Abstract
The genome of a recently identified virus, hepatitis G virus (HGV), shows considerable homology to hepatitis C virus (HCV). Two HGV proteases similar to nonstructural proteins NS2 and NS3 of HCV were identified, and their cleavage site specificity was investigated. Amino acids essential for the protease activities were determined by mutation analysis. NS4A of HGV was demonstrated to be a cofactor for NS3-mediated proteolysis, with a region critical for activity residing between Leu1561 and Ala1598.
Hepatitis G virus (HGV) is a new transfusion-transmissible agent identified in the sera of hepatitis patients (18). Recently, two nonhuman viruses, GBV-A and GBV-B, and a human virus, GBV-C, were detected (26, 28). Comparison of the polyprotein amino acid sequence from the HGV isolates with that from GBV-C showed that these viruses are nearly identical (33); therefore, the HGV nomenclature will be used throughout this paper.
The HGV genome is a 9.4-kb positive-stranded RNA molecule which contains an open reading frame (ORF) encoding a 2,900-amino-acid (aa) precursor polyprotein. The polyprotein sequence resembles those of viruses in the Flaviviridae family, especially hepatitis C virus (HCV), with which it shares 25.5% overall identity at the amino acid level. Regions of conservation cover the whole nonstructural region of the polyprotein, reaching 40 to 60% identity in parts of the putative NS3 and NS5B regions (18). Therefore, the putative HGV nonstructural region is expected to be organized similarly to the NS2-NS3-NS4A-NS4B-NS5A-NS5B structure of HCV (22). The HGV polyprotein contains consensus sequences typical of the Flaviviridae family, such as a putative serine protease and helicase in the NS3 region as well as an RNA-dependent RNA polymerase in the NS5B region. It also contains an NS2 protease motif, characteristic of HCV (18). In HCV, NS2, NS3, and NS4A activities are responsible for the proteolytic processing of the nonstructural region of the polyprotein. NS3 of HCV is a chymotrypsin-like serine protease responsible for cleavages at NS3/NS4A, NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B sites (1, 7, 9, 16, 20, 24). Processing at these sites is abolished when Ser1165 in the catalytic site of the NS3 is mutated to alanine (7, 30). NS4A of HCV is a cofactor required for the NS3-mediated cleavages at the NS3/NS4A, NS4A/NS4B, and NS4B/NS5A sites, but it is not obligatory for the NS5A/NS5B cleavage (2, 3, 13, 16, 19, 21, 27, 29). Proteolytic activity located in the NS2 region of HCV is required for the NS2/NS3 cleavage (8, 9). His952 and Cys993 (numbered as in HCV-H sequence) were shown to be essential for the NS2 activity (8).
In these studies, we used the recombinant baculovirus expression system to study the proteolytic processing of the HGV polyprotein. Using site-directed mutagenesis of the putative HGV protease motifs and deletion mapping, we demonstrate that HGV polyprotein encodes protease activities which are functionally similar to the NS2, NS3, and NS4A activities of HCV.
Construction of recombinant baculoviruses.
Plasmid p3ZHGV-6, encoding the entire HGV (PNF2161) polyprotein (18a), was used as a cloning source of the HGV genes, either as DNA restriction fragments or as a template for PCRs. Portions of the HGV genome were cloned into baculovirus plasmid transfer vectors pAcG1 and pAcG3X (5). These vectors contain the glutathione S-transferase (GST) gene, allowing the expression of HGV genes as amino-terminal fusion proteins under the control of the very late baculovirus polyhedrin promoter. The BglII-StuI fragment coding for the Ile806-Glu1658 region of the HGV polyprotein was cloned into the BamHI-SmaI-cut pAcG3X vector, which enabled expression of the part of the HGV polyprotein containing the NS2 and NS3 protease motifs and a putative NS4A region. Amino acids are numbered from the first methionine encoded by AUG461 in the polyprotein ORF of HGV (PNF2161). This construct was designated P.
