Abstract
Oesophagostomum spp. (Family: Chabertiidae) is keeping a low profile in terms of severity in Bangladesh while maintaining economic loss through disguise within sheep and goats. The study was performed to identify prevalence, confirmation of species through morphology and morphometry followed by phylogeny using ITS2 and COX1 genes. In total 384 slaughterhouse-sourced small and large intestines were pooled from Mymensingh, Kishoreganj, Netrokona, Sherpur and Tangail districts of Mymensingh division. Followed by isolation, O. columbianum and O. asperum were identified following their key morphological features. Notably, O. asperum was first time detected in Bangladesh. The overall prevalence of Oesophagostomum spp. was found 60.93%. The prevalence of O. columbianum (64.95%) was almost double than that of O. asperum (35.04%). Among several characters, only the distance between anus to tail tip showed a significant morphological disparity in female. The Neighbor-joining (NJ) phylogenic trees based on ITS2 and COX1 genes confirmed the study species. The first time identified O. asperum along with morphometry and phylogeny will add value to the fact that nematodes are invisibly present with high prevalence in this country. This study will help to draw specific attention to command a practical control strategy for intervening in economic loss.
Keywords: Oesophagostomum, ITS2, COX1, Morphology, Morphometry, Phylogeny
1. Introduction
Oesophagostomum or nodular worm is a bursate nematode parasite of large intestine belonging to the family Chabertiidae, a helminth with euryxenous host range starting from ruminants, apes, domestic and wild pigs, monkeys to human (Acevedo-Ramírez et al., 2019, Krief et al., 2010, Legesse and Erko, 2005, McCarthy and Moore, 2000, Stewart et al., 2013, Tariq et al., 2008). Among different Oesophagostomum species, Oesophagostomum columbianum, O. asperum and O. venulosum are the most dominant species in small ruminants around the world (Roeber et al., 2011). A plethora of literatures documented on the prevalence of Oesophagostomum spp. globally including in Central Mexico (4.5 % in sheep), Ethiopia (58.7 % in sheep and 50.8 % in goats), India (82.9 % in goats and 55.4 % in sheep), Cameron (90 % in both species), China (82.2 % in sheep) and Myanmar (Acevedo-Rami rez et al., 2019; Choubisa and Jaroli, 2013, Hou et al., 2022, Negasi et al., 2012, Ntonifor et al., 2013, Win et al., 2020). In Bangladesh, the reported prevalence of GI nematode infection ranged from 63 to 89.8 % and 63–92 % in sheep and goats, respectively (Chakrabortty et al., 2023, Mazid et al., 2006, Mohanta et al., 2007).
Oesophagostomum are geo-helminths and the suitable climatic ambience of Bangladesh paved their way towards high biotic potential with rapid establishment rate (Kusiluka and Kambarage, 1996, Mohanta et al., 2007). Food or soil contaminated with infective larvae (L3) is the principal route of transmission (Sweeny et al., 2012). Due to browsing feeding habit (top browser), goats are more prone to infection than sheep as larvae of this species are positively hydrotropic and available at the tip of grass-blade (Soulsby, 1982). Adult Oesophagostomum causes anemia and rapid weight loss while larvae infection induces black-green diarrhea with mucus and occasionally blood (Zhao et al., 2014).
Diagnosis of Oesophagostomum spp. observing only morphological criteria is not supportive enough as almost all nematodes show uniform outer view. However, application of former criterion along with morphometry and PCR-based tools devise solid and doubtless evidence for specific identification (Dorris et al., 1999, Nath et al., 2021, Wang et al., 2013).
Nowadays, the identification of parasite species and their phylogram analysis are validated through genetic characterization (Cerutti et al., 2010). Besides, several studies accepted that genes of both ribosomal and mitochondrial DNA provide practical knowledge regarding diagnostic probes or species identification markers (Jex et al., 2008, Nath et al., 2021).
As a specific vaccine against Oesophagostomum spp. is yet to be ascertained and commercialized, implementation of effective management strategies is the best solution for now. But, in return, these strategies demand accurate identification of hosts and parasites to the species level (Chilton, 2004). Only a few studies detected O. columbianum in Bangladesh. Still, there is a paucity of research works mentioning the morphological, morphometrical and molecular identification of adult O. asperum. In this study, morphology, morphometry and genetic diversity of adult O. columbianum and O. asperum had been explored along with differentiation between these two species based on the trinity.
2. Methodology
2.1. Sampling area and sampling period
A slaughter-house based experiment was performed in Mymensingh, Kishoreganj, Netrokona, Sherpur and Tangail districts of Mymensingh in a year-long period from December 2021 to November 2022.
2.2. Sampling technique
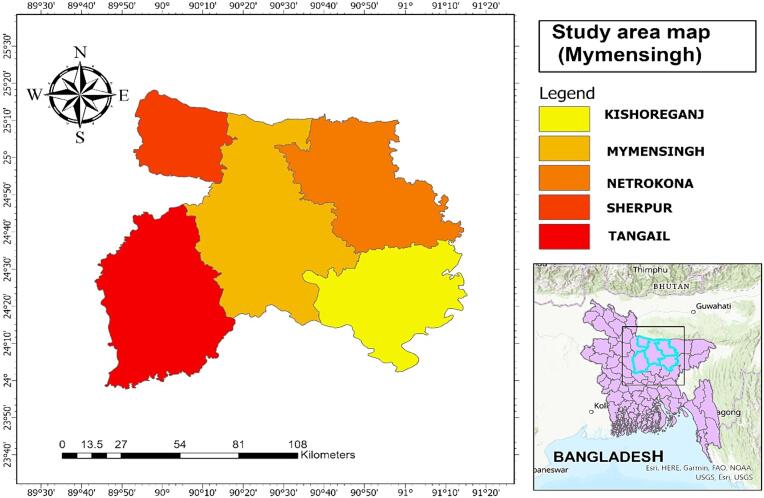
A simple random sampling method was applied to study Oesophagostomum infection in Mymensingh. Sample requirement of this study was determined using the formula generated by Thrusfield (1995). Considering the prospective prevalence of 50 % (P = 0.50) (Chakrabortty et al., 2023) and a precision of 5 % (d = 0.05), 384 samples were obtained from the study areas of Mymensingh (Fig. 1) at 95 % (i.e. 1.96) confidence interval.
Fig. 1.
Map showing the study area. Sheep and goat viscera were collected from several slaughter houses of Kishoreganj, Mymensingh, Netrokona, Sherpur and Tangail districts of Mymensingh division.
2.3. Collection of adult parasites
After collection of viscera particularly small and large intestines from sheep and goats, the samples were shifted into the laboratory and cut opened through long axis by giving longitudinal incision. The contents were processed for examination by simultaneous washing and sedimentation. Finally, the sediments were examined for intended nematode species for further processing.
2.4. Microscopic examination and identification of parasites
After isolation of adult parasites, they were washed multiple times with phosphate buffered saline (PBS) and subjected to morphological identification using lactophenol blue and identified according to keys and description given previously (Bowman, 2014, Gaddam et al., 2017, Singh, 2003, Soulsby, 1982,).
For morphometric analysis, adult nematodes were treated with warm glycerin alcohol to make them straight and convenient for measuring different body features using photomicroscope (Labomed, Los Angeles, USA). For male parasites, length of the spicules and gubernaculum were measured and calculation of the distances between the vulva to the anus, and the anus to the tip of the tail were performed for female. However, measurement of total body length, body width and esophageal length were also done.
