Abstract
Endoplasmic reticulum (ER) stress-related genes are closely related to the occurrence, development, and immunotherapy response of tumors. This study provides a comprehensive assessment of HSPA5 from a pan-cancer perspective using multi-omics data. We analyzed the function of HSPA5 in multiple tumor types using multiple databases. Finally, immunohistochemistry was used to examine the relationship between HSPA5 expression in tissue microarrays from 100 patients with bladder cancer and the prognosis of patients with bladder cancer. Using the TCGA database, we were able to determine that HSPA5 is significantly elevated in a number of common malignancies and is linked with a bad prognosis. Cox regression analysis showed that the high expression of HSPA5 was correlated with OS, progression free survival (PFS), disease free survival (DFS), and disease special survival (DSS) of adrenocortical carcinoma (ACC). In addition, we discovered significant disparities in HSPA5 methylation and phosphorylation levels between various malignancies and normal tissues. HSPA5 expression was significantly correlated with the levels of infiltrating cells and immune checkpoint genes. HSPA5 is highly expressed in bladder cancer and patients with high HSPA5 expression have a poor prognosis. Our study provides a basis for further understanding of the role of ER stress-related gene HSPA5 in different tumor genesis and development. HSPA5 has also been shown to be a prognostic biomarker for bladder cancer patients.
Keywords: HSPA5, ER stress, Prognosis, BLCA
1. Introduction
Cancer is one of the main causes of mortality and disability worldwide, making it a major public health concern [1]. Cancer may be treated in a number of ways today, some of which include cutting into the patient. Despite these medicines' successes in combating cancer, a sizable percentage of patients get relatively little from them. This serious problem emphasises the need for a comprehensive knowledge of tumour genesis pathways [2]. New targets and biomarkers for the diagnosis and treatment of cancer are urgently needed. Through their ongoing development, the TCGA and GEO databases have simplified the analysis of individual gene expression, gene connection, clinical prognosis, and signal pathway regulation.
Secretory protein synthesis, folding, and modification all take place in the endoplasmic reticulum (ER), a key organelle [3]. Even though the endoplasmic reticulum (ER) tightly regulates protein processing and synthesis, ER stress, characterized by the accumulation of misfolded or unfolded proteins, may be caused by a variety of external factors and intracellular processes [4]. Cancer cells' function, destiny, and survival are all adversely affected by ER stress, which is caused by the tumor microenvironment and abnormal transcription and metabolism [5]. Targeting HSPA5 was shown to induce endoplasmic reticulum stress in both melanoma and hepatocellular carcinoma [6,7]. HSPA5 is a crucial chaperone protein in the normal unfolded protein response (UPR). However, this effect can easily lead to resistance of cancer cells to treatment [8]. Through many molecular processes, HSPA5 promotes the proliferation and survival of cancer cells. Its overexpression leads to primary cancer cell growth and metastasis, increased medication and treatment resistance, and worse clinical outcomes overall [9]. Inhibiting HSPA5 also boosts the potency of cancer drugs like chemotherapy. For instance, mantle cell lymphoma cells were made more sensitive to the anticancer proteasome inhibitor bortezomib by the small molecule Hsp 90 inhibitor retaspimycin hydrochloride [10]. As consequently, HSPA5 is pivotal in tumour development. Recent studies have shown that HSPA5 regulates proliferation, metastasis and iron death of bladder cancer cells through the P53/SLC7A11/GPX4 pathway [11]. And our study further confirmed the important value of HSPA5 in bladder cancer through clinical samples. It has also been shown that HSPA5 should play an important role in the entry of SARS-Cov-2 into cancer patients through the lungs. In addition, HSPA5 showed a high correlation with COVID-19 [12,13]. Chimeric antigen receptor T (CAR-T) cell therapy has been successfully applied to treat hematologic malignancies but faces many challenges in solid tumors. Cell surface GRP78-targeted CAR-T cells have also been shown to be effective in treating human pancreatic cancer [14]. In addition, HSPA5 has been shown to affect the stemness profile of cancer cells in esophageal, glioblastoma, gastric and breast cancers [[15], [16], [17], [18]].
This is the first research to investigate HSPA5's possible molecular pathways in the aetiology of various malignancies by analysing its gene expression, survival, and mutation using the TCGA datasets.
2. Materials and methods
2.1. Samples and datasets
Using information from the GTEx database and the Cancer Genome Atlas (TCGA), we compared HSPA5 expression in distinct cancer types and normal tissues. In addition, between June 2012 and March 2018, the Nantong Tumour Hospital collected data from 100 patients diagnosed with bladder cancer and 41 cases of normal bladder tissue undergoing partial and radical cystectomy. After surgery, the follow-up period for each patient ranged from one to six years and lasted until August 2019. All subjects provided their informed assent in writing. The Ethics Committee of Nantong Cancer Hospital approved the study.
2.2. Prognostic analysis of HSPA5 in different tumors
Forest plots and Kaplan-Meier curves were used to examine the association between HSPA5 expression and the various survival rates of patients with multiple tumors. A univariate survival analysis was performed in order to calculate the hazards ratios (HR) and the confidence intervals for 95%.
2.3. Analysis of methylation and phosphorylation levels of HSPA5 in different tumors
The study compared HSPA5 methylation and phosphorylation levels in cancer and surrounding tissues using the UALCAN database.
2.4. Gene mutation analysis of HSPA5 in different tumors
The somatic mutation, structural variation, amplification, lack of depth, and numerous alterations in HSPA5 in malignancies were evaluated using the Cbioportal for Cancer Genomics website. On top of that, TCGA patients with and without HSPA5 gene mutations were compared for OS, DFS, PFS, and RFS.
2.5. Functional enrichment analysis of HSPA5 gene
RNAseq data (level 3) and corresponding clinical information for the eight tumors required for analysis were obtained from the TCGA dataset. Two categories, high and low HSPA5 expression, were used to classify the samples. Limma, a R tool for analysing differential gene expression, was used to analyze the HSPA5 mRNA levels (version: 3.40.2). Threshold mRNA differential expression screening was specified as “log2 (fold change) >2 or log2 (fold change)<-2.” ClusterProfiler package (version: 3.18.0) in R was employed to analyze the gene ontology (GO) function of potential targets and enrich the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [19].
2.6. Correlation analysis of HSPA5 expression with tumor immune infiltration-related cells and immunoassay site genes
Accurate evaluation of immune scores for these tumors was performed using Immuneeconv, a R programme that incorporates the most recent TIMER algorithms. Using TIMER methods, we analyzed heat maps of the Spearman connection between HSPA5 and genes associated with immune examination locations and immunological scores. TISCH was used to obtain immune cell-related single-cell data.
2.7. Correlation analysis between HSPA5 gene expression and TMB or MSI, and the efficacy of immune checkpoint blockade
The publication of the article “The Immune Landscape of Cancer” by Vesteinn Thorsteinn et al., in 2018 served as the inspiration for the development of the TMB protocol. A 2017 publication by Russell Bonneville et al., titled “Landscape of Microsatellite Instability Across 39 Cancer Types,” is the source for MSI. Two-group data have been typically analyzed using the Wilcoxon test unless otherwise specified. The TIDE method was used to provide a prediction of the potential ICB reaction [20].
2.8. Immunohistochemistry
The tissue microarrays underwent several steps for processing. First, they were baked in an oven at 85 °C for 10 min. Then, they were soaked in xylene for 15 min and hydrated in a series of ethanol concentrations: 100%, 95%, 80%, and 70%. After that, the chips were treated with citric acid solution in an autoclave for antigen repair. Once cooled, the tissue chips were washed with PBS and incubated with hydrogen peroxide for 20 min. Following this, HSPA5 antibody (1:2000, ab21685) was added and incubated for 2 h at room temperature. After the completion of the above procedure, the tissue microarrays were washed 3 times with PBS and incubated with an immunohistochemical secondary antibody for 20 min at room temperature. The microarrays were washed 3 times with PBS again, and then DAB was added for staining, followed by staining with hematoxylin. Subsequently, the microarrays were dehydrated in a series of ethanol concentrations: 70%, 80%, 90%, and 100%. Finally, they were immersed in xylene for 8 min, and the microarrays were blocked after all of the aforementioned processes. Immunostaining intensity scores ranged from 0 to 3 (0, no reaction; 1, weak reaction; 2, moderate reaction; 3, strong reaction). Scales were scored as 1 (0%–25%), 2 (26%–50%), 3 (51%–75%) and 4 (76%–100%). The final scores were obtained by multiplying the strength scores and the proportional scores. The results are as follows: 0–5: low expression; 6–12: high expression.