Mutations at the NS2 and NS3 protease motifs were made by oligonucleotide-directed mutagenesis (15). Oligonucleotides for the His849-to-Tyr and Cys890-to-Leu mutations in the NS2 protease motif were 5′-CGAATAAACAAGCTAATATACCCGAG and 5′-CCTGAAACAGGAATCCCGTCACGCAG, respectively. Nucleotides which differ from those of the HGV (PNF2161) strain are underlined. Amino acids are numbered from the first methionine of the long ORF encoded by AUG461 in the HGV (PNF2161) genome (18). Oligonucleotide 5′-GCACCGAGCCGACCGAGTGG was used to mutate Ser1062 to Ala in the NS3 serine protease putative active site. In addition, some of the constructs were provided with a carboxy-terminal tag, derived from herpes simplex virus (HSV) glycoprotein D. This allowed the carboxy-terminal part of the HGV polyprotein encoded in these constructs to be monitored during the polyprotein processing studies. A synthetic EcoRI DNA fragment encoding the 11-aa Gln-Pro-Glu-Leu-Ala-Pro-Glu-Asp-Pro-Glu tag was cloned into the unique EcoRI site of the NS2-NS3-NS4A-expressing constructs in frame with the carboxy terminus of the HGV polyprotein fragment. Constructs P23 and P234 were derived by making carboxy-terminal truncations of the NS4 region in the P construct. Construct P23 was largely devoid of the putative NS4 sequence and encoded the Ile806-Arg1550 segment of the HGV polyprotein containing the NS2 and NS3 sequences. Construct P234 spanned the beginning of the NS4 region and encoded the Ile806-Ala1598 segment of the HGV polyprotein. Constructs 4, 41, and 42 encoded the Arg1550-Lys1655, Leu1561-Lys1655, and Leu1576-Lys1655 portions of the HGV polyprotein, respectively. They were derived by cloning PCR fragments from the putative NS4 region into the pAcG1 vector.
Construct 45 encodes an Asp1806-Lys2235 segment of the HGV polyprotein, which includes the putative NS4B/NS5A junction. It was obtained by ligating a HindIII-BglII fragment of the p3ZHGV-6 plasmid into the pAcG1 vector. Construct 55 encodes a part of the HGV polyprotein containing the putative NS5A/NS5B junction. Plasmid p3ZHGV-6 was digested with the Eco47.III and EcoRI restriction endonucleases. The Eco47.III-EcoRI fragment encoding the Arg2078-Gly2873 segment of the HGV polyprotein was ligated into the EcoRI- and SmaI-digested pAcG1 vector.
The plasmid transfer vectors containing the HGV genes were cotransfected with the BaculoGold linearized baculovirus DNA (PharMingen, San Diego, Calif.) according to the manufacturer’s instructions. The resultant white plaques were selected and purified by two sequential plaque assays (11, 12).
NS2 mediates cleavage at the NS2/NS3 junction.
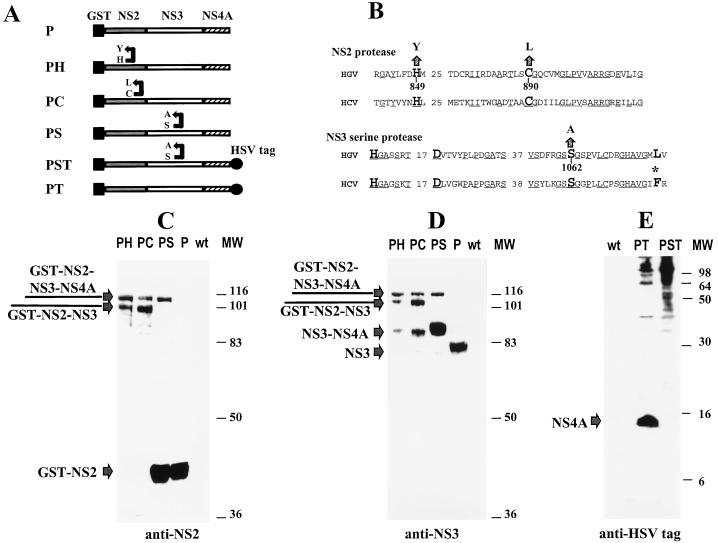
In order to elucidate the role of the putative NS2 protease active site in the HGV polyprotein cleavage, Sf21 insect (Spodoptera frugiperda) cells were infected either with the NS2-NS3-NS4A construct containing the wild-type NS2 putative protease (P) or with constructs carrying mutations of His849 or Cys890 in the NS2 active site (PH or PC) or with a construct carrying a Ser1062 mutation in the NS3 serine protease active site (PS). Cells were harvested at 60 h postinfection, and cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with rabbit polyclonal antibodies raised against the NS2 (Ser872-Ser889) or NS3 (Gln1488-Pro1504) peptides (Fig. 1). GST-NS2 was completely cleaved from NS3 in the P construct, because GST-NS2 was the only product reacting with anti-NS2 antibodies, and there was no protein-associated reactivity at a higher molecular mass (Fig. 