Abstract
Background
The cellular mechanism of the formation of abdominal aortic aneurysm (AAA) is very complicated. A series of sophisticated events eventually led to significant pathological changes in the anatomical structure and function of the arterial wall and they are still not clear nowadays.
Methods
We pooled publicly available GEO datasets (GSE57691 and GSE47472) to get a comprehensive comparisons between normal tissues and AAA tissues to try to reveal molecular mechanism underlying the disease. Total 63 AAA samples and 18 normal tissue samples were compared and we fond that there were 784 significantly different gene (DEGs, threshold set as adjusted P < 0.05 and Log FC < 1) were identified. At the same time, we validate the possible signaling factor expression of AAA by comparing the normal tissue of the human body with the AAA tissue.
Results
In the pathway enrichment, we found that FOXP3 related signaling pathways, inflammation-related cytokine signaling pathways, interleukin-8-CXCR1 related signaling pathways and VEGFA and FGFR1 related signal pathway were significantly enrichmented. In Weighted gene co-expression network analysis (WGCNA), we found that the key hub genes were significantly related to lipid catabolic metabolism, which further verified the possibility that AAA might relate to energy metabolism disorders.
Conclusion
Based on the comprehensive analysis of previous high-throughput data and the validation of basic experiments, we found that the occurrence of AAA may be related to energy metabolism disorders and local inflammation.
Keywords: Abdominal aortic aneurysm, Metabolism, inflammation, Gene expression profiling, RNA
1. Introduction
In developed countries, abdominal aortic aneurysm (AAA) is a major burden of medical care. It mainly occurs in men over 65 years old and is the 13th leading cause of death in the United States [1]. The cellular mechanism of the formation of AAA is very complicated. A series of sophisticated events eventually led to significant pathological changes in the anatomical structure and function of the arterial wall. For a long time, smooth muscle cell (SMC) apoptosis and degeneration of the aortic media have been considered as signs of AAA pathology [2]. Inflammation, reactive oxygen species (ROS) production and endoplasmic reticulum stress were also all found related to SMC apoptosis in AAA [3]. In addition to some individual studies of molecular biological processes that might occur in vascular epithelial cells or vascular smooth muscle cells, many studies also fully considered the interaction between different cell types in vascular tissues and the relationship between the occurrence and development of AAA [[4], [5], [6], [7]].
However, the molecular biological mechanism related to the occurrence of AAA was so complicated that it is difficult to draw a comprehensive conclusion from a single investigation of a certain signaling pathway or a single key molecule. With the development of technology and the increasing number of people engaged in AAA research, many high-quality basic research has been published in the last five years. In 2018, a study revealed the possible role of perivascular adipose tissue (PVAT) in AAA pathogens and suggested that AAA is an immune disease with a basic autoimmune component [8]. A separate study, which was published in 2020, discovered that following Bonferroni's adjustment, the levels of 21 proteins associated with protein analysis, oxidative stress, lipid metabolism, and inflammation in the plasma significantly increased. However, the levels of Paraoxonase 3, which is linked to the metabolism of high-density lipoproteins, decreased in patients with abdominal aneurysm [9]. The two above studies represent two directions of AAA-related basic research, namely the exploration of the mechanism of disease and the discovery of biological markers. Regrettably, more advancement in these two fields is necessary due to the absence of reliable animal models and the limited number of instances in populations.
In order to fully understand the mechanism behind AAA, many RNA high-throughput sequencing results had been published [10]. Many interesting conclusions based on high-throughput sequencing have been published, but there is still no manuscript published for the integrated analysis of all high-throughput sequencing data. Therefore, this research will be based on the published RNA sequencing data and conduct integrated analysis to obtain the target A more comprehensive understanding of the molecular mechanism of AAA. At the same time, immunohistochemistry, Western blot and qRT-PCR experiments were performed on normal and AAA tissues to further verify the further mechanisms of AAA development.
2. Methods
2.1. Bioinformatics analysis
We conducted a comprehensive data search in the GEO and EBI databases. The inclusion conditions are as follows: Abdominal aortic aneurysm appears in the abstract keywords. The exclusion conditions are as follows: data sets before 2010 are excluded, sequencing data derived from non-human tissues are excluded, and data with a sample size of less than 6 cases are excluded.
After completing data retrieval, screening and inclusion, we first use the sva (3.36.0) program package to eliminate batch effects. The annotation of the probes was conducted with idmap3 (0.1.0) package. In the data preparation stage, all data are centralized and standardized for more objective comparison. We used the limma (3.44.3) package to calculate and identify differential genes. In the process of determining differential genes, a linear fitting models were used, and the statistical significance is determined by using the T test. The FDR is used as the adjusted P value in this analysis. In this study, a significantly different gene (DEG) is defined as the absolute value of fold change greater than 2 (LogFC>1), and adjusted P is less than 0.001.
After finishing the selection of DEG, further enrichment analysis is mainly completed by three parts: kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis, gene set enrichment analysis (GSEA) enrichment analysis, and weighted correlation network analysis (WGCNA) enrichment analysis. KEGG enrichment analysis is mainly done through the clusterProfiler (3.16.1) package, and the significant enrichment pathway is defined as a P < 0.05. GSEA analysis was performed with GSEA (4.1.0) software, the detailed parameter settings are as follows: permutation method was applied and permutation number was set as 1000, weighted enrichment statistic method was applied, Signal-to-noise method was applied to rank gene, real number mode was chosen to sort gene list, descending mode was applied to order gene list, largest gene subset number was limited up to 500, and the minimum gene subset was set as 15, meandiv mode was chosen to normalize datasets. Significant enrichment results was defined as P < 0.05. In WGCNA analysis, the Hierarchical Clustering method wad used to identify outliers. Calculate soft power to determine the best co-expression network model construction parameters, mean connectivity (<100) and signed R2 (>0.8) were calculated to determine the soft power value. After the co-expression subgroup gene grouping is completed, the correlation between the expression of the gene subgroup and the risk of AAA is calculated by the Pearson correlation and the student-t test is used for the significance test. The symbols of the most closely related gene subgroups that were up-regulated in the AAA were extracted and further analyzed.