2.9. Statistical analysis
Expression levels of HSPA5 were compared in cancerous and healthy tissues using T-tests. With the use of a univariate Cox regression, we were able to calculate the HR and the p-value for the survival analysis. The p < 0.05 threshold was used in all statistical analyses. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
3. Result
3.1. Expression of HSPA5 mRNA in tumor
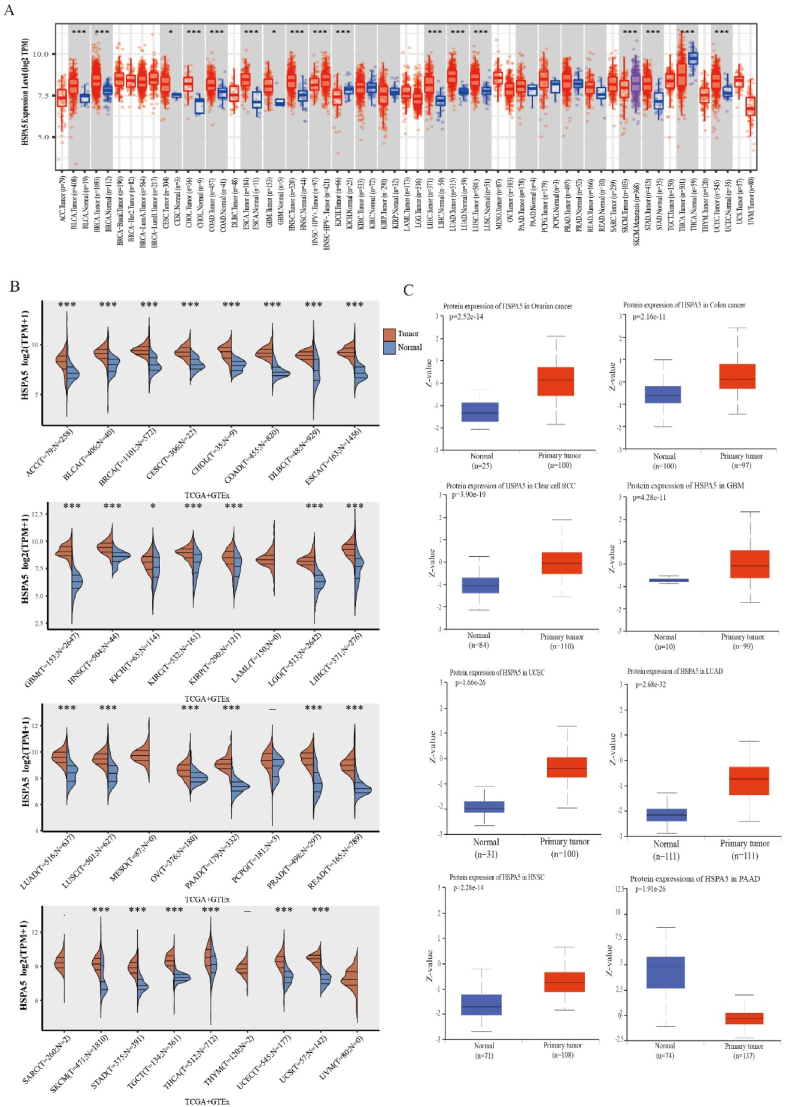
First, we investigated the frequency of HSPA5 mRNA expression in 34 different types of common human malignant tumors using data from the TIMER database [21]. Our findings revealed that BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, LIHC, LUAD, LUSC, STAD, and UCEC exhibited significantly higher HSPA5 mRNA expression levels compared to the corresponding normal samples (Fig. 1A). Furthermore, we also analyzed HSPA5 expression using TCGA + GTEx data and observed that the expression of HSPA5 mRNA in most tumors remained significantly higher than that in normal tissues, except for LAML, MESO, PCPG, SARC, THYM, and UVM (Fig. 1B). To further investigate the expression of HSPA5 protein in tumors, we examined the CPTAC database. Our analysis revealed that the expression of HSPA5 was significantly higher in OV, COAD, KIRC, GBM, UCEC, LUAD, and HNSC compared to normal tissues. However, in the case of PAAD, the expression of HSPA5 in CPTAC was significantly lower than that in the corresponding normal tissues, possibly due to the smaller sample size (Fig. 1C).
Fig. 1.
mRNA expression of HSPA5 in different tumors in general. (A) HSPA5 mRNA levels in different tumors in the TCGA database. (B) HSPA5 mRNA levels in different tumors in the TCGA + GTEx database. (C) CPTAC dataset showing significant differences in HSPA5 protein expression in normal and primary tissues of OV, COAD, KIRC, GBM, UCEC, LUAD, HNSC and PAAD.
3.2. Prognostic analysis of HSPA5 function in tumor
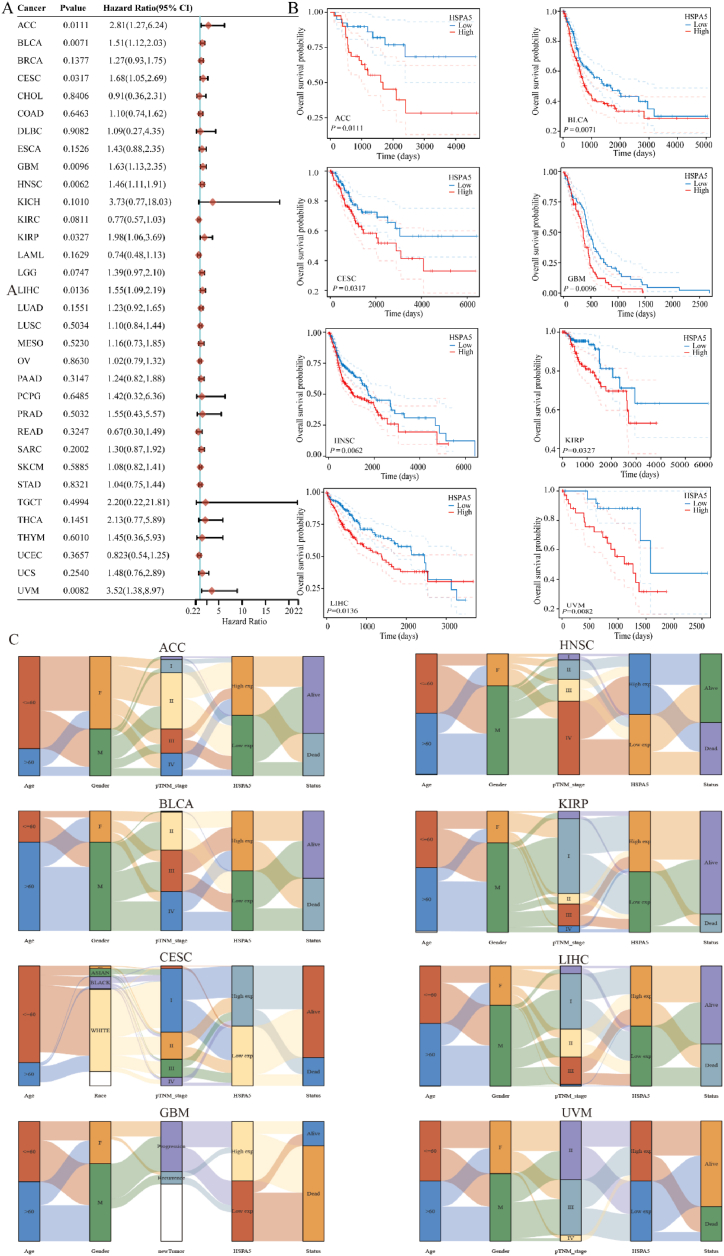
Next, we looked at how HSPA5 expression was linked to survival rates across different malignancies. Survival indicators in our analyses included OS, PFS, DFS, and DSS. Cox regression analysis showed that HSPA5 expression was significantly associated with OS in 8 of the 33 cancers, including ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and uveal melanoma (UVM). In all eight tumors, the prognosis of patients in the HSPA5 high-expression group was worse than that in the HSPA5 low-expression group. Kaplan-Meier survival curves were used to demonstrate the results (Fig. 2A and B). Based on the above conclusions, we further analyzed the correlation between HSPA5 expression and patient age, sex, race, and TNM stage in eight tumors, which revealed a significant correlation to poor prognosis in overall survival in Fig. 2A. Using Sankey diagrams, we found that in these eight tumors, there were always more deaths in samples with high HSPA5 expression than in samples with low HSPA5 expression (Fig. 2C). Finally, the correlation between HSPA5 and PFS, DFS, and DSS in these 33 tumors was analyzed by Cox regression. HSPA5 expression was significantly correlated with PFS in ACC, BLCA, CESC, HNSC, LGG, LUSC, THCA, and UVM (Supplementary Fig. 1A), and was significantly correlated with DFS in HNSC, LUSC, mesothelioma (MESO), and THCA (Supplementary Fig. 1B). Moreover, HSPA5 expression was significantly correlated with DSS in ACC, BLCA, ESCA, GBM, HNSC, KIRP, LGG, LIHC, LUSC, and UVM (Supplementary Fig. 1C).
