Abstract
Oxidative stress plays an important role in the pathogenesis of acute lung injury (ALI). As a typical post-translational modification triggered by oxidative stress, protein S-glutathionylation (PSSG) is regulated by redox signaling pathways and plays diverse roles in oxidative stress conditions. In this study, we found that GSTP downregulation exacerbated LPS-induced injury in human lung epithelial cells and in mice ALI models, confirming the protective effect of GSTP against ALI both in vitro and in vivo. Additionally, a positive correlation was observed between total PSSG level and GSTP expression level in cells and mice lung tissues. Further results demonstrated that GSTP inhibited KEAP1-NRF2 interaction by promoting PSSG process of KEAP1. By the integration of protein mass spectrometry, molecular docking, and site-mutation validation assays, we identified C434 in KEAP1 as the key PSSG site catalyzed by GSTP, which promoted the dissociation of KEAP1-NRF2 complex and activated the subsequent anti-oxidant genes. In vivo experiments with AAV-GSTP mice confirmed that GSTP inhibited LPS-induced lung inflammation by promoting PSSG of KEAP1 and activating the NRF2 downstream antioxidant pathways. Collectively, this study revealed the novel regulatory mechanism of GSTP in the anti-inflammatory function of lungs by modulating PSSG of KEAP1 and the subsequent KEAP1/NRF2 pathway. Targeting at manipulation of GSTP level or activity might be a promising therapeutic strategy for oxidative stress-induced ALI progression.
Keywords: GSTP, S-glutathionylation, Acute lung injury, Oxidative stress, KEAP1, NRF2
Graphical abstract

Highlights
-
•
GSTP promotes S-glutathionylation (PSSG) of KEAP1 in acute lung injury (ALI).
-
•
GSTP-catalyzed PSSG of KEAP1 activates NRF2 pathway and alleviates ALI.
-
•
C434 in KEAP1 is the key PSSG site in this process.
-
•
Targeting at GSTP might be a promising therapeutic strategy for ALI.
Abbreviations
- ALI
acute lung injury
- PSSG
S-glutathionylation
- ROS
reactive oxygen species
- GSTP
glutathione S-transferase isoform P
- KEAP1
kelch-like ECH-associated protein 1
- NRF2
nuclear factor-erythroid 2-related factor 2
- HO-1
heme oxygenase-1
- NQO1
NAD(P)H:quinone oxidoreductase 1
- FBS
fetal bovine serum
- LPS
lipopolysaccharide
- BALF
bronchoalveolar lavage fluid
- MDA
malondialdehyde
- MPO
myeloperoxidase
- ARDS
acute respiratory distress syndrome
1. Introduction
Acute lung injury (ALI), commonly caused by bacterial and respiratory viral infections [1,2], is a disordered severe inflammatory process in the lung that leads to substantial damages to pulmonary endothelial and epithelial barriers, with a mortality rate up to 40% [3,4]. Oxidant-mediated tissue disruption is considered to play a crucial role in ALI pathogenesis. Oxidative stress is commonly observed in both experimental ALI models and ALI patients, involving the imbalance between elevated oxidant levels and decreased endogenous antioxidant levels, increased formation of lipid peroxidation byproducts and oxidative modification of intracellular proteins [3,4]. Oxidative stress induces inflammation by activating the expression of transcription factors and the subsequent production of inflammatory mediators, ultimately leading to the development of ALI [5]. Intracellular oxidant levels are positively correlated with the severity of alveolar epithelial inflammatory damage and the outcome of ALI, while antioxidant enzymes play vital roles by eliminating intracellular oxidants and reducing inflammation thereby attenuating the severity of injury [6]. Thus, suppression of inflammation and/or oxidative stress is considered as a potential strategy for the prevention and treatment of ALI.
Reactive oxygen species (ROS) are constantly generated as products of normal cellular metabolism and participate in the maintenance of cellular redox homeostasis, whereas the overproduction and accumulation of ROS prompt oxidative stress [7]. The redox status equilibrium in cells is maintained by various mechanisms, among which the tripeptide glutathione (GSH) plays a central role. Depletion of endogenous GSH is commonly reported in ALI cases [8,9], while GSH supplementation has been used to attenuate oxidative stress and inflammation, thereby impeding disease progression [10,11]. Besides its prominent role of being a major nucleophile in cells, GSH participates in a plenty of cell signaling pathways. Among them, a protein post-translational modification process, known as protein S-glutathionylation (PSSG), is reported to implicate in many redox imbalance-related diseases [12]. PSSG is a unique redox-driven post-translational modification in which the cysteine of glutathione binds to the-SH (thiol) group of a target cysteine in the protein via disulfide bond formation. PSSG is reversible in nature and results in temporary changes in the structure and function of target proteins. PSSG occurs spontaneously upon oxidative stimuli, whereas the rate and extent of this process increase with the catalysis of enzymes [13,14].
Glutathione S-transferase P (GSTP) is the prominent enzyme recognized to catalyze PSSG process [15,16]. GSTP is widely expressed in different type of cells from various organs. Though being considered as a cytosolic enzyme, GSTP is also found in the cytoplasm, nucleus, and mitochondria [[17], [18], [19]]. Besides catalyzing GSH conjugation reactions of electrophilic substrates, GSTP is involved in regulating a variety of cellular functions and maintenance of cellular redox homeostasis. PSSG mediated by GSTP is associated with the pathogenesis of many diseases, including pulmonary impairment, neurodegenerative disorders, and cancers [[20], [21], [22]]. In addition, GSTP-mediated PSSG process of essential protein targets are reported to play antioxidant, anti-inflammatory, and anti-apoptotic functions in acute and chronic inflammatory responses. Although the significance of GSTP in modulation respiratory diseases via catalyzing specific PSSG process is continuously being revealed, the mechanism of regulation and reduction of ALI mediated by GSTP is still unclear. Moreover, the target proteins of GSTP-mediated PSSG in ALI pathogenesis have not been investigated.
Kelch-like ECH-associated protein 1 (KEAP1), the suppressor of nuclear factor-erythroid 2-related factor 2 (NRF2), is a critical chaperonin for E3 ubiquitin ligases for degrading NRF2 [23]. KEAP1 protein is a 27-cysteine enriched structure, many of which were reported to be highly redox sensitive [24]. Upon electrophilic or oxidative stress, cysteine residues of KEAP1 are easily oxidized and NRF2 was released from KEAP1, which leads to the upregulation of subsequent antioxidant genes and the activation of KEAP1-NRF2 oxidative stress response pathway [25]. In fact, over a decade ago, Holland et al. have illustrated the cysteine modification map of KEAP1 protein, including PSSG modification, under varying GSH/GSSG ratios and proposed a consequential structural change of Kelch domain in KEAP1 protein, which eventually resulted in disrupted KEAP1-NRF2 binding using computational modeling analysis [26]. KEAP1 has been reported to be S-glutathionylated in inflammation [27] and GSTP-catalyzed KEAP1 PSSG process leading to NRF2 activation is an important neuronal protection mechanism [28]. However, up until now, the information of KEAP1 modification and role of GSTP in ALI is not available.
In this study, we explored the roles of GSTP and KEAP1 PSSG process in ALI. With integration of in vitro, in sillico, and in vivo findings, it is proposed that GSTP-catalyzed PSSG of KEAP1 promoted the dissociation of KEAP1-NRF2 complex and nuclear translocation of NRF2, activated the downstream pathways, and eventually ameliorate ALI progress. In summary, GSTP-catalyzed PSSG of KEAP1 exerts anti-inflammatory effects in the oxidative stress response in ALI process, which provided a potential therapeutic target for its treatment.
2. Materials and methods
2.1. Chemicals and reagents
LPS was purchased from Sigma-Aldrich (L-2880, USA). Penicillin and streptomycin, Fetal bovine serum (FBS), Dulbecco's modified Eagle's medium (DMEM) and Ham's F–12 K (Kaighn's) Medium were acquired from Invitrogen-Gibco (Grand Island, NY). Antibodies against NRF2, KEAP1, GSTP, HO-1, NQO1, Lamin B, and β-Actin were supplied by Cell Signaling or Abcam. All other chemicals were offered by Sigma-Aldrich, if not otherwise indicated. Recombinant human GSTP was prepared in our laboratory as described previously [29].
2.2. Cell culture
HPAEpiCs were purchased from the ScienCell Research Laboratories (#3200, USA) and cultured in Alveolar Epithelial Cell Medium (Sciencell, 3201, USA) consisting of 96.15 % basal medium, 2.0 % FBS, 1.0 % epithelial cell growth supplement, and 1.0 % antibiotics. BEAS-2B cells, purchased from the cell bank of Shanghai Institutes for Biological Sciences (China Academy of Science, Shanghai), were cultured in DMEM containing 10% FBS. A549 cells, purchased from the cell bank of Shanghai Institutes for Biological Sciences (China Academy of Science, Shanghai), were cultured in Ham's F–12 K (Kaighn's) Medium containing 10% FBS. All cells were cultured in a humidified incubator at 37 °C with 5% CO2.
2.3. Plasmid or siRNA transfection
Expression plasmids containing wild type and C368, C434, and C613 mutant KEAP1 genes, and siRNA targeting at GSTP (GSTP-siRNA) gene and the negative control (NC-siRNA) were provided by Tsingke Biotech (Beijing, China). For transient transfection, cells were cultured to 60%–80% confluence and then transfected using Lipofectamine™ 3000 and P3000™ reagent (Invitrogen, L3000015, USA) according to the manufacturer's instructions.
GSTP siRNA sense (5′-3′): GGCAAGGAUGACUAUGUGATT, antisense (5′-3′): UCACAUAGUCAUCCUUGCCTT.
2.4. GSH and GSSG measurement
Intracellular GSH and GSSG levels were measured using the luminescence-based GSH/GSSG-Glo method according to the manufacturer's instructions (Promega, V6611, USA).
2.5. Fe assays
Fe2+ and total iron levels in HPAEpiCs were measured using an iron assay kit (Abcam, ab83366, UK) in accordance with the manufacturer's protocol.
