Abstract
Simian hemorrhagic fever virus (SHFV) was recently reclassified and assigned to the new virus family Arteriviridae. During replication, arteriviruses produce a 3′ coterminal, nested set of subgenomic mRNAs (sgRNAs). These sgRNAs arise by discontinuous transcription, and each contains a 5′ leader sequence which is joined to the body of the mRNA through a conserved junction sequence. Only the 5′-most open reading frame (ORF) is believed to be transcribed from each sgRNA. The SHFV genome encodes nine ORFs that are presumed to be expressed from sgRNAs. However, reverse transcription-PCR analysis with leader- and ORF-specific primers identified only eight sgRNA species. The consensus sequence 5′-UCNUUAACC-3′ was identified as the junction motif. Our data suggest that sgRNA 2 may be bicistronic, expressing both ORF 2a and ORF 2b. SHFV encodes three more ORFs on its genome than the other arteriviruses. Comparative sequence analysis suggested that SHFV ORFs 2a, 2b, and 3 are related to ORFs 2 through 4 of the other arteriviruses. Evidence which suggests that SHFV ORFs 4 through 6 are related to ORFs 2a through 3 and may have resulted from a recombination event during virus evolution is presented.
Simian hemorrhagic fever virus (SHFV), an enveloped, positive-stranded RNA virus, causes a persistent, asymptomatic infection in monkeys belonging to the genera Papio, Erythrocebus, and Cercopithecus (17, 23). However, this virus induces a fatal hemorrhagic fever in species within the genus Macaca (29, 36). SHFV, together with equine arteritis virus (EAV), murine lactate dehydrogenase-elevating virus (LDV), and porcine reproductive and respiratory syndrome virus (PRRSV), has recently been assigned to the monogeneric family Arteriviridae within the newly established order Nidovirales (4) (reviewed in references 10 and 35).
The arteriviral genomes are monopartate, capped (31), and polyadenylated (3, 30), and they vary in length from 12.7 to 15.1 kb (7, 15, 26). The 5′-terminal two-thirds of the viral genomes are taken up by two large overlapping open reading frames (ORFs) designated ORF 1a and ORF 1b. These ORFs encode polyproteins from which the viral polymerase and other nonstructural proteins are derived (reviewed in reference 10). In the case of EAV, LDV, and PRRSV, six smaller ORFs are present downstream of ORF 1b encoding four glycoproteins (GP2 through GP5), the unglycosylated membrane protein (M), and the nucleocapsid protein (N), as ordered from the 5′ → 3′ direction, according to the nomenclature of van Nieuwstadt et al. (39). These ORFs are expressed through a 3′ coterminal, nested set of subgenomic mRNAs (sgRNAs) which are assumed to be functionally monocistronic (9, 37). Each sgRNA contains a leader sequence of approximately 200 nucleotides (nt) (7, 15, 19, 26, 42) which is derived from the extreme 5′ end of the genome and is joined to the body of the RNA via a poorly understood discontinuous transcription mechanism. Fusion of the leader and body sequences occurs at a short conserved motif (6–8, 25, 42), the transcription-associated sequence (TAS) (18). The TAS is found at the 3′ end of the common leader sequence and at positions upstream of each of the ORFs (7, 15, 25).