2.5. DNA extraction and PCR amplification
The microscopically identified adult parasites were considered for DNA extraction followed by preservation of DNA at −20 °C. The ITS2 region of ribosomal DNA (rDNA) and COX1 region of mitochondrial DNA (mtDNA) were selected and amplified using the primer sets, NC1 and NC2 for ITS2 gene (350 bp) and Nemat F and Nemat R for COX1 gene (720 bp) (Stevenson et al., 1995, Bowles et al., 1992). For both primer pairs, final reaction volume (25 µl) and quantity of primers (1 μl) were exactly the same. PCR conditions for both ITS2 and COX1 genes are presented in Table 1. Extracted DNA was analyzed and examined through a gel documentation system (1.5 % agarose gel).
Table 1.
Primers and their sequences used in the study.
| Target Genes | Primers | Amplicon size | References |
|---|---|---|---|
| ITS2 | NC1 (Forward): ACGTCTGGTTCAGGGTTGTT | 350 bp | Stevenson et al., 1995 |
| NC2 (Reverse):TTAGTTTCTTTTCCTCCGCT | |||
| COX1 | Nemat F: TTTTTTGGGCATCCTGAGGTTTAT | 720 bp | Bowles et al., 1992 |
| Nemat R:TAAAGAAAGAACATAATGAAAATG |
2.6. Sequencing of DNA
The positive PCR products showing clear single band in appropriate size were considered for sequencing by applying suitable primers. To single out the exact species of recovered sequences, the reference sequences were searched out through BLAST program, all of them were aligned and edited using MEGA software (Tamura et al., 2013). The aligned studied sequences were submitted in GenBank database with the accession number of LC777640-LC777643, LC777649-LC777650, LC778201-LC778206 and LC778212-LC778221.
2.7. Genetic data analysis
The genotypes, nucleotide diversity (Nd), haplotypes and haplotype diversity (Hd) of each gene were computed applying the DnaSP v6 program (Rozas, 2009). Pairwise comparisons (%) were performed between the studied genotypes and previously published sequences applying the program Bio-Edit (Hall, 1999). Genetic evaluation was conducted by adopting NJ method grounded on the Tamura-Nei model (Tamura et al., 2013). Besides, for NJ tree, bootstrap support and other contexts were adopted from the values available in MEGA (Tamura et al., 2013) along with a 50 % threshold level. In addition, a total of 29 sequences were retrieved from GenBank (Supplementary file 1).
2.8. Statistical analysis
Prevalence was calculated using GraphPad Prism software (Agresti and Coull, 1998). An independent sample t-test was executed to compare several morphometric characters of O. asperum and O. columbianum using SPSS version 25 (Statistical Package for the Social Sciences).
3. Results
3.1. Prevalence of Oesophagostomum in sheep and goats from Mymensingh
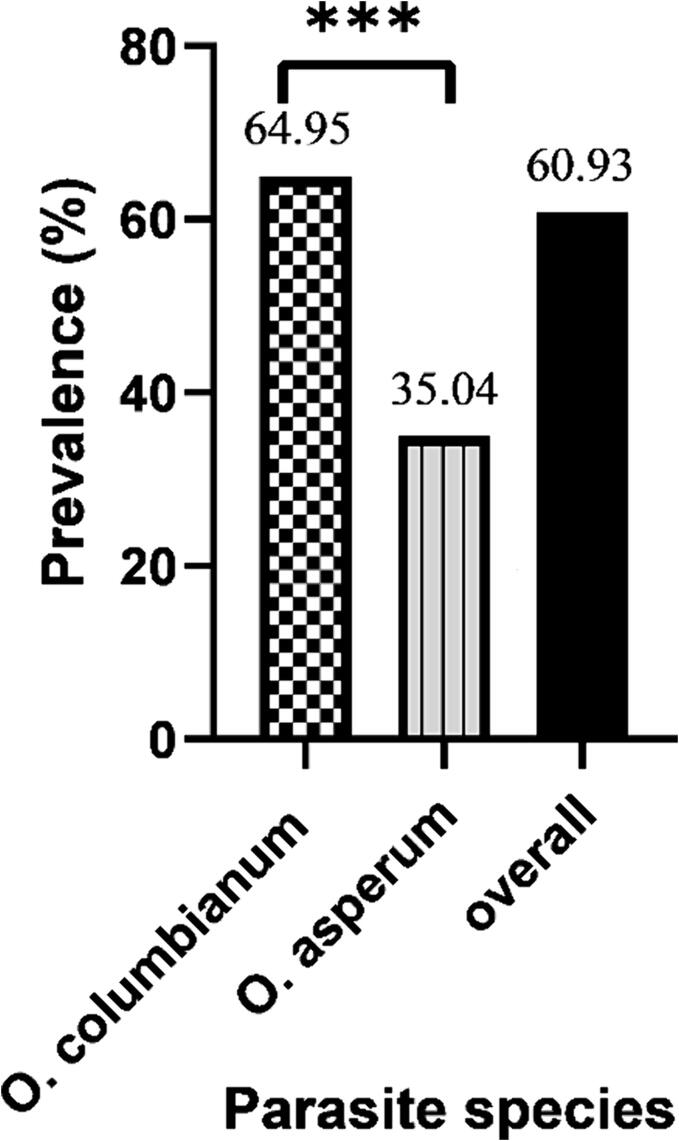
In this study, out of total 384 slaughtered viscera (small and large intestine), 60.93 % (234/384) samples manifested Oesophagostomum spp. infections in sheep (82/234) and goats (152/234). O. columbianum identified in 51 % goat and 27.28 % sheep, respectively. Also, O. asperum showed 32.89 % prevalence in goat and 17.45 % prevalence in sheep. However, irrespective of host species, the prevalence of infection was higher in O. columbianum (64.95 %) compared to O. asperum (35.04 %) (Fig. 2).
Fig. 2.
Prevalence of Oesophagostomum species in Mymensingh. The bar diagram indicates significant difference in the prevalence between O. columbianum and O. asperum. ***, p < 0.0001.
3.2. Morphological features of o. Asperum
Adult male and female O. asperum were dioecious and twinning in color (milky-white) with medium body size. Males were analogously shorter than the female worms. The adult parasite had a typical straight body with much broader anterior end, depicting as a hood and transversely striated cuticle. Buccal capsule was divided into two elements, external and internal corona radiata along with a well-developed and inflated cervical vesicle. A constriction separated the cervical groove from the rest of the body. And further down near the end of the groove, there was a depression, called the excretory canal. In the case of esophagus, they showed typical club-shaped pattern which can be characterized by initial tapering behind the esophageal duct followed by progressive inflation posteriorly (Fig. 3A).
Fig. 3.