2.6. Western blot
Lung tissue samples or cells were lysed in a RIPA buffer (Beyotime, P0013B, China) with protease inhibitor cocktail (Roche, 4693132001, USA) for 20 min. The protein concentrations were measured using a BCA protein assay kit (Beyotime, P0010, China), and 50 μg of proteins were transferred onto a PVDF membrane following separation on a 10% SDS-polyacrylamide gel. The membrane was blocked with blocking solution (5% (w/v) nonfat dry milk) for 2 h, followed by an overnight incubation at 4 °C with antibodies against KEAP1 (Cell Signaling Technology, 8047, 1:1000), NRF2 (Cell Signaling Technology, 12,721, 1:1000), GSTP (Cell Signaling Technology, 3369, 1:1000), HO-1 (Abcam, ab189491, 1:2000), NQO1 (Abcam, ab80588, 1:10,000), Lamin B (Abcam, ab32535, 1:500) and β-Actin (Cell Signaling Technology, 8457, 1:1000). The following day, the membrane was incubated for an additional 1 h with HRP-conjugated secondary antibody (1:1000 dilution) at room temperature after thoroughly washing three times with PBST. Bands were detected by ECL (Beyotime, P0018AM, China) and band intensities were quantified using Image J gel analysis software. All experiments were performed in triplicate.
2.7. Co-immunoprecipitation
Cells were lysed with IP buffer (Beyotime, P0013J, China) containing a protease inhibitor cocktail (Roche, 4693132001, USA). The lysates were incubated with the anti-KEAP1 (Cell Signaling Technology, 4678) primary antibody overnight on a rotating shaker at 4 °C, followed by the addition of Protein A/G Magnetic Beads (Thermo Fisher Scientific, 88,804, USA) for another 2 h. Proteins were eluted by sample loading buffer, followed by western blotting as described above.
2.8. Generation of Gstp knockout mice
Gstp1/p2/p3−/− mice were generated by Shanghai Model Organisms Center, Inc. (Shanghai, China). Clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) systems was used to disrupt the three Gstp genes in mice. A pair of gRNA sequences (TGCTAGAATTGGCTAGTCCCTGG and ACCATGTTCCATTGGCGTAATGG) was designed for Gstp1/p2/p3−/−. All mice were on the C57BL/6 J background and were maintained under specific-pathogen-free conditions. All mice used were male and 6–7 weeks of age.
2.9. Adeno-associated virus vectors
AAV-ZsGreen (1 × 1013 Vg/mL) and AAV-GSTP (1 × 1013 Vg/mL), prepared by Hanbio Biotechnology Co., Ltd. (Shanghai, China), were used for GST overexpression in C57BL/6 J mice. The virus vector diluted by sterile saline was instilled via intratracheally (i.t.) using microsprayer aerosolizer (YUYAN Instrument, China) after mice were anesthetized with isoflurane. The dose was 2.5 × 1010 Vg/animal. Lungs were harvested on day 28 for further analysis.
2.10. Animal treatment
Wild-type (WT) and Gstp−/− C57BL/6 J mice were deeply anesthetized with 3–5% isoflurane, and administered with sterile saline or LPS (3 mg/kg) intratracheally (i.t.) using microsprayer aerosolizer (YUYAN Instrument, China). After LPS challenge for 24 h, all mice were euthanized by CO2 asphyxiation. Lung samples were collected and the lung coefficient was calculated. Left lobes were used for hematoxylin and eosin (H&E) staining. Proportion of right lobes were homogenized in RIPA buffer for Western blot assay. The bronchoalveolar lavage fluid (BALF) were harvested for subsequent analysis of cell count, protein assay and ELISA.
2.11. Malondialdehyde (MDA) and myeloperoxidase (MPO) assays
The lung tissues were homogenized and dissolved in extraction buffer for the analysis of MDA (Nanjing Jiancheng Bioengineering Institute, A003-1-1, China) and MPO (Nanjing Jiancheng Bioengineering Institute, A044-1-1, China) levels. To examine the level of lipid peroxidation in the lung tissue and accumulation of neutrophils, MDA and MPO levels were assessed using commercially assay kits according to the respective manufacturer's instructions.
2.12. Cell counting and protein concentration assay in BALF
Cell counts in BALF samples were analyzed by the multispecies hematology analyzer (Sysmex XN-1000 V, Japan). The protein concentrations of BALF were measured.
2.13. ELISA
Cytokine concentrations of IL-1β, IL-6, and TNF-α in BALF from mice after i. t. Administration of PBS or LPS (3 mg/kg) for 24 h were quantified using mouse ELISA kits according to manufacturer's protocols (Invitrogen, 88–7013, 88–7064, 88–7324, USA). Plates were read at 450 nm on a microplate reader (BioTek Synergy Neo 2 Hybrid Multimode Reader, USA).
2.14. Histopathology evaluation
The lung tissues were fixed in 4% paraformaldehyde for 48 h, embedded in dehydrated paraffin, and sectioned into 4 μm slices. The sections were rehydrated in an alcohol gradient and stained with hematoxylin and eosin (H&E). Lung injury scores were evaluated by a blinded pathologist based on four criteria, including neutrophil infiltration, alveolar edema, alveolar hemorrhage, and lung parenchymal abnormalities [30,31]. Lung injury was rated as 0 to 4 points based on its severity: 0, no injury; 1, less than 25% injury; 2, 25–50% injury; 3, 50–75% injury; and 4, almost 100% injury.
2.15. Detection of KEAP1 S-glutathionylation
Lung tissues and cells were lysed in NP-40 lysis buffer (Beyotime, P0013F, China) containing 20 mM N-ethylmaleimide (Sigma-Aldrich, E1271, USA) and protease inhibitor cocktail (Roche, 4693132001, USA). The lysates were incubated with the anti-glutathione (Virogen, 101-A, USA) primary antibody overnight on a rotating shaker at 4 °C. Following washing, the mixture was then incubated with Protein A/G Magnetic Beads according to the manufacturer's instructions (Thermo Fisher Scientific, 88804, USA) for 2 h at 4 °C. Afterwards, co-IP was performed on a magnetic rack, and the obtained immune precipitate was washed and boiled sequentially. Supernatant was harvested by removing the magnetic beads and was resolved by non-reducing SDS-PAGE. A portion of cell lysates was incubated with 50 mM dithiothreitol (DTT) as the negative control, and the samples were purified through columns (Bio-Rad Laboratories, USA) to remove DTT before subsequent immunoprecipitation [32].
2.16. Immunofluorescence staining
HPAEpiCs or BEAS-2B cells were washed with PBS, fixed in 4% paraformaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 for 10 min. After blocked with 4% goat serum for 1 h at room temperature, cells were incubated with corresponding primary antibody overnight at 4 °C. The next day, cells were washed and incubated with Alexa Fluor® 488 goat anti-mouse IgG (Invitrogen, A-11001, 1:300) or Cyanine 3 goat anti-rabbit IgG (Invitrogen, A10520, 1:300) for 1 h at room temperature, and the nuclei were stained with DAPI. Slides were photographed under a Zeiss LSM 800 Confocal Laser Scanning Microcopy.
2.17. Nuclear and cytoplasmic fractionation
HPAEpiCs or BEAS-2B cells were first treated with or without LPS for the indicated time interval and washed with PBS, and then harvested. Nuclear and cytoplasmic fractions were separated using a Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific, 78833, USA).
2.18. Luciferase assays
HPAEpiCs were transfected with the Cignal Antioxidant Luciferase Reporter Kit (Qiagen, CCS-5020 L, USA) and Lipofectamine™ 3000 Reagent (Invitrogen, L3000015, USA). At 24 h after transfection, the cells were treated with LPS or vehicle solution. Luciferase activity was measured using Dual-Glo® Luciferase Assay system (Promega Corporation, E2920, USA).
2.19. Computational modeling
The crystal structures of the human BTB domain (PDB: 7X4X) and Kelch domain (PDB: 7K29 [33]) of KEAP1, and GSTP (PDB: 1AQW [34]), were retrieved from the PDB bank [35] (https://www.rcsb.org/). The full length of KEAP1 protein (https://alphafold.ebi.ac.uk/entry/Q14145) was predicted by AlphaFold [36]. All protein structures were pre-processed in Schrödinger 2018 [37] using the “protein preparation” module. This involved splitting unnecessary chains, deleting water molecules, metals, and ions, and adding hydrogens, among other steps. The covalent binding modes of glutathionylated KEAP1 at C368, C434, and C613 sites were manually built and refined through CHARMM27 [38] energy minimizations. During the minimizations, the systems were fixed except for GSH and the corresponding cysteines. These energy minimizations were performed using the Molecular Operating Environment (MOE) software [39]. The “Protein-Protein Docking” module of the Schrödinger software was used to predict the interactions between KEAP1 and GSTP dimer. The well-prepared crystal structures of the Kelch domain of KEAP1 and GSTP were utilized for this simulation. The “Standard” mode was chosen, while all other settings were kept as default values.
2.20. Molecular dynamics
The Molecular Dynamics (MD) were performed using the Desmond protocol module in Schrodinger, with OPLS3 force field [40] incorporated. The SPC solvent model was employed, and the orthorhombic periodic boundary [41] condition was introduced along with minimizing the box volume to prevent confusion among solvent molecules. Next, the system was recalculated and neutralized by adding Na+ ions. The sampling was conducted at a temperature of 300 K and constant pressure consumption. We take samples at an interval of 250 ps, completing the entire process within 50 ns, and collect a total of 200 samples for further systematical analysis. The other settings were kept at their default values. The quality of the MD simulation was evaluated using Simulation Interaction Diagram tools.
2.21. Statistical analysis
Statistical comparisons were performed in GraphPad Prism 9.0 (GraphPad Software, USA). Unpaired two-tailed Student's t-test was used to compare two groups. Comparisons of more than two groups were tested with one-way ANOVA. Experimental data for each quantitative analysis were replicated at least three times. Statistical significance was defined as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3. Results
3.1. Role of GSTP in LPS-induced oxidative damage and PSSG in human pulmonary cells
As shown in Fig. 1A, LPS stimulation induced substantially decreased GSH levels and GSH/GSSG ratio, whereas increased GSSG levels in HPAEpiCs, representing a typical oxidative stress stimulus. Similar changes were observed in A549 cells (Fig. S1A). Meanwhile, knockdown of GSTP further decreased GSH, GSH/GSSG ratio and increased GSSG levels, indicating a protective role of GSTP in LPS-induced oxidative stress. In addition, ferrous (Fe2+) and total iron accumulation levels, as indicators of oxidative stress and early ferroptosis, were significantly elevated in LPS-treated HPAEpiCs and A549 cells, which was exacerbated by the knockdown of GSTP (Fig. 1B and S1B). Meanwhile, the levels of intracellular PSSG, a post-translational process tightly regulated by redox status, was examined by immunoblotting. As shown in Fig. 1C and D, intracellular PSSG were significantly stimulated in both LPS-treated HPAEpiCs and BEAS-2B cells, and GSTP knockdown compellingly attenuated the LPS-induced upregulation of PSSG level. A similar upregulation of PSSG was observed in A549 cells (Fig. S1C). Moreover, LPS-induced intracellular PSSG was shown to follow time dependent manner in both WT and GSTP-silenced cells (Fig. 1D). These findings indicated the protective effect of GSTP in regulation of intracellular redox status and PSSG against LPS-induced lung cell injury.