Fig. 2.
Prognostic value of HSPA5 in different tumors. (A): Forest plot showing the prognostic value of HSPA5 in 33 tumors (B): Kaplan-Meier curves of HSPA5 in eight tumors. (C): Sankey plots of HSPA5 expression versus patient age, gender and TNM stage in eight tumors.
3.3. Clinical significance and protein expression of HSPA5 in different tumors
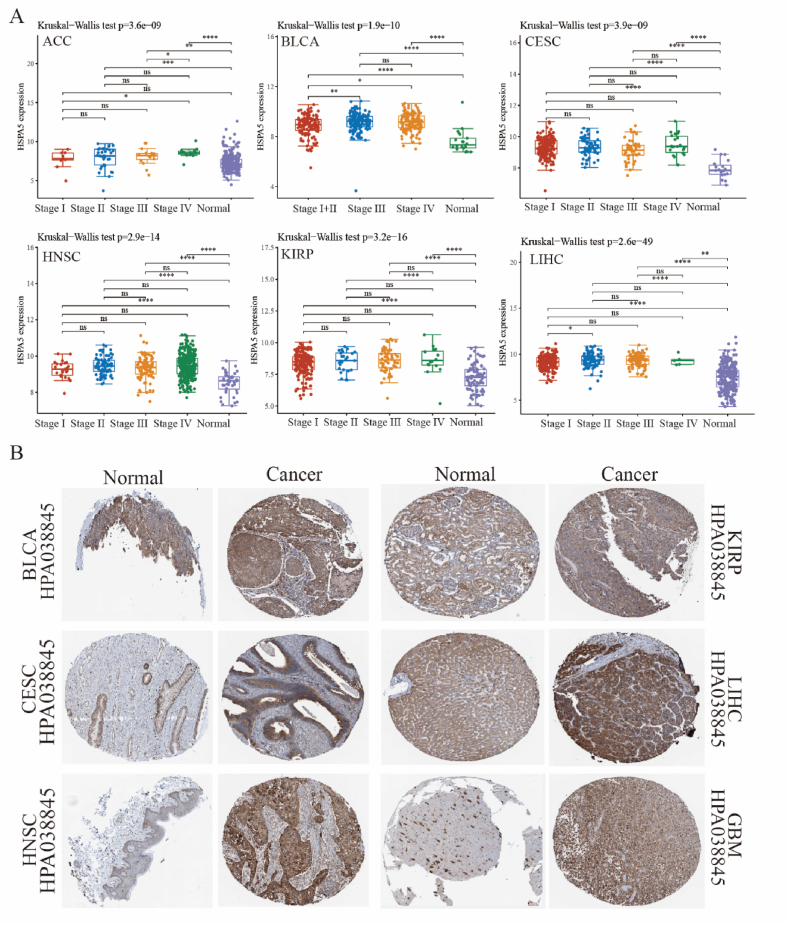
The analysis of HSPA5 mRNA expression in various tumors and its prognostic significance reveals its essential role in ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM biology. Further analysis was conducted to examine the expression of HSPA5 in different stages of these tumors. The results demonstrated significant differences in HSPA5 expression across stages of ACC, BLCA, CESC, HNSC, KIRP, and LIHC. In ACC, the expression of HSPA5 was significantly higher in Stage IV compared to Stage I and Stage III. In BLCA, the expression of HSPA5 in Stage III and Stage IV was significantly higher than that in Stage I + II. In CESC, HNSC, and KIRP, the expression of HSPA5 was higher in all stages (I, II, III, and IV) compared to normal samples. Similarly, in LIHC, HSPA5 expression was significantly higher in Stage I than in Stage II (Fig. 3A). We further clarified the expression level of HSPA5 protein in these tumors from The Human Protein Altas, showing the HSPA5 protein in BLCA, CESC, HNSC, KIRP, LIHC, and GBM tissues to be significantly higher than in the normal ones (Fig. 3B).
Fig. 3.
Expression of HSPA5 in ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM. (A): Significant differences in HSPA5 mRNA expression in ACC, BLCA, CESC, HNSC, KIRP, and LIHC across clinical stages. (B): HSPA5 protein expression levels in BLCA, CESC, HNSC, KIRP, LIHC and GBM.
3.4. Analysis of HSPA5 methylation and phosphorylation levels in different tumors
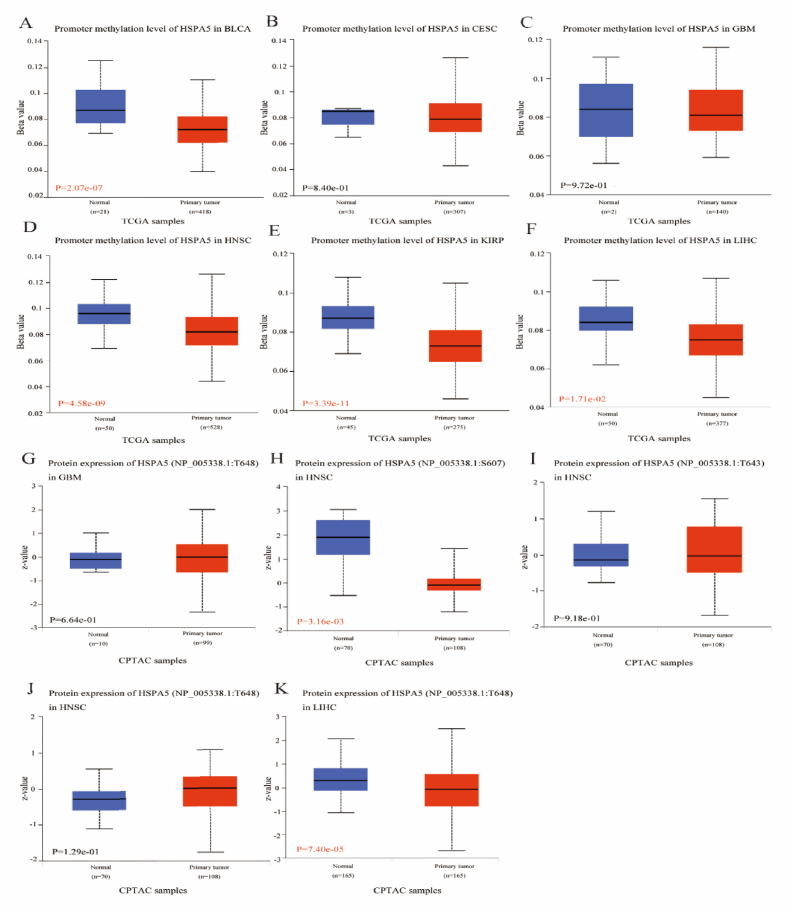
Using the UALCAN and TCGA datasets, we looked into the DNA methylation and HSPA5 phosphorylation levels in these eight tumors. BLCA, HNSC, UCEC, KIRP, and LIHC revealed substantially lower HSPA5 methylation levels compared to normal tissues in the UALCAN database, however CESC and GBM showed no significant change from normal tissues (Fig. 4A–F). Subsequently, Primary tumour tissues and normal tissues were compared for their HSPA5 phosphorylation levels. In HNSC, HSPA5 phosphorylation at S607 was considerably decreased compared to levels seen in normal tissues. In LIHC, the phosphorylation level of HSPA5 at T648 was significantly lower than that in normal tissues. Moreover, the phosphorylation level of HSPA5 in other tumors was not significantly different from that in normal tissues (Fig. 4G–K).