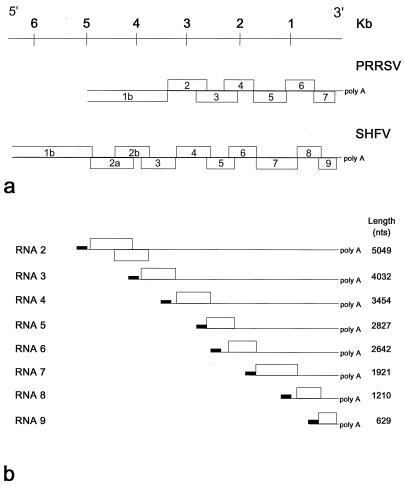
Recent analysis of the 6.3 kb, 3′ terminal SHFV genome sequence revealed the presence of nine ORFs, instead of the expected six, downstream of ORF 1b (Fig. 1) (34). The gene organization at the 3′ end of the SHFV genome is identical to that of the other arteriviruses in that ORF 7 is presumed to encode GP5 and ORFs 8 and 9 encode the M and N proteins, respectively (16, 34). The coding assignments of the remaining six SHFV ORFs are not known. Furthermore, it is not clear how these ORFs are expressed: Northern blot analysis of RNA extracted from SHFV-infected cells detected only six polyadenylated virus-specific sgRNAs, with estimated lengths of 4.7, 3.3, 2.7, 2.0, 1.2, and 0.65 kb (16, 42). Analysis of the two smallest SHFV RNAs identified a short conserved sequence, 5′-U(U/C)AACC-3′ at the junction site (42). One or more similar motifs have been found upstream of each of the SHFV ORFs (34). However, which of these potential TASs are actually used during virus replication had not been determined. Here, we have studied SHFV transcription in further detail. We have employed reverse transcription (RT)-PCR to identify all of the sgRNAs and to map each of the leader-body junction sites. In addition, we have investigated the coding assignments of SHFV ORFs 2a through 6 by computer-assisted sequence analysis.
FIG. 1.
Schematic representation of the genome organization of the PRRSV and SHFV 3′ ORFs (a) and the SHFV sgRNAs (b). ORFs are drawn approximately to scale. Solid lines represent untranslated RNA. Black rectangles represent 5′ leader sequences. The lengths of the sgRNAs, excluding the poly(A) tracts, are also shown.
To analyze the viral sgRNAs, MA104 cells were infected with the LVR 42-0/M6941 strain of SHFV at multiplicity of infection of 0.1, and total cytoplasmic RNA was isolated at 20 h postinfection according to the method of Sawicki et al. (32) with modifications (42). Leader-body junction regions of SHFV sgRNAs were amplified by RT-PCR using negative-sense ORF-specific oligonucleotide primers (Table 1) and a positive-sense primer corresponding to nt 60 through 77 of the SHFV 5′ leader sequence (42). The resulting PCR fragments were purified from 1% agarose gels, inserted into plasmid pCRII, and introduced into Escherichia coli TOP10F′ with the TA Cloning Kit, according to the manufacturer (Invitrogen Corp., San Diego, Calif.). Recombinant plasmid DNA was purified with the Quantum Prep Plasmid Miniprep kit (Bio-Rad Laboratories, Hercules, Calif.) and sequenced with M13 universal and reverse primers and the Sequenase DNA sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio). For each PCR product, at least three independent clones were analyzed.
TABLE 1.
Reverse primer sequences used for RT-PCR and locations of primer sequences downstream of initiation codons
| ORF | Reverse primer sequence (5′ → 3′) | Distance to initiation codon (nt) |
|---|---|---|
| 2a | TTGAAACCCTTCTCTGCC | 263 |
| 2b | GCAGATGAAAGCAAGCAAC | 104 |
| 3 | TTCGCCATAACCCAGAAC | 282 |
| 4 | AAAGAACTGGGTTCGCAC | 338 |
| 5 | GAAGTGCAACTTGACCTGAG | 115 |
| 6 | CGCAATCACAATGAACAAG | 506 |
| 7 | AAAGTCAAGAAAGGAGGCAG | 490 |
| 8 | GTCGCGTTAATCGCTAAAC | 313 |
| 9 | CATATGAAAGATTTCCGCC | 231 |
As shown in Fig. 2, eight distinct junction regions were identified. These were located upstream of ORF 2a and of ORFs 3 through 9. Alignment of the genomic fusion regions and comparison with the 3′ end of the leader sequence indicate that the SHFV TAS conforms to a nonanucleotide consensus sequence, 5′-UCNUUAACC-3′, in which only the residues at positions 7 and 8 are strictly conserved. The SHFV TAS resembles the TASs of PRRSV strain Lelystad [5′-U(U/C)AACC-3′] (25), LDV [5′-U(A/G)UAACC-3′] (6), and EAV (5′-UCAACU-3′) (8, 9). The distances between the SHFV TASs and the initiation codons of the 5′-most ORFs on the respective sgRNAs range from 1 to 178 nt (Fig. 2).
FIG. 2.
