Abstract
Human T-cell lymphotrophic virus type 2 (HTLV-2), a common infection of intravenous drug users and subpopulations of Native Americans, is uncommon in the general population. In contrast with the closely related HTLV-1, which is associated with both leukemia and neurologic disorders, HTLV-2 lacks a strong etiologic association with disease. HTLV-2 does shares many properties with HTLV-1, including in vitro lymphocyte transformation capability. To better assess the ability of HTLV-2 to transform lymphocytes, a limiting dilution assay was used to generate clonal, transformed lymphocyte lines. As with HTLV-1, the transformation efficiency of HTLV-2 producer cells was proportionately related to the number of lethally irradiated input cells and was comparable to HTLV-1-mediated transformation efficiency. HTLV-2-infected cells were reproducibly isolated and had markedly increased growth potential compared to uninfected cells; HTLV-2 transformants required the continued presence of exogenous interleukin 2 for growth for several months and were maintained for over 2 years in culture. All HTLV-2-transformed populations were CD2 and/or CD3 positive and B1 negative and were either CD4+ or CD8+ populations or a mixture of CD4+ and CD8+ lymphocytes. Clonality of the HTLV-2 transformants was confirmed by Southern blot analysis of T-cell receptor β chain rearrangement. Southern blot analysis revealed a range of integrated full-length genomes from one to multiple. In situ hybridization analysis of HTLV-2 integration revealed no obvious chromosomal integration pattern.
The first identified human retroviruses, the human T-cell lymphotropic virus types 1 and 2 (HTLV-1 and -2), have the ability to transform lymphocytes in vitro (10, 11, 51, 62). T lymphocytes infected with HTLV demonstrate enhanced growth potential, marked by seemingly unlimited entry into the cell cycle. The two HTLV serotypes are approximately 65% homologous on the nucleotide level (56, 59) and demonstrate a correspondingly high serologic cross-reactivity (37). Nevertheless, they have distinct seroepidemiologic and clinical profiles. HTLV-1 is associated etiologically with adult T-cell leukemia (16, 26, 63) as well as with a peripheral neuropathy known as tropical spastic paraparesis or HTLV-associated myelopathy (1, 20, 31, 48). HTLV-1 is endemic to southern Japan, the Caribbean basin, central Africa, northeastern South America, and regions of the southeastern United States (3–6, 8, 25, 38, 43, 54). HTLV-2 is occasionally found in Native American Indians (12–15, 28, 34, 39, 41) as well as in a significant proportion of intravenous drug users (IVDUs) (2, 33, 35, 53). Recently, Hall et al. defined two subtypes of HTLV-2, HTLV-2a and HTLV-2b, isolated from peripheral blood lymphocytes (PBLs) of IVDUs (23). HTLV-2 has not been associated with any disease to date, though there have been isolated reports of HTLV-2-associated neuropathy mostly from south Florida and the Caribbean (24, 27, 30, 42, 58).
Persaud et al. (50) reported a limiting dilution infectivity assay for HTLV-1 which allowed early detection of infected cultures and interleukin 2 (IL-2)-driven expansion of clonal populations. The present study extends this method to HTLV-2 and characterizes the resultant transformants. Clonal HTLV-2 populations were routinely and reproducibly generated with the HTLV-2a laboratory strain LAMP/MO. Furthermore, Southern blot analysis was used to characterize both the number and size of proviral integrations in the transformants derived from the in vitro system. These results are compared with those previously noted for HTLV-1. Additionally, chromosomal in situ hybridization was used to localize both HTLV-1 and HTLV-2 proviral integrations.
Infection-transformation efficiency and assay reproducibility.
The HTLV-2 transformation assay was performed as described by Persaud for HTLV-1 (50). Briefly, LAMP/MO cells (gift of Robert Gallo, National Cancer Institute, Bethesda, Md.) were exposed on ice to 11,700 rads (390 rads/min for 30 min) from a gamma source. Various numbers of irradiated cells (10, 100, and 1,000) were cocultivated with 104 activated peripheral blood mononuclear cells (PBMCs) in round-bottom 96-well plates in the presence of IL-2 (10 U/ml) (Boehringer-Mannheim Corp., Indianapolis, Ind.). HTLV-2-transformed cell populations were identified as cells that continued to proliferate beyond 6 weeks, the point at which most of the activated PBMCs not exposed to the HTLV-2-producing cells no longer proliferate in medium containing IL-2, and also by the continued production of HTLV p24 antigen (determined by enzyme-linked immunosorbent assay; (Coulter Immunology, Hialeah, Fl) >1,000 pg/ml) in the culture supernatant. Control wells containing only activated PBMCs or only irradiated LAMP/MO (104 cells/well) were maintained in parallel. After 6 to 9 weeks of culture, cocultures which continued to grow were expanded in growth medium supplemented with 10 U of IL-2/ml. HTLV-2-transformed clones were defined as cells exposed to HTLV-2 that continued to proliferate in the presence of IL-2 and that continuously produced p24 antigen (>1,000 pg/ml) by 12 weeks after the initial coculture. Control wells, which contained 104 phytohemagglutinin-activated PBMCs in the absence of HTLV-2-infected cells, proliferated for approximately 4 weeks and were never p24+; wells which contained only HTLV-2-irradiated cells never exhibited growth and remained positive for p24 at low levels for approximately 3 to 6 weeks after the assay set-up.
Transformation efficiency was defined as the percentage of the total cultures exposed to irradiated LAMP/MO HTLV-2 producer cells that met the criteria for transformation. The lymphocyte transformation efficiency of HTLV-2 is demonstrated in Table 1, which summarizes the results of three different experiments. Different donor PBMCs were used for each experiment. With 1,000 HTLV-2-irradiated input cells, the overall transformation efficiency was 71%, with a range of 40 to 81%. With 100 and 10 input cells, the overall transformation efficiencies dropped to 58 and 10%, respectively, demonstrating that transformation efficiency is related to the number of input infected cells.