1C). Analysis of the same lysate with anti-NS3 (Gln1488-Pro1504) antibodies indicated a major product with an apparent molecular mass of 70 kDa, which is consistent with the predicted molecular mass of NS3 (Fig. 1D). Mutation of the NS3 serine protease motif in the PS construct did not significantly affect NS2/NS3 cleavage, because the release of the GST-NS2 product was comparable in both P- and PS-infected cell lysates (Fig. 1C and D). However, mutations of the NS2 putative active site in the PH and the PC constructs were found to have a profound effect on this cleavage. Free GST-NS2B and -NS3 products were not observed on immunoblots of PH- and the PC-infected cell lysates, indicating that NS2 was the major factor responsible for the NS2/NS3 cleavage (Fig. 1C and D).
FIG. 1.
Western blot analysis of proteolytic processing of the NS2B-NS4A polyprotein expressed by recombinant baculoviruses. S. frugiperda SF21 cells were infected at a multiplicity of 5 PFU per cell with recombinant baculoviruses or wild-type (wt) Autographa californica nuclear polyhedrosis virus. Cells were harvested at 60 h postinfection. Proteins were separated by SDS-PAGE, electroblotted onto nitrocellulose membranes, and probed with either rabbit antisera raised to HGV synthetic peptides or with the monoclonal anti-HSV tag antibodies. (A) Schematic representation of constructs. (B) Mutations of HGV protease motifs. Phe154 of the HCV substrate specificity pocket is indicated by an asterisk and numbered from the N-terminal residue of NS3. (C) Western blot analysis of NS2B/NS4A polyprotein cleavage with an anti-NS2 antibody. Samples were separated on an SDS-PAGE gel (12% polyacrylamide). (D) Western blot analysis of NS2B/NS4A polyprotein cleavage with anti-NS3 polyclonal antibody. Samples were separated on an SDS-PAGE gel (12% polyacrylamide). (E) Western blot analysis of NS2B/NS4A polyprotein cleavage with anti-HSV tag antibody. Samples were separated by SDS-PAGE (18% polyacrylamide) and probed with anti-HSV tag monoclonal antibody. MW, molecular mass markers (kilodaltons).
NS3 mediates cleavages at the NS3/NS4, NS4/NS5A, and NS5A/NS5B junctions.
Cleavage at the NS3/NS4 junction was studied with the PT and PST constructs. These constructs expressed the NS2-NS3-NS4 portion of HGV polyprotein containing an easily detectable HSV tag at the carboxy terminus of NS4. Consequently, release of NS4 from the polyprotein could be studied with commercially available HSV tag monoclonal antibodies (Novagen, Madison, Wis.). The PT construct expressed wild-type NS3, while the PST construct expressed NS3 with a Ser1062 mutation in the putative serine protease active site. Cleaved NS4 product with a molecular mass of 15 kDa was observed when the cells were infected with the PT construct, whereas no cleaved NS4 product was detected on immunoblots of the cell lysates infected with the PST construct containing a mutated NS3 serine protease motif (Fig. 1E). This finding indicates that the NS3 serine protease was essential for the NS3/NS4 cleavage. In the HCV polyprotein, NS4A/NS4B cleavage occurs 54 aa downstream of the NS3/NS4 cleavage. This cleavage event was not observed in our experiments with HGV. The size of the cleavage product containing the HSV tag corresponded to the size of the intact NS4 portion expressed in cells infected with the construct PT (Fig. 1E). However, the PT construct contains an incomplete putative NS4B; therefore the possibility that the intact NS4B region is required for this cleavage has not been ruled out.