Fig. 1.
Down regulation of GSTP-mediated PSSG aggravates LPS-induced human pulmonary cells injury. Human pulmonary cells, HPAEpiCs and BEAS-2B, were transfected with NC-siRNA or GSTP-siRNA and further treated with LPS (200 ng/mL). (A) GSH, GSSG, and the GSH/GSSG ratio in HPAEpiCs. (B) Intracellular Fe2+ and total iron levels in HPAEpiCs. (C) Total PSSG in HPAEpiCs with manipulated GSTP expression levels via non-reducing Western blot. (D) Time- and GSTP-dependency of total PSSG in BEAS-2B cells via non-reducing Western blot. Data are expressed as means ± SD. Experimental data for each quantitative analysis were replicated at least three times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance.
3.2. GSTP deficiency exacerbates the oxidative damage, inflammation, and lung tissue injury in mice ALI model
Next, the role of GSTP in LPS-induced oxidative damage and inflammation in ALI mice was investigated. As markers for ROS formation and inflammatory and infiltrative processes in lungs, MDA content and MPO activity were measured. As shown in Fig. 2A and B, LPS treatment induced a remarkable increase in MDA content and MPO activity in both WT and Gstp−/− mice, while the upregulation was more pronounced in Gstp−/− mice. Total cell and neutrophils count, protein concentration, and pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α in BALF were then determined. Similar with tendency observed in MDA content and MPO activity, all these markers were significantly upregulated in the ALI condition at 24 h after LPS exposure, while the increase of markers was more prominent in Gstp−/− ALI mice compared to their WT counterparts, indicating a deteriorated lung inflammation, pulmonary edema, endothelial barrier damage, and inflammatory infiltration in Gstp−/− mice (Fig. 2C to H). Pulmonary histopathological examinations showed apparent morphological injury in WT ALI mice, including the thickened alveolar wall, aggravated inflammatory cells infiltration, alveolar hemorrhage and interstitial edema, which was exacerbated in Gstp−/− mice induced by LPS (Fig. 2J). In addition, values of lung coefficient (Fig. 2I), representing the pulmonary pathological damage during ALI development, and lung injury scores (Fig. 2K) were both significantly higher in Gstp−/− ALI mice compared with WT ALI mice. This part of the results indicated that Gstp−/− mice exhibited increased susceptibility to cell infiltration and inflammation, referring to a positive role of GSTP in alleviating ALI-induced inflammation.
Fig. 2.
GSTP alleviated the oxidative damage, inflammation and lung tissue injury in mice with ALI. Gstp−/− mice were i. t. Administrated with 3 mg/kg of LPS. After LPS challenge for 24 h, all mice were euthanized and their lungs and BALF were collected. (A–B) MDA and MPO activity in lung tissues were measured. (C–D) Total cell counts and neutrophils percentage from the BALF were measured using a multispecies hematology analyzer. (E–H) Levels of total protein and cytokines (IL-1β, IL-6 and TNF-α) secretion in BALF were measured. (I) The lung coefficient was calculated. (J) H&E staining of lung tissues. (K) Semiquantitative analysis of lung injury scores. Data are expressed as means ± SD. n = 8 or 9 mice/group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.3. GSTP promotes activation of the KEAP1/NRF2 signaling pathway
To examine how GSTP was involved in reducing oxidative stress, elevating PSSG levels, and alleviating inflammatory reactions, the role of GSTP in the oxidative stress response pathway KEAP1/NRF2 was explored. The co-immunoprecipitation examinations showed that GSTP knockdown significantly increased the affinity between KEAP1 and NRF2 (Fig. 3A), whereas GSTP overexpression reduced the affinity (Fig. 3B), suggesting an augmenting role of GSTP1 to release NRF2 from KEAP1 against the pro-inflammatory stress. Next, the subcellular location of NRF2 under oxidative stress was determined by immunostaining and western blotting. As shown in Fig. 3C, an increase in nuclear accumulation of NRF2 was detected upon LPS exposure, while in GSTP knocked-down cells the nuclear NRF2 distribution was substantially diminished. Moreover, western blotting results in Fig. 3D also showed that GSTP knockdown significantly suppressed LPS-induced upregulation of nuclear and overall NRF2 level as well as the NRF2 downstream gene HO-1, without disturbing the KEAP1 levels. As shown in Fig. 3E, the apparent LPS-exposure time dependency of NRF2 and its downstream genes in NC-siRNA cells was vanished in GSTP-siRNA cells. Nevertheless, the total KEAP1 in the two types of cells remained unchanged upon LPS treatment duration (Fig. 3D and E). Similar phenomenon was also observed in in vivo experiments. In Gstp−/− mice treated with LPS, levels of NRF2, HO-1 and NQO1 were significantly decreased compared to the WT mice (Fig. 3F). Lastly, Fig. 3G showed that LPS treatment led to an increased transcription activity of NRF2, which was notably suppressed in GSTP-siRNA treated cells compared to its NC-siRNA control. Altogether, these results showed that GSTP promoted the dissociation of KEAP1/NRF2 complexes, increased NRF2 nuclear translocation, and enhanced NRF2 downstream HO-1 and NQO1 protein expression.
Fig. 3.
Effects of GSTP on LPS-induced KEAP1/NRF2 pathway activation. (A) HPAEpiCs transfected with NC-siRNA and GSTP-siRNA (n = 3), both groups were treated with LPS (200 ng/mL) for 60 min, then co-immunoprecipitation of KEAP1 and immunoblotting analysis were performed to assess the binding of NRF2. (B) HEK293 transfected with pCMV and GSTP plasmids (n = 3), both groups were treated with LPS (200 ng/mL) for 60 min, and co-immunoprecipitation of KEAP1 and immunoblotting analysis were performed to assess the binding of NRF2. (C) Immunofluorescence staining and confocal microscopy imaging of BEAS-2B cells transfected with GSTP-siRNA after exposure to LPS (200 ng/mL) for 2 h. (D) Expression levels of KEAP1, HO-1, total and nuclear NRF2 in GSTP-knocked down and control HPAEpiCs were measured after LPS (200 ng/mL) treatment for 30 min, n = 3. (E) Time- and GSTP-dependency of KEAP1, HO-1, NQO1, total and nuclear NRF2 expression levels in GSTP-knocked down and control BEAS-2B were measured after LPS treatment (200 ng/mL), n = 3. (F) The lung tissues were collected and lysed for protein analyzed of HO-1, NQO1 and NRF2 levels. Gstp−/− mice were i. t. Administrated with 3 mg/kg of LPS. n = 8 or 9 mice/group. (G) HPAEpiCs cells were co-transfected with the antioxidant response element reporter plasmid and the indicated siRNA for 48 h and were harvested for luciferase assay, n = 4. Data are expressed as means ± SDs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance.
3.4. GSTP upregulates S-glutathionylation of KEAP1 in response to ROS
To further explore the mechanism behind GSTP-mediated KEAP1/NRF2 pathway activation, the role of GSTP1 in PSSG of KEAP1 was determined. In GSTP knocked-down HPAEpiCs and A549 cells, the PSSG level of KEAP1 was remarkably suppressed (Fig. 4A and S2A). In addition, as shown in Fig. 4B and C, overexpression of GSTP in HEK293 cells significantly upregulated the PSSG levels of total protein and KEAP1, with the overall KEAP1 level being constant, as well as in A549 GSTP-overexpressed cells (Fig. S2B). Moreover, the addition of DTT in co-IP samples basically erased the bands in Fig. 4A and C, S2A and S2B, confirming the binding of GSH to KEAP1 was via disulfide bonds. Furthermore, KEAP1 was significantly S-glutathionylated in the lungs of both WT and Gstp−/− mice after LPS dosing while the extent of S-glutathionylated KEAP1 was largely lower in Gstp−/− mice than in WT mice, while the expression of KEAP1 was unaffected (Fig. 4D). The overlapped signals observed by immunofluorescence confocal microscopy demonstrated that GSTP and KEAP1 were co-localized in the cytoplasm of HPAEpiCs, indicating possible direct interactions between GSTP and KEAP1 (Fig. 4E). Lastly, by incubating recombinant KEAP1 protein with varying concentrations of GSTP upon H2O2 oxidative insult, it is proved that GSTP catalyzed the S-glutathionylation of KEAP1 protein in both monomer and dimer forms following a clear GSTP concentration-dependent manner (Fig. 4F). Overall, these data indicated that KEAP1 was substantially S-glutathionylated in response to oxidative stress and GSTP played an essential catalytic role in this process.
Fig. 4.
KEAP1 is S-glutathionylated and catalyzed by GSTP under oxidative stress conditions. (A) Co-IP showing PSSG of KEAP1 in HPAEpiCs was reduced by GSTP-siRNA transfection via non-reducing Western blot, after exposure to LPS (200 ng/mL) for 30 min, n = 3. Whole cell lysates confirmed the expression of GSTP, KEAP1, and β-Actin via reducing Western blot. (B) Total PSSG level via non-reducing Western blot in HEK293 transfected with pCMV and GSTP expression plasmids, after exposure to LPS (200 ng/mL) for 30 min, n = 3. Whole cell lysates confirmed the expression of GSTP and β-Actin via reducing Western blot. (C) Co-IP showing PSSG of KEAP1 in HEK293 was enhanced by GSTP overexpression via non-reducing Western blot, after exposure to LPS (200 ng/mL) for 30 min, n = 3. Whole cell lysates confirmed the expression of GSTP, KEAP1, and β-Actin via reducing Western blot. (D) Non-reducing Western blot of PSSG of KEAP1 in lung tissues from Gstp−/− and WT mice following i. t. Administration of PBS or LPS. n = 8 or 9 mice/group. (E) Immunofluorescence staining and confocal microscopy imaging of HPAEpiCs showing the colocalization of GSTP and KEAP1. (F) GSTP concentration dependency of human recombinant KEAP1 PSSG in dimer and monomer status via non-reducing Western blot. The level of KEAP1 was conformed via reducing Western blot. Data are expressed as means ± SDs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance.