Sequences of the leader-body junction regions of the SHFV sgRNAs. The sequences are aligned with the 5′ ends of the SHFV genome (5′ Leader [42]) and with the genomic sequences surrounding the TAS (Genome). Colons indicate identical nucleotides. The initiation codons for ORFs 2a through 9 are underlined. Nucleotides from the leader and genome sequences which unambiguously contribute to the sgRNA are shaded. The overall junction region consensus sequence is shown at the bottom of the figure with the most common nucleotides represented in the middle line.
Previous studies have shown that during arteriviral transcription, fusion of the leader and body sequences can occur at different positions within the TAS, resulting in sgRNA sequence heterogeneity (6, 25). In this study, such heterogeneity was not observed between cDNA clones derived from the same leader-body junction region. However, only a limited number of SHFV cDNA clones were studied per sgRNA species. Furthermore, for sgRNAs 2a, 4, 7, and 9, it was not possible to pinpoint leader-body fusion sites because of the extensive sequence identity between the leader sequences and the TASs. The results shown in Fig. 2 suggest that in the case of the ORF 8 sgRNA, leader-body fusion preferentially occurred at position 4, with residues 1 through 3 being derived from the leader and 5 through 9 from the genomic sequence. Similarly, for the sgRNAs of ORFs 3 and 6, leader-body fusion took place at either position 4 or 5. In contrast, on the ORF 5 sgRNA at least the 3′-most 7 nt were derived from the genome, and leader-body fusion may have occurred at position 1 or 2 or possibly even at position −1, i.e., upstream of the TAS (Fig. 2).
Based upon our present results, we propose that SHFV produces eight sgRNA species during replication. According to conventional nomenclature (5), these sgRNAs are designated RNAs 2 through 9, with calculated lengths of 5.049, 4.032, 3.454, 2.827, 2.642, 1.921, 1.210, and 0.629 kb, respectively; these sizes exclude the 3′ poly(A) tail but include the 208-nt 5′ leader sequence (Fig. 1) (42). The lengths of sgRNAs 2, 4, 6, 7, 8, and 9 correspond to those found for the SHFV RNAs in infected cells as detected by Northern blot analysis (16, 42). Amplification of sgRNAs 3 and 5 consistently yielded reduced amounts of RT-PCR products, compared to those obtained for the other six sgRNAs. This suggests that sgRNAs 3 and 5 may be produced in small amounts during virus replication, which could explain why these transcripts were previously overlooked by Northern blot hybridization. Also, sgRNAs 5 and 6 are likely to comigrate in agarose gels, which may have precluded detection of the individual transcripts.
Surprisingly, we did not detect an ORF 2b-specific sgRNA. A potential TAS was previously identified 141 nt upstream of the ORF 2b initiation codon (34). However, RT-PCR performed with a combination of the ORF 2b- and leader-specific primers did not yield the anticipated product of 413 nt but rather a single, much larger product of 893 nt. Sequence analysis showed that the latter product had not been derived from an ORF 2b-specific sgRNA but rather from sgRNA 2. The apparent absence of a separate sgRNA for ORF 2b may indicate that this gene is silent. However, we currently favor the hypothesis that sgRNA 2 may be functionally bicistronic, directing the synthesis of both the ORF 2a and ORF 2b products. Experiments to test this hypothesis are currently under way. The expression of multiple ORFs from a single sgRNA species has not yet been reported for any other arterivirus, but it has been well documented for the closely related coronaviruses (21, 22, 33), i.e., other members of the order Nidovirales which employ a transcription scheme similar to that of the arteriviruses (reviewed in references 24 and 38).
EAV, LDV, and PRRSV carry the same complement of genes, and their genomes are essentially collinear. SHFV possesses three more ORFs than the other arteriviruses. Whereas ORFs 8 and 9 have been found to encode the SHFV homologs of M and N, respectively (Fig. 1) (16), and ORF 7 presumably encodes GP5 (34), the genes for GP2 through GP4 remain to be identified. Little is known about the characteristics and functions of these glycoproteins. GP2 is a class I membrane glycoprotein which has been found in the viral envelopes of EAV, LDV, and PRRSV (11, 13, 27, 28). Studies of LDV and PRRSV suggest that GP3, a soluble glycoprotein, and GP4, an integral membrane protein of unknown topography, are also part of the virion (12, 14, 39, 41). SHFV virions contain at least two envelope glycoprotein species, of 42 and 54 kDa (16), the latter of which presumably represents GP5 (34).