After the careful screening, there were final two RNA sequencing datasets were included in this analysis [10,11]. Total 63 AAA samples and 18 normal tissue samples were compared. of them, 14 were AAA neck tissues, 20 were small AAA tissues (mean maximum aortic diameter = 54.3 ± 2.3 mm), 29 were large AAA sample tissues (mean maximum aortic diameter = 68.4 ± 14.3 mm), all 18 health control tissues were obtained from organ donors.
2.2. Experimental validation
2.2.1. Experimental procedure
The study was approved by the Ethics Committee of Shanghai Tenth People's Hospital (approval number: 22KN258, March 9, 2022), and all participants provided written informed consent after fully understanding the study. During the study, we collected a total of 4 surgical patients, so we grouped the tissue saved after surgical resection of abdominal aortic aneurysms, labeling the tumor tissue as the “AAA group" and the paratumor tissue as the normal group, which was labeled as the “control group ".
2.3. Immunohistochemical staining
After fixing the tissue specimens with 4% paraformaldehyde and making 4 μm paraffin sections, the experimental procedure strictly followed the immunohistochemistry SP two-step method (zsbio, Beijing, China), in which paraffin sections were dewaxed, hydrated and microwave antigen repaired, incubated with 3% H2O2, and incubated at 4 °C with primary Antibodies MRPS15 (1 : 200, Thermo Fisher Scientific, #17006-1-AP), MRPS24 (1 : 200, ABclonal, #A12123) were incubated overnight. Secondary antibody incubation was performed after washing with PBS, followed by color development in DAB solution, followed by microscopic (Olympus, Tokyo, Japan) observation and image acquisition, and finally data analysis using Image J software (National Institutes of Health, Bethesda, MD, USA).
2.4. Western blot
A 100 mg tissue sample was taken, ground with 500 μLRIPA lysate, and the supernatant was centrifuged to obtain the BCA protein quantification kit (Beyotime, Shanghai, China) to detect protein concentration. Add 1.5x loading buffer, boil for 5 min at 99 °C, separate the protein by SDS-PAGE gel electrophoresis and transfer to PVDF membrane, close the membrane with 5% skimmed milk for 1 h at room temperature, wash the membrane 3 times with 1xTBST, add primary antibodies β-actin (1:2000, Abcam, ab115777), MRPS15 (1:500, Thermo Fisher Scientific, #17006-1-AP), MRPS24 (1:500,ABclonal, #A12123) at 4 °C.The bands were analyzed by ImageJ software, and the relative expression of MRPS15 and MRPS24 proteins was calculated using GAPDH as the internal reference.
2.4.1. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from each group of tissue samples by TRIzol method (Thermo Fisher Scientific, Wilmington, USA), and the RNA content and concentration were detected by spectrophotometer (Thermo Fisher Scientific, Wilmington, USA) and calculated. All primers are designed and synthesized by Shanghai Shenggong Engineering Technology Co., Ltd., as detailed in Table 4 cDNA Synthesis Kit (Thermo, #K1622) reverse transcription synthesis cDNA, procedures and reaction systems are strictly in accordance with the relevant kit specifications. The cycling conditions were 95 °C for 30 s; 95 °C for 15 s, 55 °C for 15s, 45 cycles in total and 60 °C for 30 s. Comparing the expression of the target gene to the GAPDH level, the sample Ct values were recorded, the relative expression of the target gene was calculated by 2−ΔΔCt method and repeated three times.
Table 4.
Primer sequences for RT-qPCR.
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| NHP2 | CCTCCCTGGGACTAGGTTTCA | CGATGGGGTTCTGGTTGAC |
| RPS3A | AGGGTCGTGTGTTTGAAGTGA | CATGGAAGTTAGTCAGGCAGTTT |
| RPS24 | ATGAACGACACCGTAACTATCCG | CCGAATTTCTGTCTTAGGCACTG |
| MRPS15 | ATGTCCCTGGAATTGAGAAGGTT | CTCCAGGGATCTGGTGTCCT |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
Note: RT–qPCR, real-time quantitative polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RPS24: ribosomal protein S24; RPS3A:ribosomal protein S3A. MRPS15: Mitochondrial Ribosomal Protein S15.
2.5. Statistical analysis
GraphPad Prism 8 software was used for graphing and statistical analysis. All data were expressed as mean ± SEM. Student's t-test was used to compare data between two groups. P < 0.05 was considered statistically significant. (*P value < 0.05, **P value < 0.01.)
3. Results
After the careful screening, there were final two RNA sequencing datasets were included in this analysis [10,11]. Total 63 AAA samples and 18 normal tissue samples were compared. of them, 14 were AAA neck tissues, 20 were small AAA tissues (mean maximum aortic diameter = 54.3 ± 2.3 mm), 29 were large AAA sample tissues (mean maximum aortic diameter = 68.4 ± 14.3 mm), all 18 health control tissues were obtained from organ donors. Detailed information was offered in Table 1. When set threshold adjusted P value as 0.05 and threshold Log FC as 1, total 784 DEGs were identified (Fig. 1A). A heat map based on the top 50 representative DEGs is provided (Fig. 1B), and it could be found that the signature based on the differential gene can significantly distinguish AAA samples and the control samples. In the further KEGG enrichment analysis, we found that molecular pathways related to energy metabolism play an important role in the formation of AAA, especially galactose metabolism (Fig. 2A). In further screening of hub genes based on string database and cytospace software, we found that RPS24, RPS3A, MRPS15, MRPS12, and NHP2 may be key molecules in the formation of AAA (Fig. 2B). Using the Genemania database to perform PPI network analysis based on the key molecule, it was found that the hub molecules that were screened out were closely related to mitochondrial protein complexes (adjusted P = 9.08*10^-6) (see Table 2).