3.5. C434 is essential for NRF2 nuclear accumulation in responding to oxidative stress
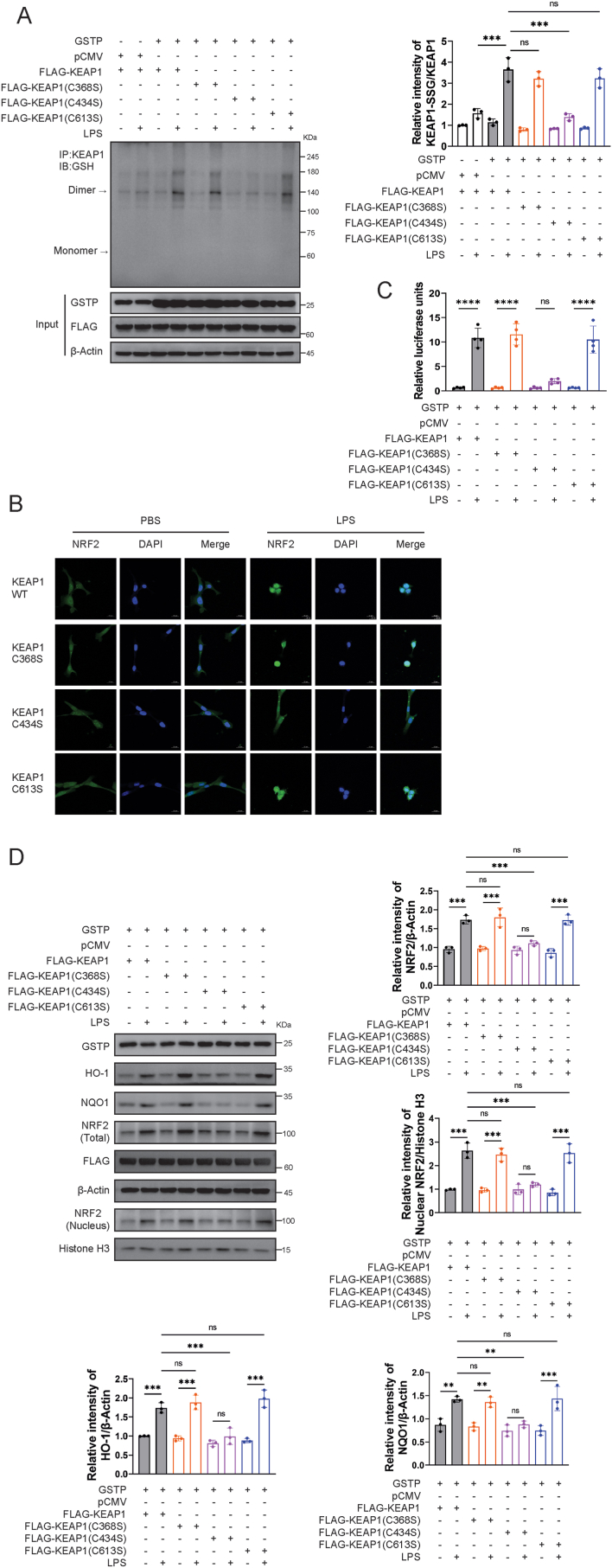
The PSSG site of KEAP1 in the presence of GSTP was determined by LC-MS/MS from recombinant KEAP1 protein samples. Fig. S3A showed the MS/MS spectra and identified sequences of the three detected peptides in KEAP1 molecule. By alignment with the published KEAP1 sequence, C368, C434, and C613 were identified as the S-glutathionylated sites of KEAP1 catalyzed by GSTP. In addition, the binding modes of GSH to C368, C434, and C613 in KEAP1 were visualized by molecular docking approach, as shown in Fig. S3B. Notably, these three cysteine residues were all located in the Kelch domain of the KEAP1 protein. The three identified S-glutathionylated cysteine sites were mutated to serine, respectively, to evaluate the biological significance of KEAP1 PSSG. As shown in Fig. 5A and B, the difference in KEAP1 PSSG levels in mutated KEAP1-transfected HPAEpiCs was evaluated. Co-immunoprecipitation results showed that LPS-induced PSSG of KEAP1 in HPAEpiCs transfected with C434S mutant plasmids was significantly reduced, but not in cells transfected with C368S or C613S mutant plasmids. Next, we validated the subcellular localization of NRF2 in cells transfected with mutant plasmids in the presence or absence of oxidative stress. When exposed to LPS, NRF2 accumulated in the nucleus of HPAEpiCs transfected with wild type plasmid as well as C368S and C613S mutant plasmids, but not in C434S counterparts (Fig. 5B). Consistently, NRF2 transcriptional activity was significantly suppressed in C434S mutant expressed cells (Fig. 5C). Furthermore, it was found that expression of nuclear NRF2 and activation of its downstream enzymes HO-1 and NQO-1 were largely reduced in C434S mutant expressed HPAEpiCs with LPS treatment (Fig. 5D).
Fig. 5.
C434 PSSG of KEAP1 enhances NRF2 nuclear translocation and activated downstream pathways. (A) Co-immunoprecipitation of KEAP1 and GSH was performed, and relative KEAP1 PSSG levels were measured, n = 3. (B) Immunofluorescence staining and confocal microscopy imaging of HPAEpiCs cells with overexpression of KEAP1 WT, C368S, C434S and C613S after exposure to LPS (200 ng/mL) for 1 h. (C) HPAEpiCs were co-transfected with the antioxidant response element reporter plasmid and the indicated cDNA for 48 h and were harvested for luciferase assay, n = 4. (D) Levels of HO-1, NQO1, total and nuclear NRF2 in BEAS-2B were measured and calculated LPS (200 ng/mL), n = 3. Data are expressed as means ± SD. Experimental data for each quantitative analysis were replicated at least three times. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance.
3.6. Computational analysis of GSTP-catalyzed KEAP1 PSSG at C434 and the impact on NRF2 binding
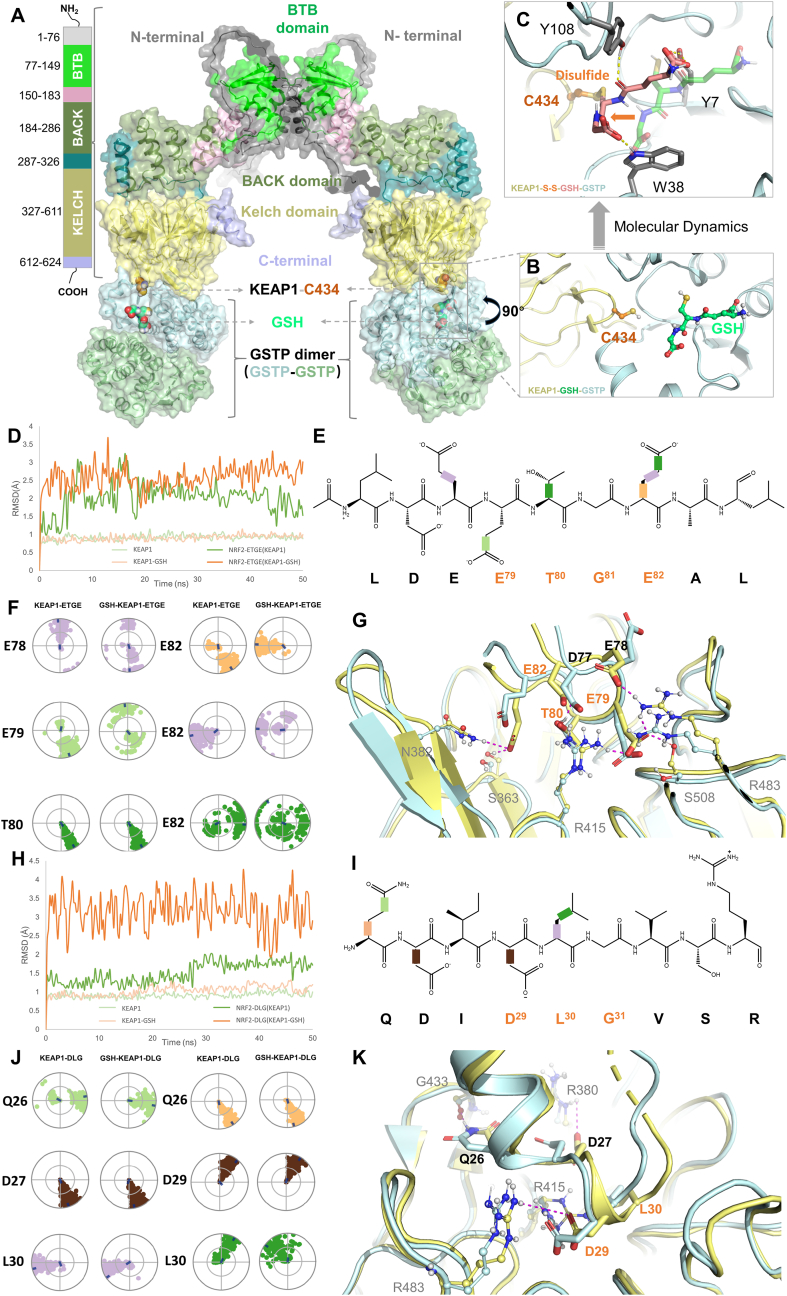
To illustrate the binding mode between KEAP1 and GSTP in context of C434 PSSG, a protein-protein docking simulation was performed. Since functional KEAP1 and GSTP proteins both exist as homodimers, the stoichiometry of KEAP1 dimer to GSTP dimer was illustrated as 1 to 2. As shown in Fig. 6A, C434 was located in the outer loop connecting the β-sheets in Kelch domain of KEAP1 structure, which was able to contact GSTP and forms interaction. The very close distance between KEAP1-C434 and G-site of GSTP, as depicted in Fig. 6B, suggested the possibility of a disulfide bond formation between GSH and KEAP1-C434. By performing a molecular dynamic simulation, the formation of KEAP-SSG at C434 was visualized in Fig. 6C, where Y7, W38, and Y108 in GSTP were identified as the key interacting residues in this complex.
Fig. 6.