TABLE 1.
HTLV-2 transformation efficiency and reproducibility
| Expt. | No. of antigen-positive culturesa/no. of wells (%) with LAMP/MO input cell number of:
|
||
|---|---|---|---|
| 10 | 100 | 1,000 | |
| A | 1/10 (10) | 2/10 (20) | 4/10 (40) |
| B | 2/8 (25) | 4/8 (50) | 6/8 (75) |
| C | 3/40 (8) | 22/30 (73) | 22/27 (81) |
| Total | 6/58 (10) | 28/48 (58) | 32/45 (71) |
Results are expressed as the number (%) of antigen-positive (p24 concentration >1,000 pg/ml) cultures over the total number of wells seeded that could be established as cell lines after cocultivation of 104 PBMCs with different cell concentrations (10, 100, and 1,000) of irradiated LAMP/MO.
By linear regression analysis, the log input number of LAMP/MO cells correlated with the efficiency of transformation with an r value of 0.88 and a P value of <0.0001. There was no evidence of a significant difference in donor lymphocytes relative to lymphocyte transformation. All continuous lymphocyte growth was HTLV related: there were no cultures which were positive for growth and negative for p24.
Time course of HTLV-2-mediated T-cell transformation.
In one representative experiment (Table 1, experiment C), at 9 weeks after coculture with 104 PBMCs (the earliest time point tested, to allow residual p24 to reach undetectable levels), 67% of the wells with 100 LAMP/MO cells/well were p24+, indicating infection. By week 10, 70% of the wells with 103 LAMP/MO cells/well were p24 positive. By week 12, all cultures which demonstrated growth had detectable p24, and no additional positive cultures were noted. Supernatant from cells in control wells containing only irradiated LAMP/MO cells were p24 negative at the earliest time point tested. Most of the wells that continued to proliferate could be readily expanded and continued to produce p24. These cultures have been maintained in culture for over 2 years with the addition of IL-2. Abrupt removal of IL-2 results in cessation of cell growth, but it is possible to generate IL-2 independence of HTLV-2-transformed cells (data not shown). We conclude that these HTLV-2 lymphocyte lines are IL-2 dependent in the initial phases of transformation.
Cell surface phenotypes of HTLV-2 transformants.
The cell surface phenotypes of 41 cultures from experiment C (Table 1) were determined at the earliest time possible after coculture (approximately 12 weeks). As shown in Table 2, all HTLV-2-transformed cultures were positive for the T-lymphocyte markers CD2 and/or CD3 and negative for the B1 cell surface antigen. Forty-four percent of the HTLV-2 transformants generated consisted of pure (>95%) populations of CD4+ or CD8+ T lymphocytes, and the remainder consisted of mixtures of the two subsets. A greater percentage of the transformants (39 versus 5%) were CD4+ than CD8+ in this system. In contrast, analyses of lymphocytes in vivo indicate that HTLV-1 is detected primarily in the CD4+ subset (52) and HTLV-2 is found in the CD8+ population (29). The findings reported here indicate that in vitro, the CD4+ cell is at least as susceptible to infection and transformation with HTLV-2 as the CD8+ T cell. One possible explanation for the difference between the in vitro and in vivo observations is that HTLV-2-infected CD4+ T lymphocytes may be eliminated in vivo (45) or suppressed by the host immune system.
TABLE 2.
Cell surface phenotypes of HTLV-2 transformants
| Phenotypea | No. of cultures with indicated phenotype at LAMP/MO input cell no. of:
|
Total no. (%) of cultures with phenotype | ||
|---|---|---|---|---|
| 10 | 100 | 1,000 | ||
| CD2+ or CD3+ | 2b | 19c | 20d | 41 |
| CD4+ | 1 | 13 | 2 | 16 (39) |
| CD8+ | 0 | 0 | 2 | 2 (5) |
| CD4+ and CD8+ | 1 | 6 | 16 | 23 (56) |
Cell lines were designated as CD4+ or CD8+ if >95% of the cells were positive for the subtype by flow cytometric analysis as described in the text. All HTLV-2-positive transformants in these experiments were B1 negative. The cell lines were phenotyped by one- or two-color fluorescence by using a FACScan (Becton-Dickinson and Co., Mountain View, Calif.) and were analyzed using the Lysys 2.0 software package. The following antibodies were used: CD2, CD3, CD4, CD8, B1, KC56 (T200 Ag), and MsIgG (mouse isotypic control containing immunoglobulin subtypes G1, G2a, G2b, G3) (Coulter Immunology).
From three HTLV-2-transformed cultures.
From 22 HTLV-2-transformed cultures.
From 24 HTLV-2-transformed cultures.
T-cell receptor rearrangement of HTLV-2 transformants.
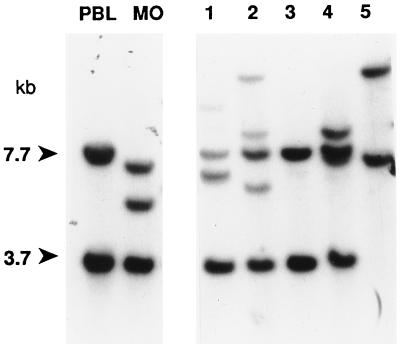
The cell surface analyses indicated that several of the HTLV-2 transformants were homogenous T-lymphocyte populations. Southern blot analysis for T-cell receptor beta chain (TCRβ) gene rearrangement was performed to confirm lineage and to establish clonality (44, 60). A representative blot is shown in Fig. 1. All 15 cultures analyzed were confirmed to be T cells by virtue of rearrangement. Ten were determined to be clonal populations, and 5 were oligoclonal. Notably, the TCRβ gene pattern demonstrated by LAMP/MO (Fig. 1, lane MO) was distinct from that of the other samples analyzed, indicating that the cultures generated in the infectivity assay were in fact new transformants and not the result of outgrowth of insufficiently irradiated LAMP/MO.