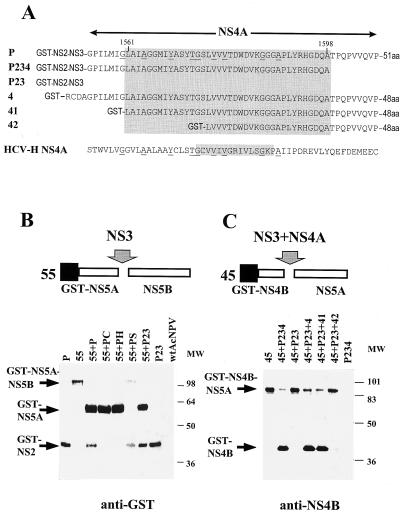
NS3-mediated cleavage at the NS4B/NS5A and NS5A/NS5B junctions was studied in trans and not in cis, because cytotoxicity associated with the middle part of the NS4 region precluded efficient expression of the NS3-NS4-NS5 constructs. For this, constructs 45 and 55 containing NS4B/NS5A and NS5A/NS5B junctions, respectively, were coinfected with constructs expressing wild-type or mutated NS3 and NS2B, with or without NS4A. As in HCV, these cleavages were found to be dependent on the NS3 serine protease activity, but not on the NS2 protease activity. Mutations in the NS2 protease active site (constructs PC and PH) or deletion of the NS4A region (construct P23) did not impair NS5A/NS5B cleavage (Fig. 2B). Therefore, the NS3 protease was sufficient in mediating NS5A/NS5B cleavage. However, as in HCV, NS4B/NS5A cleavage required expression of both NS3 and NS4A. Deletion of the NS4A region (construct P23) abolished the cleavage, but this was restored when construct P23 was coinfected with construct P4, which expresses the NS4A region (Fig. 2C).
FIG. 2.
Analysis of cleavage at the NS4B/NS5A and NS5A/NS5B sites. (A) Schematic representation of the recombinant baculovirus constructs. NS4A sequences are shown and compared with those of HCV-H NS4A. Borders of the putative NS4A region are indicated by arrows. Identical amino acids are underlined. HCV-H NS4A (17) and HGV activity domains are indicated inside shaded rectangles. (B) Western blot analysis of NS5A/NS5B cleavage products. S. frugiperda SF21 cells were infected, at a multiplicity of 5 PFU per cell, with each type of recombinant baculovirus or wild-type Autographa californica nuclear polyhedrosis virus (wtAcNPV). Cells were harvested at 60 h postinfection. Proteins containing the GST tag were purified by affinity chromatography on glutathione-Sepharose, separated by SDS-PAGE (12% polyacrylamide), and electroblotted onto nitrocellulose membranes, which were reacted with rabbit anti-GST tag antibodies (Sierra Biosource). (C) Western blot analysis of NS4B/NS5A cleavage. Cells were infected with recombinant baculoviruses, and proteins containing the GST tag were purified and reacted with NS4B(Ala1884-Tyr1899) antibodies. MW, molecular mass markers (kilodaltons).
Identification of the NS4A active region.