Computational modeling of GSTP-catalyzed KEAP1 PSSG at C434 and molecular dynamic analysis of C434 PSSG on KEAP1-NRF2 binding. (A) The binding mode of the full-length structure of KEAP1 (dimer) bound to GSTP dimers. Major domains of KEAP1 were shown in different colors. The BTB domain (PDB: 7X4X) and Kelch domain (PDB: 7K29) are crystal structures, while the other domains are predicted by Alphafold. The GSTP dimers are shown in pale blue and pale orange, respectively. Key residue C434 and GSH are shown in orange and green-cyan spheres. The whole complex is shown as a cartoon as well as transparent surfaces. (B) The detailed binding information of KEAP1 (pale yellow cartoon) bound to the G-site of GSTP (pale blue cartoon) with GSH (green-cyan ball and sticks). (C) The binding modes of GSH (pink-cyan sticks) to KEAP1 by forming a disulfide bond with C434 after molecular dynamics simulations. The un-bounded conformation of GSH is shown in transparent green-cyan sticks. Other key residues are shown in grey sticks, and key interactions are depicted as dotted lines. (D) The root-mean-square deviation (RMSD) values of the backbone (αC) of KEAP1 (pale green), KEAP1-GSH (pale orange), and the corresponding bound NRF2 ETGE motif (green and orange, respectively) were determined. The RMSD is used to measure the average displacement change of a group of atoms within a specific frame, relative to a reference frame, which was calculated for each frame in the trajectory. (E) The chemical structure of the ETGE motif is presented. 2D schematic NRF2 ETGE with color-coded rotatable bonds is presented, and each rotatable bond torsion is accompanied by a dial plot and bar plot of the same color in panel F. (F) The NRF2 ETGE motif torsion plot summarizes the conformational changes of every rotatable bond (RB) in NRF2 ETGE throughout the simulation trajectory (0.00–50.00 ns). The dial plots describe the torsion over the course of the simulation, with the center representing the beginning and the radial direction representing the time evolution. The dial plots on the left and right refer to the KEAP1-NRF2-ETGE and GSH-KEAP1-NRF2-ETGE system, respectively. (G) Comparative binding modes of ETGE motif of NRF2 with the KEAP1 Kelch domain before (pale-yellow) and after (pale-blue) molecular dynamics. Key residues are highlighted in corresponding colors, and key interactions are indicated by magenta dotted lines. The residues that correspond to the residues of KEAP1 are depicted as balls and sticks, and labelled in grey to differentiate them from the orange-labelled residues in ETGE motif. (H) The RMSD values of the backbone (αC) of KEAP1 (pale green), KEAP1-GSH (pale orange), and the corresponding bound NRF2 DLG motif (green and orange, respectively) were determined. (I) The chemical structure of the DLG motif is presented. 2D schematic of DLG motif with color-coded rotatable bonds is presented, and each rotatable bond torsion is accompanied by a dial plot and bar plot of the same color in panel J. (J) The DLG motif torsion plot summarizes the conformational changes of every RB in DLG motif throughout the simulation trajectory (0.00–50.00 ns). The dial plots on the left and right refer to the KEAP1-DLG and GSH-KEAP1-DLG system, respectively. (K) Comparative binding modes of DLG motif of NRF2 with the KEAP1 Kelch domain before (pale-yellow) and after (pale-blue) molecular dynamics is shown. Key residues are highlighted in corresponding colors, and key interactions are indicated by magenta dotted lines. The residues that correspond to the residues of KEAP1 are depicted as balls and sticks, and labelled in grey to differentiate them from the orange-labelled residues in DLG motif. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To elucidate the molecular mechanism of KEAP1 PSSG at C434 on the binding of NRF2, the 50 ns MD simulations were performed on the high-affinity binding ETGE and low-affinity binding DLG motifs of NRF2, respectively, to compare KEAP1-NRF2 and C434-glutathionylated KEAP1-NRF2 systems. Fig. 6D and H indicated that the KEAP1-ETGE, KEAP1-DLG, GSH-KEAP1-ETGE, and GSH-KEAP1-DLG systems all achieved equilibrium after 50 ns. Apparently, both ETGE and DLG motifs of NRF2 bound to C434-glutathionylated KEAP1 exhibited higher root-mean-square deviation (RMSD) values, indicating a lowered stability of binding (Fig. 6D and H). Afterwards, a systematic analysis on the conformations of the rotatable bonds of both ETGE and DLG motifs were conducted. As depicted in Fig. 6F and J, significant changes of rotation angle patterns for GSH-KEAP1-ETGE system were observed at E78, E79, and E82 and for GSH-KEAP1-DLG system were observed at Q26 and L30, indicating less stable conformations of both GSH-KEAP1-ETGE and GSH-KEAP1-DLG complexes. It is of interesting to note that although L30 did not directly interact with KEAP1 in a polar manner, it did influence the interaction between DLG and KEAP1 by transiting the α-helix around L30 into a more flexible loop conformation (Fig. 6K). Comparative binding modes before and after PSSG of KEAP1 at C434 (Fig. 6G) showed that multiple interactions between ETGE-KEAP1 were split, including ETGE-E78 and KEAP1-R483, ETGE-E79 and KEAP1-R483/S508, ETGE-E82 and KEAP1-S363/N382, whereas an intramolecular hydrogen bond formed between ETGE-T80 and ETGE-D77 remained conserved. Similar results were observed in DLG-KEAP1 system, including a decrease in the occurrence of H-bond/ionic interactions between DLG-D27 and KEAP1-R380 from 68% to less than 30% and a decrease in the occurrence of H-bond/ionic interactions between DLG-D29 and KEAP1-R483 from 100% to 86%. However, interactions between DLG-Q26 and KEAP1-G433, as well as DLG-Q29 and KEAP1-G415, remained conserved.
The observations, altogether, provided molecular details of GSTP1 binding to KEAP1 for the catalysis of PSSG at C434 and revealed the mechanism of promoting NRF2 disassociation from computational simulation view.
3.7. Antioxidant pathway activation of GSTP in the model of LPS–induced acute lung injury
To further ascertain whether and how GSTP plays a protective role in LPS-induced ALI, multiple endpoints related to inflammation and lung injury were compared in AAV-GSTP treated (high expression of GSTP protein as evidenced in Fig. S2F) and AAV-ZsGreen mice. As shown in Fig. 7A, the PSSG level of KEAP1 versus total KEAP1 in AAV-GSTP pretreated mice was significantly higher than that in AAV-ZsGreen mice in ALI, while the total KEAP1 level remained unchanged. Meanwhile, notable elevated levels of NRF2 and its downstream antioxidant proteins HO-1 and NQO1 were detected in AAV-GSTP ALI mice compared to AAV-ZsGreen ALI mice (Fig. 7B). Furthermore, GSTP suppressed LPS-induced MDA (Fig. 7C) expression and MPO (Fig. 7D) activity upregulation in lungs, indicating reduced inflammatory damage. The total number of cells, neutrophils infiltration, and protein exudation in BALF were reduced in AAV-GSTP ALI mice (Fig. 7E–G). Moreover, the IL-1β, IL-6 and TNF-α levels in BALF significantly decreased in GSTP overexpressed mice after LPS treatment (Fig. 7H–J). Consistently, the lung coefficient, lung histological examinations and corresponding lung injury scores all demonstrated a significant reduction of inflammation by GSTP overexpression in lungs (Fig. 7K-M). Taken together, this part of the results confirmed that extrinsic administration of GSTP attenuated LPS-induced oxidative stress and inflammation damage of lungs via promoting PSSG of KEAP1 to disturb KEAP1-NRF2 complex and activating the NRF2 downstream pathways.
Fig. 7.
GSTP alleviated the oxidative damage, inflammation and lung tissue injury in mice with ALI via promoting KEAP1 PSSG. Mice pretreated with AAV-GSTP were i. t. Administrated with 3 mg/kg of LPS. After LPS challenge for 24 h, all mice were euthanized and their lungs and BALF were collected. (A) PSSG of KEAP1 in lung tissues from mice pretreated with AAV-GSTP. (B) The lung tissues were collected and lysed for the immunoblotting of HO-1, NQO-1 and NRF2 levels. (C–D) MDA and MPO activity in lung tissues were measured. (E–F) The total cell counts and neutrophils percentage from the BALF were measured counted using a multispecies hematology analyzer. (G–J) Levels of total protein and cytokines (IL-1β, IL-6 and TNF-α) secretion in BALF were measured. (K) The lung coefficient was calculated. (L) H&E staining of sections of lungs. (M) Semiquantitative analysis of lung injury scores. Data are expressed as means ± SD. n = 8 or 9 mice/group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = no significance.
4. Discussion
The redox-activated factor NRF2 regulates transcription of around 250 genes, including those encoding proteins involved in cellular redox homeostasis and inflammation [[42], [43], [44]]. NRF2 activity is predominantly regulated by its repressor, KEAP1. In the present study, it is found that GSTP promoted KEAP1/NRF2 signaling pathway activation in ALI. Moreover, GSTP-catalyzed PSSG of KEAP1 at C434 residue was shown to play a key role in KEAP1/NRF2 pathway activation, which reversed pulmonary edema and inflammation progression. By elucidating the catalytic role of GSTP in KEAP1 PSSG in ALI, our study proposed a novel underlying mechanism for oxidative stress-induced ALI, as well as a potential strategy for the treatment of ALI.
GSH is the core of redox defenses during oxidative stress. Upregulation of inflammatory response in lung cells are usually influenced by the cellular redox status (GSH/GSSG ratio) [[42], [43], [44], [45], [46]]. Decreased GSH/GSSG ratio under oxidative stress conditions represents a significant conversion from GSH to GSSG. The elevated level of intracellular GSSG promoted the extent of PSSG level through the disulfide bond formation between the protein thiol group and GSH. Dysregulation of PSSG has been suggested to substantially influence the progression of diseases, most of which are results of redox imbalance. The roles of PSSG in various diseases, particularly the respiratory related syndromes, have been widely explored. For example, PSSG levels were found to be downregulated in sputum from eosinophilic and neutrophilic asthmatics [47]. While in contrast, the overall PSSG levels in the lungs from mice with lung fibrosis and from patients with idiopathic pulmonary fibrosis were elevated [21,48]. GSTP is the major enzyme catalyzing the forward reaction of PSSG, besides the well-known roles in catalyzing GSH conjugation of small molecules [49]. Previous studies have showed that the consequences of GSTP-mediated PSSG in different respiratory diseases were diverse, suggesting the complexity of mechanisms behind [21,[50], [51], [52]]. The details of mechanisms by which GSTP regulates the pathophysiology of ALI and the target proteins of S-glutathionylation are to be clarified.
Up to date, the exact redox mechanisms by which oxidative stress-induced modifications leading to ALI are still unclear. In this study, we found that deficiency of GSTP in human alveolar epithelial cells significantly exacerbated ALI by modulating KEAP1/NRF2 signaling pathway. Moreover, we showed that KEAP1 was prone to PSSG under oxidative stress conditions and this process was substantially catalyzed by GSTP. Further experiments showed that S-glutathionylation at C434 residue of KEAP1 promoted the KEAP1-NRF2 complex disassociation and NRF2 nuclear translocation, thereby activating NRF2 antioxidant signaling pathway and ameliorating lung cell inflammation.
Afterwards we used LPS as the oxidative stress inducer, which is one of the most commonly utilized models in ALI research and shares high similarities in pathophysiology to human ARDS [53]. LPS exposure to the lung causes activation of macrophages and leakage of inflammatory cells, especially the neutrophils, into the lungs, which not only leads to the release of inflammatory cytokines, but also induces the formation of ROS and oxidative stress [54,55]. In our work, LPS exposure increased GSH/GSSG ratio, Fe2+ and total Fe levels, and total PSSG levels in human lung cells, while the loss of GSTP significantly aggravated LPS-induced lung cell damage, confirming the protective effect of GSTP-regulated PSSG in ALI. This is consistent with recent studies that GSTP plays key protective roles in oxidative stress [56,57].