Table 1.
Data Sources and relative information. AAA = Abdominal Aortic Aneurysm.
| Dataset | Year | Organism | Experiment type | Sample number | Control source | Platform |
|---|---|---|---|---|---|---|
| GSE57691 | 2015 | Homo sapiens | Expression profiling by array | 49 AAA samples; 10 normal controls. |
Organ donors | GPL10558 |
| GSE47472 | 2013 | Homo sapiens | Expression profiling by array | 14 AAA samples; 8 normal controls |
Organ donors | GPL10558 |
Fig. 1.
Volcano plot and heatmap of presentative DEGs. A: Volcano plot of all genes, threshold was set as adjusted P < 0.05, LogFC>1. B: heatmap of presentative DEGs. DEGs = Differentially expressed genes.
Fig. 2.
Enrichment analysis of KEGG signaling pathway, hub gene screening and PPI network analysis based on Genemania database. A:Enrichment analysis of KEGG signaling pathway based on DEGs. B:hub gene screening results. C: PPI analysis based on Genemania database. DEGs: Differentially expressed genes. PPI: Protein-protein interaction.
Table 2.
GSEA analysis between abdominal aortic aneurysm and normal tissue. GSEA = Gene Set Enrichment Analysis. AAA = Abdominal Aortic Aneurysm.
| Gene Set | Upregulated in class | Normalized Enrichment Score | p-value |
|---|---|---|---|
| FOXP3 targets | AAA | 1.97 | 0.006 |
| Cytokines and Inflammatory Response | AAA | 1.87 | 0.006 |
| Interleukin 10 Signaling | AAA | 1.83 | 0.018 |
| IL8 CXCR1 Pathway | AAA | 1.81 | 0.012 |
| VEGFA targets | AAA | 1.82 | 0.032 |
| FGFR1 Mutant Receptor Activation | AAA | 1.80 | 0.004 |
In order to obtain a more comprehensive signal pathway enrichment analysis result, we conducted a GSEA analysis. Through GSEA analysis, we found that FOXP3 related signaling pathways in AAA samples showed a significant enrichment state (Enrichment Score, ES = 0.64, P = 0.006, Fig. 3A). Inflammation-related cytokine signaling pathways (ES = 0.66, P = 0.006, Fig. 3B), interleukin-10 related signaling pathways (ES = 0.64, P = 0.018, Fig. 3C), interleukin-8-CXCR1 related signaling pathways (ES = 1.81, P = 0.012, Fig. 3D), VEGFA and FGFR1 related signal pathway were also significantly related to AAA (Fig. 3E/F).
Fig. 3.
GSEA analysis results. GSEA: Gene Set Enrichment Analysis.
Weighted gene co-expression network analysis (WGCNA), this analysis method aims to find co-expressed gene modules, and to explore the relationship between gene networks and the phenotype of interest, as well as the core genes in the network. In this analysis, we first confirmed that there is no strong heterogeneity between the samples included in this study after data cleaning and standardization (Fig. 4A). By calculating the average connection degree and signed R2, it was determined that the soft power value that should be used was 8 (Fig. 4B). When the soft power value was set to 8, the maximum number of gene subgroups was set as 5000, and the minimum number of gene subgroups was set as 25, a total of 20 interrelated gene subgroups are obtained (Fig. 4C). Pearson correlation test was used to test the correlation between the identified gene subgroups and the AAA phenotype, and a total of 6 subgroups were found to be significantly correlated with AAA (Table 3). Among them, the gene subgroup labeled Green was significantly positively correlated with AAA, and the r coefficient was as high as 0.44. We extracted the gene subgroups labeled Green, screened the hub genes, and performed molecular pathway enrichment analysis based on the Genemania database. We found that the key hub genes were still significantly related to lipid catabolic metabolism, which further verified the possibility that AAA might relate to energy metabolism disorders.
Fig. 4.
WGCNA analysis results. A: Outlier identification conducted by Hierarchical Clustering method. B: Soft power value confirmation. C: Schematic diagram of gene co-expression network. D: PPI plot based on Genemania database. Hub genes were obtained from Green subgroup in co-expression network.
Table 3.
WGCNA results between abdominal aortic aneurysm and normal tissue. WGCNA= Weighted Correlation Network Analysis. r = Correlation coefficient. *: Calculated by Pearson correlation. **: Calculated by Student asymptotic method.