In the HCV polyprotein, the NS3 protease is followed by NS4A, which serves as a cofactor for the NS3-mediated cleavages (22). Identification of the NS4A active region was performed by making truncations of the NS4 region in constructs P and 4. Construct P encoded the NS2, NS3, and NS4 portions of the HGV polyprotein, and construct 4 encoded only the NS4 portion. The activity of the truncated constructs P234, 41, and 42 was investigated by determining the ability of these constructs to complement the NS3-mediated cleavage at the NS4B/NS5A junction following coinfection with construct 45 (Fig. 2A and C). Cells were harvested at 60 h postinfection, and the GST-tagged proteins were purified by affinity chromatography on glutathione-Sepharose (7). The purified proteins were separated by SDS-PAGE, and the GST-NS4B cleavage product was identified by Western blot analysis with polyclonal antibody raised against the NS4B (Ala1884-Tyr1899) peptide (Fig. 2C). Construct 4, covering all of the putative NS4A region; construct 41, with an amino-terminal truncation; and construct P234, with a carboxy-terminal truncation in the region, retained full NS4A activity. Construct 42 exhibited very little residual NS4A activity, indicating that the NS4A active domain was impaired in this construct. Therefore, the NS4A active domain was located within the Leu1561-Ala1598 HGV polyprotein sequence. Amino acid sequence comparison between the HCV NS4A and HGV NS4A active domains indicated limited homology between these sequences; however, there was a considerable conservation on the secondary structure level, as predicted by the Chou-Fasman algorithm (4) (data not shown).
Analysis of the HGV polyprotein cleavage sites.
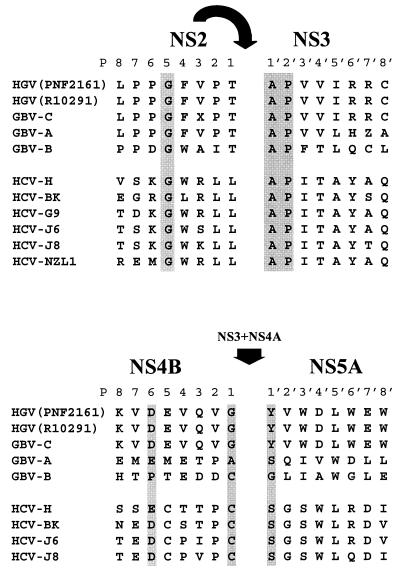
The sequence at the NS2 protease-mediated NS2/NS3 cleavage site was established by amino-terminal sequence analysis of the NS3 cleavage product, which was identified in cell lysates of the insect cells infected with the P construct (Fig. 1D). Amino-terminal sequence analysis of the NS3 product yielded the Ala-Pro-Val-Val-Ile sequence, which indicates that the NS2/NS3 cleavage occurred within the Gly-Phe-Val-Pro-Thr/Ala926-Pro-Val-Val-Ile sequence. Alignment of the HCV, HGV, GBV-A, and GBV-B sequences at the NS2/NS3 junction shows strict conservation of amino acids at positions P5, P1′, and P2′ (Fig. 3). The reason for such strict conservation remains unclear and could reflect other functions distinct from proteolytic processing. As previously demonstrated for HCV, NS2B/NS3 cleavage was not strictly dependent on the Gly, Ala, and Pro in these positions; conservative, as well as some nonconservative P5, P1′, and P2′ amino acid substitutions were tolerated (24).
FIG. 3.
Alignment of the HCV, HGV, GBV-A, and GBV-B sequences at the NS2B/NS3 and NS4B/NS5A junctions. The cleavages have been experimentally established for HGV (this study) and HCV only (7, 8). The amino acids demonstrated to be conserved in cleavage sites found in HCV are shaded.