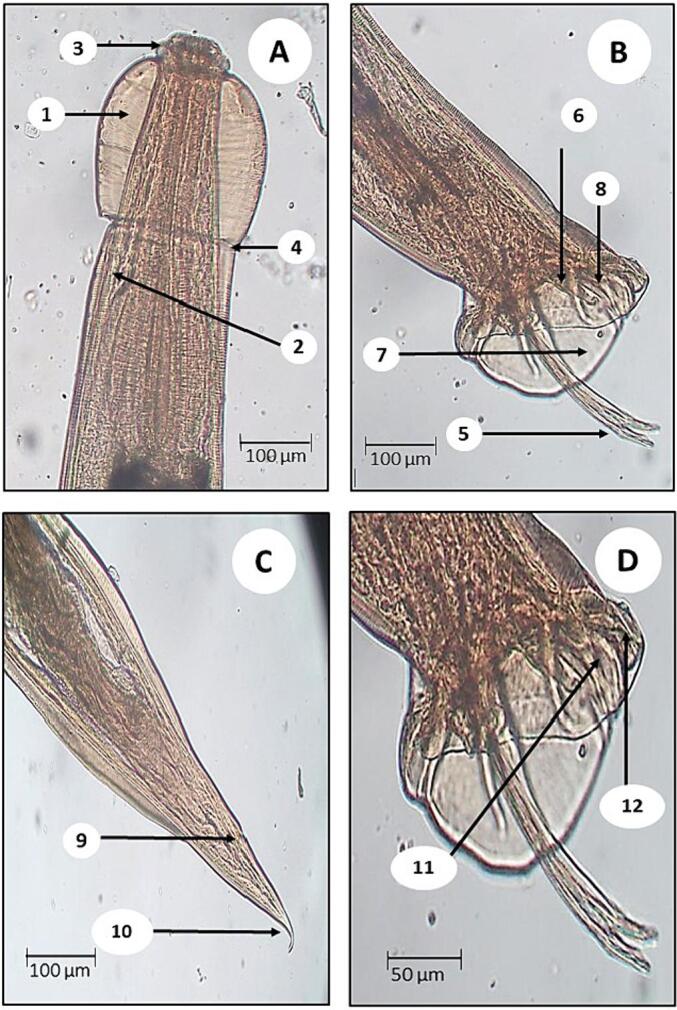
Microscopic identification of O. asperum collected from sheep and goat viscera from Mymensingh. A) Anterior part (both male and female): 1- cervical vesicle, 2- excretory pore, 3- mouth collar, 4- cervical groove; B and D) Posterior part of male: 5- spicule, 6- ventral ray, 7- dorsal lobe of bursa, 8- lateroventral ray, 11- mediolateral ray, 12- posterior-lateral ray; C) Posterior part of female: 9- anus, 10- tail.
Male had a convex-shaped bursa with bursal rays. The medio and posterio-lateral rays were fused proximally, and discernible from the latero-ventral ray. Dorsal rays had 2 short lateral branches along with externo-dorsal ray and spicule. Spicules were found paired and equal (Fig. 3B, 3D).
In female, tail was suddenly tapered from vagina to the tip of the tail. The constriction marked off a conical terminal posterior end where vulva and less prominent anus were located. However, in this nematode, vagina was long and opened into relatively anteriorly placed ovejector (Fig. 3C).
3.3. Morphological features of o. Columbianum
Despite being originated from the same genus, O. columbianum had some morphological disparity with O. asperum. At the anterior end, well-developed striated lateral alae, cephalic vesicle and cervical papillae (Fig. 4A) are those three striking features which display the discreteness of this species. Leaf-like projections known as external and internal corona radiata wrap the margin of small buccal capsule. On top of that, hook-like curvature of the upper portion of the body was also visible. However, inspite of housing several unique characters, inflated cervical vesicle and excretory canal were little to nowhere to be found.
Fig. 4.
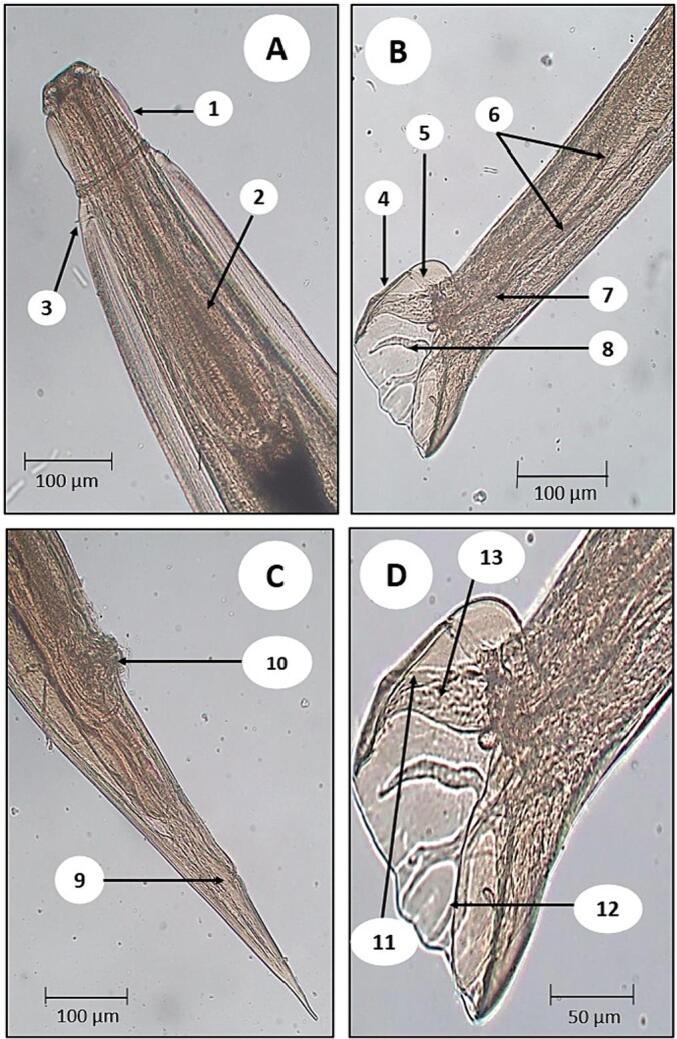
Microscopic identification of O. columbianum collected from sheep and goat viscera from Mymensingh. A) Anterior part (both male and female): 1- cervical vesicle, 2- esophagus, 3- cervical papillae; B and D) Posterior part of male: 4- bursa, 5- ventral ray, 6- spicules, 7- gubernaculum, 8- externo-dorsal ray, 11- mediolateral ray, 12- dorsal ray, 13- posterio-lateral ray; C) Posterior part of female: 9- anus, 10- pars-ejectrix.
In male, O. columbianum endorsed morphological uniformity with O. asperum in parameters like bell-shaped bursa, dorsal rays, lateral rays and spicule (Fig. 4B, 4D).
In female, the tail was gradually diminished along with distinct kidney-shaped pars-ejectrix (Fig. 4C).
3.4. Morphometric measurements of o. Asperum and o. Columbianum
A total of 200 adult parasites (50 male and 50 female for each species) were undergone through morphometry. Except for the length of spicules and gubernaculum for male and distance from vulva to anus for female; numerous morphometric features were common for both sexes. Measurements have been shown in Table 2. Oesophagostomum columbianum evidently stated morphometric deviation from O. asperum. Females displayed longer distances ranging 0.24–1.04 mm from vulva and 0.10–1.46 mm to tail tip depositing anus as the middle point within these two features. While numerous parameters complied similar ranges with O. asperum, length of spicules (0.20–1.44 mm) of O. columbianum showed variation in the sense of elongation (Table 2).
Table 2.
Morphometric measurements of Oesophagostomum species.
| Parameters (millimeter) |
Oesophagostomum asperum |
Oesophagostomum columbianum |
||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| Total body length | 7.60–10.0 | 9.8–13.48 | 6.0–10.0 | 9.0–14.4 |
| Total body width | 0.34–0.38 | 0.42–0.48 | 0.28–0.40 | 0.28–0.50 |
| Length of the esophagus | 0.62–0.64 | 0.60–0.68 | 0.60–0.70 | 0.60–0.76 |
| Length of spicules | 0.90–1.33 | – | 0.20–1.44 | – |
| Length of the gubernaculum | 0.07–0.12 | – | 0.07–0.16 | – |
| Distance from vulva to anus | – | 0.28–0.55 | – | 0.24–1.04 |
| Distance from anus to tail tip | – | 0.10–0.14 | – | 0.10–0.46 |
3.5. Genetic analysis of o. Columbianum and o. Asperum
3.5.1. Species identification and genotyping
For the identification of species, amplification of ITS2 gene was followed by sequencing of 23 representative samples from different areas of Mymensingh. A 320 base pair consensus length was obtained for all the samples. From 23 ITS2 sequences, 6 distinct genotypes were obtained. The sequence similarities were 98.1–99.6 % and 99.6–100 %, when contrasted with one another, or with 2 ITS2 BLAST sequences (1 for each) of O. columbianum (JX188470.1) and O. asperum (KM200806.1), respectively. Following the comparison of 6 study genotypes with the one ITS2 sequence of Oesophagostomum bifurcum (HQ283349.1), the nucleotide identities ranged from 21.2 % to 26.2 % involving both study species (Table 3).