| Gene module | Gene number | r* | p-value** |
|---|---|---|---|
| Brown | 455 | −0.45087 | 2.40E-05 |
| Green | 297 | 0.441624 | 3.67E-05 |
| Light Yellow | 310 | −0.40682 | 0.000164 |
| Light Green | 1992 | −0.36431 | 0.000827 |
| Turquoise | 238 | −0.33224 | 0.002444 |
| Cyan | 64 | 0.221934 | 0.046451 |
| Light Cyan | 42 | 0.187156 | 0.094319 |
| Black | 238 | 0.184431 | 0.099301 |
| Midnight Blue | 43 | 0.175472 | 0.117142 |
| Yellow | 310 | −0.16659 | 0.137158 |
| Purple | 98 | −0.16093 | 0.151215 |
| Magenta | 202 | 0.138919 | 0.216146 |
| Red | 245 | 0.127562 | 0.256434 |
| Pink | 229 | 0.081005 | 0.472205 |
| Grey | 236 | 0.080284 | 0.476171 |
| Salmon | 66 | 0.075709 | 0.501739 |
| Green Yellow | 80 | −0.0452 | 0.688677 |
| Tan | 72 | −0.02765 | 0.806426 |
| Blue | 632 | −0.00811 | 0.942717 |
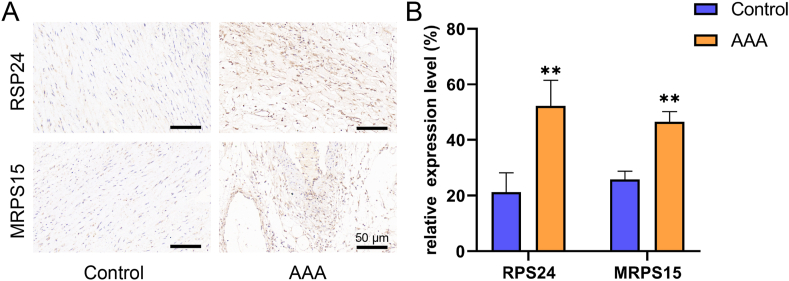
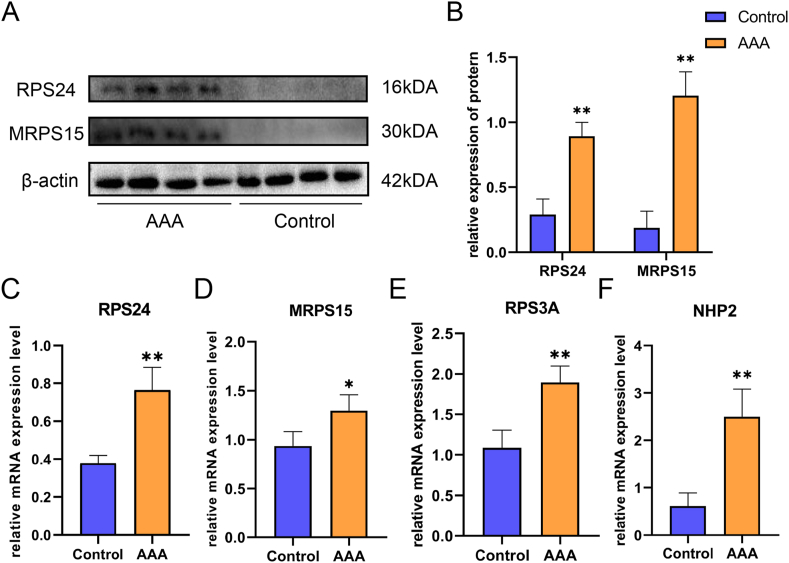
Further, we verified the above results and signaling factors by basic experiments. Firstly, we found that the expression levels of RPS24 and MRPS15 were significantly higher in the AAA group compared with the control group by immunohistochemistry (Fig. 5A), and the difference was statistically significant (P < 0.01, Fig. 5B). Secondly, in Western blot, the expression levels of RPS24 and MRPS15 showed the same trend as in immunohistochemistry (Fig. 6A/B). Finally, we again verified by RT-qPCR that the expression of RPS24 was significantly higher in the AAA group than in the control group (Fig. 6C), while the expression levels of MRPS15 (Fig. 6D, P < 0.05) RPS3A (Fig. 6E) and NHP2 (Fig. 6F) were also elevated in the AAA group (P < 0.01).
Fig. 5.
A: Immunohistochemical detection of RPS24 and MRPS15 expression in 2 groups of specimens (scale bar = 50 μm). B: Quantification of RPS24 and MRPS15 in immunohistochemical staining (n = 4). All data are expressed as the mean ± SEM, *P value < 0.05, **P value < 0.01.
Fig. 6.
A: Representative Western blot protein bands of RPS24, MRPS15 in each group. B: Statistical analysis of Western blot results (n = 4). C-E:Analysis of mRNA expression of RPS24, MRPS15, RPS3A and NHP2 in each group by RT–qPCR (n = 4). All data are expressed as the mean ± SEM, *P value < 0.05, **P value < 0.01.
4. Discussion
Abdominal aortic aneurysm (AAA) represents a complex and multifactorial vascular pathology, the elucidation of which necessitates a nuanced understanding of its molecular underpinnings. Central to its pathogenesis is the orchestration of chronic inflammation within the arterial wall [12]. Infiltration of immune cells, particularly macrophages and T lymphocytes, is a hallmark event, with these cells releasing potent inflammatory mediators, including interleukin-6 (IL-6), IL-8 and tumor necrosis factor-alpha (TNF-α) [[13], [14], [15], [16]]. The intricate interplay between these cytokines contributes to the perpetuation of a pro-inflammatory microenvironment, fostering deleterious cellular responses [17].
Matrix metalloproteinases (MMPs), specifically MMP-2 and MMP-9, emerge as key effectors in the dysregulated matrix remodeling characterizing AAA [18]. This involves the proteolytic degradation of crucial extracellular matrix (ECM) components, notably elastin and collagen, leading to the structural weakening of the aortic wall. The imbalance between MMPs and their endogenous regulators, tissue inhibitors of metalloproteinases (TIMPs), assumes paramount importance in dictating the pathological trajectory. Excessive ECM degradation, compounded by impaired matrix synthesis, underscores the vulnerability of the aortic wall to aneurysmal dilation [19].
