Summary
Extracellular vesicles (EVs) play a crucial role in facilitating communication between cancer cells and their immediate or remote microenvironments, thereby promoting the extensive spread of cancer throughout the body. In this context, we present a protocol for the isolation of tumor cell-derived EVs followed by in vivo metastasis assessment in a murine ovarian cancer model. We describe steps for the isolation and characterization of EVs from ID8 cells, development of a metastatic mouse model, and sample preparation for flow cytometry.
For complete details on the use and execution of this protocol, please refer to Gupta et al.1
Subject areas: Cancer, Immunology
Graphical abstract

Highlights
-
•
Generation of stable cell lines using lentivirus
-
•
Isolation of tumor cell-derived EVs using ultracentrifugation
-
•
Evaluation of EVs-specific biomarkers to ascertain EVs enrichment
-
•
Generation of preclinical model to evaluate role of EVs in cancer progression
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Extracellular vesicles (EVs) play a crucial role in facilitating communication between cancer cells and their immediate or remote microenvironments, thereby promoting the extensive spread of cancer throughout the body. In this context, we present a protocol for the isolation of tumor cell-derived EVs followed by in vivo metastasis assessment in a murine ovarian cancer model. We describe steps for the isolation and characterization of EVs from ID8 cells, development of a metastatic mouse model, and sample preparation for flow cytometry.
Before you begin
The protocol described here outlines the isolation and utilization of EVs originating from ovarian cancer cells within the context of a metastatic tumor model. Additionally, it is worth noting that this protocol is adaptable for the isolation of EVs from the culture medium of various other cancer cell types. For EVs isolation we followed guidelines established by the International Society for Extracellular Vesicles (ISEV).2
Institutional permissions
All experiments complied with protocols approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin.
All mouse experiments must be performed with the approval of the animal care committee relevant to your research institution.
Lentiviral transduction
Timing: 2 weeks
This section describes stable cell lines generation using lentivirus.
-
1.Packaging of lentivirus.
-
a.Preparation and transfection of 293T cells. Split 293T cells at 8×105 293T cells in 5 mL of complete DMEM without antibiotics (no penicillin or streptomycin) in a 6 cm tissue culture plate for each lentiviral expression vector.
-
b.Incubate cells at 37°C, 5% CO 2 for 16 h.Note: We recommend that cells should reach 70% confluence on the day of transfection
-
c.In microfuge tubes, prepare a 20 μL mixture of the 3 transfection plasmids in Opti-MEM Carefully pipette up and down or vortex briefly to mix.
Reagent Amount per 6 cm dish∗ pLCP-GFP or pLCP-GFP-SPHK1 1 μg psPAX2 750 ng pMD2.G 250 ng Optional: To enhance viral production, you can adjust the ratio of lentiviral plasmid, packaging plasmid, and envelope plasmid. When conducting the transfection process, we suggest the use of FuGENE Transfection Reagent (Roche Applied Science) or Lipofectamine 2000 (Invitrogen).-
i.In a 1 mL tube, make 80 μL of Lipofectamine master mix by adding 6 μL Lipofectamine to 74 μL Opti-MEM.Note: Do not allow Lipofectamine 2000 reagent to touch the walls of the tube. Blend the mixture by gently swirling or flicking the tube. Allow it to incubate for 5 min at 25°C–37°C.
-
ii.Add 80 μL of Lipofectamine master mix to 20 μL mixture of the 3 transfection plasmids, making a total volume of 100 μL.
-
iii.Dispense the master mix directly into the liquid, avoiding contact with the tube walls. Swirl the tube to ensure thorough mixing.
-
iv.Incubate the mixture for 20–30 min at 25°C–37°C.
-
v.Slowly add the transfection mixture prepared in the previous step (step iii) drop by drop onto the cells, taking care not to touch the sides of the dish.
-
vi.Carefully swirl the plate back and forth and side to side to evenly distribute the complex.
-
i.
-
d.Incubate cells at 37°C with 5% CO2 for a period of 12–18 h.
-
e.Remove the media and add fresh media to eliminate the transfection reagent.
-
f.Wash cells once with Mg2+/Ca2+free PBS.
-
g.Subsequently, substitute the medium with 5 mL fresh DMEM supplemented with 10% FBS and penicillin/streptomycin.
-
h.Incubate the cells at 37°C with 5% CO2 for an additional 24 h.
-
i.Collect the first batch of media containing lentiviral particles from transfected 293T cells and transfer it to a 15 mL tube.
-
j.Store the medium at 4°C.
-
k.Add 5 mL of fresh media containing antibiotics to the cells and incubate at 37°C with 5% CO2 for another 24 h.
-
l.Collect the second batch of media from cells and combine it with the first batch.
-
m.Centrifuge the media at 200 × g for 5 min to pellet any inadvertent collection of 293T during the harvesting process.Optional: Filter the media through a 0.45 μm filter to remove cellular debris. Avoid using a 0.2 μm filter, as this may damage the virus envelope.Note: All procedures involving lentivirus must be carried out within a Biosafety Level 2 (BSL-2) facility. Supernatants can be stored for a maximum of one week at 4°C or in separate aliquots for extended storage at −80°C.
-
a.
-
2.Lentiviral transduction to generate stably transfected cells.
-
a.Seed 1×105 ID8 cells (or the specific target cells of your experiment) in 6cm dishes.
-
b.Allow them to grow at 37°C with 5% CO2 until they reach approximately 60% confluency for transduction.
-
i.Remove the media and add polybrene (8 μg/mL) containing media.