Fig. 4.
Analysis of HSPA5 methylation and phosphorylation levels in various cancers. (A–F): Plots showing methylation levels of HSPA5 in BLCA, CESC, GBM, HNSC, KIRP, and LIHC. (G–K): Plots showing phosphorylation levels of HSPA5 in GBM, HNSC, and LIHC.
3.5. Mutation analysis of HSPA5 in different tumors
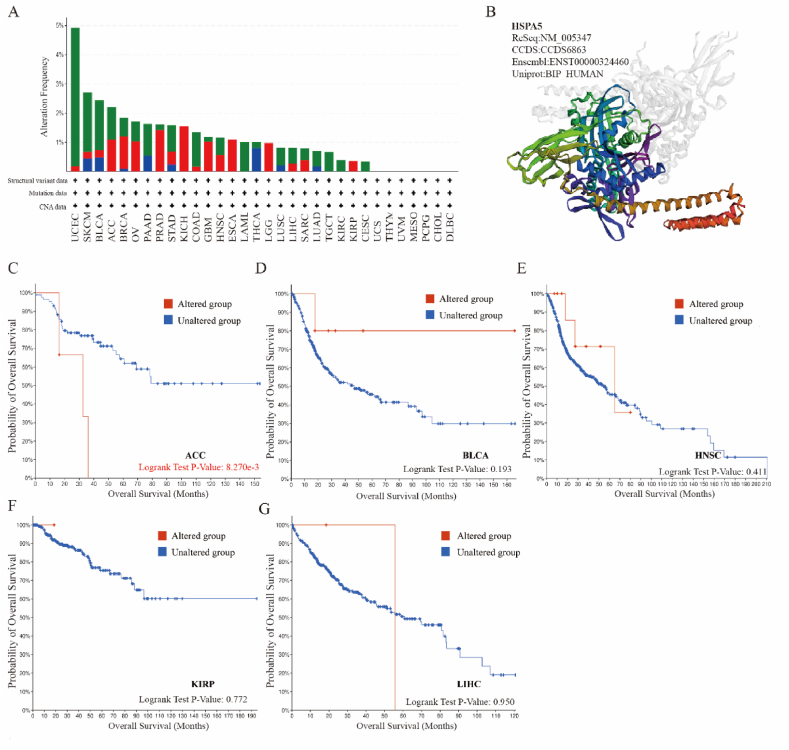
We investigated HSPA5 mutations in different tumors using the cBioPortal database. In Fig. 5A, Mutation is shown in green, amplification in red, and a large deletion in blue. Among ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM, HSPA5 had the highest mutation frequency in BLCA. Mutations were also predominant in CESC, and LIHC. HSPA5 was mainly amplified in ACC, GBM, HNSC, and KIRP. Subsequently, we performed a three-dimensional structural analysis of HSPA5, depicted by a schematic (Fig. 5B). Finally, an analysis of the impact of HSPA5 mutations on the overall survival of patients with these tumors showed them to be significantly related to the poor prognosis of ACC patients, despite no significant difference in other tumors (Fig. 5C–G).
Fig. 5.
Mutation analysis of the HSPA5 gene. (A): Outputs from cBioPortal database, showing the modification frequency with various mutations in the HSPA5 gene. (B): Plots showing the 3D analysis result of HSPA5. (C–G): Effect of HSPA5 gene mutation on overall survival in ACC, BLCA, HNSC, KIRP, and LIHC.
3.6. GSEA analysis of the potential role of HSPA5 in tumors
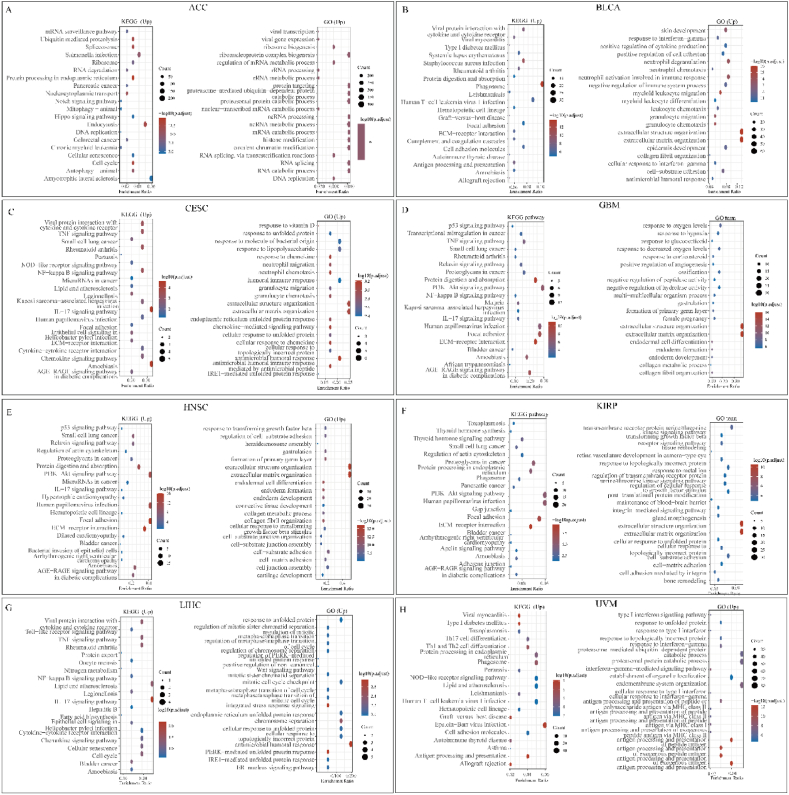
To further analyze the pathogenic mechanism of HSPA5 in different tumors, we used KEGG analysis and GO analysis. In the ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM, we divided the samples into two groups depending on HSPA5 high and low expression, for differential analysis, and the obtained differentially expressed genes were analyzed for gene enrichment analysis. In ACC, several cell cycles signaling pathways related to HSPA5 were significantly enriched with upregulated gene sets, suggesting that this gene may affect ACC progression by affecting ACC cell cycle. (Fig. 6A). GSEA results of down-regulated gene sets were also enriched in various signaling pathways, such as PPAR signaling pathway (Supplementary Fig. 2A). In BLCA, GSEA results of up-regulated gene sets indicate that HSPA5 regulates Phagosome, Focal adhesion, and immune-related signaling pathways (Fig. 6B), the results of the down-regulated gene set enrichment analysis were also enriched for the PPAR signaling pathway (Supplementary Fig. 2B). In CESC, the enrichment analysis of up-regulated gene sets revealed several well-known signaling pathways, such as TNF, NF-Kappa B signaling pathway (Fig. 6C). The results of down-regulated gene set enrichment analysis indicated that the Estrogen signaling pathway may be regulated by HSPA5 (Supplementary Fig. 2C). In GBM, due to the low number of down-regulated genes, we only performed functional analysis on up-regulated genes. The HSPA5-mediated tumor mechanisms were similar to those in CESC (Fig. 6D). In HNSC, enrichment analysis showed the probability of HSPA5 affecting the PI3K-Akt and P53 signaling pathway, and the results of the enrichment analysis of down-regulated gene sets all indicated that the IL17 signaling pathway plays an important role (Fig. 6E, Supplementary Fig. 2D). In KIRP, the number of down-regulated genes was similarly low. GSEA showed significant enrichment of various well-known signaling pathways, such as PI3K-Akt and AGE-RAGE signaling pathway (Fig. 6F). Similarly, we hypothesized that HSPA5 may affect LIHC progression by affecting the cell cycle and immune microenvironment (Fig. 6G, Supplementary Fig. 2E). Finally, in UVM, enrichment analysis showed that HSPA5 could affect disease progression through NOD-like receptor, JAK-STAT signaling pathways (Fig. 6H, Supplementary Fig. 2F). In addition, we show a volcano map of HSPA5-related differential genes in these eight tumors (Supplementary Fig. 2G).