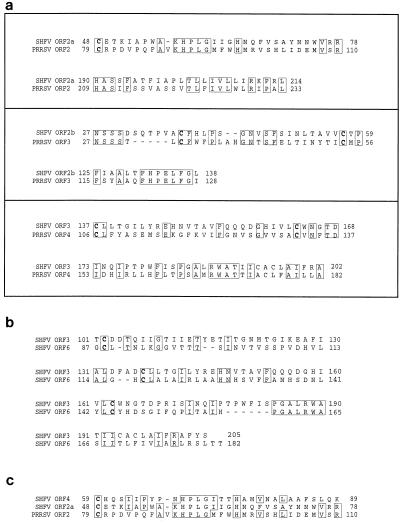
The products encoded by SHFV ORFs 2a through 6 all show characteristics of glycoproteins. To determine the relationship of these proteins to those encoded by other arteriviruses, the deduced amino acid sequences were used as a query to search the nonredundant protein database at the National Center for Biotechnology Information (NCBI) (Bethesda, Md.) by using the software program BLASTP (1). Regions of significant amino acid sequence similarity were detected between the products of SHFV ORFs 2a, 2b, and 3 and those of ORFs 2, 3, and 4 of the Lelystad isolate of PRRSV (Fig. 3a). These results suggest that ORFs 2a, 2b, and 3 encode homologs of GP2, GP3, and GP4, respectively.
FIG. 3.
(a) Alignment of conserved regions in the predicted translation products of SHFV ORFs 2a, 2b, and 3 with those of ORFs 2 through 4, respectively, of the Lelystad isolate of PRRSV (26). (b) Alignment of the carboxy-terminal halves of the predicted translation products of SHFV ORFs 3 and 6. (c) Alignment of parts of the deduced amino acid sequences of SHFV ORFs 2a and 4 with that of the ORF 2 product of the Lelystad isolate of PRRSV. Cysteine residues are indicated in boldface type. The alignments were based on database searches using version 1.4.9MP of BLASTP (1) in combination with pairwise comparisons and multiple sequence alignments generated with the Genetics Computer Group software programs GAP and PILEUP, respectively.
A search using the products of ORFs 4 through 6 as a query did not reveal significant sequence similarity to any protein sequence in the NCBI data library. Surprisingly, however, a comparison with SHFV sequences revealed that the ORF 6 product is closely related to the protein encoded by ORF 3, having 27% sequence identity and 46% sequence similarity in the carboxy-terminal 105 residues (Fig. 3b). This observation suggests that ORF 6 resulted from a gene duplication event and led us to speculate that ORFs 4 and 5 may have arisen similarly. Indeed, a short region with 30% sequence identity and 42% sequence similarity was detected in the proteins encoded by ORFs 2a and 4 (Fig. 3c). However, sequence similarity between the products encoded by ORFs 2b and 5 was virtually absent.