Oxidative stress further intricately weaves into the narrative of AAA pathogenesis. Reactive oxygen species (ROS), generated through diverse cellular processes, not only inflict direct damage upon cellular structures but also actively participate in redox-sensitive signaling pathways [20]. This results in a perpetuating cycle of inflammation and oxidative stress, amplifying the vascular damage observed in AAA [21]. The upregulation of NADPH oxidases, responsible for ROS generation, signifies a pivotal event in this oxidative milieu, further underscoring the complexity of the molecular landscape [22].
Genetic factors contribute significantly to the predisposition to AAA, with specific polymorphisms in genes associated with MMPs, TIMPs, and inflammatory pathways influencing individual susceptibility [23,24]. The familial clustering of AAA cases attests to the heritable component of this vascular disorder. Exploring the genetic landscape offers promising avenues for discerning potential biomarkers and therapeutic targets, thereby affording opportunities for personalized interventions [25].
The transforming growth factor-beta (TGF-β) signaling pathway, traditionally recognized for its pivotal role in maintaining vascular homeostasis, undergoes a dichotomous transformation in AAA pathogenesis [26]. TGF-β, conventionally an anti-inflammatory and anti-proteolytic factor, paradoxically shifts to a pro-inflammatory phenotype in AAA [27]. Dysregulated TGF-β signaling manifests in increased MMP production, culminating in aberrant ECM turnover and vascular remodeling [28]. Unraveling the nuances of TGF-β signaling dynamics is imperative for devising interventions that rectify its divergent roles and restore its protective functions.
Notch signaling, a highly conserved pathway regulating cell fate decisions, emerges as a discerning factor in AAA [29]. The dysregulation of Notch receptors and ligands within vascular cells contributes to endothelial dysfunction and vascular smooth muscle cell (VSMC) phenotypic alterations [30]. These disruptions further fuel the pathological cascade, prompting a reevaluation of Notch signaling as a potential therapeutic target in modulating cellular behaviors in AAA.
In terms of molecular mechanisms related to the pathogenesis of AAA, many signal pathways worthy of attention have been proposed. First of all, the role of local inflammation was a factor that is difficult to ignore [31]. A sign of the formation of AAA was a strong inflammatory response, which basically involved all the classical cellular components of inflammation, as well as the local inflammatory response in the arterial wall [[32], [33], [34]]. Neutrophil infiltration occurs in the early stages of AAA, but it is short-lived. Observations in the elastase model showed that treatment with neutrophil neutralizing antibodies slowed the expansion of AAA, indicating that neutrophils have functionally related effects [35,36]. Although there were still some controversies, some studies had also pointed out that AAA might be closely related to autophagy-related processes, and this correlation is reasonable in the arterial wall damage-repair cycle [37,38].
Since diseases such as AAA involve large-scale extracellular matrix remodeling, matrix metalloproteinases (MMP) have received extensive attention in basic research related to AAA. In AAA, many studies have confirmed that compared with normal aorta, the amount of elastin, collagen and glycosaminoglycan is reduced, and the imbalance between active MMP and its inhibitor is one of the main reasons for these changes [39,40]. Evidence of up-regulation of MMP-1 expression has been found in AAA [41]. Along with increased MMP-1 expression, there is a concurrent decrease in the levels of MMP-1 inhibitors [42]. However, analysis data based on human samples failed to correlate genetic polymorphisms in the MMP-1 promoter region with clinical outcomes [43]. MMP-13 expression was increased in AAA too, especially in symptomatic AAA and those at high risk for rupture [44]. MMP-3 was also highly expressed in the AAA wall. This enzyme usually produced by fibroblasts and epithelial cells might be produced by macrophages in the AAA environment. Polymorphism in the promoter region of the MMP-3 gene called 5A/6A (5 adenine vs. 6 adenine at −1612) enhances transcriptional activity and serves as an independent risk factor for the formation of AAA [42,45]. The renin-angiotensin system was also considered to be closely related to the occurrence of AAA. The proof of the induction of AAA by Ang II infusion in apoE48 and LDL (low density lipoprotein) 49 receptor KO mice was undoubtedly the most direct data to record the causal relationship between the renin-angiotensin system and the formation of AAA [46,47].
In this study, through a comprehensive analysis of the existing RNA sequencing data, we found that the formation and development of AAA was related to energy metabolism and local inflammation. In this study, we found that the formation and development of AAA are related to energy metabolism and local inflammation through comprehensive analysis of available RNA sequencing data. By reviewing the literature we found that RPS24, MRPS15, RPS3A and NHP2 have been rarely reported in the formation and development of AAA, and our biosignature analysis and experimental validation provide research directions for the mechanistic study of AAA disease development. In addition, the galactose metabolism-related pathway, the IL8-CXCR pathway, was the first time we have discovered in AAA. In terms of energy metabolism, there was currently a published study based on plasma metabolomics that had reached a similar conclusion to this study, that the disturbance of energy metabolism might be closely related to the occurrence of AAA [48]. Cell inflammatory factor related pathways represented by IL8 and IL10 were screened again in this study, which further shows that local inflammation is still closely related to the occurrence of AAA. At the same time, we experimentally verified the key factors of AAA formation in human specimens, and showed that RPS24, RPS3A, MRPS15 and NHP2 are key regulators of AAA formation and development through basic experiments.
5. Conclusion
Based on the integrated analysis of previous high-throughput data and experimental validation of human specimens, we found that the occurrence of AAA may be related to energy metabolism disorders and local inflammation.