-
ii.Add 0.5–1 mL of lentiviral particle solution to target cells.Note: Modify the amount of virus added based on the size of your target plate. For a 6cm plate, consider adding between 0.05-1 mL virus. It is advisable to experiment with a range of multiplicity of infections (MOIs) to determine the optimal MOI for infection. MOI refers to the number of viral particles infecting each cell.
-
iii.Incubate cells at 37°C with 5% CO2 for 48 h.
-
iv.Replace the media and supplement it with G418 at a concentration of 1000 μg/mL. This will facilitate the selection of cells that have achieved stable integration of the lentiviral vector.
-
v.Continue growing the cells under standard culture conditions, supplementing the medium with G418 (1000 μg/mL).
-
vi.Replace the culture medium every 2 days with fresh medium containing G418.Note: The optimal G418 concentration may vary depending upon the specific cell type. It is crucial to determine the optimal G418 concentration preliminary dose-response experiments when working with different cell lines.After 7 days, most untransduced cells will be eliminated, and individual colonies will be apparent.
-
i.
-
c.Perform serial dilution of bulk culture in a 96 wells plate to isolate and expand individual colonies.
-
d.Evaluate the infected cells for overexpression using suitable methods such as RT-PCR or western blotting.Note: It is advisable to maintain one plate of uninfected cells in parallel. This plate will serve as a positive control during G418 selection process.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| TSG101 | Proteintech | Dilution (1:1,000); Clone: N/A; Cat# 14497-I-AP; RRID:AB_2208090 |
| CD63 | Proteintech | Dilution (1:1,000); Clone: N/A; Cat# 25682-I-AP; RRID:AB_2208090 |
| Alix | Santa Cruz Biotechnology | Dilution (1:1000); Clone: 1A12; Cat# sc-53540; RRID:AB_673819 |
| TSG101 | Thermo Fisher Scientific | Dilution (1:1,000); Clone: 4A10; Cat# MA1-23296; RRID:AB_2208088 |
| Anti-rabbit HRP | Cell Signaling Technology | Dilution (1:1,000);; Cat#: 7074S: RRID:AB_2099233 |
| Anti-mouse HRP | Cell Signaling Technology | Dilution (1:1,000); Cat#: 7076P2: RRID:AB_330924 |
| GFP | Santa Cruz Biotechnology | Dilution (1:1,000); Clone: NA; Cat# Sc-9996; RRID:AB_627695 |
| SPHK1 | Proteintech | Dilution (1:1,000); Clone: N/A; Cat# 10670-I-AP; RRID:AB_2195809 |
| Zombie UV fixable viability kit | BioLegend | Dilution (1:1,000); Cat#: 423107 |
| Anti-mouse CD4-BV711 | BioLegend | Dilution (1:100); Clone: GK1.5; Cat# 100447; RRID: AB_2564586 |
| Anti-mouse CD8-BV605 | BioLegend | Dilution (1:100); Clone: 53-6.7; Cat# 100744; RRID:AB_2562609 |
| Anti-mouse CD45-BV510 | BioLegend | Dilution (1:100); Clone: 30-F11; Cat# 103138; RRID:AB_2563061 |
| Anti-mouse CD3-BUV395 | BD Horizon | Dilution (1:100); Clone; 145-2C11 (RUO) Cat# 563565; RRID:AB_2738278 |
| Anti-mouse PD1-PE | BD Pharmingen | Dilution (1:100); Clone: N/A; Cat# 561788; RRID:AB_10895570 |
| Anti-mouse TIM3-BV421 | BD Biosciences | Dilution (1:100); Clone: 5D12; Cat# 747626; RRID:AB_2744192 |
| Chemicals, peptides, and recombinant proteins | ||
| RIPA buffer | Thermo Fisher Scientific | Cat# 89901 |
| Halt protease and phosphatase inhibitor cocktail | Thermo Fisher Scientific | Cat# 78444 |
| Pierce BCA assay protein kit | Thermo Fisher Scientific | Cat# 23225 |
| ECL Prime western blotting detection reagent | Thermo Fisher Scientific | Cat# 34580 |
| G418 | Thermo Fisher Scientific | Cat# J63871-AB |
| ACK lysis buffer (RBC lysis buffer) | Quality Biological | Cat# 118-156-101 |
| FACS buffer | Tonbo Biosciences | Cat# 4222-L500 |
| Cyto-Fast Fix/Perm buffer | BioLegend | Cat# 426803 |
| TF Perm/Wash buffer (5X) | BD | Cat# 562725 |
| Polybrene | Sigma | Cat# 107689 |
| Bovine serum albumin (BSA) | Sigma | Cat# A3059 |
| Opti-MEM (1X) | Gibco | Cat#31985-070 |
| Dulbecco’s modified Eagle’s medium (DMEM) | Thermo Fisher Scientific | Cat# 10569010 |
| ITS | Thermo Fisher Scientific | Cat# 41400045 |
| Fetal bovine serum (FBS) | Atlanta Biologicals | Cat# H17112 |
| Hanks' balanced salt solution (HBSS) | Atlanta Biologicals | Cat# 14170120 |
| Antibiotic (penicillin/streptomycin) | Thermo Fisher Scientific | Cat# 15140122 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668027 |
| Vybrant DiI cell-labeling solution | Thermo Fisher Scientific | Cat# V22885 |
| DPBS | Sigma | Cat#D8662 |
| psPAX2 | Addgene | Cat#12260 |
| pMD2.G | Addgene | Cat#12259 |
| pLCP-GFP | Addgene | Cat#17448 |
| pLCP-GFP-SPHK1 | Genecopoeia | Cat#EX-Mm05392-M68 |
| Experimental models: Cell lines | ||
| 293T | ATCC | Cat# CRL-3216 |
| ID8 | Sigma | Cat# SCC145 |
| Experimental models: Organisms/strains | ||
| 4–6-weeks-old female C57BL/6 mice | Envigo | - |
| Software and algorithms | ||
| GraphPad Prism 9.5 | GraphPad | https://www.graphpad.com/ |
| FlowJo v10.10 | FlowJo LLC | https://www.flowjo.com/ |
| NTA software 3.1 | Malvern, Worcestershire, United Kingdom | https://www.malvernpanalytical.com |
| Others | ||
| Insulin syringe (31G) | BD | Cat# 328820 |
| Centrifuge tube polypropylene (38.5 mL) | Beckman Coulter | Cat# 355642 |
| PVDF | Bio-Rad | Cat #1620177 |
| 40 μM strainer | Greiner | Cat#542040 |
| 27G x 1/2 needle | BD | Cat# 305109 |
| 18G needle | BD | Cat#305136 |
| NanoSight LM10 | Malvern, Worcestershire, United Kingdom | https://www.malvernpanalytical.com |
Step-by-step method details
Isolation and characterization of extracellular vesicles from conditioned media
Timing: 1 week
This section describes the steps of isolating EVs from cancer cell-conditioned media. Specifically, we aim to isolate EVs from the culture medium of ID8-Control and SPHK1 ectopically expressed in ID8 cell lines.