The sequence at the NS3-mediated NS4B/NS5A cleavage site was established by amino-terminal sequence analysis of the NS5A cleavage product present in the extracts of the cells coinfected with construct 45, which contained the putative NS4B/NS5A junction, and construct P, which had HGV protease activities. The NS5A cleavage product was identified in the cell extracts by Western blot analysis with anti-NS5A (Ser2196-Gly2211) peptide antibodies (data not shown). Amino-terminal sequence analysis of the NS5A product yielded the sequence Tyr-Val-Trp-Asp-Leu-Trp-Glu-Trp-Ile-Met, indicating that the NS4B/NS5A cleavage occurs within the Glu-Val-Gln-Val-Gly/Tyr1899-Val-Trp-Asp-Leu sequence. Unlike NS2/NS3 cleavage, HCV and HGV NS4B/NS5A cleavage sites have little in common apart from the conserved acidic amino acid residue at position P6 and the presence of small residues at positions P1 and P1′ (Fig. 3). The acidic amino acid at P6 was conserved in all NS3-mediated HCV cleavage sites; however, it was not essential for efficient cleavage (14). The P1 position was shown to be more important for this cleavage (14, 31, 32). P1 Cys is conserved in all trans-cleaved HCV sites, and a recent study demonstrated that it was critical for the trans cleavage at the NS5A/NS5B site (32). P1 Cys is likely to interact with the S1 substrate pocket of HCV NS3 protease, with a cysteine sulfhydryl group probably playing a critical role in substrate binding (10, 19, 23, 32). However, Gly instead of Cys was found in the P1 position of the trans-cleaved NS4B/NS5A site of HGV (Fig. 3). Moreover, Phe154, which determines the preference of the HCV NS3 for cysteine in the P1 substrate position (6, 32), is substituted for Leu154 in HGV NS3, probably determining the different substrate specificity of HGV NS3 (Fig. 1B). Interestingly, GBV-B NS3 protease, which shares substrate specificity with HCV NS3, has both Phe154 in its substrate binding pocket (25) and Cys in the P1 position of the putative NS4B/NS5A cleavage site (Fig. 3).
In summary, the mode of the HGV polyprotein cleavage in the nonstructural protein region appears to be very similar to that of HCV, except for the substrate specificity of the NS3 protease. HCV NS3 serine protease activity is required for all of the cleavages downstream of NS3, while the NS4A activity is absolutely required for all of the cleavages except for NS5A/NS5B (2, 16). Similarly, HGV NS3, but not NS2B, was found to be required for the NS4B/NS5A and NS5A/NS5B cleavages, and NS4A was found to be absolutely required for the NS4B/NS5A cleavage but not for the NS5A/NS5B cleavage. As in the case of HCV, the HGV NS4B/NS5A and NS5A/NS5B cleavages could occur in trans. Similarity between HGV and HCV is not surprising, because these viruses share considerable amino acid sequence similarity in the nonstructural regions of their polyproteins (18). In both viruses, processing of the nonstructural part of the polyprotein is mediated by the NS2 and NS3 proteases and the NS4A cofactor, which are encoded at corresponding parts of the genomes of these viruses, have similar active domains, and can effect cleavages at analogous sites of the polyprotein in a similar fashion.
Acknowledgments
We express our gratitude to Kirk Fry, Jeffrey Linnen, and Patrice Yarbough for helpful discussions during the course of this study and to Nancy Alexi and Jane Bardwell for comments during the preparation of the manuscript.
REFERENCES
- 1.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V, Wilkinson T, Koch J O. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou P Y, Fasman G D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Davies A H, Jowett J B M, Jones I M. Recombinant baculovirus vectors expressing glutathione-S-transferase fusion proteins. Bio/Technology. 1993;11:933–936. doi: 10.1038/nbt0893-933. [DOI] [PubMed] [Google Scholar]
- 6.Failla C M, Pizzi E, De Francesco R, Tramontano A. Redesigning the substrate specificity of the hepatitis C virus NS3 protease. Folding Design. 1996;1:35–42. [PubMed] [Google Scholar]
- 7.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4655–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J L, Morgenstern K A, Lin C, Fox T, Dwyer M D, Landro J A, Chambers S P, Markland W, Lepre C A, O’Malley E T, Sharbeson S L, Rice C M, Murcko M A, Caron P R, Thomson J A. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 11.King L A, Possee R D. The baculovirus expression system: a laboratory guide. London, United Kingdom: Chapman & Hall; 1992. [Google Scholar]
- 12.Kitts P A, Possee R D. A method for producing recombinant baculovirus expression vectors at high frequency. BioTechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 13.Koch J O, Lohmann V, Herian U, Bartenschlager R. In vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. Virology. 1996;221:54–66. doi: 10.1006/viro.1996.0352. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov A A, Agapov E V, Rice C M. Specificity of the hepatitis C virus serine protease: effects of substitutions at the 3/4A, 4A/4B, 4B/5A, and 5A/5B cleavage sites on polyprotein processing. J Virol. 1994;68:7525–7533. doi: 10.1128/jvi.68.11.7525-7533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel T A. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Prágai B M, Grakoui A, Xu J, Rice C M. The hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin C, Thomson J A, Rice C M. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine protease complex in vivo and in vitro. J Virol. 1995;69:4373–4380. doi: 10.1128/jvi.69.7.4373-4380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linnen J, Wages J, Zhang-Keck Z-Y, Fry K, Krawczynski K Z, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih J, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a new transfusion transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 18a.Linnen, J., et al. Personal communication.