Table 3.
Pairwise identities (%) among 6 ITS2 genotypes of O. columbianum and O. asperum from Mymensingh using reference sequences of different Oesophagostomum spp. from GenBank.
| Sample ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| 1.OA1 | |||||||||
| 2.OA2 | 99.6 | ||||||||
| 3.OC1 | 79.9 | 80.2 | |||||||
| 4.OC2 | 79.9 | 80.2 | 99.3 | ||||||
| 5.OC3 | 79.6 | 79.9 | 99.6 | 99.6 | |||||
| 6.OC4 | 80.2 | 80.5 | 99.6 | 99.0 | 99.3 | ||||
| 7. O. asperum (KM200806.1) | 99.6 | 100 | 80.2 | 80.2 | 79.9 | 80.5 | |||
| 8.O. columbianum (JX188470.1) | 80.2 | 80.5 | 98.7 | 98.1 | 98.4 | 98.4 | 80.5 | ||
| 9.O. bifurcum (HQ283349.1) | 26.2 | 26.2 | 21.6 | 22.2 | 21.9 | 21.2 | 26.2 | 22.5 |
OC, O. columbianum; OA, O.asperum; Black boxes indicate 100%sequence similarity
Four ITS2 genotypes of O. columbianum were aligned with the reference sequence (JX188470.1) and nucleotide positions 68, 106, 121, 156, 157 and 238 displayed polymorphisms, each depicted as single nucleotide polymorphism (SNP). There were two transversions (G<->A) and four transitions (T<->C) among those 6 polymorphisms (Table 4). Besides, O. columbianum also showed the genetic divergence of 0.00343 (nucleotide diversity) and 0.8492 (genotype diversity). Alignment of 2 ITS2 genotypes of O. asperum with the recovered GenBank sequence (KM200806.1) detected only a single nucleotide polymorphism (SNP) at nucleotide position 140 which was a transversion (G<->A). ITS2 sequences of O. asperum showed the nucleotide diversity of 0.00081 along with the genotype diversity of 0.261.
Table 4.
Nucleotide details and distribution of 4 genotypes from 9O. columbianum worms isolated from sheep and goats.
| Genotypes | Nucleotide position |
|||||
|---|---|---|---|---|---|---|
| 68 | 106 | 121 | 156 | 157 | 238 | |
| JX188470.1 | T | G | C | T | G | T |
| OC1 | . | . | . | C | A | C |
| OC2 | C | . | T | C | A | C |
| OC3 | C | . | . | C | A | C |
| OC4 | . | A | . | C | A | C |
OC, O. columbianum; Dot (.) represents similar position with JX188470.1
3.5.2. Phylogenetic analysis using ITS2 gene
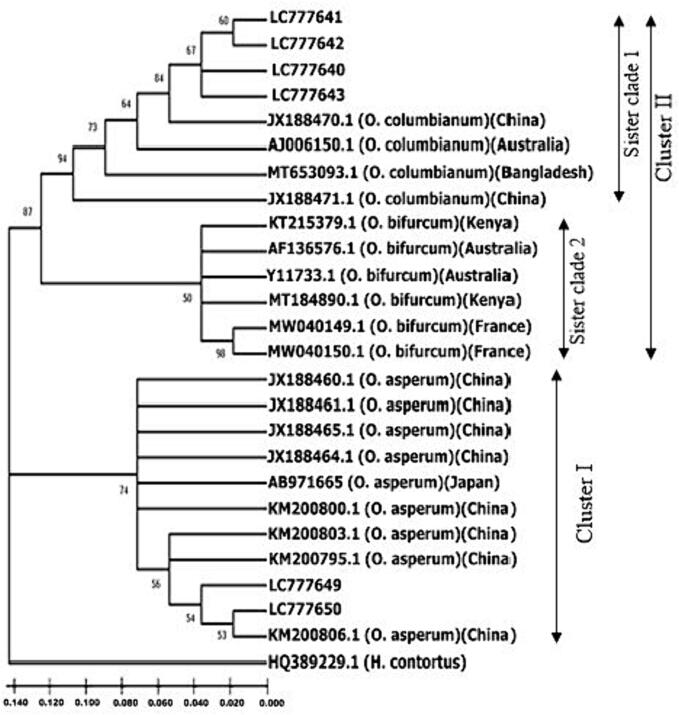
Following the identification of species, the phylogram was created using 6 studied sequences of O. columbianum and O. asperum combining with 19 ITS2 sequences of Oesophagostomum from different countries (Supplementary file 1) where Haemonchus contortus used as out-group (Fig. 5). The analysis separated the NJ phylogram into two main clusters depicting two study species. Cluster I was unique with the studied and reference sequences of O. asperum with strong nodal support (74 %). But cluster II divided into two sister clades where sister clade 1 clustered with the sequences of O. columbianum and sister clade 2 with the sequences of O. bifurcum. The unique clustering pattern of studied species with the reference sequences of O. columbianum and O. asperum confirming the species of the studied sequences.
Fig. 5.
The Neighbor-joining phylogenetic analysis was based on 25 nucleotide sequences of ITS2 gene of O. columbianum and O. asperum. Among them nineteen ITS2 sequences were retrieved from GenBank databases and applied for analysis with six studied sequences (Accession nos. LC777641-LC777643, LC777649-LC777650). Only > 50 % of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown. The sequence of Haemonchus contortus (Accession no. HQ389229.1) was used as out group.
3.5.3. COX1 gene: Genetic diversity
Total 25 COX1 sequences of O. columbianum and O. asperum were analyzed. A 695 base pair gene fragment was considered for making a phylogenetic tree. From the 25 COX1 amplicons, 16 distinct haplotypes were identified (6 from O. columbianum and 10 from O. asperum). For O. columbianum (6 isolates), the nucleotide diversity was 0.00534 and haplotype diversity was 0.8364. Besides, for 10 haplotypes of O. asperum, haplotype diversity was noted 0.9231 with nucleotide diversity, 0.0315.
3.5.4. COX1 gene: Phylogenetic analysis
All recovered sequences were harmonized with other available COX1 reference sequences of both study species with 97 %-100 % similarity.