Note: ID8 cells can survive in serum-free media for a duration of 2–3 days, due to their limited serum components dependency for growth.
CRITICAL: For in vivo injection, it is recommended to isolate fresh EVs each time.
-
1.
Seed 1×106 control and SPHK1 overexpressed cells in 15 cm tissue culture plates, and culture them in DMEM supplemented with 4% FBS and 1X ITS.
-
2.
Incubate the cells at 37°C with 5% CO2.
-
3.
Once the cells reach a confluency of 70%, wash them with 1X PBS followed by serum free DMEM.
-
4.
Incubate cells in serum free media at 37°C with 5% CO2. After 48 h, collect the supernatant enriched with EVs in 50 mL conical tube and keep it on ice.
Note: We collected 30 mL serum free media for EVs isolation from two 15 cm tissue culture plates. It is important to optimize the initial number of cells and volume of serum free media required for EVs isolation due to variations in EVs complexity.
-
5.
Proceed with the isolation of EVs using a serial ultracentrifugation method.
-
6.
Centrifuge the supernatant from step 3 at 2,000 × g at 4°C for 20 min to eliminate cell debris.
-
7.Transfer the resulting supernatant to a new 50 mL conical tube for subsequent EVs isolation.
 Pause point: The EVs enriched supernatant can be stored at −80°C for long-term storage (up to 6 months) or 4°C for short-term storage prior to ultracentrifugation.Note: It is imperative that all steps are conducted at 4°C to prevent the degradation of EVs.
Pause point: The EVs enriched supernatant can be stored at −80°C for long-term storage (up to 6 months) or 4°C for short-term storage prior to ultracentrifugation.Note: It is imperative that all steps are conducted at 4°C to prevent the degradation of EVs.-
a.Carefully transfer the conditioned medium into 38.5 mL open top ultracentrifuge tubes and centrifuge at 16,500 × g for 50 min at 4°C.
-
b.Transfer the resulting supernatant into a fresh ultracentrifuge tube.
-
c.To ensure the proper balance, weigh the tubes containing supernatant and adjust their balance using sterile ice-cold PBS.
-
d.Ultracentrifuge the supernatant at 100,000 × g for 2 h.
 CRITICAL: It is vital to ensure that all the tubes are properly balanced before ultracentrifuge. An imbalance rotor may lead to spillage or operational failure, and potential damage to the roto or ultracentrifuge machine.
CRITICAL: It is vital to ensure that all the tubes are properly balanced before ultracentrifuge. An imbalance rotor may lead to spillage or operational failure, and potential damage to the roto or ultracentrifuge machine. -
e.Retrieve the EVs from pellet and resuspend in cold PBS. Gently pipet up and down along the wall of the tube to thoroughly dislodge and disperse the pellet.Note: During this step, decant the supernatant gently to avoid disturbing the pellet. Leave approximately 500 μL of supernatant in the tube.
-
f.Again, subject the tubes for ultracentrifugation at 100,000 × g for 2 h at 4°C to collect the washed EVs pellet (Figure 1).
-
g.Carefully discard the PBS by decanting and remove any remaining liquid by gently inverting the tubes onto a paper towel.
-
h.Resuspend the EVs pellet in 100–1000 μL cold PBS and transfer into low protein-bound microcentrifuge tubes.Note: We recommend allocating a portion to sterile tubes for subsequent quantification and characterization processes such as nanoparticle tracking analysis (NTA), protein quantification, western blotting, and transmission electron microscopy (TEM).
-
i.Maintain the tube on ice or store it at −80°C for preservation.
-
j.Quantify EVs through total protein quantification using a BCA protein assay kit.Note: In our experimental setup, conducted multiple times, we consistently yield 80–100 μg of EVs from 30 mL of supernatant from ovarian cancer cells
-
a.
-
8.Determine EVs concentration and size distribution using NTA.Note: In this protocol, we employ NanoSight LM10 instrument (Malvern Panalytical, Malvern, UK) equipped with a 488 nm laser and NTA3.1 software for analysis.
 CRITICAL: Always keep the EVs on ice throughout this process. NTA measurement should be performed within 24–48 h of EVs collection to reduce EVs aggregation and lysis.
CRITICAL: Always keep the EVs on ice throughout this process. NTA measurement should be performed within 24–48 h of EVs collection to reduce EVs aggregation and lysis.-
a.Dilute the EVs sample in sterile ice-cold PBS and keep it on ice.Note: The optimal particle concentration should fall within 0.05–0.1 μg /μL (typically 108–109 particles/mL).