To further explore the roles of GSTP in ALI, a Gstp1/2/3 triple-knockout mice model was constructed. Humans only have one isoform, GSTP1, while mice have three isoforms, Gstp1, Gstp2 and Gstp3 [58]. The human GSTP1 gene has high homology with mouse Gstp1 and 2 of 83%, and slightly lower homology with mouse Gstp3 of 75% [17]. In humans, GSTP1 is mainly expressed in the lung, liver, heart, and brain. In particular, GSTP1 is highly expressed in all types of respiratory epithelial cells, including type I and type II alveolar epithelial cells, in human lungs [59]. Similarly, murine Gstp is highly expressed in lungs [60,61]. In our study, it is clearly seen that the absence of GSTP in mice exacerbated LPS-induced ALI by increasing lipid peroxidation and pro-inflammatory cytokines production, which is in accordance with in vitro data. These results suggested that the beneficial effects of GSTP against ALI may be closely related to its antioxidant and anti-inflammatory activities.
Next, in human lung epithelial cells, the potential molecular target of GSTP-catalyzed PSSG under ALI-mimicking conditions was identified as KEAP1, which is susceptible to redox-dependent regulation. As the predominant oxidative stress response signaling pathway, KEAP1/NRF2 activation is commonly implicated in the ALI process [62,63]. This study showed that GSTP promoted NRF2 activation under oxidative stress conditions without influencing total KEAP1 level, while it induced significant intracellular PSSG. Following experiments showed that KEAP1 is a glutathionylation target protein in ALI while its glutathionylation was essentially catalyzed by GSTP. KEAP1 structure is enriched with reactive cysteine residues, which act as sensors for endogenously produced and exogenously encountered chemical inducers that readily react with sulfhydryl groups. The reactivity of KEAP1 cysteines has been comprehensively reviewed before [24], among which C77, C297, C319, C368, and C434 were proposed as the most sensitive ones to form protein-Cys-SG type of disulfides [26]. Besides, various types of chemical inducers were reported to target at different cysteine sites in KEAP1 to regulate NRF2 [64,65]. For example, KEAP1 C151 and C288 act as the functional sensors for the electrophile DEM and 15 d-PGJ2, respectively [66,67].
Covalent modification and oxidation of KEAP1 cysteine residues can lead to dissociation of NRF2 and facilitate its nuclear translocation, eventually leading to activation of NRF2-dependent antioxidant genes [68]. Previous studies have reported various cases of direct modification of specific cysteine sensors in KEAP1 by electrophiles as the initiation event of oxidative stress response [[68], [69], [70]]. In fact, PSSG of KEAP1 was implicated in several biological processes including neutral protection, immune response activation, and anti-cancer treatment [27,28,71]. Nevertheless, the GSTP-catalyzed KEAP1 PSSG in ALI has never been investigated before. To unravel the underlying mechanism by which GSTP activated the KEAP1/NRF2 signaling pathway under ALI condition, effects of PSSG of KEAP1 on KEAP1-NRF2 interaction were evaluated. The KEAP1-NRF2 interaction is essential for the activation of the ARE-controlled antioxidant enzyme system, and also has been implicated as therapeutic targets for many diseases prevention and treatment [72,73]. Our in vivo and in vitro experimental results showed that GSTP could further significantly promote weakening of KEAP1-NRF2 interaction, enhance NRF2 expression and nuclear translocation, and increase the expression of its downstream antioxidant proteins, including HO-1 and NQO1. Notably, we identified that GSTP induced PSSG of KEAP1 at the C368, C434, and C613 sites, with the modification at C434 proved to be the pivotal PSSG site which promoted NRF2 nuclear translocation, binding to ARE and subsequent activation of downstream pathways. In fact, C434 has been known as one of the most sensitive cysteine sites in KEAP1, which was prone to chemical-induced modifications [26,28,74,75]. Holland et al. have proposed C434 and C368 as the critical sites for KEAP1-NRF2 binding, while the detailed evidences were computational-based energy minimization analysis and citation of earlier publications, in which the single mutation of C368 or C434 neighboring residues, e.g. G364C, G340C, or H436A in KEAP1, resulted in weakened NRF2 affinity or repressed NRF2 activity [26]. Following studies like Fujii et al. [75] and Cheng et al. [74] showed that PSSG or chemical modification of KEAP1 C434 in cellular experiments using co-IP method and suggested a mechanism of PSSG- or chemical covalent binding-mediated inactivation of Cul 3-KEAP1 complex ubiquitin ligase activity, which resulted the activation of NRF2 pathway. Additionally, Carvalho et al. reported a protective role of GSTP-catalyzed KEAP1 PSSG against a neurotoxicant and predicted C434 as the plausible modification site without wet lab evidence [28]. In the present study, we combined site-specific mutation experiment with computational simulation approaches to fully elucidate the molecular mechanisms of PSSG at C434 on KEAP1/NRF2 pathway.
As shown in Fig. 6, the protein-protein interaction of KEAP1 and GSTP to form C434-GSH disulfide was clearly visualized and the changes in chemical bond rotation leading to an unstable binding between C434-glutathionylated KEAP1 and ETGE and DLG motifs of NRF2 were proposed. Although it is generally accepted that DLG motif binds to KEAP1 with a much lower affinity than that of ETGE [76], the detailed KEAP1 binding mechanisms to two motifs of NRF2 and how that controls NRF2 ubiquitination were very complicated and to be fully validated [24]. Originally, ETGE was considered as the essential motif for the KEAP1-NRF2 interaction and NRF2 activation [77]. Later on, Fukutomi et al. reported that KEAP1 C434 located near the DLG binding surface [78], and suggested the previously-observed S-guanylation of C434 with 8-nitro-cGMP may specifically disrupted the interaction between DLG motif and KEAP1 [75]. Consistent with previous understanding that ETGE and DLG motifs binding modes to KEAP1 were distinct from each other [78], our MD simulation and rotatable bonds conformation analysis results showed that the binding of both motifs to KEAP1 were remarkably influenced by C434 PSSG, which supported the observation of subsequential NRF2 pathway activation.
Lastly, a GSTP overexpression mice model was used to validate the previous observations and proposed hypothesis in this study. Consistent with the rest of results, overexpression of GSTP in lungs increased the PSSG level of KEAP1, promoted the upregulation of antioxidant signaling factors after LPS stimulation, and alleviated the levels of pro-inflammatory cytokines and oxidative damage in lung tissue. Therefore, it is suggested that GSTP is able to reduce oxidative damage in lung tissue and exhibit a protective effect against LPS-induced ALI and inflammation in vivo. Currently, selective GSTP inhibition by small-molecule chemical inhibitors (Telintra) [79] or siRNA (NBF-006) [80] has been developed for the treatment of myelodysplastic syndrome or cancers, respectively, whereas the strategies of targeted and specific activation of GSTP in vivo is to be explored. Based on our research findings, pulmonary administration of GSTP recombinant protein preparations loaded by advanced biopharmaceuticals delivery system might be a potential strategy for the treatment of ALI in the future.
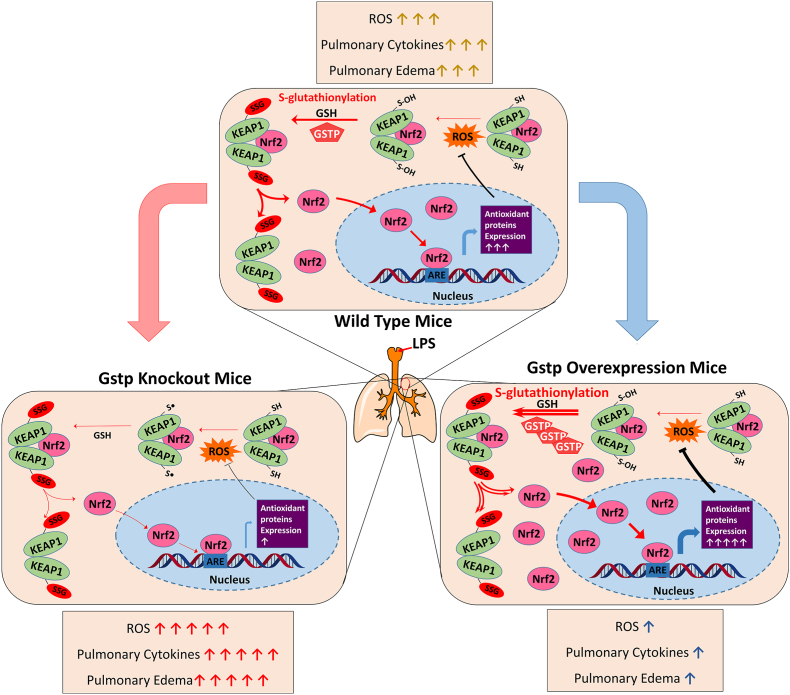
In conclusion, as presented in Fig. 8, this study elucidated the GSTP-catalyzed KEAP1 PSSG process in lung epithelia under oxidative conditions and proposed the biological mechanisms and therapeutic significances behind. The GSTP-mediated enhancement of KEAP1 PSSG process disassociate KEAP1-NRF2 interactions, facilitate the dissociation of KEAP1-NRF2 complex, thereby promote NRF2 nuclear translocation and activate downstream proteins expression, eventually leading to ameliorated lung inflammation and injury. Our results indicate that GSTP is a potential target for anti-inflammatory drug development and suggest that PSSG of KEAP1 at C434 plays an anti-inflammatory role in lung epithelial cells in ALI. This study reveals the underlying mechanism by which lung epithelial cells prevent self-overactivation under oxidative stress.
Fig. 8.
Proposed mechanism for alleviating inflammation in acute lung injury via GSTP-mediated KEAP1/NRF2 pathway activation. GSTP increased the dissociation of the KEAP1-NRF2 by catalyzing KEAP1 PSSG process, subsequently activated NRF2 downstream genes including HO-1 and NQO1 and alleviated the progression of inflammation in acute lung injury.
Funding
This study was supported by the Natural Science Foundation of China (No. 82003877 and No. 82200510) and the Natural Science Foundation of Jiangsu Province (No. BK20190557).