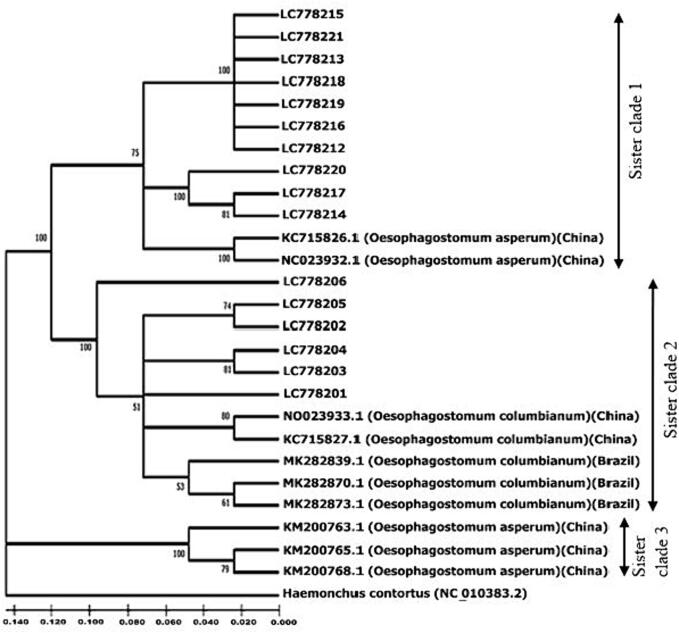
To genetically compare all recovered COX1 sequences from Mymensingh district with foreign countries, a NJ phylogenic tree was constructed. For this, 16 studied haplotype data set of O. columbianum and O. asperum along with 7 sequences from China, 3 from Brazil and as out-group, Haemonchus contortus was employed (Supplementary file 1). Here, two crucial clusters were viewed by bootstrap analysis with 1000 replicates. The large group divided into two sister clades with strong bootstrap support (100 %). Sister clade 1 and 2 uniquely grouped with the studied and reference sequences. But the smaller group (Sister clade 3) of Chinese reference sequences did not grouped with the study sequences. The clade forming nature of studied sequences confirmed species identification. The isolates of O. columbianum from Brazil and China formed monophyletic clade with the studied isolates of O. columbianum (Accession no: LC778201- LC778206) from Bangladesh. Besides, the isolates of O. asperum from China formed cluster with Bangladeshi isolates (LC778212-LC778221). However, another small group is found occupying only Chinese isolates of O. asperum. In this tree, it is evident that, the isolates did not follow any specific relation in terms of host and geographical origin (Fig. 6).
Fig. 6.
The Neighbor-joining phylogenetic analysis was based on 26 nucleotide sequences of COX1 gene of O. columbianum and O. asperum. Only > 50 % of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) was represented in the dendogram. Ten COX1 sequences were retrieved from GenBank databases and used for analysis with sixteen studied sequences (Accession nos. LC778201-LC778206, LC778212-LC778221). The sequence of Haemonchus contortus (Accession no. NC_010383.2) was used as out group.
4. Discussion
Oesophagostomum, a nodular worm inhabiting in small and large intestine of cattle, sheep, pig and primates as a serious pathogen and causing severe interference in absorption of nutrients, movement of bowel and digestion (Soulsby, 1982). This parasite is prevalent worldwide but more commonly in tropical and subtropical areas (Nath et al., 2014, Ntonifor et al., 2013, Win et al., 2020).
The frequency of Oesophagostomum worm encountered in this experiment (60.93 %) is higher but not as much as Mazid et al., (2006) (89.89 %) and Nath et al., (2014) (92 %) documented. Besides, Islam et al., (2017) (15.9 %), Rahman et al., (2017) (10.8 %), Hassan et al., (2011) (10.9–12.9 %) and Dagnachew et al., (2011) (37.6 %) reported comparatively lower frequency. Both of the study species belong to the same genus as well as share common location and food resources which might lead to ‘interspecies competition’. Higher prevalence of O. columbianum may be an indication of triumph over O. asperum. Besides parasite species, there is also variation in prevalence between two host species. As a top browser, goats are more prone to infection (Soulsby, 1982) and in sheep, presence of putative quantitative trait loci (QTL) responsible for resistance against internal nematode infection is already reported (Dominik et al., 2010). Besides, lack of sufficient available data concerning prevalence of only O. asperum in Bangladesh pairing with smaller and scattered study areas might be the reasons behind the variation in prevalence. However, the noticeable contrasts amid the present and previous findings may be as a result of variation in location, climate, sample size, nature and population size of studied animals together with methods of examinations.
This study announced with certainty the morphological elements of O. asperum describing dioecious nature and sexual dimorphism are in agreement with Nath et al., (2021). Unlike O. columbianum, O. asperum depicted inflated cervical vesicle which mimics the explanation of Gaddam et al., (2017) and Singh (2003). However, there was absence of lateral alae in this study which is in coincidence with the statement of Soulsby (1982). Yadav and Tandon (1992) illustrated similar features of this study regarding mouth collar and cervical groove. In male O. asperum, the statement of this study explaining developed bursa along with lateral and dorsal rays expressed similarity with Gaddam et al., (2017) and Singh (2003). Spicules were equal in length just like Soulsby (1982) detected. In female, Gaddam et al., (2017) and Singh (2003) reported that the tail tapered abruptly with shorter distance between vulva and tail tip which is coherent with the present study findings. In addition, the length and position of vagina endorse the descriptions of Singh (2003).
In morphometry of male O. asperum, total body length and esophageal length were found 7.60–10.0 and 0.62–0.64 mm, respectively which are relatively shorter than the features described by Soulsby (1982). Body width, length of the gubernaculum and spicules were recorded 0.34–0.38, 0.07–0.12 and 0.90–1.33 mm, sequentially. In female, only total body width of the study O. asperum (0.42–0.48 mm) were broader than the features illustrated by Soulsby (1982). As per our knowledge, still today, there is scarcity of articles mentioning measurements of other morphometric features of adult O. asperum, namely, esophageal length, distance between vulva to anus and anus to tip of the tail, length of the gubernaculum and spicules.
All adult male and female O. columbianum were pointed out by observing their morphological characters marking both anterior and posterior ends which displayed analogy with the specimens described previously (Gaddam et al., 2017, Nath et al., 2021). Interestingly, anterior end was identical irrespective of sex of the parasites. Even if there was presence of well-developed lateral alae, cephalic vesicle was merely present in the current study. Nath et al., (2021) agreed with the statement while Goodey (1924) and Soota (1981) outlined highly developed cephalic vesicle. Oesophagostomum columbianum was occupied with two pear-shaped cervical papillae just behind the constriction. All of these observations are compatible with the description of Nath et al., (2021) and Zhao et al., (2014). Yadav and Tendon (1992) addressed bending of the anterior end like a hook which also represent one of the findings of the present study. Descriptions of these inquiry namely, moderate sized worm with sexual dimorphism, females being larger than the male, straight body with tapering at both ends are also in agreement with Gaddam et al., (2017) and Nath et al., (2021). Striated cuticle with full-length lateral alae matched with the characteristics reported by Goodey, 1924, Nath et al., 2021 and Zhao et al., (2014). The Scanning Electron Microscopy findings of Yadav and Tandon, 1992, Gaddam et al., 2017 and Soulsby (1982) supported the present study inferences. In male O. columbianum, present study reported dorsal lobe with several ventral and lateral rays. All of these are in coincidence with the findings of Gaddam et al., 2017, Singh, 2003 and Soulsby (1982). Spicules were equal and alate which are identical to the statement of Singh (2003) and Soulsby (1982). In females, anus appeared as a fissure and tail was tapered following gradual reduction manner with slightly prominent vulva. Vagina was shorter which opened posteriorly in a kidney-like structure, Pars-ejectrix which are coherent with Khanmohammadi et al., (2013) and Soulsby (1982).
Unlike anterior end, during morphometry, lower part of O. columbianum showed more versatility and represented several elements appropriate not only for identification but also to sort sexuality. In male, length of the body was found relatively shorter than the reports previously published by Goodey, 1924, Nath et al., 2021 and Soota et al., (1981). Yet, total body width and length of gubernaculum were in harmony with Goodey (1924) and Nath et al., (2021). However, longer esophageal (0.60–0.70 mm) and spicule length (0.07–0.16 mm) were outlined in multiple literatures (Nath et al., 2021, Ransom, 1911; Soota et al., 1981) than those in the present study. In female O. columbianum, higher range of body length (12–18 mm) had been documented in former experiments (Goodey, 1924, Nath et al., 2021; Soota et. al., 1981) compared to this research work (9.0–14.4 mm). Interestingly, total body width and length of the esophagus were also coherent with the results of Nath et al., (2021) and Soota et al., (1981). Besides, distance between vulva to anus and anus to tip of the tail supported the statements of the previously published data (Goodey, 1924, Nath et al., 2021). Both for morphology and morphometry, inspite of having same origin, due to genetic changes over time, variation (based on physical and reproductive parts) may exist between species within the same genus.