-
b.Use the diluted EVs sample for measuring the size of particles with the NanoSight LM10 using NTA software 3.1 (Malvern) following the manufacturer’s instructions.
-
c.Use deionized-water-dampened cleaning tissue designed for optical lenses to gently cleanse the LM10 viewing unit.
-
d.Gently wipe the inner surface of the top-plate window and the optical plate using the dampened tissue.
-
e.To remove any moisture, softly wipe the surfaces with the tissue and use an air compressor’s air stream for drying.
-
f.Turn on the power and start the NTA v.3.1 program.Note: we recommend performing a double rinse of the beam path. This can be achieved by gradually injecting 1 mL of PBS into the chamber using a syringe. This procedure will help maintain an impurity-free beam path.
-
g.Vortex the sample tube containing the EVs at full power.
-
h.Using 1 mL syringe, withdraw 500 μL of sample, ensuring no air bubbles are trapped inside the syringe.
-
i.Insert the syringe into Malvern Panalytical NanoSight syringe pump and press the load button.
-
j.Once the sample is loaded, make necessary adjustments to the imaging position, focus, and camera level on the capture setting to achieve a clear particle image on the screen and press the ‘Start Camera’ button.Note: The presence of EVs aggregates can affect the NTA analysis. To eliminate aggregates, dilute the EVs with ice cold PBS and vortex the solution for 10–15 s.
-
k.Fine tune the focus and record a video of the particles for 30–60 s.
-
l.Repeat steps f-i to capture a total of 5 different videos.
-
m.After video capture, set the detection threshold to the level where most of the particles are accurately identified.
-
n.Proceed with NTA analysis to determine the particle number and size distribution.
-
o.Upon completing the experiment, ensure to thoroughly clean the fluidic system.
-
a.
-
9.Detection of protein markers in EVs by western blotting.Note: We have used a BCA protein assay kit to quantify EVs.
-
a.Add 100 μL RIPA buffer (with protease inhibitor) to the EVs suspension and vortex for 30 s to lyse the EVs.
-
b.Place on ice for 20–30 min to allow complete lysis.
-
c.Add 2X SDS loading dye and heat the mixture at 95°C for 5 min.
-
d.Cool the sample on ice for 5 min before loading it onto the gel.
-
e.Load 30–50 μg of EVs protein and run the standard SDS-PAGE followed by western blotting onto a PVDF membrane.
-
f.Block the membrane in 5% non-fatty acid milk /1XPBS-0.05% Tween 20 (PBS-T) solution.
-
g.Place the membrane on a rocker shaker for 1 h at 25°C–37°C.
-
h.Wash the membrane two times with 1X PBST for 10 min each.
-
i.Incubate the membrane in primary antibody diluted in 1X PBST at 4°C for 16 h.
-
j.Wash the membrane thrice with 1X PBST for 10 min each.
-
k.Incubate the membrane with an HRP- labeled secondary antibody diluted in 1X PBST for 1 h at 25°C–37°C.
-
l.Wash the membrane thrice with 1X PBST for 10 min each.
-
m.For HRP detection, incubate membrane with 5 mL of ECL and visualize using the image detector.Note: To prepare for TEM, resuspend the EVs in 100 μL of cold PBS and follow the protocol and guidelines provided by the electron microscopy facility.
-
a.
Figure 1.
Isolation of EVs from cancer cell conditioned medium
(A) The cleared media is carefully transferred to an ultracentrifuge tube and the presence of the EVs pellet is observed following ultracentrifugation.
Generation of ovarian cancer mice model
Timing: 11 weeks
This section describes the development of an ovarian cancer model using C57BL/6 female mice. We developed this protocol based on previous ref.1
-
10.Preparation of cancer cells for injection
-
a.Grow RAB27a−/−ID8 cells in DMEM supplemented with 1X ITS, 4% FBS, and 1% antibiotics.Note: We used RAB27a−/−ID8 cells for tumor generation to prevent the secretion of EVs by tumor cells in mice. Rab27a is a critical regulatory protein essential for EVs secretion from the plasma membrane3.
-
b.Discard the culture medium and wash the cells once with sterile PBS.
-
c.Now add 2 mL of trypsin/EDTA solution to the cells and incubate at 37°C until the cells detach from the surface of the culture plate.
-
d.Inactivate the trypsin by adding 2 mL of culture medium to the plate.
-
e.Resuspend the cells and collect in a 50 mL tube.
-
f.Gently resuspend the cells a few times. Take 100 μL cells to a 1.5 mL centrifuge tube and mix them with an equal volume of trypan blue.
-
g.Determine the total number of cells with a hemocytometer or automatic cell counter.
-
h.Centrifuge the cells at 200 × g for 5 min to form a pellet.
-
i.Gently aspirate the supernatant without disturbing the cell pellet and wash the cells twice with 20–30 mL sterile PBS.Note: This step is crucial as it effectively eliminates trypsin and FBS residue from the cell pellet. These substances can interfere with tumor growth in vivo.
-
j.Discard the PBS and resuspend the cell pellet in HBSS.
-
k.Centrifuge the cells at 200 × g for 5 min.
-
l.Gently aspirate the supernatant without disturbing the cell pellet.
-
m.Add a calculated volume of HBSS to the cell pellet to attain a cell density of 1 × 106 cells per 20 μL.Note: The number of cells required for tumor formation, which may vary depending on the cell line, can be determined empirically from the literature or by performing a preliminary in vivo experiment.