Ethics statement
All the animal experiments are complied with ethical standards of the Animal Care and Welfare Committee of China Pharmaceutical University. The animal experiments were approved by the Ethics Committee of China Pharmaceutical University (No. 2022-03-048), and all the experimental processes were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
CRediT authorship contribution statement
Xiaolin Sun: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation, Conceptualization. Chaorui Guo: Writing – review & editing, Visualization, Validation, Resources, Methodology, Investigation, Funding acquisition, Formal analysis. Chunyan Huang: Validation, Resources, Methodology, Investigation, Formal analysis. Ning Lv: Validation, Resources, Methodology, Investigation, Formal analysis. Huili Chen: Writing – review & editing, Visualization, Validation, Methodology. Haoyan Huang: Visualization, Validation, Resources, Methodology. Yulin Zhao: Visualization, Validation, Methodology. Shanliang Sun: Writing – original draft, Visualization, Validation, Methodology, Investigation. Di Zhao: Writing – review & editing, Validation, Supervision, Project administration. Jingwei Tian: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Xijing Chen: Writing – review & editing, Supervision, Resources, Funding acquisition, Conceptualization. Yongjie Zhang: Writing – review & editing, Visualization, Supervision, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Haiteng Deng and Ms. Xiaolin Tian (Tsinghua University) for their technical assistance in protein MS analysis, and Ms. Jie Zhao (Animal Experimental Center of China Pharmaceutical University) for her kind help with animal experiment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103116.
Contributor Information
Jingwei Tian, Email: tianjingwei618@163.com.
Xijing Chen, Email: chenxj-lab@hotmail.com.
Yongjie Zhang, Email: zhangyongjie@cpu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Gallelli L., Zhang L., Wang T., Fu F. Severe acute lung injury related to COVID-19 infection: a review and the possible role for escin. J. Clin. Pharmacol. 2020;60:815–825. doi: 10.1002/jcph.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long M.E., Mallampalli R.K., Horowitz J.C. Pathogenesis of pneumonia and acute lung injury. Clin Sci (Lond). 2022;136:747–769. doi: 10.1042/CS20210879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubenfeld G.D., Caldwell E., Peabody E., Weaver J., Martin D.P., Neff M., Stern E.J., Hudson L.D. Incidence and outcomes of acute lung injury. N. Engl. J. Med. 2005;353:1685–1693. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 4.Schouten L.R., Veltkamp F., Bos A.P., Van Woensel J.B., Neto A.S., Schultz M.J., Wösten R.M., Van Asperen Incidence and mortality of acute respiratory distress syndrome in children: a systematic review and meta-analysis. Crit. Care Med. 2016;44:819–829. doi: 10.1097/CCM.0000000000001388. [DOI] [PubMed] [Google Scholar]
- 5.Bezerra F.S., Lanzetti M., Nesi R.T., Nagato A.C., Silva C.P.E., Kennedy-Feitosa E., Melo A.C., Cattani-Cavalieri I., Porto L.C., Valenca S.S. Oxidative stress and inflammation in acute and chronic lung injuries. Antioxidants. 2023;12:548. doi: 10.3390/antiox12030548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang J.D., Mcardle P.J., O'reilly P.J., Matalon S. Oxidant-antioxidant balance in acute lung injury. Chest. 2002;122:314S–320S. doi: 10.1378/chest.122.6_suppl.314s. [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y., Uys J.D., Tew K.D., Townsend D.M., S-glutathionylation From molecular mechanisms to health outcomes. Antioxidants Redox Signal. 2011;15:233–270. doi: 10.1089/ars.2010.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghezzi P. Role of glutathione in immunity and inflammation in the lung. Int. J. Gen. Med. 2011;4:105–113. doi: 10.2147/IJGM.S15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biswas S.K., Rahman I. Environmental toxicity, redox signaling and lung inflammation: the role of glutathione. Mol. Aspect. Med. 2009;30:60–76. doi: 10.1016/j.mam.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prousky J. The treatment of pulmonary diseases and respiratory-related conditions with inhaled (nebulized or aerosolized) glutathione. Evid. Based. Complement. Alternat. Med. 2008;5:27–35. doi: 10.1093/ecam/nem040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lana J.F.S.D., Lana A.V.S.D., Rodrigues Q.S., Santos G.S., Navani R., Navani A., Da Fonseca L.F., Azzini G.O.M., Setti T., Mosaner T., Simplicio C.L., Setti T.M. Nebulization of glutathione and N-Acetylcysteine as an adjuvant therapy for COVID-19 onset. Adv. Redox. Res. 2021;3 doi: 10.1016/j.arres.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musaogullari A., Chai Y.C. Redox regulation by protein S-glutathionylation: from molecular mechanisms to implications in health and disease. Int. J. Mol. Sci. 2020;21:8113. doi: 10.3390/ijms21218113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corteselli E., Aboushousha R., Janssen-Heininger Y. S-glutathionylation-controlled apoptosis of lung epithelial cells; potential implications for lung fibrosis. Antioxidants. 2022;11:1789. doi: 10.3390/antiox11091789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalinina E., Novichkova M. Glutathione in protein redox modulation through S-glutathionylation and S-nitrosylation. Molecules. 2021;26:435. doi: 10.3390/molecules26020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tew K.D., Townsend D.M. Regulatory functions of glutathione S-transferase P1-1 unrelated to detoxification. Drug Metab. Rev. 2011;43:179–193. doi: 10.3109/03602532.2011.552912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tew K.D., Manevich Y., Grek C., Xiong Y., Uys J., Townsend D.M. The role of glutathione S-transferase P in signaling pathways and S-glutathionylation in cancer. Free Radic. Biol. Med. 2011;51:299–313. doi: 10.1016/j.freeradbiomed.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Blazejewski S.M., Bennison S.A., Toyo-Oka K. Glutathione S-transferase Pi (Gstp) proteins regulate neuritogenesis in the developing cerebral cortex. Hum. Mol. Genet. 2021;30:30–45. doi: 10.1093/hmg/ddab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Z.W., Zhang J., Ancrum T., Manevich Y., Townsend D.M., Tew K.D. Glutathione S-transferase P-mediated protein S-glutathionylation of resident endoplasmic reticulum proteins influences sensitivity to drug-induced unfolded protein response. Antioxidants Redox Signal. 2017;26:247–261. doi: 10.1089/ars.2015.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamada K., Goto S., Okunaga T., Ihara Y., Tsuji K., Kawai Y., Uchida K., Osawa T., Matsuo T., Nagata I. Nuclear glutathione S-transferase π prevents apoptosis by reducing the oxidative stress-induced formation of exocyclic DNA products. Free Radic. Biol. Med. 2004;37:1875–1884. doi: 10.1016/j.freeradbiomed.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., Ye Z.W., Chen W., Manevich Y., Mehrotra S., Ball L., Janssen-Heininger Y., Tew K.D., Townsend D.M. S-Glutathionylation of estrogen receptor α affects dendritic cell function. J. Biol. Chem. 2018;293:4366–4380. doi: 10.1074/jbc.m117.814327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mcmillan D.H., Van Der Velden J.L., Lahue K.G., Qian X., Schneider R.W., Iberg M.S., Nolin J.D., Abdalla S., Casey D.T., Tew K.D. Attenuation of lung fibrosis in mice with a clinically relevant inhibitor of glutathione-S-transferase π. JCI insight. 2016;1 doi: 10.1172/jci.insight.85717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J., Ye Z.W., Chen W., Culpepper J., Jiang H., Ball L.E., Mehrotra S., Blumental-Perry A., Tew K.D., Townsend D.M. Altered redox regulation and S-glutathionylation of BiP contribute to bortezomib resistance in multiple myeloma. Free Radic. Biol. Med. 2020;160:755–767. doi: 10.1016/j.freeradbiomed.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D.D., Lo S.C., Cross J.V., Templeton D.J., Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul 3-dependent ubiquitin ligase complex. Mol. Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baird L., Yamamoto M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 2020;40 doi: 10.1128/MCB.00099-20. 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Gu, Zhao K., Ju Y., Mani S., Cao Q., Puukila S., Khaper N., Wu L., Wang R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxidants Redox Signal. 2013;18:1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]
- 26.Holland R., Hawkins A.E., Eggler A.L., Mesecar A.D., Fabris D., Fishbein J.C. Prospective type 1 and type 2 disulfides of Keap1 protein. Chem. Res. Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambhir L., Checker R., Thoh M., Patwardhan R., Sharma D., Kumar M., Sandur S.K. 1, 4-Naphthoquinone, a pro-oxidant, suppresses immune responses via KEAP-1 glutathionylation. Biochem. Pharmacol. 2014;88:95–105. doi: 10.1016/j.bcp.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho A.N., Marques C., Guedes R.C., Castro‐Caldas M., Rodrigues E., Van Horssen J., Gama M.J. S‐Glutathionylation of Keap1: a new role for glutathione S‐transferase pi in neuronal protection. FEBS Lett. 2016;590:1455–1466. doi: 10.1002/1873-3468.12177. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y., Huang H., Lv N., Huang C., Chen H., Xing H., Guo C., Li N., Zhao D., Chen X. Glutathione s-transferases mediate in vitro and in vivo inactivation of genipin: implications for an underlying detoxification mechanism. J. Agric. Food Chem. 2023;71:2399–2410. doi: 10.1021/acs.jafc.2c08175. [DOI] [PubMed] [Google Scholar]
- 30.Shi J., Yu T., Song K., Du S., He S., Hu X., Li X., Li H., Dong S., Zhang Y. Dexmedetomidine ameliorates endotoxin-induced acute lung injury in vivo and in vitro by preserving mitochondrial dynamic equilibrium through the HIF-1a/HO-1 signaling pathway. Redox Biol. 2021;41 doi: 10.1016/j.redox.2021.101954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amatullah H., Maron-Gutierrez T., Shan Y., Gupta S., Tsoporis J.N., Varkouhi A.K., Monteiro A.P.T., He X., Yin J., Marshall J.C. Protective function of DJ-1/PARK7 in lipopolysaccharide and ventilator-induced acute lung injury. Redox Biol. 2021;38 doi: 10.1016/j.redox.2020.101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anathy V., Aesif S.W., Guala A.S., Havermans M., Reynaert N.L., Ho Y.S., Budd R.C., Janssen-Heininger Y. Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J. Cell Biol. 2009;184:241–252. doi: 10.1083/jcb.200807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortet P.C., Muellers S.N., Viarengo-Baker L.A., Streu K., Szymczyna B.R., Beeler A.B., Allen K.N., Whitty A. Recapitulating the binding affinity of Nrf2 for KEAP1 in a cyclic heptapeptide, guided by NMR, X-ray crystallography, and machine learning. J. Am. Chem. Soc. 2021;143:3779–3793. doi: 10.1021/jacs.0c09799. [DOI] [PubMed] [Google Scholar]