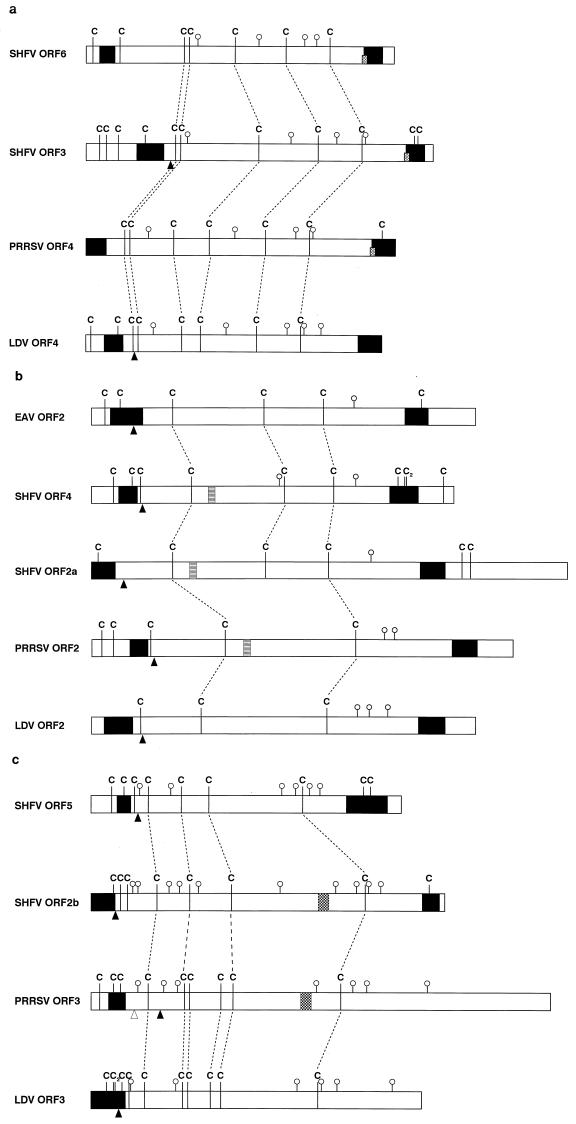
During divergent evolution, conservation primarily acts at the level of overall protein structure whereas the primary sequence may vary considerably. Cysteine residues, critically involved in glycoprotein folding through the formation of disulfide bonds, are generally more strictly conserved than other amino acid residues. As shown in Fig. 4a, the distribution of cysteine residues in the SHFV ORF 6 product resembles that of the SHFV ORF 3 product and the ORF 4 products (GP4) of PRRSV and LDV. Similarly, the cysteine pattern in the presumptive ectodomain of the SHFV ORF 4 product is similar to that of the ORF 2a product of SHFV and the ORF 2 products (GP2) of PRRSV, LDV, and EAV (Fig. 4b). Finally, the cysteine patterns of the products encoded by SHFV ORFs 2b and 5 can also be matched (Fig. 4c). The combined data suggest that SHFV ORFs 4 through 6 may have arisen from a heterologous RNA recombination event during which the genes for GP2, GP3, and GP4 were duplicated. Further support for this scenario can be found in the ORF overlap regions. All but two of the SHFV ORFs overlap the adjacent 5′ ORF; there is a 42-nt gap between ORFs 3 and 4 and a 5-nt gap between ORFs 6 and 7 (34). These gaps suggest that an RNA segment carrying ORFs 4 through 6 may have been placed between existing overlapping ORFs. Such a recombination event may have involved closely related SHFV genomes, in which case the considerable sequence differences between ORFs 2a through 3 and their duplicates resulted from subsequent divergence. Alternatively, ORFs 4 through 6 may have been acquired from a more distantly related arterivirus. A recombinant virus can only become established if the newly gained genetic information provides a selective advantage. At present, we do not know how the acquired genes contribute to SHFV fitness. Future studies of the biosynthesis and function of the various SHFV proteins and characterization of additional SHFV isolates may shed light on this issue.
FIG. 4.
Schematic representations of the ORF 4 products of LDV and PRRSV with the ORF 3 and ORF 6 products of SHFV (a), the ORF 2 products of EAV, LDV, and PRRSV with the ORF 2a and ORF 4 products of SHFV (b), and the ORF 3 products of LDV and PRRSV with the ORF 2b and ORF 5 products of SHFV (c). Cysteine residues are indicated by C, and potential N-linked glycosylation sites are denoted by open circles. The fine broken lines identify aligned cysteine residues, and the coarse broken lines in panel c identify arbitrarily aligned residues. Black triangles indicate the positions at which the signal sequences are cleaved from the peptide as predicted by von Heijne’s algorithm (40), and the open triangle symbolizes an alternative signal sequence cleavage site. The black boxes represent N- and C-terminal hydrophobic domains as determined by Kyte and Doolittle (20). The short diamond-squared boxes in panel a represent the core sequence, RWA. The striped boxes in panel b symbolize the core residues, HPLG, of a highly conserved amino acid sequence. The checkered boxes in panel c indicate the locations of the conserved seven-amino-acid sequence, FHPELFG. Nucleotide sequences were taken from den Boon et al. (7) for the Utrecht strain of EAV, Chen et al. (6) for LDV strain P, Meulenberg et al. (26) for the Lelystad isolate of PRRSV, and Smith et al. (34) for the LVR 42-0/M6941 strain of SHFV.