-
n.Gently mix the cells and place them on ice. The cells are now ready for intraovarian injection, and this injection should be executed within 1 h of cell collection.
-
o.Keep the cell preparation on ice throughout this process.
-
a.
-
11.Orthotopic (Intraovarian) injection of cancer cells into mice.Note: One day prior to surgery, conduct a visual assessment of all mice to evaluate their general health. Shave the mice on one side of the abdominal cavity.
-
a.Slowly swirl the cells to ensure uniform suspension and load the syringe by drawing up the desired volume using an 18G needle.
-
b.Afterward, replace this needle with 27½G needle. Place the syringe on ice until it is used for injection.
-
c.Place the mice in the isoflurane chamber.
-
d.Once the animals are anaesthetized, administer the pain reliever buprenorphine-SR (10 μL/mouse, Stock 1 mg/ml) subcutaneously using a 22½G syringe.
-
e.Keep the syringe on ice until it is used for injection.
-
f.Position the anesthetized mice laterally (left or right depending on the ovary to be injected) on a sterile gauze pad connected to an isoflurane tube with a nose cone.
-
g.The mouse head should face away from the investigator, while the tail toward the investigator (Figure 2).
-
h.Thoroughly disinfect the shaved area by applying chlorhexidine-soaked sterile cotton swab followed by a sterile surgical alcohol pad three times.
 CRITICAL: Proper sterile draping is essential to minimize contamination of the surgical site. The drapes should cover the entire animal while exposing the surgical site.
CRITICAL: Proper sterile draping is essential to minimize contamination of the surgical site. The drapes should cover the entire animal while exposing the surgical site. -
i.Carefully lift the peritoneal wall lining and create a small vertical incision.
-
j.While keeping the incision open with one pair of forceps, employ another sterile pair of forceps to carefully draw the ovarian fat pad through the incision, directing it towards the midline.
-
k.Adjust the fat pad slightly to expose the ovary.
-
l.Carefully and slowly inject the cells into the ovaries, taking care to avoid any leakages.
 CRITICAL: With utmost care and precision, perform this procedure to avoid any damage to the oviduct, and bursa, or the risk of rupturing the bursa due to excessive pressure or overfilling.
CRITICAL: With utmost care and precision, perform this procedure to avoid any damage to the oviduct, and bursa, or the risk of rupturing the bursa due to excessive pressure or overfilling. -
m.Carefully reposition the reproductive tract and fat pad to their original locations beneath the body wall.
-
n.Suture the incision in the peritoneal wall with 2–4 sutures, using 4-0 sterilized surgical silk thread and needles.
-
o.Seal the skin with surgical staples or wound clips.Note: To prevent hypothermia and facilitate a speedy recovery, return the mice to a cage placed on a heating pad until they have completely regained consciousness from the anesthesia. The surgical staples can be taken out from the skin seven days after the operation.
 CRITICAL: Following the surgery, observe the mice at 30 min intervals for a duration of 2 h, paying close attention to indications such as choking, bleeding, rapid or shallow breathing, vocalizations, head tilting, or any other sustained unusual behaviors. Additionally, monitor the mice's daily behavior and mobility for a period of 10 days following the surgery. If an infection is suspected, promptly seek veterinary assistance.
CRITICAL: Following the surgery, observe the mice at 30 min intervals for a duration of 2 h, paying close attention to indications such as choking, bleeding, rapid or shallow breathing, vocalizations, head tilting, or any other sustained unusual behaviors. Additionally, monitor the mice's daily behavior and mobility for a period of 10 days following the surgery. If an infection is suspected, promptly seek veterinary assistance.
-
a.
-
12.Intravenous EVs injection.Note: We recommend initiating EVs treatment one week after cancer cell injections in mice.
-
a.Before the injection, position the mouse cages at least 15 cm from a heat lamp. Allow them to remain in this position for 5 min, to facilitate tail vein dilation.
 CRITICAL: Prevent the mice from becoming overheated.
CRITICAL: Prevent the mice from becoming overheated. -
b.Load a 1 mL syringe with the EVs or PBS using a 31-gauge sterile needle. Make sure not to trap any air bubbles in the syringe.
-
c.Inject 200 μL EV or PBS (as a control) into the lateral tail vein of tumor-cells bearing mice.
-
d.Administer tumor cell derived EVs through the tail vein three times a week for a duration of twelve weeks or until any of the mice exhibit moribund symptoms.
-
e.During necropsy, document the tumor weight, ascites volume and distribution of tumor nodules.Note: Investigators should standardize the EVs concentration for tail vein injections.
 CRITICAL: Before injections, vortex the EVs to ensure there are no aggregates. The presence of aggregated EVs could result in immediate adverse effects or mouse mortality.
CRITICAL: Before injections, vortex the EVs to ensure there are no aggregates. The presence of aggregated EVs could result in immediate adverse effects or mouse mortality.
-
a.
Figure 2.
Orthotopic (intraovarian) injection of tumor cells
(A) Anesthetized mouse on a sterile gauze pad connected to an isoflurane tube with a nose cone.
(B) Ovarian fat pad with ovary.
(C) Reproductive tract with normal ovary and ovarian tumor.
Ascites and tissue sample collection
Timing: 2 days
This section describes the harvesting of tumor samples including ascites and tumor tissue.
-
13.
Before dissection, measure the mice weight and euthanize the mice by CO2 inhalation (might interfere with some of the parameters) and/or cervical dislocation.
-
14.
Proceed to harvest the tumor tissue and ascites.
-
15.
Collect ascites through abdominal aspiration.
-
16.
For mouse dissection, delicately elevate the skin and create an anterior skin incision along the ventral midline of the mouse, starting from the upper thighs and extending to the lower rib cage.
