ligation assembly. Nature. 2020;578:461–466. doi: 10.1038/s41586-020-2000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott D.C., Rhee D.Y., Duda D.M., Kelsall I.R., Olszewski J.L., Paulo J.A., et al. Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell. 2016;166:1198–1214.e24. doi: 10.1016/j.cell.2016.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horn-Ghetko D., Krist D.T., Prabu J.R., Baek K., Mulder M.P.C., Klügel M., et al. Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly. Nature. 2021;590:671–676. doi: 10.1038/s41586-021-03197-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Z.Q., Kentsis A., Dias D.C., Yamoah K., Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 13.Unk I., Hajdú I., Fátyol K., Szakál B., Blastyák A., Bermudez V., et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13 Dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen D.E., Brzovic P.S., Klevit R.E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 15.Krönke J., Fink E.C., Hollenbach P.W., MacBeth K.J., Hurst S.N., Udeshi N.D., et al. Lenalidomide induces ubiquitination and degradation of CK1α indel(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petzold, G G., Fischer E.S., Thomä N.H. Structural basis of lenalidomide-induced CK1α degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- 17.Wu K., Huynh K.Q., Lu I., Moustakim M., Miao H., Yu C., et al. Inhibitors of cullin-RING E3 ubiquitin ligase 4 with antitumor potential. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2007328118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu G., Weng S., Matyskiela M., Zheng X., Fang W., Wood S., et al. UBE2G1 governs the destruction of cereblon neomorphic substrates. Elife. 2018;7 doi: 10.7554/eLife.40958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sievers Q.L., Gasser J.A., Cowley G.S., Fischer E.S., Ebert B.L. Genome-wide screen identifies cullin-RING ligase machinery required for lenalidomide-dependent CRL4CRBN activity. Blood. 2018;132:1293–1303. doi: 10.1182/blood-2018-01-821769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill S., Reichermeier K., Scott D.C., Samentar L., Coulombe-Huntington J., Izzi L., et al. Robust cullin-RING ligase function is established by a multiplicity of poly-ubiquitylation pathways. Elife. 2019;8 doi: 10.7554/eLife.51163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G., Xu G., Schulman B.A., Jeffrey P.D., Harper J.W., Pavletich N.P. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol. Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 22.Saha A., Deshaies R.J. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce N.W., Kleiger G., Shan S.O., Deshaies R.J. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z.Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dikic I., Schulman B.A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 2023;24:273–287. doi: 10.1038/s41580-022-00543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y., Wang W., Kirschner M.W. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015;348 doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown N.G., VanderLinden R., Watson E.R., Weissmann F., Ordureau A., Wu K.P., et al. Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell. 2016;165:1440–1453. doi: 10.1016/j.cell.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonetta K.R., Taygerly J., Boyle K., Basham S.E., Padovani C., Lou Y., et al. Prospective discovery of small molecule enhancers of an E3 ligase-substrate interaction. Nat. Commun. 2019;10:1402. doi: 10.1038/s41467-019-09358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao S., Kang S., Mao H., Yao J., Gu L., Zheng N. Defining molecular glues with a dual-nanobody cannabidiol sensor. Nat. Commun. 2022;13:815. doi: 10.1038/s41467-022-28507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K., Ching K., Chong R.A., Pan Z.Q. A new Forster resonance energy transfer-based platform to track substrate ubiquitination by fluorescence. J. Biol. Chem. 2020;296 doi: 10.1074/jbc.RA120.016858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.