FIG. 1.
TCRβ rearrangement of HTLV-2 transformants. DNAs from selected HTLV-2 lines were digested with HindIII, subjected to Southern blotting as previously described (50), and hybridized with a 0.42-kb probe to the constant regions, CTβ1 and CTβ2, of the β chain of the T-cell receptor (Oncor, Gaithersburg, Md.). CTβ DNA was labeled with 32P by using a random-priming kit (GIBCO/BRL) and was purified on a Sephacryl G-50 column (Pharmacia, Piscataway, N.J.). The germline control (lane PBL) consisted of DNA from normal PBLs and gave the predicted bands of 3.7 and 7.7 kb. LAMP/MO (lane MO) demonstrated a clonal pattern distinct from the HTLV-2 transformants. Lanes 3, 4, and 5 are clonal populations by this analysis, and lanes 1 and 2 are oligoclonal.
Proviral copy number in HTLV-2-transformed cell lines.
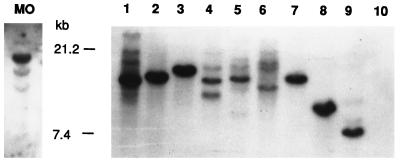
LAMP/MO and the 15 HTLV-2 transformants studied above were next analyzed for viral copy number by Southern blot analysis. Digestion of DNA from the HTLV-2 transformants with the enzyme HindIII, which does not cut in the proviral sequence, was utilized to determine the number of integrated proviruses. Each band generated by HindIII digest on Southern blot analysis thus represents a distinct proviral integration site. A representative blot is shown in Fig. 2. LAMP/MO contained a single major band and several (at least three) other, much fainter appearing bands. Of the 10 cultures determined to be clonal by TCRβ rearrangement, 5 had single integration sites, 3 had a single strong band and several faint bands, and 2 had multiple bands. The five samples which were oligoclonal by TCRβ rearrangement all had multiple integration sites. No difference in growth properties in cells with single or multiple integration sites was noted (with regard to IL-2 requirements and generation time).
FIG. 2.
Detection of HTLV-2 proviral integration sites in transformed populations of T lymphocytes. Southern blot analyses of DNA from HTLV-2 transformants determined to be clonal by TCRβ rearrangement analysis were digested with HindIII and hybridized with HTLV-2 probe pMO-4, which consisted of inserts of the entire HTLV-2 genome (57) (kindly provided by George Shaw, University of Alabama, Birmingham, Ala.). Lane MO, LAMP/MO; lanes 1 to 9, HTLV-2 transformants; lane 10, negative control (PBLs from HTLV-negative donor).
Defective proviral forms associated with HTLV-2.
Clonal populations of HTLV-1-infected lymphocytes have been shown to contain both full-length and subgenomic, or defective, proviruses. The presence of three proviral forms (full-length, 8.8 kb; defective, 6.5 and 3.5 kb) in LAMP/MO has been described previously (9). However, the transmissibility of these forms has not been investigated qualitatively in vitro. The HTLV-2 transformants derived from this transformation assay provided the opportunity to determine whether defective proviral forms are associated with T lymphocytes transformed in vitro with HTLV-2. For these analyses, a combination of the enzymes AseI and EcoRV, which each cut once in the 5′ (bp 1032) and 3′ (bp 8035) long terminal repeat, respectively, were used. A full-length provirus digested with the combination EcoRV/AseI would be visualized at 7.0 kb; a defective with internal deletions would be detected as a smaller band.
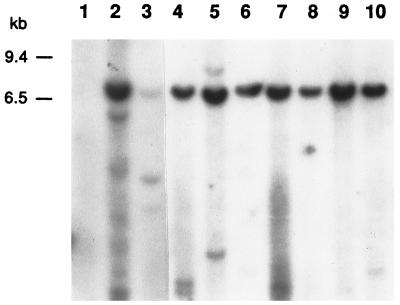
LAMP/MO and a total of 18 HTLV-2 transformants were analyzed for proviral structure. As demonstrated in Fig. 3, all transformants contained a full-length proviral copy and no transformant contained only a subgenomic fragment. LAMP/MO (Fig. 3, lane 2) contains, in addition to the full-length provirus (visualized here as a band of 7.0 kb) one major defective form of approximately 6.0 kb. One sample (Fig. 3, lane 5) contained, in addition to the full-length form, a band slightly larger than the full-length form. Two other HTLV-2 transformants harbored a smaller HTLV-2 proviral form in addition to the full-length provirus (Fig. 3, lane 3, and data not shown). With these three exceptions, all of the other 15 samples examined contained exclusively a full-length provirus.
FIG. 3.
Detection of HTLV-2 proviral integration forms in clonal populations of T lymphocytes. Southern Blot analyses of HTLV-2 transformants digested with a combination of AseI/EcoRV as described in the text and hybridized with probe pMO-4. Lane 1, negative control; lane 2, LAMP/MO; lanes 3 to 10, HTLV-2 transformants. Low-molecular-weight bands (<1 kb) seen in lanes 2, 4, 5, 7, and 10 most likely represent cellular-probe degradation products.
Production of subgenomic, or defective, provirus by HTLV-1 has been demonstrated in clinical samples (47). Similarly, analyses of HTLV-1-transformed T lymphocytes demonstrated that most transformants contained defective proviral forms in addition to the full-length provirus (data not shown). In contrast to the data for HTLV-1 transformants, our studies present little evidence for the existence of defective HTLV-2 provirus. The findings reported herein suggest either that defectives are not efficiently generated in HTLV-2 replication, and therefore play little role in HTLV-2-mediated viral transformation, or that HTLV-2 defectives that are generated in vitro are “lethal-dominants” and cannot support replication.