-
17.
Then, produce lateral incisions at the upper and lower ends of the skin, effectively detaching it from the peritoneal tissue.
-
18.
Collect metastatic tumor, remove tumor nodules from each peritoneal organ, and record the weight of the tumor tissue.
Note: Be cautious not to harm any organs during this process.
CRITICAL: Ascites and tissue harvesting should be expeditious.
Note: The tumor tissue can be used for histopathological assessment. First, fix the tissue in 4% paraformaldehyde for 24–48 h, followed by paraffin embedding. Alternatively, you can also rapidly freeze the tissue by immersing it in OCT and liquid nitrogen. Both types of tissue, the formalin-fixed paraffin-embedded (FFPE) sections and the flash-frozen sections, are amenable to staining with markers related to proliferation, apoptosis, or pertinent signaling proteins. It is recommended to set aside a portion of the frozen tumor tissue for RNA or protein-related applications.
Optional: Depending upon your experimental need, also collect other tissues such as the lung, liver, spleen, or kidney
Sample preparation for flow cytometry
Timing: 2 days
Note: The presence of cancer cells and immune cells in ascites offers a means to monitor ovarian cancer progression. Therefore, in our study, ascites samples were utilized to investigate EVs-mediated T cell inactivation and dysfunction.
-
19.Preparation of cells.
-
a.Collect the ascites, as mentioned in step 12, into 50 mL conical tubes.
-
b.Centrifuge ascites at 200 × g for 5 min at 4°C. There should be a visible cell pellet at the bottom of the tube.
-
c.Carefully remove the supernatant and store at −80°C for later analysis.
-
d.Add red blood cell lysis solution (ACK lysis buffer) to the cell pellet and incubate for 10–15 min at 37°C. Do not incubate for more than 15 min.
-
e.Add cold PBS to neutralize the lysis buffer.
-
f.Centrifuge the cells at 200 × g for 5 min at 4°C. Discard the supernatant.Note: Repeat RBS lysis until a clear white pellet is visible based on manufactures protocol.
-
g.Resuspend the cells in ice cold PBS.
-
h.Filter the cells through 40 μm cells strainer.
-
i.Centrifuge the cells at 200 × g for 5 min at 4°C. Discard the supernatant.
-
a.
-
20.Staining for flow cytometry.Note: Ascites contain a variety of cells such as tumor cells, immune cells, and mesothelial cells. Analyzing the composition of this cellular fraction can significantly enhance our understanding of tumor cell proliferation, progression, chemoresistance, and immune evasion in tumor microenvironment. Flow cytometry provides a platform for profiling individual cell types present in ascites. In this context, we profiled T-cells present in ascites sample collected earlier.
-
a.Resuspend the cells in PBS and adjust number to 1 × 106 cells per well of round bottom 96 wells plate.Note: For applying this protocol to other cells, careful titration on cell number is recommended
-
b.Centrifuge the cells at 200 × g for 5 min at 4°C and discard the supernatant.
 CRITICAL: Carefully remove the supernatant without disrupting the cell pellet.
CRITICAL: Carefully remove the supernatant without disrupting the cell pellet. -
c.Thaw zombie live-dead stain and prepare a 1/1000 dilution in PBS.
-
d.Resuspend cells with 100 μL of live-dead stain and incubate them at 4°C for 30 min in dark.
-
e.Centrifuge the cells at 200 × g for 5 min at 4°C and remove the supernatant.
-
f.Resuspend cells in PBS and spin them at 200 × g for 5 min at 4°C and remove the supernatant.
-
g.Prepare 1/100 dilution of FC block in PBS and add 100 μL of this to cell pellet. Mix well and incubate for 30 min on ice.Note: During FC blocking step, prepare antibody mixtures in stain buffer (1:100 dilution). After spinning down cells, remove supernatant.
 CRITICAL: To prevent non-specific binding of antibodies to cells, it is advisable to add an FC blocker to your sample.
CRITICAL: To prevent non-specific binding of antibodies to cells, it is advisable to add an FC blocker to your sample. -
h.Add 100 μL of the antibody mixture to the cells and incubate for 30 min on ice.
-
i.Wash cells one time with 200 μL FACS Buffer.
-
j.Fix cells by incubating them for 30 min on ice or for 16 h at 4°C with 100 μL fixation/perm buffer.
-
k.Wash cells one time with 200 μL FACS Buffer.
-
l.Finally, add 200 μL FACS Buffer and store cells in the dark at 4°C until they are ready for analysis.
-
a.
-
21.Intracellular staining.
-
a.Following fixation, remove the supernatant, and then incubate with a working dilution of Perm/Wash buffer for 30 min on ice.
-
b.Centrifuge the cells at 200 × g for 5 min at 4°C and carefully discard the supernatant.
-
c.Add intracellular antibody appropriately diluted in 1X permeabilization buffer and incubate for 30 min on ice.
-
d.Wash the cells once with 1X perm/wash buffer.
-
e.Add 200 μL FACS Buffer and store sample until analysis.
-
a.
CRITICAL: Ensure that plates or tubes are sealed to prevent evaporation. Fixed cells should be stable for up to 1 week prior to analysis. Note that conjugated antibodies may degrade over time once exposed to fixative.
FACS analysis
Timing: 2 h
Note: FACS analysis was performed using FlowJo_v10.9.0 software.
-
22.
Initially, ensure there are no instances of clogging or acquisition-related issues.
-
23.
Create a gate to isolate the acquired cells according to their cytoplasmic granularity (Side Scatter, SSC-A) and size (Forward Scatter, FSC-A), aiming to exclude debris and non-viable cells.
-
24.
Continue creating the following gates as shown in Figure 3.