- 34.Prade L., Huber R., Manoharan T.H., Fahl W.E., Reuter W. Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor. Structure. 1997;5:1287–1295. doi: 10.1016/s0969-2126(97)00281-5. [DOI] [PubMed] [Google Scholar]
- 35.Burley S.K., Berman H.M., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. Protein Data Bank (PDB): the single global macromolecular structure archive. Methods Mol. Biol. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maestro Schrödinger. Schrödinger, Inc.; Portland: 2018. Version 11.5. [Google Scholar]
- 38.Mackerell A.D., Jr., Feig M., Brooks C.L. Improved treatment of the protein backbone in empirical force fields. J. Am. Chem. Soc. 2004;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 39.Chemical Computing Group . Chemical Computing Group Inc.; Montreal: 2014. MOE® Version 2014.0901. [Google Scholar]
- 40.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theor. Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 41.Nam K. Acceleration of ab initio QM/MM calculations under periodic boundary conditions by multiscale and multiple time step approaches. J. Chem. Theor. Comput. 2014;10:4175–4183. doi: 10.1021/ct5005643. [DOI] [PubMed] [Google Scholar]
- 42.Liu S., Pi J., Zhang Q. Signal amplification in the KEAP1-NRF2-ARE antioxidant response pathway. Redox Biol. 2022;54 doi: 10.1016/j.redox.2022.102389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patinen T., Adinolfi S., Cortés C.C., Härkönen J., Deen A.J., Levonen A.L. Regulation of stress signaling pathways by protein lipoxidation. Redox Biol. 2019;23 doi: 10.1016/j.redox.2019.101114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghunath A., Sundarraj K., Nagarajan R., Arfuso F., Bian J., Kumar A.P., Sethi G., Perumal E. Antioxidant response elements: discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297–314. doi: 10.1016/j.redox.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moldoveanu B., Otmishi P., Jani P., Walker J., Sarmiento X., Guardiola J., Saad M., Yu J. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2008;2:1–11. doi: 10.2147/jir.S4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahman I., Yang S.R., Biswas S.K. Current concepts of redox signaling in the lungs. Antioxidants Redox Signal. 2006;8:681–689. doi: 10.1089/ars.2006.8.681. [DOI] [PubMed] [Google Scholar]
- 47.Kuipers I., Louis R., Manise M., Dentener M.A., Irvin C.G., Janssen-Heininger Y.M., Brightling C.E., Wouters E.F., Reynaert N.L. Increased glutaredoxin-1 and decreased protein S-glutathionylation in sputum of asthmatics. Eur. Respir. J. 2013;41:469–472. doi: 10.1183/09031936.00115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anathy V., Lahue K.G., Chapman D.G., Chia S.B., Casey D.T., Aboushousha R., Van Der Velden J.L., Elko E., Hoffman S.M., Mcmillan D.H. Reducing protein oxidation reverses lung fibrosis. Nat. Med. 2018;24:1128–1135. doi: 10.1038/s41591-018-0090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Ye Z.W., Singh S., Townsend D.M., Tew K.D. An evolving understanding of the S-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 2018;120:204–216. doi: 10.1016/j.freeradbiomed.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van De Wetering C., Manuel A.M., Sharafi M., Aboushousha R., Qian X., Erickson C., Macpherson M., Chan G., Adcock I.M., Zounematkermani N. Glutathione-S-transferase P promotes glycolysis in asthma in association with oxidation of pyruvate kinase M2. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones J.T., Qian X., Van Der Velden J.L., Chia S.B., Mcmillan D.H., Flemer S., Hoffman S.M., Lahue K.G., Schneider R.W., Nolin J.D. Glutathione S-transferase pi modulates NF-κB activation and pro-inflammatory responses in lung epithelial cells. Redox Biol. 2016;8:375–382. doi: 10.1016/j.redox.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anathy V., Roberson E., Cunniff B., Nolin J.D., Hoffman S., Spiess P., Guala A.S., Lahue K.G., Goldman D., Flemer S. Oxidative processing of latent Fas in the endoplasmic reticulum controls the strength of apoptosis. Mol. Cell Biol. 2012;32:3464–3478. doi: 10.1128/MCB.00125-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matute-Bello G., Frevert C.W., Martin T.R. Animal models of acute lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 2008;295:L379–L399. doi: 10.1152/ajplung.00010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhatia M., Zemans R.L., Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;46:566–572. doi: 10.1165/rcmb.2011-0392tr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abraham E. Neutrophils and acute lung injury. Crit. Care Med. 2003;31:S195–S199. doi: 10.1007/978-3-642-57210-4_15. [DOI] [PubMed] [Google Scholar]
- 56.Carvalho A.N., Marques C., Rodrigues E., Henderson C.J., Wolf C.R., Pereira P., Gama M.J. Ubiquitin–proteasome system impairment and MPTP-induced oxidative stress in the brain of C57BL/6 wild-type and GSTP knockout mice. Mol. Neurobiol. 2013;47:662–672. doi: 10.1007/s12035-012-8368-4. [DOI] [PubMed] [Google Scholar]
- 57.Conklin D.J., Guo Y., Jagatheesan G., Kilfoil P.J., Haberzettl P., Hill B.G., Baba S.P., Guo L., Wetzelberger K., Obal D. Genetic deficiency of glutathione s-transferase p increases myocardial sensitivity to ischemia–reperfusion injury. Circ. Res. 2015;117:437–449. doi: 10.1161/CIRCRESAHA.114.305518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang Z., Snouwaert J.N., Kovarova M., Nguyen M., Repenning P.W., Latour A.M., Cyphert J.M., Koller B.H. Mice lacking three loci encoding 14 glutathione transferase genes: a novel tool for assigning function to the GSTP, GSTM, and GSTT families. Drug Metab. Dispos. 2014;42:1074–1083. doi: 10.1124/dmd.113.056481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van De Wetering C., Elko E., Berg M., Schiffers C.H., Stylianidis V., Van Den Berge M., Nawijn M.C., Wouters E.F., Janssen-Heininger Y.M., Reynaert N.L. Glutathione S-transferases and their implications in the lung diseases asthma and chronic obstructive pulmonary disease: early life susceptibility? Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habig W.H., Pabst M.J., Jakoby W.B. Glutathione S-transferases: the first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. doi: 10.1016/S0021-9258(19)42083-8. [DOI] [PubMed] [Google Scholar]
- 61.Conklin D.J., Haberzettl P., Lesgards J.F., Prough R.A., Srivastava S., Bhatnagar A. Increased sensitivity of glutathione S-transferase P-null mice to cyclophosphamide-induced urinary bladder toxicity. J. Pharmacol. Exp. Therapeut. 2009;331:456–469. doi: 10.1124/jpet.109.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou J., Zheng Q., Chen Z. The Nrf2 pathway in liver diseases. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.826204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen Z., Wang Y., Su Z., Kou R., Xie K., Song F. Activation of p62-keap1-Nrf2 antioxidant pathway in the early stage of acetaminophen-induced acute liver injury in mice. Chem. Biol. Interact. 2018;282:22–28. doi: 10.1016/j.cbi.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 64.Canning P., Cooper C.D., Krojer T., Murray J.W., Pike A.C., Chaikuad A., Keates T., Thangaratnarajah C., Hojzan V., Marsden B.D. Structural basis for Cul 3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J. Biol. Chem. 2013;288:7803–7814. doi: 10.1074/jbc.a112.437996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hourihan J.M., Kenna J.G., Hayes J.D. The gasotransmitter hydrogen sulfide induces nrf2-target genes by inactivating the keap1 ubiquitin ligase substrate adaptor through formation of a disulfide bond between cys-226 and cys-613, Antioxid. Redox Signal. 2013;19:465–481. doi: 10.1089/ars.2012.4944. [DOI] [PubMed] [Google Scholar]
- 66.Saito R., Suzuki T., Hiramoto K., Asami S., Naganuma E., Suda H., Iso T., Yamamoto H., Morita M., Baird L. Characterizations of three major cysteine sensors of Keap1 in stress response. Mol. Cell Biol. 2016;36:271–284. doi: 10.1128/MCB.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takaya K., Suzuki T., Motohashi H., Onodera K., Satomi S., Kensler T.W., Yamamoto M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012;53:817–827. doi: 10.1016/j.freeradbiomed.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eggler A.L., Liu G., Pezzuto J.M., Van Breemen R.B., Mesecar A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong F., Sekhar K.R., Freeman M.L., Liebler D.C. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J. Biol. Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 71.Wang L., Qu G., Gao Y., Su L., Ye Q., Jiang F., Zhao B., Miao J. A small molecule targeting glutathione activates Nrf2 and inhibits cancer cell growth through promoting Keap-1 S-glutathionylation and inducing apoptosis. RSC Adv. 2018;8:792–804. doi: 10.1039/c7ra11935f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 73.Lu M.C., Ji J.A., Jiang Z.Y., You Q.D. The Keap1–Nrf2–ARE pathway as a potential preventive and therapeutic target: an update. Med. Res. Rev. 2016;36:924–963. doi: 10.1002/med.21396. [DOI] [PubMed] [Google Scholar]
- 74.Cheng Y., Cheng L., Gao X., Chen S., Wu P., Wang C., Liu Z. Covalent modification of Keap1 at Cys 77 and Cys 434 by pubescenoside a suppresses oxidative stress-induced NLRP3 inflammasome activation in myocardial ischemia-reperfusion injury. Theranostics. 2021;11:861–877. doi: 10.7150/thno.48436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujii S., Sawa T., Ihara H., Tong K.I., Ida T., Okamoto T., Ahtesham A.K., Ishima Y., Motohashi H., Yamamoto M. The critical role of nitric oxide signaling, via protein S-guanylation and nitrated cyclic GMP, in the antioxidant adaptive response. J. Biol. Chem. 2010;285:23970–23984. doi: 10.1074/jbc.M110.145441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Canning P., Sorrell F.J., Bullock A.N. Structural basis of Keap1 interactions with Nrf2. Free Radic. Biol. Med. 2015;88:101–107. doi: 10.1016/j.freeradbiomed.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Gene Cell. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. https://doi:10.1046/j.1365-2443.2002.00561.x [DOI] [PubMed] [Google Scholar]
- 78.Fukutomi T., Takagi K., Mizushima T., Ohuchi N., Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell Biol. 2014;34:832–846. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tew K.D. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 2016;76:7–9. doi: 10.1158/0008-5472.CAN-15-3143. [DOI] [PubMed] [Google Scholar]
- 80.Yin G., Huang J., Petela J., Jiang H., Zhang Y., Gong S., Wu J., Liu B., Shi J., Gao Y. Targeting small GTPases: emerging grasps on previously untamable targets. pioneered by KRAS. 2023;8:212. doi: 10.1038/s41392-023-01441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.