Chromosomal localization of HTLV integration.
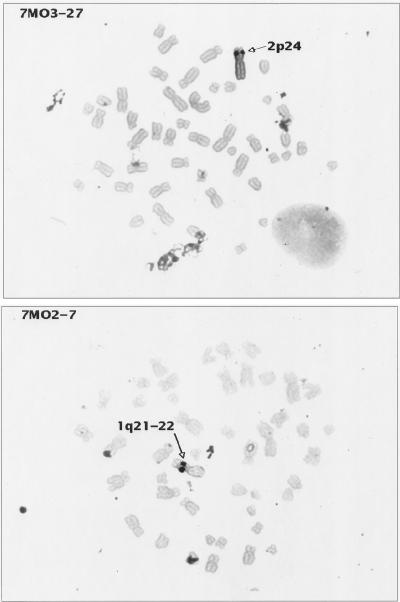
Several studies have addressed the site specificity of HTLV-1 proviral chromosomal integration (40, 55, 65). No obvious clustering of proviral integration has been observed, indicating that HTLV-1 probably integrates randomly into chromosomal DNA. The localization of HTLV-2 proviral integration has not been previously addressed. In this series of experiments, chromosomal in situ hybridization of several HTLV-1 and HTLV-2 transformants was used to determine the proviral chromosomal localization. If there were a preferential site(s) of proviral integration, the transformants derived from this assay could facilitate its localization because multiple transformants from a single donor can be analyzed. Figure 4 shows an example of in situ hybridization of metaphase spreads of two HTLV-2-transformed cell lines with probe pMO4: proviral integration at 2p24 was noted for cell line 7MO3-27, and proviral integration at 1q21-22 was noted for cell line 7MO2-7. A minimum of 20 cells were analyzed for each cell line. Signals detected more than three times at the same chromosome locations were scored as specific proviral integration sites. Eight HTLV-2 transformants were examined, and a total of 17 integration locations were detected; 10 HTLV-1 transformants were also examined, and a total of 25 integration locations were noted (data not shown). No specific chromosomal integration site or pattern was identified with transformants from either virus.
FIG. 4.
In situ hybridization of HTLV-2-transformed cell lines. Metaphase preparations of two HTLV-2 transformants (7MO2-7 and 7MO3-27) were hybridized with probe pMO-4. Cell cultures were harvested for metaphase preparation with colcemid by standard cytogenetic laboratory procedures. Probes were labelled by nick translation with either biotin-dUTP or digoxigenin-dUTP (Boehringer-Mannheim Corp.) and detected by nonfluorescent methods which use peroxidase-tagged avidin (Vector Lab Inc., Burlington, Calif.) or peroxidase-tagged anti-digoxigenin (Boehringer-Mannheim), followed by diaminobenzidine and silver amplification (Amersham Corp., Arlington Heights, Ill.). This method was originally described by Burns et al. (7), and hybridization and signal detection were performed as described by Lee et al. (36) with modifications (61). The probe concentration used was 10 μg/ml, and hybridization was performed overnight by standard in situ hybridization protocols (Oncor). For each probe and transformant cell line, several amplification times were tested: the best signal localization was achieved with the minimal amplification time (ranging from 20 to 40 min) sufficient to observe specific signal while minimizing the background. Cells were chosen for further analysis when the silver signals were confined to a single small dot on each chromatid. Images were recorded and analyzed with Cytovision software (Applied Imaging, Pittsburgh, Pa.). These results are representative of the results obtained with the other HTLV-2-transformed cell lines analyzed. Similar results were also achieved when HTLV-1-transformed cell lines were hybridized with the HTLV-1 probe pMT2, a full-length (8.25 kb) HTLV-1 probe as previously described (57).
The chromosomal localization studies of the transformants derived from this in vitro transformation system confirm and extend the conclusions of previous studies for HTLV-1. Analysis of multiple transformants, including several from a single donor, have failed to reveal a conserved integration locus. However, the possibility of HTLV integration adjacent to conserved, genome-wide repetitive sequences has not been eliminated. Recent work indicating HTLV proviral integration in the GC-rich fraction of the genome (65) supports the concept that HTLV-1 integration may be governed by yet undefined rules. Further studies are necessary to elucidate completely the relationship between HTLV integration and T-lymphocyte transformation.
The data presented in this report show that clonal populations of lymphocytes transformed with HTLV-2 can be reproducibly generated by using a modified limiting dilution cocultivation assay. Clonal transformants are typically CD3+, B1−, CD4+, or CD8+ T lymphocytes demonstrating IL-2-dependent long-term growth and continuous p24 production. The time course of transformation and transformation efficiency, as well as the phenotypic profile of the resultant HTLV-2 clones, are comparable to the results obtained when HTLV-1 is used to transform PBMCs (50). One notable difference is the frequent detection of defective proviral formation with HTLV-1 transformants and the rarity of defective proviruses noted with HTLV-2 transformants.
The present results do not illuminate one of the more intriguing biologic questions about HTLV-1 and HTLV-2, namely, the viral properties that account for the markedly different ecologic niches. In recent years, the HTLV seroprevalence among IVDUs has been attributed predominantly to HTLV-2, indicating a possible quantitative or qualitative difference in infectivity between the two viruses. Since in vitro transformation with HTLV-1 and HTLV-2 are apparently equivalent, perhaps other factors are important determinants of transmission and explain the seeming differences in the niches occupied by these closely related retroviruses. Particularly puzzling from a seroepidemiologic perspective is the relative absence of HTLV-2 in the general population and, in spite of a low level of HTLV-1 in the general population, the relative absence of HTLV-1 in IVDUs (2, 33, 35, 53). It is clear that HTLV-1 is etiologically associated with clinical disease; it seems possible that HTLV-2 may be as virulent as HTLV-1 but the high mortality of infected patients from other causes (49) may obscure HTLV-2-specific disease. Alternatively, HTLV-2 may produce only subclinical infections in the vast majority of HTLV-2-infected patients and differ qualitatively from HTLV-1 in disease potential.