Funding statement
Supported by theFoundation for the Talents by the Tenth People's Hospital of Tongji University(Grant No: 2021SYPDRC028); Supported by the Fundamental Research Funds for the Central Universities.
Data availability statement
Data included in article. material/referenced in article.
CRediT authorship contribution statement
Jun Li: Writing – original draft, Formal analysis, Data curation, Conceptualization. Yang Liu: Writing – review & editing, Funding acquisition, Formal analysis, Data curation. Zhitao Wei: Visualization, Validation, Supervision, Investigation, Formal analysis, Data curation. Jie Cheng: Visualization, Validation, Supervision, Software, Project administration, Investigation, Conceptualization. Yongfa Wu: Visualization, Validation, Supervision, Resources, Methodology, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27912.
Contributor Information
Zhitao Wei, Email: dr_sirius@163.com.
Jie Cheng, Email: jiechg@163.com.
Yongfa Wu, Email: slkyongfa@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Abdulameer H., Al Taii H., Al-Kindi S.G., Milner R. Epidemiology of fatal ruptured aortic aneurysms in the United States (1999-2016) J. Vasc. Surg. 2019;69(2):378–384.e372. doi: 10.1016/j.jvs.2018.03.435. [DOI] [PubMed] [Google Scholar]
- 2.López-Candales A., Holmes D.R., Liao S., Scott M.J., Wickline S.A., Thompson R.W. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am. J. Pathol. 1997;150(3):993–1007. [PMC free article] [PubMed] [Google Scholar]
- 3.Qin Y., Wang Y., Liu O., Jia L., Fang W., Du J., Wei Y. Tauroursodeoxycholic Acid attenuates angiotensin II induced abdominal aortic aneurysm formation in apolipoprotein E-deficient mice by inhibiting endoplasmic reticulum stress. Eur. J. Vasc. Endovasc. Surg. 2017;53(3):337–345. doi: 10.1016/j.ejvs.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Sun J., Deng H., Zhou Z., Xiong X., Gao L. Endothelium as a potential target for treatment of abdominal aortic aneurysm. Oxid. Med. Cell. Longev. 2018;2018 doi: 10.1155/2018/6306542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okuno K., Cicalese S., Eguchi S. Depletion of CD11c+ cell attenuates progression of abdominal aortic aneurysm. Clin. Sci. (Lond.) 2020;134(1):33–37. doi: 10.1042/CS20191083. [DOI] [PubMed] [Google Scholar]
- 6.Hannawa K.K., Eliason J.L., Woodrum D.T., Pearce C.G., Roelofs K.J., Grigoryants V., Eagleton M.J., Henke P.K., Wakefield T.W., Myers D.D., et al. L-selectin-mediated neutrophil recruitment in experimental rodent aneurysm formation. Circulation. 2005;112(2):241–247. doi: 10.1161/CIRCULATIONAHA.105.535625. [DOI] [PubMed] [Google Scholar]
- 7.Rateri D.L., Howatt D.A., Moorleghen J.J., Charnigo R., Cassis L.A., Daugherty A. Prolonged infusion of angiotensin II in apoE(-/-) mice promotes macrophage recruitment with continued expansion of abdominal aortic aneurysm. Am. J. Pathol. 2011;179(3):1542–1548. doi: 10.1016/j.ajpath.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piacentini L., Werba J.P., Bono E., Saccu C., Tremoli E., Spirito R., Colombo G.I. Genome-wide expression Profiling Unveils autoimmune response signatures in the perivascular adipose tissue of abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 2019;39(2):237–249. doi: 10.1161/ATVBAHA.118.311803. [DOI] [PubMed] [Google Scholar]
- 9.Memon A.A., Zarrouk M., Ågren-Witteschus S., Sundquist J., Gottsäter A., Sundquist K. Identification of novel diagnostic and prognostic biomarkers for abdominal aortic aneurysm. Eur J Prev Cardiol. 2020;27(2):132–142. doi: 10.1177/2047487319873062. [DOI] [PubMed] [Google Scholar]
- 10.Biros E., Gäbel G., Moran C.S., Schreurs C., Lindeman J.H., Walker P.J., Nataatmadja M., West M., Holdt L.M., Hinterseher I., et al. Differential gene expression in human abdominal aortic aneurysm and aortic occlusive disease. Oncotarget. 2015;6(15):12984–12996. doi: 10.18632/oncotarget.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biros E., Moran C.S., Rush C.M., Gäbel G., Schreurs C., Lindeman J.H., Walker P.J., Nataatmadja M., West M., Holdt L.M., et al. Differential gene expression in the proximal neck of human abdominal aortic aneurysm. Atherosclerosis. 2014;233(1):211–218. doi: 10.1016/j.atherosclerosis.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Abe J., Berk B.C. Atheroprone flow activation of the sterol regulatory element binding protein 2 and nod-like receptor protein 3 inflammasome mediates focal atherosclerosis. Circulation. 2013;128(6):579–582. doi: 10.1161/CIRCULATIONAHA.113.004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raffort J., Lareyre F., Clément M., Hassen-Khodja R., Chinetti G., Mallat Z. Monocytes and macrophages in abdominal aortic aneurysm. Nat. Rev. Cardiol. 2017;14(8):457–471. doi: 10.1038/nrcardio.2017.52. [DOI] [PubMed] [Google Scholar]