-
25.
Following the completion of the gating strategy, calculate the percentage of T cell subtype relative to the total CD45-positive cells (CD45+), representing the entire population of immune cells infiltrating the tumor.
Figure 3.
Representative plots demonstrating the overall gating strategy employed for the identification of CD8+ and CD4+ T cells in the mouse samples
Expected outcomes
Building upon the ultracentrifuge approach, this protocol enables users to isolate both high yield and purity of EVs for in vivo functional experiments4,5. We used ID8 cells to isolate EVs which can approximately yield 80–100 μg/1 × 107 cells of EVs proteins. The quantities of EVs can vary based on the cell lines and the conditions of the culture. Therefore, it is imperative to empirically determine the appropriate culture scale for each experiment, depending on the desired amount of EVs for subsequent applications.
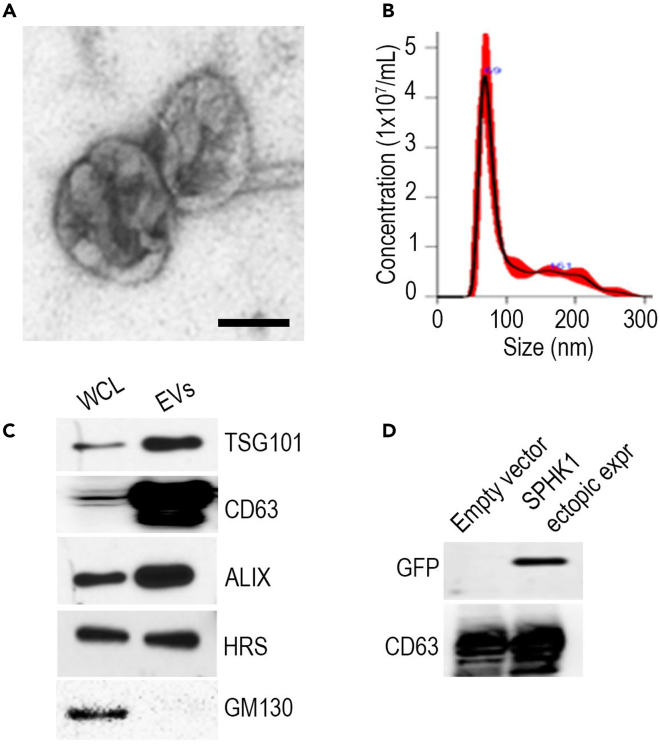
The EVs that were isolated underwent subsequent characterization utilizing TEM and NTA, which confirmed their size to be within the range of approximately 50–200 nm (as shown in Figures 4A and 4B). Furthermore, we confirmed the successful isolation of EVs by detecting the presence of positive markers (CD63, TSG101, HRS, and ALIX) as well as a negative marker (GM130) using Western blot analysis (Figure 4C). We also generated ovarian cancer cells that overexpress SPHK-GFP plasmid. We further validated the presence of GFP tagged SPHK within EVs of ovarian cancer cells through the expression of green fluorescent protein (GFP) (Figure 4D).
Figure 4.
Size distribution and western blot analysis of EVs
Adapted from Gupta et al. (2022).1
(A) Representative TEM image of purified EVs from ovarian cancer cells. Scale bar, 50 nm.
(B) Characterization of purified EVs using nanoparticle tracking (NTA).
(C) Western blotting for EV positive markers (TSG101, CD63, ALIX, HRS) and negative marker GM130.
(D) GFP-tagged SPHK1 was ectopically expressed in ovarian cancer cells, and its presence in EVs was observed by western blotting.
Afterward, we examined whether EVs packaged with SPHK1 trigger immune suppression while simultaneously promoting enhanced tumor growth and metastasis in an in vivo setting. We injected RAB27a−/− ID8 cells into the ovaries of C57BL/6 mice. We observed that the administration of EVs obtained from SPHK1 overexpressed tumor cells led to increased tumor weight and ascites volume, in comparison to EVs derived from parental cells (see Figures 5A–5C).
Figure 5.
Effect of SPHK1-enriched EVs on tumor progression
Adapted from Gupta et al. (2022).1
(A) General appearance of mice at the end of treatment. The mice treated with EVs carrying ectopically expressed SPHK1 displayed increased ascites volume and tumor nodules.
(B and C) Bar graph represents average tumor weight and ascites volume with SEM. ∗∗∗∗p<0.0001 compared to control by Student’s t-test.