The present studies are the first qualitative analyses of the HTLV-2 proviral structure in transformed T lymphocytes. Clonal T-cell populations were shown to contain either single or multiple integration sites. HTLV-1 proviral integration has been characterized by numerous studies: adult T-cell leukemia cells contain oligo- or monoclonal viral integration (64), while in cells from patients with tropical spastic paraparesis/HTLV-associated myelopathy, there is a notably polyclonal pattern (21, 22). Though the demonstration of immortalization with a single proviral integration in both HTLV-1 and HTLV-2 suggests that one integration site, or virus by itself, is sufficient for transformation, additional cellular events are most likely involved as well in HTLV-mediated T-cell transformation. Transformation associated with multiple integrants could be due to successive integration cycles until one favorable integration event occurs; alternatively, the transformed phenotype may represent the cumulative effect of multiple integrations. The in vitro data presented above provide support for the former hypothesis.
HTLV transformation confers enhanced growth potential on the infected lymphocyte. This transforming capacity has been exploited in the laboratory as a method of immortalizing lymphocytes for further study, in a manner similar to Epstein-Barr virus transformation of B lymphocytes. PBLs from patients with adenosine deaminase deficiency (32) and paroxysmal nocturnal hemoglobinura (46) have been immortalized with HTLV-1, and a series of papers have examined hormone responsiveness in Pygmy and normal HTLV-2-transformed T-cell lines (17–19). The transforming ability of HTLV-2, the relative frequency of monoclonal integrants, and the absence of disease association render HTLV-2 a suitable candidate for the immortalization of T lymphocytes from patients for multiple purposes.
REFERENCES
- 1.Bhagavati S, Ehrlich G, Kula R W, Kwok S, Sninsky J, Udani V, Poiesz B J. Detection of human T-cell lymphoma/leukemia virus type I DNA and antigen in spinal fluid and blood of patients with chronic progressive myelopathy. N Engl J Med. 1988;318:1141–1147. doi: 10.1056/NEJM198805053181801. [DOI] [PubMed] [Google Scholar]
- 2.Biggar R J, Buskell-Bales Z, Yakshe P N, Caussy D, Gridley G, Seeff L. Antibody to human retroviruses among drug users in three east coast American cities, 1972–1976. J Infect Dis. 1991;163:57–63. doi: 10.1093/infdis/163.1.57. [DOI] [PubMed] [Google Scholar]
- 3.Biggar R J, Johnson B K, Oster C, Sarin P S, Ocheng D, Tukei P, Nsanze H, Alexander S, Bodner A J, Siongok T A, et al. Regional variation in prevalence of antibody against human T-lymphotropic virus types I and III in Kenya, East Africa. Int J Cancer. 1985;35:763–767. doi: 10.1002/ijc.2910350611. [DOI] [PubMed] [Google Scholar]
- 4.Biggar R J, Saxinger C, Gardiner C, Collins W E, Levine P H, Clark J W, Nkrumah F K, Blattner W A. Type-I HTLV antibody in urban and rural Ghana, West Africa. Int J Cancer. 1984;34:215–219. doi: 10.1002/ijc.2910340212. [DOI] [PubMed] [Google Scholar]
- 5.Blattner W A, Kalyanaraman V S, Robert-Guroff M, Lister T A, Galton D A, Sarin P S, Crawford M H, Catovsky D, Greaves M, Gallo R C. The human type-C retrovirus, HTLV, in blacks from the Caribbean region, and relationship to adult T-cell leukemia/lymphoma. Int J Cancer. 1982;30:257–264. doi: 10.1002/ijc.2910300302. [DOI] [PubMed] [Google Scholar]
- 6.Blayney D W, Blattner W A, Robert-Guroff M, Jaffe E S, Fisher R I, Bunn P, Jr, Patton M G, Rarick H R, Gallo R C. The human T-cell leukemia-lymphoma virus in the southeastern United States. JAMA. 1983;250:1048–1052. [PubMed] [Google Scholar]
- 7.Burns J, Chan V, Jonasson J, Fleming K, Taylor S, McGee J. Sensitive system for visualising biotinylated DNA probes hybridised in situ: rapid sex determination of intact cells. J Clin Pathol. 1985;38:1085–1092. doi: 10.1136/jcp.38.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catovsky D, Greaves M F, Rose M, Galton D A, Goolden A W, McCluskey D R, White J M, Lampert I, Bourikas G, Ireland R, Brownell A I, Bridges J M, Blattner W A, Gallo R C. Adult T-cell lymphoma-leukaemia in blacks from the West Indies. Lancet. 1982;1:639–643. doi: 10.1016/s0140-6736(82)92200-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen I S, McLaughlin J, Gasson J C, Clark S C, Golde D W. Molecular characterization of genome of a novel human T-cell leukaemia virus. Nature. 1983;305:502–505. doi: 10.1038/305502a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen I S, Quan S G, Golde D W. Human T-cell leukemia virus type II transforms normal human lymphocytes. Proc Natl Acad Sci USA. 1983;80:7006–7009. doi: 10.1073/pnas.80.22.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chosa T, Yamamoto N, Tanaka Y, Koyanagi Y, Hinuma Y. Infectivity dissociated from transforming activity in a human retrovirus, adult T-cell leukemia virus. Gann. 1982;73:844–847. [PubMed] [Google Scholar]