Acknowledgments
This work was supported by Public Health Service grant RR06841 from NCRR.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1989. [Google Scholar]
- 3.Brinton M A, Gavin E I, Fernandez A V. Genotypic variations among six isolates of lactate dehydrogenase-elevating virus (LDV) J Gen Virol. 1986;67:2673–2684. doi: 10.1099/0022-1317-67-12-2673. [DOI] [PubMed] [Google Scholar]
- 4.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 5.Cavanagh D, Brian D A, Enjuanes L, Holmes K V, Lai M M C, Laude H, Siddell S G, Spaan W, Taguchi F, Talbot P J. Recommendations of the coronavirus study group for the nomenclature of the structural proteins, mRNAs, and genes of coronaviruses. Virology. 1990;176:306–307. doi: 10.1016/0042-6822(90)90259-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z, Kuo L, Rowland R R R, Even C, Faaberg K S, Plagemann P G W. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J Gen Virol. 1993;74:643–660. doi: 10.1099/0022-1317-74-4-643. [DOI] [PubMed] [Google Scholar]
- 7.den Boon J, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Boon J A, Kleijnen M F, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative form RNAs. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales. Sem Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vries A A F, Raamsman M J B, van Dijk H A, Horzinek M C, Rottier P J M. The small envelope glycoprotein (GS) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J Virol. 1995;69:3441–3448. doi: 10.1128/jvi.69.6.3441-3448.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drew T W, Meulenberg J J M, Sands J J, Paton D J. Production, characterization and reactivity of monoclonal antibodies to porcine reproductive and respiratory syndrome virus. J Gen Virol. 1995;76:1361–1369. doi: 10.1099/0022-1317-76-6-1361. [DOI] [PubMed] [Google Scholar]
- 13.Faaberg K S, Plagemann P G W. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology. 1995;212:512–525. doi: 10.1006/viro.1995.1509. [DOI] [PubMed] [Google Scholar]
- 14.Faaberg K S, Plagemann P G W. ORF 3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology. 1997;227:245–251. doi: 10.1006/viro.1996.8310. [DOI] [PubMed] [Google Scholar]
- 15.Godeny E K, Chen L, Kumar S N, Methven S L, Koonin E V, Brinton M A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godeny E K, Zeng L, Smith S L, Brinton M A. Molecular characterization of the 3′ terminus of the simian hemorrhagic fever virus genome. J Virol. 1995;69:2679–2683. doi: 10.1128/jvi.69.4.2679-2683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gravell M, London W T, Leon M E, Palmer A E, Hamilton R S. Differences among isolates of simian hemorrhagic fever (SHF) virus (42231) Proc Soc Exp Biol Med. 1986;181:112–119. doi: 10.3181/00379727-181-42231. [DOI] [PubMed] [Google Scholar]
- 18.Hiscox J A, Mawditt K L, Cavanagh D, Britton P. Investigation of the control of coronavirus subgenomic mRNA transcription by using T7-generated negative-sense RNA transcripts. J Virol. 1995;69:6219–6227. doi: 10.1128/jvi.69.10.6219-6227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kheyar A, St.-Laurent G, Archambault D. Sequence determination of the extreme 5′ end of equine arteritis virus leader region. Virus Genes. 1996;12:291–295. doi: 10.1007/BF00284650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Liu D X, Cavanagh D, Green P, Inglis S C. A polycistronic mRNA specified by the coronavirus infectious bronchitis virus. Virology. 1991;184:531–544. doi: 10.1016/0042-6822(91)90423-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D X, Inglis S C. Identification of two new polypeptides encoded by mRNA 5 of the coronavirus infectious bronchitis virus. Virology. 1992;186:342–347. doi: 10.1016/0042-6822(92)90094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.London W T. Epizootiology, transmission and approach to prevention of fatal simian hemorrhagic fever virus in rhesus monkeys. Nature (London) 1977;268:344–345. doi: 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- 24.Luytjes W. Coronavirus gene expression: genome organization and protein synthesis. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 33–54. [Google Scholar]