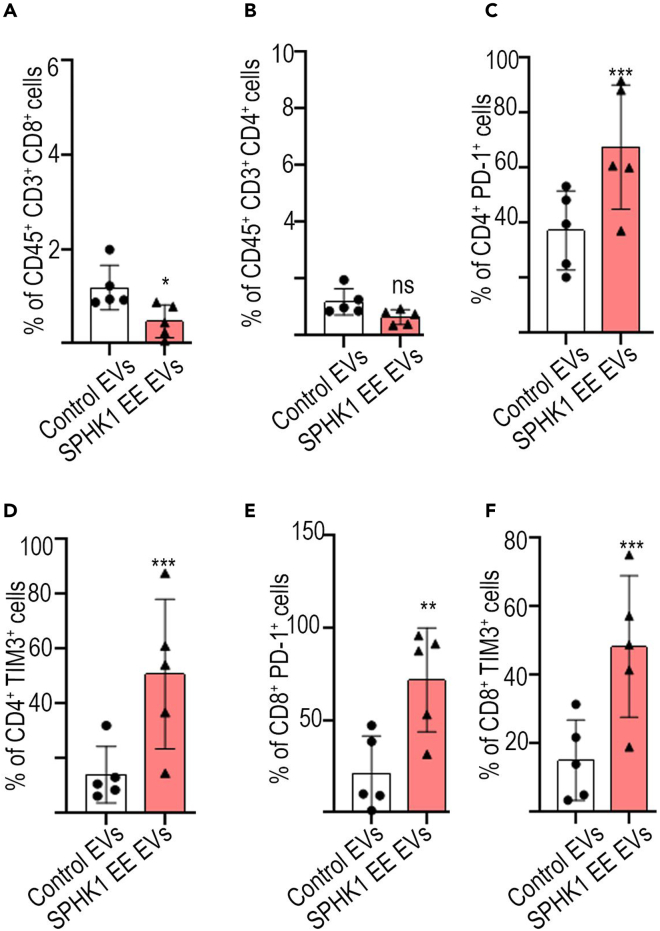
Next, we profiled the T cells from ascites samples. Immune profiling of T cell populations within the ascites from EV-treated mice demonstrated decreased infiltration of CD8 and CD4 positive T lymphocytes (TILs) in the group with ectopically expressed SPHK1, in contrast to the control EVs group (as depicted in Figures 6A and 6B). In addition, the marker of T cell exhaustion such as PD-1 and TIM3 were increased in TILs in ascites obtained from mice treated with SPHK1-overexpressed EVs in comparison to control EVs (Figures 6C–6F). Altogether, these findings collectively suggest that SPHK1enriched in EVs, plays a crucial role in the progression of ovarian tumors.
Figure 6.
Inhibition of T cell cytotoxicity by SPHK1-enriched EVs
Adapted from Gupta et al. (2022).1
(A and B) Percentage of CD8+ and CD4+ cells in ascites of tumor bearing mice.
(C–F) Expression of PD-1+ and TIM3+ cells among CD8+ cells and CD4+ T cells in ascites of mice. Error bars indicate mean ± SEM, ∗p<0.05, ∗∗p<0.01 ∗∗∗p<0.001, (Student's t-test).
Our method carries substantial importance in elucidating the impact of EVs composition on both cancer progression and immunotherapy. Additionally, our protocol can be used to study the therapeutic potential of engineered EVs carrying anti-tumor molecules such as proteins, nucleic acid, or drugs.
Limitations
While the differential ultracentrifugation (UC) protocol provides a robust method for processing and analyzing a wide range of specimens from different sources and varying sample volumes, it does have some inherent limitations. UC segregates particles based on their hydrodynamic size and density, resulting in limited resolution and purity of the extracted EVs. The isolation method primarily captures various categories of large vesicles, microparticles, and small EVs, with an enrichment of exomeres and exosomes. It’s important to note that the EVs within each of these categories exhibit heterogeneity, and there is a possibility of contamination by non-EVs proteins4,5
Another limitation of this protocol pertains to accurately determining the appropriate quantity of EVs for in vivo injections. The quantity of EVs may vary depending on the specific cancer cell type, which can subsequently affect tumor growth and metastasis. As a result, the dosage and duration of EV administration may require adjustments based on the tumor type being studied.
Troubleshooting
Problem 1
No EVs pellet after ultracentrifugation (see step 5e).
Potential solution
Examine the lower section of the tube under adequate lighting against a dark background. When removing the tube from the rotor, avoid abrupt movements. As insufficient yield of EVs may make the pellet difficult to detect, it is worth noting that the number of cells significantly influences the yield of EVs.
Problem 2
Death of mouse during intraovarian injection (see step 9).
Potential solution
Adjust the concentration of tumor cells and make single cell suspension. An excess of cells or cell clumps might lead to mouse fatalities during surgery. Additionally, ensure a sufficient and consistent supply of isoflurane and oxygen throughout the entire surgical procedure.
Problem 3
Death of mouse during intravenous EVs injection (see step 10).
Potential solution
Adjust the EVs concentration accordingly for intravenous injection. An excessive EVs injection can result in immediate fatality. It is recommended to gently vortex the EVs before injection to eliminate any aggregated EVs. Also, inject the EVs as slowly as possible and hold the syringe in place for a short time after injecting.
Resource availability
Lead contact
Further requests for information should be directed to the lead contact, Sunila Pradeep (spradeep@mcw.edu).
Technical contact
Requests for technical information should be directed to smittal@mcw.edu.
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate any codes.
Acknowledgments
This work was supported in part to S.P. by MCW Cancer Center Institutional Research Grants, the Department of Defense (W81XWH-21-1-0361, W81XWH-21-1-0138, and W81XWH-21-1-0413), and NCI R01CA258433.
Author contributions
S.P. and S.M. conceived the study and generated hypotheses. S.M. wrote the manuscript. S.M., S.K., P.G., and M.S. prepared figures for the manuscript. S.P. and P.C.-R. edited the manuscript. S.P. allocated funding and finalized the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Gupta P., Kadamberi I.P., Mittal S., Tsaih S.W., George J., Kumar S., Vijayan D.K., Geethadevi A., Parashar D., Topchyan P., et al. Tumor derived extracellular vesicles drive T cell exhaustion in tumor microenvironment through sphingosine mediated signaling and impacting immunotherapy outcomes in ovarian cancer. Adv. Sci. 2022;9 doi: 10.1002/advs.202104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc L., Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. 2018;9:95–106. doi: 10.1080/21541248.2016.1264352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3–22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 5.Tauro B.J., Greening D.W., Mathias R.A., Mathivanan S., Ji H., Simpson R.J. Two distinct populations of exosomes are released from LIM1863 colon carcinoma cell-derived organoids. Mol. Cell. Proteomics. 2013;12:587–598. doi: 10.1074/mcp.M112.021303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any codes.