- 12.Duenas-Barajas E, Bernal J E, Vaught D R, Nerurkar V R, Sarmiento P, Yanagihara R, Gajdusek D C. Human retroviruses in Amerindians of Colombia: high prevalence of human T cell lymphotropic virus type II infection among the Tunebo Indians. Am J Trop Med Hyg. 1993;49:657–663. doi: 10.4269/ajtmh.1993.49.657. [DOI] [PubMed] [Google Scholar]
- 13.Echeverria de Perez G, Leon-Ponte M, Noya O, Botto C, Gallo D, Bianco N. First description of endemic HTLV-II infection among Venezuelan Amerindians. J Acquired Immune Defic Syndr. 1993;6:1368–1372. [PubMed] [Google Scholar]
- 14.Feigenbaum F, Fang C, Sandler S G. Human T-lymphotropic virus type II in Panamanian Guaymi Indians. Transfusion. 1994;34:158–161. doi: 10.1046/j.1537-2995.1994.34294143946.x. [DOI] [PubMed] [Google Scholar]
- 15.Ferrer J F, Del Pino N, Esteban E, Sherman M P, Dube S, Dube D K, Basombrio M A, Pimentel E, Segovia A, Quirulas S, et al. High rate of infection with the human T-cell leukemia retrovirus type II in four Indian populations of Argentina. Virology. 1993;197:576–584. doi: 10.1006/viro.1993.1631. [DOI] [PubMed] [Google Scholar]
- 16.Gallo R C, Kalyanaraman V S, Sarngadharan M G, Sliski A, Vonderheid E C, Maeda M, Nakao Y, Yamada K, Ito Y, Gutensohn N, Murphy S, Bunn P, Jr, Catovsky D, Greaves M F, Blayney D W, Blattner W, Jarrett W F, zur Hausen H, Seligmann M, Brouet J C, Haynes B F, Jegasothy B V, Jaffe E, Cossman J, Broder S, Fisher R I, Golde D W, Robert-Guroff M. Association of the human type C retrovirus with a subset of adult T-cell cancers. Cancer Res. 1983;43:3892–3899. [PubMed] [Google Scholar]
- 17.Geffner M E, Bailey R C, Bersch N, Vera J C, Golde D W. Insulin-like growth factor-I unresponsiveness in an Efe Pygmy. Biochem Biophys Res Commun. 1993;193:1216–1223. doi: 10.1006/bbrc.1993.1755. [DOI] [PubMed] [Google Scholar]
- 18.Geffner M E, Bersch N, Bailey R C, Golde D W. Growth hormone induces resistance to the mitogenic action of insulin through local IGF-I. Studies in normal and Pygmy T-cell lines. Diabetes. 1994;43:68–72. doi: 10.2337/diab.43.1.68. [DOI] [PubMed] [Google Scholar]
- 19.Geffner M E, Bersch N, Golde D W. Insulin and IGF-I stimulate normal and virally transformed T-lymphocyte cell growth in vitro. Brain Behav Immun. 1992;6:377–386. doi: 10.1016/0889-1591(92)90036-n. [DOI] [PubMed] [Google Scholar]
- 20.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;2:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 21.Gessain A, Saal F, Gout O, Daniel M T, Flandrin G, de The G, Peries J, Sigaux F. High human T-cell lymphotropic virus type I proviral DNA load with polyclonal integration in peripheral blood mononuclear cells of French West Indian, Guianese, and African patients with tropical spastic paraparesis. Blood. 1990;75:428–433. [PubMed] [Google Scholar]
- 22.Greenberg S J, Jacobson S, Waldmann T A, McFarlin D E. Molecular analysis of HTLV-I proviral integration and T cell receptor arrangement indicates that T cells in tropical spastic paraparesis are polyclonal. J Infect Dis. 1989;159:741–744. doi: 10.1093/infdis/159.4.741. [DOI] [PubMed] [Google Scholar]
- 23.Hall W W, Takahashi H, Liu C, Kaplan M H, Scheewind O, Ijichi S, Nagashima K, Gallo R C. Multiple isolates and characteristics of human T-cell leukemia virus type II. J Virol. 1992;66:2456–2463. doi: 10.1128/jvi.66.4.2456-2463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington W, Jr, Sheremata W, Hjelle B, Dube D K, Bradshaw P, Foung S K, Snodgrass S, Toedter G, Cabral L, Poiesz B. Spastic ataxia associated with human T-cell lymphotropic virus type II infection. Ann Neurol. 1993;33:411–414. doi: 10.1002/ana.410330416. [DOI] [PubMed] [Google Scholar]
- 25.Hinuma Y, Komoda H, Chosa T, Kondo T, Kohakura M, Takenaka T, Kikuchi M, Ichimaru M, Yunoki K, Sato I, Matsuo R, Takiuchi Y, Uchino H, Hanaoka M. Antibodies to adult T-cell leukemia-virus-associated antigen (ATLA) in sera from patients with ATL and controls in Japan: a nation-wide sero-epidemiologic study. Int J Cancer. 1982;29:631–635. doi: 10.1002/ijc.2910290606. [DOI] [PubMed] [Google Scholar]
- 26.Hinuma Y, Nagata K, Hanaoka M, Nakai M, Matsumoto T, Kinoshita K I, Shirakawa S, Miyoshi I. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc Natl Acad Sci USA. 1981;78:6476–6480. doi: 10.1073/pnas.78.10.6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hjelle B, Appenzeller O, Mills R, Alexander S, Torrez-Martinez N, Jahnke R, Ross G. Chronic neurodegenerative disease associated with HTLV-II infection. Lancet. 1992;339:645–646. doi: 10.1016/0140-6736(92)90797-7. [DOI] [PubMed] [Google Scholar]