- 19.Love R A, Parge H E, Wickersham J A, Hostomsky Z, Habuka N, Moomaw E W, Adachi T, Hostomska Z. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 20.Manabe S, Fuke I, Tanishita O, Kaji C, Gomi Y, Yoshida S, Mori C, Takamizawa A, Yoshida I, Okayama H. Production of nonstructural proteins of hepatitis C virus requires a putative viral protease encoded by NS3. Virology. 1994;198:636–644. doi: 10.1006/viro.1994.1075. [DOI] [PubMed] [Google Scholar]
- 21.Mori A, Yamada K, Kimura J, Koide T, Yausa S, Yamada E, Miyamura T. Enzymatic characterization of purified NS3 serine proteinase of hepatitis C virus expressed in Escherichia coli. FEBS Lett. 1996;378:37–42. doi: 10.1016/0014-5793(95)01423-3. [DOI] [PubMed] [Google Scholar]
- 22.Neddermann P, Tomei L, Steinkuhler C, Gallinari P, Tramontano A, De Francesco R. The nonstructural proteins of the hepatitis C virus: structure and functions. Biol Chem. 1997;378:469–476. [PubMed] [Google Scholar]
- 23.Pizzi E, Tramontano A, Tomei L, La Monica N, Failla C, Sardana M, Wood T, de Francesco R. Molecular model of the specificity pocket of the hepatitis C virus protease: implication for substrate recognition. Proc Natl Acad Sci USA. 1994;91:888–892. doi: 10.1073/pnas.91.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed K E, Grakoui A, Rice C M. Hepatitis C virus-encoded NS2-3 protease: cleavage site mutagenesis and requirements for bimolecular cleavage. J Gen Virol. 1995;69:4127–4136. doi: 10.1128/jvi.69.7.4127-4136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scarselli E, Urbani A, Sbardellati A, Tomei L, De Francesco R, Traboni C. GB virus B and hepatitis C virus NS3 serine proteases share substrate specificity. J Virol. 1997;71:4985–4989. doi: 10.1128/jvi.71.7.4985-4989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlauder G G, Dawson G, Simons J N, Pilot-Matias T J, Gutierrez R A, Heynen C A, Knigge M F, Kurpiewski G S, Buijk S L, Leary T P, Muerhoff A S, Desai S M, Mushahwar I K. Molecular and serologic analysis in the transmission of the GB hepatitis agents. J Med Virol. 1995;46:81–90. doi: 10.1002/jmv.1890460117. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu Y, Yamaji K, Masuho Y, Yokota T, Inoue H, Sudo K, Satoh S, Shimotohno K. Identification of the sequence on NS4A required for enhanced cleavage of the NS5A/5B site by hepatitis C virus NS3 protease. J Virol. 1996;70:127–132. doi: 10.1128/jvi.70.1.127-132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 29.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urbani A, Bianchi E, Narjes F, Tramontano A, De Francesco R, Steinkuhler C, Pessi A. Substrate specificity of the hepatitis C virus serine protease NS3. J Biol Chem. 1997;272:9204–9209. doi: 10.1074/jbc.272.14.9204. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R, Durkin J, Windsor W T, McNemar C, Ramanathan L, Le H V. Probing the substrate specificity of hepatitis C virus NS3 serine protease by using synthetic peptides. J Virol. 1997;71:6208–6213. doi: 10.1128/jvi.71.8.6208-6213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuckerman A J. Alphabet of hepatitis viruses. Lancet. 1996;347:558–559. doi: 10.1016/s0140-6736(96)91267-2. [DOI] [PubMed] [Google Scholar]