Unfortunately, there are several articles where both of the species were studied for morphological analysis without involving any morphometrical comparison between them which justifies lack of referencing data regarding the difference between adult O. asperum and O. columbianum in case of morphometry (Gaddam et al., 2017, Zhao et al., 2013, Zhao et al., 2014).
When the study sequences were compared with reference sequences from China, Japan and Australia from BLAST search, sequence identities ranged from 97 to 100 % (NCBI, 2022). Distinguished morphological characters of these two study species is proved by very high intra specific variation (19.5–20.1 %) between them which is supported by Hu et al., (2014) and Li et al., (2016). Furthermore, slightly higher sequence diversity has also been reported by Jia et al., (2014) and Zhao et al., (2014). In this research work, 0.7–1 % and 0.4 % variation in sequence identities were observed in ITS2 sequences among O. columbianum and O. asperum isolates, respectively. The value of disparity exactly coincides with the variation of reference sequences from China and Brazil (Bacelar et al., 2022, Li et al., 2016, Zhao et al., 2014). However, divergence (0–1.5 %) was also reported among O. asperum isolates from Shaanxi province and Hunan province, China (Li et al., 2016, Yu et al., 2012). Among 23 sequences of ITS2 gene, we determined 6 unique genotypes. Of them, recovered 4 isolates of O. columbianum outnumbered the previously reported articles including a single isolate from Bangladesh and two isolates from China (Nath et al., 2021, Zhao et al., 2013). Among 6 isolates, 2 of them belong to O. asperum but the number varied in several studies experimented with this species including Makouloutou et al., 2014, Yu et al., 2012 and Zhao et al., (2013).
Among several mtDNA, cytochrome oxidase subunit 1 (COX1) gene was more suitable genetic markers than that of nuclear rDNA due to maternal origin and higher substitution rates, thus making it proficient to differentiate among closely connected groups (Anderson et al., 1998, Liu et al., 2012, Troell et al., 2006). Here, the COX1 gene was adopted to ascertain the presence and level of genetic diversity of O. columbianum and O. asperum in Mymensingh. Comparatively lower nucleotide diversity (0.009) has been reported in China (Li et al., 2013) than that of O. columbianum (0.00534) and O. asperum (0.0315) of this study. This variation of nucleotide diversity may be due to scarce availability of relevant sequences in GenBank. Here, 25 COX1 sequences from Mymensingh denoted 16 unique haplotypes with high haplotype diversity (Hd) of both species (O. columbianum: 0.8364 and O. asperum: 0.9231). These significant values are supported by Li et al., (2013) who found nine haplotypes for pCOX1 gene (Hd = 0.81) and fifteen haplotypes for pnad1 gene (Hd = 0.939). High degrees of haplotype diversity have also been documented in other nematode species including Haemonchus contortus isolates originated from Bangladesh (77 haplotypes), Thailand (122 haplotypes) and Pakistan (73 haplotypes) (Dey et al., 2019, Hussain et al., 2014, Pitaksakulrat et al., 2021). Disparity of genetic diversity may resurface when a species undergoes mutations and genetic differentiation over an extended period of time since their time of origin.
As a nematode, Oesophagostomum spp. is difficult to identify and differentiate according to their morphological features. To serve the purposes of detection and distinction, the revolutionary molecular technique has evolved and is accepted worldwide (Yu et al., 2012). The nuclear (ITS2) and mitochondrial (COX1) genes play an effective role in performing accurate genetic analysis of the parasites. The ITS2 gene of rDNA is an important marker in identifying helminths (Králová-Hromadová et al., 2012, Luton et al., 1992, Stevenson et al., 1995, Zhao et al., 2014) because of its easy amplification, enough conserved regions and large variation to discriminate the inter and intra species (Ghobashy and Taeleb, 2015). In the study, within the phylogenetic tree, O. columbianum and O. asperum formed distinct monophyletic clusters with sequences from several countries mostly China, Japan and Australia. Bootstrap support (87 % and 74 %) of O. columbianum and O. asperum expressed noteworthy harmony with Yu et al., (2012) and Zhao et al., (2014).
Besides ITS2, mtDNA is also practiced as a marker because of its greater variation rate and uniparental inheritance (Liu et al., 2012, Nath et al., 2021). The phylogeny of COX1 followed the same pattern of ITS2. COX1 gene sequences also formed a distinctive group of both study parasites and GenBank references of O. columbianum and O. asperum with strong bootstrap value (100 %). This finding is consistent with Zhao et al., (2013). Besides, Bangladeshi isolates of Haemonchus contortus and Mecistocirrus digitatus also formed cluster with China and Japan in dendograms made with ITS2 and nad4 genes (Dey et al., 2019, Parvin et al., 2021). In these phylogenic trees, the reason behind considering sequences from China, Japan and Australia as a sister clade by study sequences might be owing to importation of a great quantity of animals and by products from those countries.
5. Conclusions
Oesophagostomum is highly prevalent and O. columbianum is more common compared to O. asperum in Mymensingh, Bangladesh. Morphology of both species showed distinct morphological features especially in cephalic vesicle and excretory canal at the anterior end as well as shape of spicule in male and visibility of anus in female at the posterior part. In morphometry, female features of O. columbianum showed larger distance ranges than those in O. asperum. Among all the morphometric features of O. asperum female, only the distance between anus to tail tip showed significant difference with O. columbianum female. The phylogenetic analysis of the sequence data belonging to both ITS2 and COX1 genes confirmed two distinct species as O. columbianum and O. asperum while O. asperum was first time detected in Bangladesh.
Author’s contribution
NNS received the thesis grant. NNS and MZA were involved in designing the present study and collection of data. NNS, BCK and MZA performed laboratory experiments. NNS and ARD conducted data analyses. NNS, SA and MZA interpreted the data. NNS, SAR and MZA wrote the draft of the manuscript. All authors checked and approved the final manuscript.
Ethical approval
The present study was approved by Animal Welfare and Experimentation Ethics Committee (AWEEC) of Bangladesh Agricultural University, Mymensingh, Bangladesh (AWEEC/BAU/2021 (49)).
CRediT authorship contribution statement
Nusrat Nowrin Shohana: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review and editing, Formal analysis, Software. Anita Rani Dey: Writing – review and editing, Visualization, Validation, Software. Sharmin Aqter Rony: Methodology, Supervision, Validation. Shirin Akter: Methodology, Writing – review and editing, Supervision. Bimal Chandra Karmakar: Data curation. Mohammad Zahangir Alam: Conceptualization, Funding acquisition, Visualization, Investigation, Supervision, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors greatly acknowledge Bangladesh Agricultural University for funding the research project (2020/957/BAU) and Ministry of Science and Technology, The Government of Bangladesh for National Science and Technology grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2024.103980.
Contributor Information
Nusrat Nowrin Shohana, Email: shohana.vpar@bau.edu.bd.
Anita Rani Dey, Email: anitadey@bau.edu.bd.
Sharmin Aqter Rony, Email: s.a.rony@bau.edu.bd.
Shirin Akter, Email: shirin.akter@bau.edu.bd.
Bimal Chandra Karmakar, Email: bimal.231105901@bau.edu.bd.