- 28.Hjelle B, Scalf R, Swenson S. High frequency of human T-cell leukemia-lymphoma virus type II infection in New Mexico blood donors: determination by sequence-specific oligonucleotide hybridization. Blood. 1990;76:450–454. [PubMed] [Google Scholar]
- 29.Ijichi S, Ramundo M B, Takahashi H, Hall W W. In vivo cellular tropism of human T cell leukemia virus type II (HTLV-II) J Exp Med. 1992;176:293–296. doi: 10.1084/jem.176.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson S, Lehky T, Nishimura M, Robinson S, McFarlin D E, Dhib-Jalbut S. Isolation of HTLV-II from a patient with chronic, progressive neurological disease clinically indistinguishable from HTLV-I-associated myelopathy/tropical spastic paraparesis. Ann Neurol. 1993;33:392–396. doi: 10.1002/ana.410330411. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson S, Raine C S, Mingioli E S, McFarlin D E. Isolation of an HTLV-1-like retrovirus from patients with tropical spastic paraparesis. Nature. 1988;331:540–543. doi: 10.1038/331540a0. [DOI] [PubMed] [Google Scholar]
- 32.Kohn D B, Mitsuya H, Ballow M, Selegue J E, Barankiewicz J, Cohen A, Gelfand E, Anderson W F, Blaese R M. Establishment and characterization of adenosine deaminase-deficient human T cell lines. J Immunol. 1989;142:3971–3977. [PubMed] [Google Scholar]
- 33.Kwok S, Gallo D, Hanson C, McKinney N, Poiesz B, Sninsky J J. High prevalence of HTLV-II among intravenous drug abusers: PCR confirmation and typing. Aids Res Hum Retroviruses. 1990;6:561–565. doi: 10.1089/aid.1990.6.561. [DOI] [PubMed] [Google Scholar]
- 34.Lairmore M D, Jacobson S, Gracia F, De B K, Castillo L, Larreategui M, Roberts B D, Levine P H, Blattner W A, Kaplan J E. Isolation of human T-cell lymphotropic virus type 2 from Guaymi Indians in Panama. Proc Natl Acad Sci USA. 1990;87:8840–8844. doi: 10.1073/pnas.87.22.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Swanson P, Shorty V S, Zack J A, Rosenblatt J D, Chen I S. High rate of HTLV-II infection in seropositive I.V. drug abusers in New Orleans. Science. 1989;244:471–475. doi: 10.1126/science.2655084. [DOI] [PubMed] [Google Scholar]
- 36.Lee J J, Warburton D, Robertson E J. Cytogenetic methods in the mouse: preparation of chromosomes, karyotyping, and in situ hybridization. Anal Biochem. 1990;189:1–17. doi: 10.1016/0003-2697(90)90036-9. [DOI] [PubMed] [Google Scholar]
- 37.Lee T H, Coligan J E, McLane M F, Sodroski J G, Popovic M, Wong-Staal F, Gallo R C, Haseltine W, Essex M. Serological cross-reactivity between envelope gene products of type I and type II human T-cell leukemia virus. Proc Natl Acad Sci USA. 1984;81:7579–7583. doi: 10.1073/pnas.81.23.7579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine P H, Blattner W A, Clark J, Tarone R, Maloney E M, Murphy E M, Gallo R C, Robert-Guroff M, Saxinger W C. Geographic distribution of HTLV-I and identification of a new high-risk population. Int J Cancer. 1988;42:7–12. doi: 10.1002/ijc.2910420103. [DOI] [PubMed] [Google Scholar]
- 39.Levine P H, Jacobson S, Elliott R, Cavallero A, Colclough G, Dorry C, Stephenson C, Knigge R M, Drummond J, Nishimura M, et al. HTLV-II infection in Florida Indians. Aids Res Hum Retroviruses. 1993;9:123–127. doi: 10.1089/aid.1993.9.123. [DOI] [PubMed] [Google Scholar]
- 40.Macera M J, Szabo P, Verma R S. Chromosomal localization of HTLV-1 viral integration sites using in situ hybridization: detection of a novel IL2R fragment. Mol Gen Genet. 1992;234:466–474. doi: 10.1007/BF00538707. [DOI] [PubMed] [Google Scholar]
- 41.Maloney E M, Biggar R J, Neel J V, Taylor M E, Hahn B H, Shaw G M, Blattner W A. Endemic human T cell lymphotropic virus type II infection among isolated Brazilian Amerindians. J Infect Dis. 1992;166:100–107. doi: 10.1093/infdis/166.1.100. [DOI] [PubMed] [Google Scholar]
- 42.McFarlin D E. Neurological disorders related to HTLV-I and HTLV-II. J Acquired Immune Defic Syndr. 1993;6:640–644. [PubMed] [Google Scholar]
- 43.Merino F, Robert-Guroff M, Clark J, Biondo-Bracho M, Blattner W A, Gallo R C. Natural antibodies to human T-cell leukemia/lymphoma virus in healthy Venezuelan populations. Int J Cancer. 1984;34:501–506. doi: 10.1002/ijc.2910340412. [DOI] [PubMed] [Google Scholar]
- 44.Minden M D, Toyonaga B, Ha K, Yanagi B, Chin B, Gelfand E, Mak T. Somatic rearrangement of T-cell antigen receptor gene in human T-cell malignancies. Proc Natl Acad Sci USA. 1985;82:1224–1227. doi: 10.1073/pnas.82.4.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyamoto K, Kamiya T, Minowada J, Tomita N, Kitajima K. Transformation of CD8+ T-cells producing a strong cytopathic effect on CD4+ T-cells through syncytium formation by HTLV-II. Jpn J Cancer Res. 1991;82:1178–1183. doi: 10.1111/j.1349-7006.1991.tb01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakakuma H, Nagakura S, Horikawa K, Hidaka M, Kawaguchi T, Iwamoto N, Sanada I, Kagimoto T, Takatsuki K. Interleukin-2-dependent T-cell lines established from paroxysmal nocturnal hemoglobinuria patients. Blood. 1994;84:309–314. [PubMed] [Google Scholar]