Fig. 6.
Gene Set Enrichment Analysis (GSEA) of HSPA5 in various cancers. (A–H): Enrichment plots of KEGG and GO analysis in ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM using HSPA5 high and low expression groups.
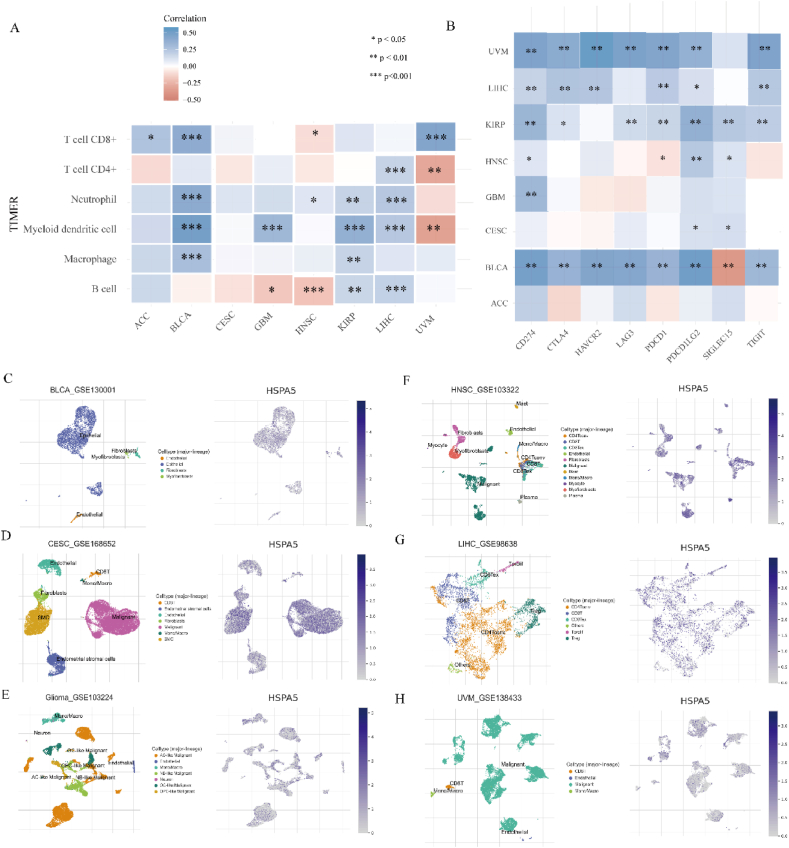
3.7. Correlation analysis of HSPA5 expression with immune cell infiltration and immunoassay sites
The association between HSPA5 expression and immune infiltration was investigated using the TIMER algorithm. HSPA5 expression showed significant correlation with immune infiltration in all tumor cells except for CESC. In ACC, HSPA5 was positively correlated with T cell CD8+ infiltration. In BLCA, HSPA5 was positively correlated with T cell CD8+, Neutrophil, Myeloid dendritic cell, and Macrophage infiltration. In GBM, HSPA5 was negatively correlated with B cell infiltration and positively correlated with Myeloid dendritic cell infiltration. In HNSC, HSPA5 was negatively correlated with T cell CD8+ and B cell infiltration, and positively correlated with Neutrophil infiltration. In KIRP, HSPA5 was positively correlated with Neutrophil, Myeloid dendritic cell, Macrophage, and B cell infiltration. In LIHC, HSPA5 was positively correlated with T cell CD4+, Neutrophil, Myeloid dendritic cell, and B cell infiltration. In UVM, HSPA5 was positively correlated with T cell CD8+ infiltration and negatively correlated with T cell CD4+ and Myeloid dendritic cell infiltration (Fig. 7A). Furthermore, we conducted an analysis on the correlation between HSPA5 and immune checkpoints. Our findings revealed a significant correlation between HSPA5 expression in BLCA and the expression of all immune checkpoint-associated genes. However, in the case of ACC, the opposite was observed (Fig. 7B). To explore the correlation of immune cell distribution and ratio with HSPA5 expression levels at the single-cell level, we obtained relevant data for BLCA, CESC, GBM, HNSC, LIHC, and UVM from the Tumor Immune Single-cell Hub 2 (TISCH2). This database does not contain ACC and KIRP-related data, so we only analyzed the other six tumors (Fig. 7C–H). In Summary, these results indicated that HSPA5 may be involved in immune regulation, which may influence immunotherapy response.
Fig. 7.
The HSPA5 expression correlated with immune infiltration. (A): The TIMER algorithm was used to analyze the correlation between HSPA5 and the level of immune infiltration of different tumor cells. (B): Analysis of HSPA5 correlation with different immune checkpoint genes. (C): Single-cell analysis of HSPA5 correlates with the level of immune infiltration of different tumor cells.
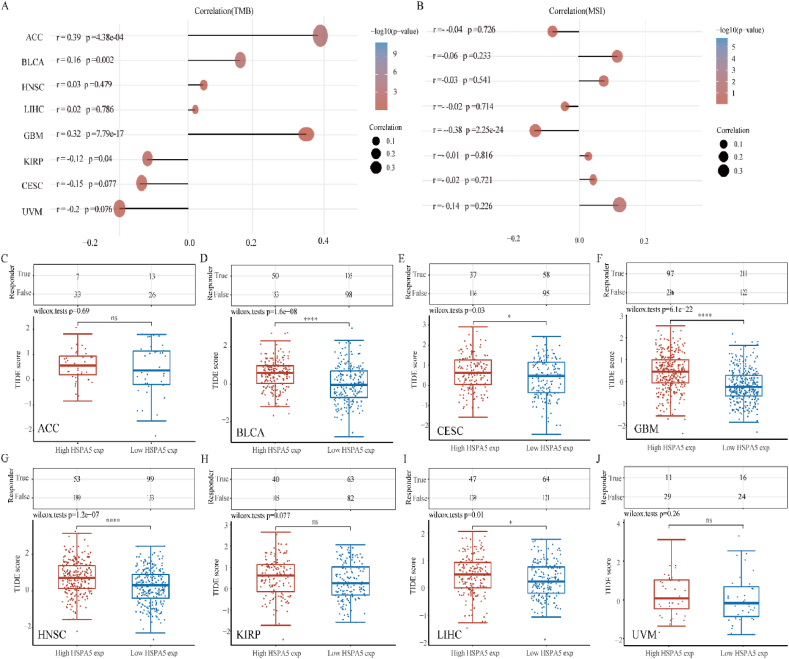
3.8. Analysis of HSPA5 correlation with TMB, MSI while predicting potential immunotherapeutic response using TIDE algorithm
TMB and MSI are two new biomarkers for assessing the effectiveness of immunotherapy. First, we investigated the relationship between HSPA5 expression and TMB, and we found that HSPA5 expression and TMB were positively correlated in ACC, BLCA, and GBM, while HSPA5 expression and TMB were negatively correlated in KIRP (Fig. 8A). We then analyzed the correlation between HSPA5 expression and MSI, and we found that only in GBM HSPA5 expression was negatively correlated with MSI. (Fig. 8B). High TIDE score, ineffective ICB therapy, and a short survival time are all indicators of tumor immune escape via the two mechanisms of dysfunction of tumor-infiltrating cytotoxic T lymphocytes (CTLS) and rejection of CTLS by immunosuppressors used in the tumor Immune Dysfunction and Exclusion (TIDE) algorithm [22]. On the basis of HSPA5 mRNA expression profile data, the TIDE algorithm was used to provide predictions about the degree to which individual samples will respond to immune checkpoint inhibitors. In BLCA, CESC, GBM, HNSC, and LIHC, the TIDE score of the HSPA5 high expression group was significantly higher than that of the HSPA5 low expression group. This suggests that patients with high HSPA5 expression have a worse outcome when treated with immune checkpoint inhibitors compared to patients in the low HSPA5 expression group. However, in ACC, KIRP and UVM, there was no significant difference in direct TIDE scores between the HSPA5 high and HSPA5 low expression groups (Fig. 8C–J). In conclusion, our results suggested that in most tumors, when HSPA5 is highly expressed, immune checkpoint blockade therapy has poor efficacy and survival is short after receiving ICB therapy.