Mohammad Zahangir Alam, Email: mzalam@bau.edu.bd.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Acevedo-Ramírez P.M.D.C., Hallal-Calleros C., Flores-Pérez I., Alba-Hurtado F., Mendoza-Garfías M.B., Del Campo N.C., Barajas R. Anthelmintic effect and tissue alterations induced in vitro by hydrolysable tannins on the adult stage of the gastrointestinal nematode Haemonchus contortus. Vet. Parasitol. 2019;266:1–6. doi: 10.1016/j.vetpar.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Agresti A., Coull B.A. Approximate is better than ‘‘exact” for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126. [Google Scholar]
- Anderson T.J., Blouin M.S., Beech R.N. Population biology of parasitic nematodes: applications of genetic markers. Adv. Parasitol. 1998;41:219–283. doi: 10.1016/s0065-308x(08)60425-x. [DOI] [PubMed] [Google Scholar]
- Bacelar P.A.A., Monteiro K.J.L., Calegar D.A., Santos J.P., Coronato-Nunes B., Carvalho-Costa F.A. Cytochrome c oxidase subunit 1 gene reveals species composition and phylogenetic relationships of Oesophagostomum spp. infecting pigs in northeastern Brazil. Rev. Bras. Parasitol. Vet. 2022;31 doi: 10.1590/S1984-29612022016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J., Blair D., McManus D.P. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 1992;54:165–173. doi: 10.1016/0166-6851(92)90109-w. [DOI] [PubMed] [Google Scholar]
- Bowman D.D. 10th ed. Elsevier Health Sciences; 2014. Georgis’ parasitology for veterinarians-e-book. [Google Scholar]
- Cerutti M.C., Citterio C.V., Bazzocchi C., Epis S., D'Amelio S., Lanfranchi P. Genetic variability of Haemonchus contortus (Nematoda: Trichostrongyloidea) in alpine ruminant host species. J. Helminthol. 2010;84:276–283. doi: 10.1017/S0022149X09990587. [DOI] [PubMed] [Google Scholar]
- Chakrabortty M., Shohana N.N., Begum N., Dey A.R., Rony S.A., Akter S., Alam M.Z. Diversity and prevalence of gastrointestinal parasites of Black Bengal goats in Natore. Bangladesh. J. Adv. Vet. Anim. Res. 2023;10:80–87. doi: 10.5455/javar.2023.j655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilton N.B. The use of nuclear ribosomal DNA markers for the identification of bursate nematodes (order Strongylida) and for the diagnosis of infections. Anim. Health Res. Rev. 2004;5:173–187. doi: 10.1079/ahr200497. [DOI] [PubMed] [Google Scholar]
- Choubisa S., Jaroli V. Gastrointestinal parasitic infection in diverse species of domestic ruminants inhabiting tribal rural areas of southern Rajasthan. India. J. Parasit. Dis. 2013;37:271–275. doi: 10.1007/s12639-012-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnachew S., Amamute A., Jemberu W.T. Epidemiology of gastrointestinal helminthiasis of small ruminants in selected sites of North Gondar zone, Northwest Ethiopia. Ethiop. Vet. J. 2011;15:57–68. [Google Scholar]
- Dey A.R., Zhang Z., Begum N., Alim M.A., Hu M., Alam M.Z. Genetic diversity patterns of Haemonchus contortus isolated from sheep and goats in Bangladesh. Infect. Genet. Evol. 2019;68:177–184. doi: 10.1016/j.meegid.2018.12.021. [DOI] [PubMed] [Google Scholar]
- Dominik S., Hunt P.W., Mcnally J., Murrell A., Hall A., Purvis I.W. Detection of quantitative trait loci for internal parasite resistance in sheep. I. Linkage analysis in a Romney×Merino sheep backcross population. Parasitology. 2010;137:1275–1282. doi: 10.1017/S003118201000020X. [DOI] [PubMed] [Google Scholar]
- Dorris M., Ley P.D., Blaxter M.L. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol. Today. 1999;15:188–193. doi: 10.1016/s0169-4758(99)01439-8. [DOI] [PubMed] [Google Scholar]
- Gaddam R., Murthy G.S.S., Kommu S. Occurrence of Oesophagostomum species in slaughtered sheep in area of Hyderabad, Telangana State. J. Parasit. Dis. 2017;41:809–813. doi: 10.1007/s12639-017-0893-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghobashy M.A., Taeleb A.A. Molecular characterization of Raillietina (R.) spp. Ortlepp, 1938 (Cestoda: Cyclophyllidea: Davaineidae) infecting domestic and wild birds (Columba livia and Columba livia domestica) World J. Zool. 2015;10(2):136–141. [Google Scholar]
- Goodey T. Oesophagostome of goats, sheep, and cattle. J. Helminthol. 1924;3:97–110. [Google Scholar]
- Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series [London]: Information Retrieval Limited c1979-c2000 95-98.
- Hassan M.M., Hoque M.A., Islam S.K.M.A., Khan S.A., Roy K., Banu Q. A prevalence of parasites in Black Bengal goats in Chittagong. Bangladesh. Int. J. Livest. Prod. 2011;2:40–44. [Google Scholar]
- Hou B., Yong R., Wuen J., Zhang Y., Buyin B., Subu D., Zha H., Li H., Hasi S. Positivity rate investigation and anthelmintic resistance analysis of gastrointestinal nematodes in sheep and cattle in ordos China. Animals. 2022;12:891. doi: 10.3390/ani12070891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T., Periasamy K., Nadeem A., Babar M.E., Pichler R., Diallo A. Sympatric species distribution, genetic diversity and population structure of Haemonchus isolates from domestic ruminants in Pakistan. Vet. Parasitol. 2014;206:188–199. doi: 10.1016/j.vetpar.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Islam M.S., Hossain M.S., Dey A.R., Alim M.A., Akter S., Alam M.Z. Epidemiology of gastrointestinal parasites of small ruminants in Mymensingh. Bangladesh J. Adv. Vet. Anim. Res. 2017;4:356–362. [Google Scholar]
- Jex A.R., Hu M., Littlewood D.T.J., Waeschenbach A., Gasser R.B. Using 454 technology for long-PCR based sequencing of the complete mitochondrial genome from single Haemonchus contortus (Nematoda) BMC Genomics. 2008;9:11. doi: 10.1186/1471-2164-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y.Q., Zhao G.H., Hu B., Cheng W.Y., Du S.Z., Bian Q.Q., Ma X.T., Yu S.K. Genetic variability in the three mitochondrial genes among Oesophagostomum asperum isolates from different regions in Shaanxi and Hunan Provinces. China. Mitochondrial DNA. 2014;25:298–302. doi: 10.3109/19401736.2013.800508. [DOI] [PubMed] [Google Scholar]
- Khanmohammadi M., Halajian A., Ganji S. First scanning electron microscope observation on adult Oesophagostomum venulosum (rudolphi, 1809) (nematoda: strongylida, chabertiidae) Veterinarija Ir Zootechnika. 2013;62:56–61. [Google Scholar]
- Králová-Hromadová I., Bazsalovicsová E., Oros M., Scholz T. Sequence structure and intragenomic variability of ribosomal ITS2 in monozoic tapeworms of the genus Khawia (Cestoda: Caryophyllidea), parasites of cyprinid fish. Parasitol. Res. 2012;111:1621–1627. doi: 10.1007/s00436-012-3001-z. [DOI] [PubMed] [Google Scholar]
- Krief S., Vermeulen B., Lafosse S., Kasenene J.M., NieguITSila A., Berthelemy M., L‘Hostis M., Bain O., Guillot J. Nodular worm infection in wild chimpanzees in western Uganda: a risk for human health? PLOS Negl. Trop. Dis. 2010;4:e630. doi: 10.1371/journal.pntd.0000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusiluka, L., Kambarage, D., 1996. Diseases of small ruminants: a handbook. In, Common Diseases of sheep and goats in Sub-Saharan Africa. edn. VETAID, Scotland Kuznetsov D, Kuznetsova N 2007: Sequences of the second internal transcribed spacer of ribosomal DNA for three species of Trichostrongylus (Nematoda: Trichostrongylidae from sheep in Russia. Helminthologia. 44, 43-46.