- 47.Ohshima K, Kikuchi M, Masuda Y, Kobari S, Sumiyoshi Y, Eguchi F, Mohtai H, Yoshida T, Takeshita M, Kimura N. Defective provirus form of human T-cell leukemia virus type I in adult T-cell leukemia/lymphoma: clinicopathological features. Cancer Res. 1991;51:4639–4642. [PubMed] [Google Scholar]
- 48.Osame M, Matsumoto M, Usuku K, Izumo S, Ijichi N, Amitani H, Tara M, Igata A. Chronic progressive myelopathy associated with elevated antibodies to human T-lymphotropic virus type I and adult T-cell leukemia-like cells. Ann Neurol. 1987;21:117–122. doi: 10.1002/ana.410210203. [DOI] [PubMed] [Google Scholar]
- 49.Page J B, Lai S H, Chitwood D D, Klimas N G, Smith P C, Fletcher M A. HTLV-I/II seropositivity and death from AIDS among HIV-1 seropositive intravenous drug users. Lancet. 1990;335:1439–1441. doi: 10.1016/0140-6736(90)91456-k. . (Comments.) [DOI] [PubMed] [Google Scholar]
- 50.Persaud D, Munoz J, Tarsis S, Parks E, Parks W. Time course and cytokine dependence of human T-cell lymphotropic virus type 1 T-lymphocyte transformation as revealed by a microtiter infectivity assay. J Virol. 1995;69:6297–6303. doi: 10.1128/jvi.69.10.6297-6303.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Popovic M, Lange-Wantzin G, Sarin P S, Mann D, Gallo R C. Transformation of human umbilical cord blood T cells by human T-cell leukemia/lymphoma virus. Proc Natl Acad Sci USA. 1983;80:5402–5406. doi: 10.1073/pnas.80.17.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson J H, Edwards A J, Cruickshank J K, Rudge P, Dalgleish A G. In vivo cellular tropism of human T-cell leukemia virus type 1. J Virol. 1990;64:5682–5687. doi: 10.1128/jvi.64.11.5682-5687.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robert-Guroff M, Weiss S H, Giron J A, Jennings A M, Ginzburg H M, Margolis I B, Blattner W A, Gallo R C. Prevalence of antibodies to HTLV-I, -II, and -III in intravenous drug abusers from an AIDS endemic region. JAMA. 1986;255:3133–3137. [PubMed] [Google Scholar]
- 54.Saxinger W, Blattner W A, Levine P H, Clark J, Biggar R, Hoh M, Moghissi J, Jacobs P, Wilson L, Jacobson R, et al. Human T-cell leukemia virus (HTLV-I) antibodies in Africa. Science. 1984;225:1473–1476. doi: 10.1126/science.6089348. [DOI] [PubMed] [Google Scholar]
- 55.Seiki M, Eddy R, Shows T B, Yoshida M. Nonspecific integration of the HTLV provirus genome into adult T-cell leukaemia cells. Nature. 1984;309:640–642. doi: 10.1038/309640a0. [DOI] [PubMed] [Google Scholar]
- 56.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw G M, Broder S, Essex M, Gallo R C. Human T-cell leukemia virus: its discovery and role in leukemogenesis and immunosuppression. Adv Intern Med. 1984;30:1–27. [PubMed] [Google Scholar]
- 58.Sheremata W A, Harrington W, Jr, Bradshaw P A, Foung S K, Raffanti S P, Berger J R, Snodgrass S, Resnick L, Poiesz B J. Association of ‘(tropical) ataxic neuropathy’ with HTLV-II. Virus Res. 1993;29:71–77. doi: 10.1016/0168-1702(93)90126-8. [DOI] [PubMed] [Google Scholar]
- 59.Shimotohno K, Takahashi Y, Shimizu N, Gojobori T, Golde D W, Chen I S, Miwa M, Sugimura T. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc Natl Acad Sci USA. 1985;82:3101–3105. doi: 10.1073/pnas.82.10.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siu G, Clark S P, Yoshikai Y, Malissen M, Yanagi Y, Strauss E, Mak T W, Hood L. The human T cell antigen receptor is encoded by variable, diversity joining gene segments that rearrange to generate a complete V gene. Cell. 1984;37:393–401. doi: 10.1016/0092-8674(84)90369-6. [DOI] [PubMed] [Google Scholar]
- 61.Warburton D, Yu M T, Tantravahi U, Lee C, Cayanis E, Russo J, Fischer S. Regional localization of 32 NotI-HindIII fragments from a human chromosome 13 library by a somatic cell hybrid panel and in situ hybridization. Genomics. 1993;16:355–360. doi: 10.1006/geno.1993.1197. [DOI] [PubMed] [Google Scholar]
- 62.Yamamoto N, Okada M, Koyanagi Y, Kannagi M, Hinuma Y. Transformation of human leukocytes by cocultivation with an adult T cell leukemia virus producer cell line. Science. 1982;217:737–739. doi: 10.1126/science.6980467. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci USA. 1984;81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zoubak S, Richardson J H, Rynditch A, Hollsberg P, Hafler D A, Boeri E, Lever A M, Bernardi G. Regional specificity of HTLV-I proviral integration in the human genome. Gene. 1994;143:155–163. doi: 10.1016/0378-1119(94)90091-4. [DOI] [PubMed] [Google Scholar]