Fig. 8.
The relationship between HSPA5 and tumor immunotherapy. (A): Stick chart showing HSPA5 gene expression with TMB in several malignancies. (B): A stick chart depicting the association between HSPA5 gene expression and MSI in various cancers. (C–J) Prediction of potential immunotherapeutic responses in patients with high HSPA5 expression and patients with low HSPA5 expression in different tumors using the TIDE algorithm.
3.9. Prognostic value of HSPA5 in bladder cancer patients by tissue microarray analysis
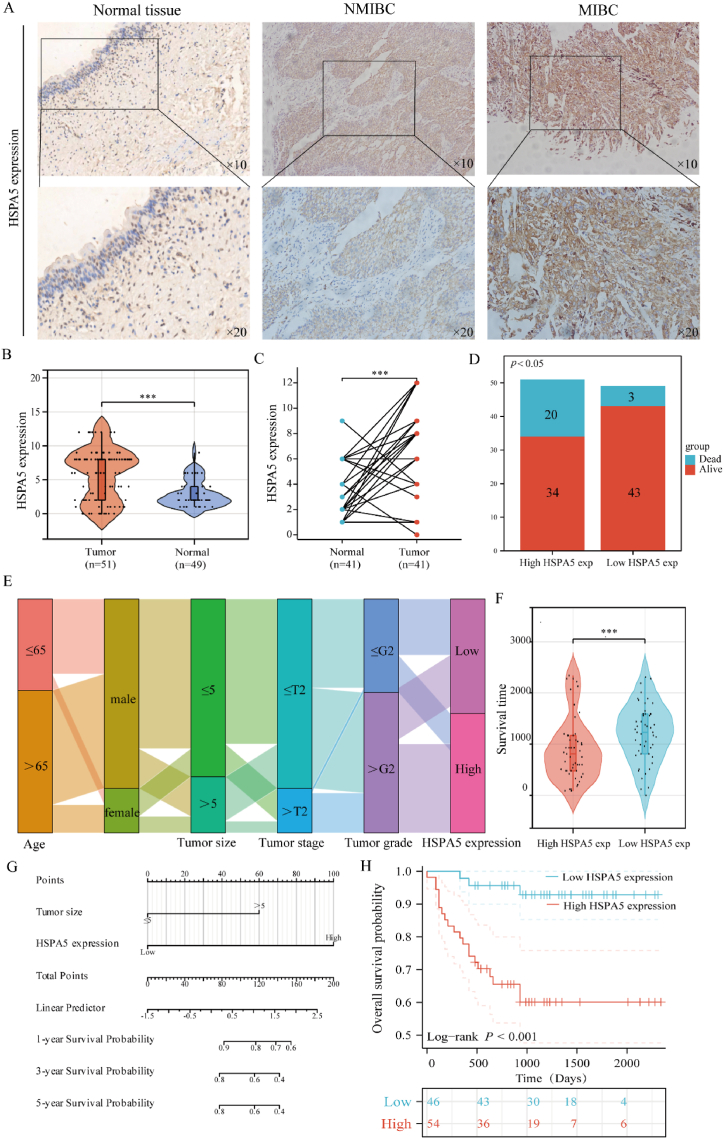
Based on these studies, we found that among the many tumors with significant differences in HSPA5 expression. It has been shown that HSPA5 has a significant impact on the prognosis of bladder cancer as well as the immune microenvironment. Furthermore, in addition to its prognostic value for bladder cancer, HSPA5 correlated with all of the immune checkpoint genes included in the study, and after grouping based on HSPA5 expression, we found that the difference in responsiveness scores to receiving immune checkpoint inhibitor therapy was most significant between the HSPA5 high-expression group and the HSPA5 low-expression group. Therefore, we chose to further validate its role in bladder cancer. We collected samples from 100 bladder cancer patients to further validate the prognosis and expression differences of HSPA5 in bladder cancer samples. Expression of HSPA5 was shown to be considerably greater in bladder cancer compared to normal bladder tissue using immunohistochemical staining, and expression of HSPA5 was further found to be significantly higher in muscle-infiltrating bladder cancer compared to non-muscle-infiltrating bladder cancer using the same method. (Fig. 9A–B). Furthermore, we compared the levels of HSPA5 expression in 41 pairs of bladder cancer and normal bladder tissues and found that, once again, the cancerous tissues expressed much more of this gene (Fig. 9C). In addition, we found that the ratio of dead to surviving population was significantly higher in the HSPA5 high expression group than in the HSPA5 low expression group (Fig. 9D). Patient survival is shown in a Sankey diagram alongside the frequency of high and low HSPA5 protein expression in bladder cancer samples of varying stage grades, ages, and other clinical features (Fig. 9E). Patient survival was also considerably lower in the high HSPA5 expression group compared to the low HSPA5 expression group (Fig. 9F). Univariate analysis showed that HSPA5 expression (p<0.001) was linked with overall survival, as were tumor size (p = 0.023), tumor stage (p = 0.004), vascular invasion (p = 0.032), and recurrence (p = 0.032). Multivariate study of bladder cancer patients revealed that tumor size (p = 0.002) and HSPA5 expression (p = 0.002) were independently associated with prognosis (Table 1). Based on the results of the multi-factor analysis we have drawn the line graphs (Fig. 9G). Finally, we subdivided individuals with bladder cancer based on HSPA5 expression, finding that those with high HSPA5 expression were more likely to have a worse prognosis than those with low HSPA5 expression (Fig. 9H). We have demonstrated that HSPA5 is highly expressed in bladder cancer and that its high expression is substantially associated with a poor prognosis.
Fig. 9.
Expression and prognostic value of HSPA5 in bladder cancer. (A): Immunohistochemistry assessed bladder carcinoma HSPA5 expression. (B): HSPA5 protein was expressed in unmatched samples of bladder cancer. (C): HSPA5 protein was expressed in paired samples of BLCA. (D): Survival and mortality rates were compared between individuals with high and low HSPA5 expression levels who were diagnosed with bladder cancer. (E): Sankey diagram showing trends in the distribution of bladder cancer samples. (F): Comparison of bladder cancer patients with high and low HSPA5 expression in terms of survival analysis. (G): Columnar plots were drawn to predict the impact of HSPA5 and tumour size on the prognosis of bladder cancer patients at 1, 3 and 5 years. (H): Analysis of the impact on the prognosis of bladder cancer patients in groups with high and low HSPA5 expression.
Table 1.