- 14.Paige E., Clément M., Lareyre F., Sweeting M., Raffort J., Grenier C., Finigan A., Harrison J., Peters J.E., Sun B.B., et al. Interleukin-6 receptor signaling and abdominal aortic aneurysm growth Rates. Circ Genom Precis Med. 2019;12(2) doi: 10.1161/CIRCGEN.118.002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X., Yao H., Yang Y., Jin L., Wang Y., Wu L., Yang S., Cheng K. miR-195 suppresses abdominal aortic aneurysm through the TNF-α/NF-κB and VEGF/PI3K/Akt pathway. Int. J. Mol. Med. 2018;41(4):2350–2358. doi: 10.3892/ijmm.2018.3426. [DOI] [PubMed] [Google Scholar]
- 16.Koch A.E., Kunkel S.L., Pearce W.H., Shah M.R., Parikh D., Evanoff H.L., Haines G.K., Burdick M.D., Strieter R.M. Enhanced production of the chemotactic cytokines interleukin-8 and monocyte chemoattractant protein-1 in human abdominal aortic aneurysms. Am. J. Pathol. 1993;142(5):1423–1431. [PMC free article] [PubMed] [Google Scholar]
- 17.Middleton R.K., Lloyd G.M., Bown M.J., Cooper N.J., London N.J., Sayers R.D. The pro-inflammatory and chemotactic cytokine microenvironment of the abdominal aortic aneurysm wall: a protein array study. J. Vasc. Surg. 2007;45(3):574–580. doi: 10.1016/j.jvs.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Maguire E.M., Pearce S.W.A., Xiao R., Oo A.Y., Xiao Q. Matrix metalloproteinase in abdominal aortic aneurysm and aortic Dissection. Pharmaceuticals. 2019;12(3) doi: 10.3390/ph12030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Wang W., Li L., Khalil R.A. MMPs and ADAMs/ADAMTS inhibition therapy of abdominal aortic aneurysm. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sánchez-Infantes D., Nus M., Navas-Madroñal M., Fité J., Pérez B., Barros-Membrilla A.J., Soto B., Martínez-González J., Camacho M., Rodriguez C., et al. Oxidative stress and inflammatory markers in abdominal aortic aneurysm. Antioxidants. 2021;10(4) doi: 10.3390/antiox10040602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emeto T.I., Moxon J.V., Au M., Golledge J. Oxidative stress and abdominal aortic aneurysm: potential treatment targets. Clin. Sci. (Lond.) 2016;130(5):301–315. doi: 10.1042/CS20150547. [DOI] [PubMed] [Google Scholar]
- 22.Salmon M. NADPH oxidases in aortic aneurysms. Antioxidants. 2022;11(9) doi: 10.3390/antiox11091830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belsley S.J., Tilson M.D. Two decades of research on etiology and genetic factors in the abdominal aortic aneurysm (AAA)--with a glimpse into the 21st century. Acta Chir. Belg. 2003;103(2):187–196. doi: 10.1080/00015458.2003.11679405. [DOI] [PubMed] [Google Scholar]
- 24.Jones G.T., Tromp G., Kuivaniemi H., Gretarsdottir S., Baas A.F., Giusti B., Strauss E., Van't Hof F.N., Webb T.R., Erdman R., et al. Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circ. Res. 2017;120(2):341–353. doi: 10.1161/CIRCRESAHA.116.308765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakalihasan N., Michel J.B., Katsargyris A., Kuivaniemi H., Defraigne J.O., Nchimi A., Powell J.T., Yoshimura K., Hultgren R. Abdominal aortic aneurysms. Nat Rev Dis Primers. 2018;4(1):34. doi: 10.1038/s41572-018-0030-7. [DOI] [PubMed] [Google Scholar]
- 26.Lareyre F., Clément M., Raffort J., Pohlod S., Patel M., Esposito B., Master L., Finigan A., Vandestienne M., Stergiopulos N., et al. TGFβ (transforming growth factor-β) blockade induces a human-like disease in a nondissecting mouse model of abdominal aortic aneurysm. Arterioscler. Thromb. Vasc. Biol. 2017;37(11):2171–2181. doi: 10.1161/ATVBAHA.117.309999. [DOI] [PubMed] [Google Scholar]
- 27.Gillis E., Van Laer L., Loeys B.L. Genetics of thoracic aortic aneurysm: at the crossroad of transforming growth factor-β signaling and vascular smooth muscle cell contractility. Circ. Res. 2013;113(3):327–340. doi: 10.1161/CIRCRESAHA.113.300675. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z., Zhou H., Zou X., Wang X. RUNX3 is up-regulated in abdominal aortic aneurysm and regulates the function of vascular smooth muscle cells by regulating TGF-β1. J. Mol. Histol. 2022;53(1):1–11. doi: 10.1007/s10735-021-10035-9. [DOI] [PubMed] [Google Scholar]
- 29.Hans C.P., Sharma N., Dev R., Blain J.M., Tonniges J., Agarwal G. DAPT, a potent Notch inhibitor regresses actively growing abdominal aortic aneurysm via divergent pathways. Clin. Sci. (Lond.) 2020;134(12):1555–1572. doi: 10.1042/CS20200456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu H., Sun J., Liang W., Chang Z., Rom O., Zhao Y., Zhao G., Xiong W., Wang H., Zhu T., et al. Cyclodextrin prevents abdominal aortic aneurysm via activation of vascular smooth muscle cell transcription factor EB. Circulation. 2020;142(5):483–498. doi: 10.1161/CIRCULATIONAHA.119.044803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peshkova I.O., Schaefer G., Koltsova E.K. Atherosclerosis and aortic aneurysm - is inflammation a common denominator? FEBS J. 2016;283(9):1636–1652. doi: 10.1111/febs.13634. [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Lindholt J.S., Sukhova G.K., Shi M.A., Xia M., Chen H., Xiang M., He A., Wang Y., Xiong N., et al. IgE actions on CD4+ T cells, mast cells, and macrophages participate in the pathogenesis of experimental abdominal aortic aneurysms. EMBO Mol. Med. 2014;6(7):952–969. doi: 10.15252/emmm.201303811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuruda T., Kato J., Hatakeyama K., Kojima K., Yano M., Yano Y., Nakamura K., Nakamura-Uchiyama F., Matsushima Y., Imamura T., et al. Adventitial mast cells contribute to pathogenesis in the progression of abdominal aortic aneurysm. Circ. Res. 2008;102(11):1368–1377. doi: 10.1161/CIRCRESAHA.108.173682. [DOI] [PubMed] [Google Scholar]