- 25.Meulenberg J J M, de Meijer E J, Moormann R J M. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993;74:1693–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- 26.Meulenberg J J M, Hulst M M, de Meijer E J, Moonen P L J M, den Besten A, de Kluyver E P, Wensvoort G, Moormann R J M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS) is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meulenberg J J M, Petersen-den Besten A. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF 2 is incorporated in virus particles. Virology. 1996;225:44–51. doi: 10.1006/viro.1996.0573. [DOI] [PubMed] [Google Scholar]
- 28.Meulenberg J J M, Petersen-den Besten A, de Kluyver E P, Moormann R J M, Schaaper W M M, Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer A E, Allen A M, Tauraso N M, Shelokov A. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am J Trop Med Hyg. 1968;17:404–412. [PubMed] [Google Scholar]
- 30.Sagripanti J L. Polyadenylic acid sequences in the genomic RNA of the togavirus of simian hemorrhagic fever. Virology. 1985;145:350–355. doi: 10.1016/0042-6822(85)90171-0. [DOI] [PubMed] [Google Scholar]
- 31.Sagripanti J L, Zandomeni R O, Weinmann R. The cap structure of simian hemorrhagic fever virion RNA. Virology. 1986;151:146–150. doi: 10.1016/0042-6822(86)90113-3. [DOI] [PubMed] [Google Scholar]
- 32.Sawicki S G, Sawicki D L, Kääriänen L, Keranen S. A Sindbis virus mutant temperature sensitive in the regulation of minus strand RNA synthesis. Virology. 1981;115:161–172. doi: 10.1016/0042-6822(81)90098-2. [DOI] [PubMed] [Google Scholar]
- 33.Senanayake S D, Hofmann M A, Maki J L, Brian D A. The nucleocapsid gene of bovine coronavirus is bicistronic. J Virol. 1992;66:5277–5283. doi: 10.1128/jvi.66.9.5277-5283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith S L, Wang X, Godeny E K. Sequence of the 3′ end of the simian hemorrhagic fever virus genome. Gene. 1997;191:205–210. doi: 10.1016/S0378-1119(97)00061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snijder E J, Spaan W J M. The coronaviruslike superfamily. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 239–255. [Google Scholar]
- 36.Tauraso N M, Shelokov A, Palmer A E, Allen A M. Simian hemorrhagic fever virus. III. Isolation and characterization of a viral agent. Am J Trop Med Hyg. 1968;17:422–431. [PubMed] [Google Scholar]
- 37.van Berlo M F, Rottier P J M, Spaan W J M, Horzinek M C. Equine arteritis virus-induced polypeptide synthesis. J Gen Virol. 1986;67:1543–1549. doi: 10.1099/0022-1317-67-8-1543. [DOI] [PubMed] [Google Scholar]
- 38.van der Most R G, Spaan W J M. Coronavirus gene expression: genome organization and protein synthesis. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 39.van Nieuwstadt A P, Meulenberg J J M, van Essen-Zandbergen A, Petersen-den Besten A, Bende R J, Moormann R J M, Wensvoort G. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J Virol. 1996;70:4767–4772. doi: 10.1128/jvi.70.7.4767-4772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieczorek-Krohmer M, Weiland F, Conzelmann K, Kohl D, Visser N, van Woensel P, Thiel H-J, Weiland E. Porcine reproductive and respiratory syndrome virus (PRRSV): monoclonal antibodies detect common epitopes on two viral proteins of European and U.S. isolates. Vet Microbiol. 1996;51:257–266. doi: 10.1016/0378-1135(96)00047-8. [DOI] [PubMed] [Google Scholar]
- 42.Zeng L, Godeny E K, Methven S L, Brinton M A. Analysis of simian hemorrhagic fever virus (SHFV) subgenomic RNAs, junction sequences, and 5′ leader. Virology. 1995;207:543–548. doi: 10.1006/viro.1995.1114. [DOI] [PubMed] [Google Scholar]