Prognostic value of HSPA5 in bladder cancer analyzed by univariate and multifactorial COX regression.
| Characteristics | Total(N) | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Age | 100 | 0.278 | |||
| ≤65 | 39 | Reference | |||
| >65 | 61 | 0.633 (0.279–1.436) | 0.274 | ||
| Gender | 100 | 0.880 | |||
| male | 81 | Reference | |||
| female | 19 | 0.921 (0.313–2.707) | 0.881 | ||
| Tumor size | 100 | 0.023 | |||
| ≤5 | 76 | Reference | Reference | ||
| >5 | 24 | 2.713 (1.188–6.196) | 0.018 | 4.010 (1.666–9.654) | 0.002 |
| Tumor stage | 100 | 0.004 | |||
| ≤T2 | 81 | Reference | Reference | ||
| >T2 | 19 | 3.820 (1.634–8.933) | 0.002 | 1.446 (0.483–4.324) | 0.510 |
| Tumor grade | 100 | 0.112 | |||
| ≤G2 | 40 | Reference | |||
| >G2 | 60 | 2.052 (0.808–5.207) | 0.130 | ||
| Vascular invasion | 100 | 0.032 | |||
| No | 87 | Reference | Reference | ||
| Yes | 13 | 3.101 (1.215–7.917) | 0.018 | 1.187 (0.391–3.600) | 0.762 |
| Lymph node metastasis | 100 | 0.820 | |||
| No | 66 | Reference | |||
| Yes | 34 | 1.105 (0.468–2.610) | 0.819 | ||
| Recurrence | 100 | 0.032 | |||
| No | 62 | Reference | Reference | ||
| Yes | 38 | 2.465 (1.078–5.638) | 0.033 | 1.834 (0.682–4.935) | 0.230 |
| Distant metastasis | 100 | 0.153 | |||
| No | 90 | Reference | |||
| Yes | 10 | 2.383 (0.807–7.037) | 0.116 | ||
| HSPA5 expression | 100 | < 0.001 | |||
| Low | 46 | Reference | Reference | ||
| High | 54 | 7.240 (2.147–24.413) | 0.001 | 7.804 (2.148–28.360) | 0.002 |
4. Discussion
One of the most perilous illnesses endangering human health is cancer, the disease burden of which is increasing worldwide [23]. Understanding the molecular process of tumour genesis and progression and looking into possible new biomarkers for cancer diagnosis and prediction of cancer treatment outcomes is essential for cancer prevention and control methods. Malignant cells and invading immune cells are subjected to persistent ER stress caused by a number of metabolic and carcinogenic abnormalities in the tumour microenvironment (TME), which in turn constitutes a sexually active ER stress response that allows malignant cells to adapt to these stresses. At the same time, Cancer development is promoted by a coordinated set of immune regulatory systems [24]. HSPA5, as an ER stress-related gene, is highly expressed in various malignant tumor tissues such as colorectal [25], liver [26], and breast cancers [27], which may lead to malignant biological behaviors such as proliferation, invasion, induction of chemotherapy resistance and immune escape of tumor cells. However, there is a lack of thorough investigations of the HSPA5 functional role to highlight the similarities and variations between various malignancies that would provide key insights to bolster cancer prevention and develop individualized treatment plans. Recent research has placed a premium on genome-wide carcinomatosis analysis for early cancer identification and treatment in an effort to pinpoint gene mutations, RNA changes, and cancer drivers involved in cancer genesis and progression.
The expression and function of HSPA5 in all malignancies were analyzed for the first time in this work. Based on our findings, HSPA5 expression was much greater in tumors than in normal tissues (Fig. 1). In some cancers, the upregulation of HSPA5 was significantly associated with poor OS, PFS, DFS, and DSS (Fig. 2). Through the above analyses, we identified a vital oncogenic role of HSPA5 in ACC, BLCA, CESC, GBM, HNSC, KIRP, LIHC, and UVM, which also confirmed these eight tumors are our choices for carrying out the follow-up work. The methylation and phosphorylation levels of HSPA5 were significantly down-regulated in LIHC, suggesting an active HSPA5 status that may affect the progression of LIHC (Fig. 4). In addition, Gene enrichment analysis showed that HSPA5 might affect a variety of signaling pathways, leading to the malignant progression of tumor pairs, with the P53, TNF, NF-kappa B, PI3k-Akt signaling-related and immune-related gene sets being significantly enriched in multiple tumors. This observation indicates that HSPA5 can not only regulate the development of malignant tumors but also affect the tumor immune microenvironment to create a more conducive niche for tumor growth (Fig. 6). P53 is a major regulator of multiple cellular biological behaviors, including apoptosis, senescence, and autophagy [28]. The study by Kamil M et al. demonstrated that HSPA5 can regulate autophagy in lung adenocarcinoma cells by affecting p53 localization [29]. One of the most investigated routes in relation to inflammation, metastasis, cell proliferation, and cellular senescence is the nuclear factor kappa B (NF-κB) signalling system [30,31]. Interleukin-17 is also a factor closely related to inflammation. This indicates that HSPA5 may promote the progression of various malignant tumors by regulating inflammatory factors. In addition, GRP78 has been shown to improve the therapeutic efficacy of MSCs against hemorrhagic shock-induced liver injury via the NF-кB pathway [32]. Through PI3K/Akt signaling, HSPA5 has also been shown to improve the sensitivity of ovarian cancer cells to paclitaxel [33]. Moreover, several immune-related signaling pathways were enriched, demonstrating HSPA5's crucial function in the immunological milieu of cancerous tumors. The enrichment analysis of down-regulated genes demonstrated, in addition, that HSPA5 has the ability to govern the appearance of malignant tumors as well as their progression via a variety of signaling pathways. For example, JAK-STAT, PPAR signaling pathway. The role of immune-related cells such as cancer-related fibroblasts (CAFs) in cancer development, treatment resistance, and disease transmission has been well-studied [34]. Recent research has indicated that activation of the UPR is necessary for the formation of a favorable tumor microenvironment, which includes the differentiation of cancer-associated fibroblasts (CAFs) [35]. Meanwhile, CAFs have been demonstrated in prior research to raise the expression of HSPA5, which is a factor that leads to the invasion of non-small cell lung cancer cells [36]. It is imperative that more research be conducted into the role and function of the UPR as a regulator of the tumor microenvironment and the actions of immune cells in order to ameliorate the immunological dysfunctions associated with cancer. Therefore, we investigated how HSPA5 regulated immune infiltration and immune checkpoint markers, which yielded beneficial results (Fig. 7). Our study showed that HSPA5 affected TMB and MSI in a variety of tumors, which led us to further use TIDE analysis to determine the effect of HSPA5 on the responders of ICB. The results collectively showed that HSPA5 can also affect ICB responders in various tumors. In conclusion, as a result of our research, HSPA5 has been identified as a candidate for a new prognostic biomarker in several types of cancer. It does this by modulating the immune milieu inside tumors. Our findings highlight the necessity of the development of therapeutic regimens targeting HSPA5.
Together, our findings suggest a critical function for HSPA5 in the immunological microenvironment and prognosis of bladder cancer. As a result, we choose to verify HSPA5's expression and prognostic importance in samples from patients with bladder cancer. Based on our findings, HSPA5 is a promising predictive biomarker for individuals with bladder cancer. However, there are definitely caveats to our work, and further tests are needed to confirm HSPA5's efficacy in large tumors.
5. Conclusions
HSPA5 expression upregulation correlates with increased immune cell infiltration and poor prognosis in a variety of malignancies. We also observed reduced levels of phosphorylation and methylation of HSPA5 in many types of cancer. HSPA5 expression was shown to be substantially linked with immunological checkpoint-related gene expression. Tissue microarray research verified that HSPA5 is substantially expressed in bladder cancer and has potential as a predictive biomarker for individuals with this disease. Future experimental investigations of HSPA5 expression and function-mediated immune cell infiltration in various cancer populations may give more significant insights to decipher the underlying tumor development mechanisms and develop novel treatments targeting HSPA5 to boost the therapeutic efficacy of immunotherapies.
Availability of data and materials
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics approval and consent to participate
The studies involving human participants were reviewed and approved by the Ethics Committee of Nantong Tumour Hospital. Ethics approval number: 2022–053. The patients provided their written informed consent to participate in this study.
CRediT authorship contribution statement
YaXuan Wang: Writing – original draft, Formal analysis, Data curation. Jinfeng Wang: Writing – original draft, Conceptualization. Yang Liu: Writing – original draft, Data curation. XiaoLin Wang: Writing – review & editing, Funding acquisition. MingHua Ren: Writing – review & editing, Validation, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by the Scientific research project of Jiangsu Provincial Health Commission (M2021005) and the Natural Science Foundation of Heilongjiang Province (No. LH2019H030).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27184.
Contributor Information
YaXuan Wang, Email: 17805050201@163.com.
Jinfeng Wang, Email: 277723534@qq.com.
Yang Liu, Email: 202101233@harbmu.edu.cn.
XiaoLin Wang, Email: cxhwyc2010@163.com.