- 34.Aortic wall inflammation predicts abdominal aortic aneurysm expansion, rupture, and need for surgical repair. Circulation. 2017;136(9):787–797. doi: 10.1161/CIRCULATIONAHA.117.028433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ricci M.A., Strindberg G., Slaiby J.M., Guibord R., Bergersen L.J., Nichols P., Hendley E.D., Pilcher D.B. Anti-CD 18 monoclonal antibody slows experimental aortic aneurysm expansion. J. Vasc. Surg. 1996;23(2):301–307. doi: 10.1016/s0741-5214(96)70274-4. [DOI] [PubMed] [Google Scholar]
- 36.Eliason J.L., Hannawa K.K., Ailawadi G., Sinha I., Ford J.W., Deogracias M.P., Roelofs K.J., Woodrum D.T., Ennis T.L., Henke P.K., et al. Neutrophil depletion inhibits experimental abdominal aortic aneurysm formation. Circulation. 2005;112(2):232–240. doi: 10.1161/CIRCULATIONAHA.104.517391. [DOI] [PubMed] [Google Scholar]
- 37.Wu Q.Y., Cheng Z., Zhou Y.Z., Zhao Y., Li J.M., Zhou X.M., Peng H.L., Zhang G.S., Liao X.B., Fu X.M. A novel STAT3 inhibitor attenuates angiotensin II-induced abdominal aortic aneurysm progression in mice through modulating vascular inflammation and autophagy. Cell Death Dis. 2020;11(2):131. doi: 10.1038/s41419-020-2326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Liu S., Pan B., Cai H., Zhou H., Yang P., Wang W. The role of autophagy in abdominal aortic aneurysm: protective but dysfunctional. Cell Cycle. 2020;19(21):2749–2759. doi: 10.1080/15384101.2020.1823731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fanjul-Fernández M., Folgueras A.R., Cabrera S., López-Otín C. Matrix metalloproteinases: evolution, gene regulation and functional analysis in mouse models. Biochim. Biophys. Acta. 2010;1803(1):3–19. doi: 10.1016/j.bbamcr.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Kadoglou N.P., Liapis C.D. Matrix metalloproteinases: contribution to pathogenesis, diagnosis, surveillance and treatment of abdominal aortic aneurysms. Curr. Med. Res. Opin. 2004;20(4):419–432. doi: 10.1185/030079904125003143. [DOI] [PubMed] [Google Scholar]
- 41.Knox J.B., Sukhova G.K., Whittemore A.D., Libby P. Evidence for altered balance between matrix metalloproteinases and their inhibitors in human aortic diseases. Circulation. 1997;95(1):205–212. doi: 10.1161/01.cir.95.1.205. [DOI] [PubMed] [Google Scholar]
- 42.Saracini C., Bolli P., Sticchi E., Pratesi G., Pulli R., Sofi F., Pratesi C., Gensini G.F., Abbate R., Giusti B. Polymorphisms of genes involved in extracellular matrix remodeling and abdominal aortic aneurysm. J. Vasc. Surg. 2012;55(1):171–179.e172. doi: 10.1016/j.jvs.2011.07.051. [DOI] [PubMed] [Google Scholar]
- 43.Saratzis A., Abbas A.A., Kiskinis D., Melas N., Saratzis N., Kitas G.D. Abdominal aortic aneurysm: a review of the genetic basis. Angiology. 2011;62(1):18–32. doi: 10.1177/0003319710373092. [DOI] [PubMed] [Google Scholar]
- 44.Mao D., Lee J.K., VanVickle S.J., Thompson R.W. Expression of collagenase-3 (MMP-13) in human abdominal aortic aneurysms and vascular smooth muscle cells in culture. Biochem. Biophys. Res. Commun. 1999;261(3):904–910. doi: 10.1006/bbrc.1999.1142. [DOI] [PubMed] [Google Scholar]
- 45.Carrell T.W., Burnand K.G., Wells G.M., Clements J.M., Smith A. Stromelysin-1 (matrix metalloproteinase-3) and tissue inhibitor of metalloproteinase-3 are overexpressed in the wall of abdominal aortic aneurysms. Circulation. 2002;105(4):477–482. doi: 10.1161/hc0402.102621. [DOI] [PubMed] [Google Scholar]
- 46.Daugherty A., Manning M.W., Cassis L.A. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J. Clin. Invest. 2000;105(11):1605–1612. doi: 10.1172/JCI7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daugherty A., Cassis L. Chronic angiotensin II infusion promotes atherogenesis in low density lipoprotein receptor -/- mice. Ann. N. Y. Acad. Sci. 1999;892:108–118. doi: 10.1111/j.1749-6632.1999.tb07789.x. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y., Wan S., Han M., Zhao Y., Li C., Cai G., Zhang S., Sun Z., Hu X., Cao H., et al. Plasma metabolomics analysis identifies abnormal energy, lipid, and amino acid metabolism in abdominal aortic aneurysms. Med Sci Monit. 2020;26 doi: 10.12659/MSM.926766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article. material/referenced in article.