MingHua Ren, Email: renminghua1972@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., et al. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Graf R.P., Fisher V., Mateo J., et al. Predictive genomic biomarkers of hormonal therapy versus chemotherapy benefit in metastatic castration-resistant prostate cancer. Eur. Urol. 2022;81(1):37–47. doi: 10.1016/j.eururo.2021.09.030. [DOI] [PubMed] [Google Scholar]
- 3.Parashar S., Ferro-Novick S. Architecture of the endoplasmic reticulum plays a role in proteostasis. Autophagy. 2022;18(4):937–938. doi: 10.1080/15548627.2022.2030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang W., Chen L., Guo X., et al. Combating multidrug resistance and metastasis of breast cancer by endoplasmic reticulum stress and cell-nucleus penetration enhanced immunochemotherapy. Theranostics. 2022;12(6):2987–3006. doi: 10.7150/thno.71693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wadgaonkar P., Chen F. Connections between endoplasmic reticulum stress-associated unfolded protein response, mitochondria, and autophagy in arsenic-induced carcinogenesis. Semin. Cancer Biol. 2021;76:258–266. doi: 10.1016/j.semcancer.2021.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerezo M., Rocchi S. New anti-cancer molecules targeting HSPA5/BIP to induce endoplasmic reticulum stress, autophagy and apoptosis. Autophagy. 2017;13(1):216–217. doi: 10.1080/15548627.2016.1246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai M.N., Chen D.M., Xiao L.X., et al. COLEC10 induces endoplasmic reticulum stress by occupying GRP78 and inhibits hepatocellular carcinoma. Lab. Invest. 2023;103(7) doi: 10.1016/j.labinv.2023.100130. [DOI] [PubMed] [Google Scholar]
- 8.Wang J., Lee J., Liem D., et al. HSPA5 Gene encoding Hsp 70 chaperone BiP in the endoplasmic reticulum. Gene. 2017;30(618):14–23. doi: 10.1016/j.gene.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W.-T., Zhu G., Pfaffenbach K., et al. GRP78 as a regulator of liver steatosis and cancer progression mediated by loss of the tumor suppressor PTEN. Oncogene. 2014;33(42):4997–5005. doi: 10.1038/onc.2013.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uckun F.M., Qazi S., Ozer Z., et al. Inducing apoptosis in chemotherapy resistant B lineage acute lymphoblastic leukaemia cells by targeting HSPA5, a master regulator of the anti apoptotic unfolded protein response signalling network. Br. J. Haematol. 2011;153(6):741–752. doi: 10.1111/j.1365-2141.2011.08671.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q., Ke S., Liu Z., et al. HSPA5 promotes the proliferation, metastasis and regulates ferroptosis of bladder cancer. Int. J. Mol. Sci. 2023;24(6):5144. doi: 10.3390/ijms24065144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T., Fu J., Cheng J., et al. New progresses on cell surface protein HSPA5/BiP/GRP78 in cancers and COVID-19. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1166680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu J., Wei C., He J., et al. Evaluation and characterization of HSPA5 (GRP78) expression profiles in normal individuals and cancer patients with COVID-19. Int. J. Biol. Sci. 2021;17(3):897–910. doi: 10.7150/ijbs.54055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Y., Fan J., Liang D., et al. Cell surface GRP78-directed CAR-T cells are effective at treating human pancreatic cancer in preclinical models. Transl Oncol. 2024;39 doi: 10.1016/j.tranon.2023.101803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin R., Ma M., Han B., et al. Esophageal cancer stem cells reduce hypoxia-induced apoptosis by inhibiting the GRP78-perk-eIF2α-ATF4-CHOP pathway in vitro. J. Gastrointest. Oncol. 2023;14(4):1669–1693. doi: 10.21037/jgo-23-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S., Wei W., Yuan Y., et al. Chimeric antigen receptor T cells targeting cell surface GRP78 efficiently kill glioblastoma and cancer stem cells. J. Transl. Med. 2023;21(1):493. doi: 10.1186/s12967-023-04330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J.L., Tai Y.S., Tsai H.Y., et al. Betulinic acid inhibits the stemness of gastric cancer cells by regulating the GRP78-TGF-β1 signaling pathway and macrophage polarization. Molecules. 2023;28(4):1725. doi: 10.3390/molecules28041725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng Y., Wang N., Wang S., et al. Cefoselis enhances breast cancer chemosensitivity by directly targeting GRP78/LRP5 signalling of cancer stem cells. Clin. Transl. Med. 2023;13(2):e1119. doi: 10.1002/ctm2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012 May;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen F., Fan Y., Cao P., et al. Pan-cancer analysis of the prognostic and immunological role of HSF1: a potential target for survival and immunotherapy. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/5551036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Li M., Yang M., et al. Analysis of immune-related signatures of lung adenocarcinoma identified two distinct subtypes: implications for immune checkpoint blockade therapy. Aging (Albany NY) 2020;12(4):3312–3339. doi: 10.18632/aging.102814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 24.Chen X., Cubillos-Ruiz J.R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer. 2021;21(2):71–88. doi: 10.1038/s41568-020-00312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahmani-Kukia N., Zamani M., Mokaram P. ERMP1 facilitates the malignant characteristics of colorectal cancer cells through modulating PI3K/AKT/β-Catenin pathway and localization of GRP78. Cell J. 2023;25(7):470–482. doi: 10.22074/CELLJ.2023.1982707.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou Y., Shi H., Lin H., et al. The abrogation of GRP78 sensitizes liver cancer cells to lysionotin by enhancing ER stress-mediated pro-apoptotic pathway. Cell Stress Chaperones. 2023;28(4):409–422. doi: 10.1007/s12192-023-01358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y., Wang N., Wang S., et al. Cefoselis enhances breast cancer chemosensitivity by directly targeting GRP78/LRP5 signalling of cancer stem cells. Clin. Transl. Med. 2023;13(2):e1119. doi: 10.1002/ctm2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein Y., Rotter V., Aloni-Grinstein R. Gain-of-Function mutant p53: all the roads lead to tumorigenesis. Int. J. Mol. Sci. 2019;20(24):6197. doi: 10.3390/ijms20246197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012 May;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haga M., Okada M. Systems approaches to investigate the role of NF-κB signaling in aging. Biochem. J. 2022;479(2):161–183. doi: 10.1042/BCJ20210547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirzaei S., Saghari S., Bassiri F., et al. NF-κB as a regulator of cancer metastasis and therapy response: a focus on epithelial-mesenchymal transition. J. Cell. Physiol. 2022;237(7):2770–2795. doi: 10.1002/jcp.30759. [DOI] [PubMed] [Google Scholar]
- 32.Han J., Jia D., Yao H., et al. GRP78 improves the therapeutic effect of mesenchymal stem cells on hemorrhagic shock-induced liver injury: involvement of the NF-кB and HO-1/Nrf-2 pathways. FASEB J. 2024;38(1) doi: 10.1096/fj.202301456RRR. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L.Y., Yu J.Y., Leng Y.L., et al. MiR-181c sensitizes ovarian cancer cells to paclitaxel by targeting GRP78 through the PI3K/Akt pathway. Cancer Gene Ther. 2022;29(6):770–783. doi: 10.1038/s41417-021-00356-y. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y., Li X., Wang L., et al. Metabolic reprogramming and crosstalk of cancer-related fibroblasts and immune cells in the tumor microenvironment. Front. Endocrinol. 2022;13 doi: 10.3389/fendo.2022.988295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y., Li Z., Li Z. GRP78 secreted by tumor cells stimulates differentiation of bone marrow mesenchymal stem cells to cancer-associated fibroblasts. Biochem. Biophys. Res. Commun. 2013;440(4):558–563. doi: 10.1016/j.bbrc.2013.09.108. [DOI] [PubMed] [Google Scholar]
- 36.Yu T., Guo Z., Fan H., et al. Cancer-associated fibroblasts promote non-small cell lung cancer cell invasion by upregulation of glucose-regulated protein 78 (GRP78) expression in an integrated bionic microfluidic device. Oncotarget. 2016;7(18):25593–25603. doi: 10.18632/oncotarget.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.