- Legesse M., Erko B. “Prevalence of intestinal parasites among school children in a rural area close to the southeast of Lake Langano, Ethiopia”.Ethiop. J. Health Dev. 2005;18:117–120. [Google Scholar]
- Li F., Hu T., Duan N., Li W., Teng Q., Li H., Cheng T.Y. Sequence variation in two mitochondrial DNA regions and internal transcribed spacer among isolates of the nematode Oesophagostomum asperum originating from goats in Hunan Province. China. J. Helminthol. 2016;90:1–6. doi: 10.1017/S0022149X14000650. [DOI] [PubMed] [Google Scholar]
- Li J.Y., Liu G.H., Wang Y., Song H.Q., Lin R.Q., Zou F.C., Liu W., Xu M.J., Zhu X.Q. Sequence variation in three mitochondrial DNA genes among isolates of Ascaridia galli originating from Guangdong, Hunan and Yunnan provinces. China. J. Helminthol. 2013;87:371–375. doi: 10.1017/S0022149X12000326. [DOI] [PubMed] [Google Scholar]
- Liu G.H., Li B., Li J.Y., Song H.Q., Lin R.Q., Cai X.Q., Zou F.C., Yan H.K., Yuan Z.G., Zhou D.H., Zhu X.Q. Genetic variation among Clonorchis sinensis isolates from different geographic regions in China revealed by sequence analyses of four mitochondrial genes. J. Helminthol. 2012;86:479–484. doi: 10.1017/S0022149X11000757. [DOI] [PubMed] [Google Scholar]
- Luton K., Walker D., Blair D. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea) Mol. Biochem. Parasitol. 1992;56(2):323–327. doi: 10.1016/0166-6851(92)90181-i. [DOI] [PubMed] [Google Scholar]
- Makouloutou P., Nguema P.P., Fujita S., Takenoshita Y., Hasegawa H., Yanagida T., Sato H. Prevalence and genetic diversity of oesophagostomum stephanostomum in wild lowland gorillas at moukalaba-doudou national park. Gabon. Helminthologia. 2014;51:83–93. [Google Scholar]
- Mazid M.A., Bhattacharjee J., Begum N., Rahman M. Helminth parasites of the digestive system of sheep in Mymensingh. Bangladesh. Bangl. J. Vet. Med. 2006;4:117–122. [Google Scholar]
- McCarthy J.S., Moore T.A. Emerging helminth zoonoses. Int. J. Parasitol. 2000;30:1351–1360. doi: 10.1016/s0020-7519(00)00122-3. [DOI] [PubMed] [Google Scholar]
- Mohanta U.K., Anisuzzaman Farjana T., Das P.M., Majumder S., Mondal M.M.H. Prevalence, population dynamics and pathological effects of intestinal helminths in black bengal goats. Bangl. J. Vet. Med. Anim. Health. 2007;5:63–69. [Google Scholar]
- Nath T.C., Bhuiyan M.J.U., Mamun M.A., Datta R., Chowdhury S.K., Hossain M., Alam M.S. Common infectious diseases of goats in chittagong district of Bangladesh. Int. J. Agric. Res. 2014;1:43–49. [Google Scholar]
- Nath T.C., Park D.L.H., Choe S.S., Ndosi B.A., Kang Y., Bia M.M., Eamudomkarn C., Mohanta U.K., Islam K.M., Bhuiyan J.U., Jeon H.K. Morphometrical and molecular characterization of oesophagostomum columbianum (Chabertiidae: Oesophagostominae) and haemonchus contortus (Trichostrongylidae: Haemonchinae) Isolated from Goat (Capra hircus) in Sylhet, Bangladesh. J Parasitol. Res. 2021 doi: 10.1155/2021/8863283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Nucleotide. 2022. National Center for Biotechnology Information. online. Available from: https://www.ncbi.nlm.nih.gov .
- Negasi W., Bogale B., Chanie M. Prevalence, species composition and associated risk factors in and around mekelle town, Northern Ethiopia. Eur. J. Biol. Sci. 2012;4:91–95. [Google Scholar]
- Ntonifor H., Shei S., Ndaleh N., Mbunkur G.N. Epidemiological studies of gastrointestinal parasitic infections in ruminants in Jakiri, Bui Division, North West Region of Cameroon. J. Vet. Med. Anim. Health. 2013;5:344–352. [Google Scholar]
- Parvin S., Dey A.R., Akter S., Anisuzzaman Talukder M.H., Alam M.Z. Molecular identification of mecistocirrus digitatus in cattle from mymensingh region of Bangladesh. J. Bangladesh Agril. Univ. 2021;19(1):67–72. [Google Scholar]
- Pitaksakulrat O., Chaiyasaeng M., Artchayasawat A., Eamudomkarn C., Boonmars T., Kopolrat K.Y., Prasopdee S., Petney T.N., Blair D., Sithithawon P. Genetic diversity and population structure of Haemonchus contortus in goats from Thailand. Infect. Genet. Evol. 2021;95 doi: 10.1016/j.meegid.2021.105021. [DOI] [PubMed] [Google Scholar]
- Rahman M.A., Labony S.S., Dey A.R., Alam M.Z. An epidemiological investigation of gastrointestinal parasites of small ruminants in Tangail. Bangladesh. J. Bangladesh Agric. Univ. 2017;15:255–259. [Google Scholar]
- Ransom, B.H., 1911. The nematodes parasitic in the alimentary tract of cattle, sheep and other ruminants, vol 71, US, Dept of Agric Bureau of Animal btd Bull.
- Roeber F., Jex A.R., Campbell A.J.D., Campbell B.E., Anderson G.A., Gasser R.B. Evaluation and application of a molecular method to assess the composition of strongylid nematode populations in sheep with naturally acquired infections. Infect. Genet. Evol. 2011;11:849–854. doi: 10.1016/j.meegid.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Bioinformatics for DNA Sequence Analysis. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Singh K.S. Veterinary helminthology ICAR. New Delhi. 2003:354–366. [Google Scholar]
- Soota T.D. On some nematodes from the unnamed collection of the zoological survey of India, along with the description of a new species. Rec. Zool. Surv. India. 1981;79:55–71. [Google Scholar]
- Soulsby E.J.L. Helminths, arthropods and protozoa of domesticated animals. Bailliere Tindall, London. 1982;7:186–191. [Google Scholar]
- Stevenson L.A., Chilton N.B., Gasser R. Differentiation of Haemonchus placei from H contortus (Nematoda: Trichostrongylidae) by the Ribosomal DNA Second Internal Transcribed Spacer. Int. J. Parasitol. 1995;25:483–488. doi: 10.1016/0020-7519(94)00156-i. [DOI] [PubMed] [Google Scholar]
- Stewart C.P., Iannotti L., Dewey K.G., Michaelsen K.F., Onyango A.W. Contextualising complementary feeding in a broader framework for stunting prevention. Matern. Child Nutr. 2013:27–45. doi: 10.1111/mcn.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny J.P., Robertson I.D., Ryan U.M., Jacobson C., Woodgate R.G. Impacts of naturally acquired protozoa and strongylid nematode infections on growth and faecal attributes in lambs. Vet. Parasitol. 2012;184:298–308. doi: 10.1016/j.vetpar.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 60. Mol Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq K., Chishti M., Ahmad F., Shawl A.S. Epidemiology of gastrointestinal nematodes of sheep managed under traditional husbandry system in Kashmir valley. Vet. Parasitol. 2008;158:138–143. doi: 10.1016/j.vetpar.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Thrusfield M. 2nd edn. Blackwell Science Ltd; Elsevier, Cambridge: 1995. Veterinary Epidemiology. [Google Scholar]
- Troell K., Engström A., Morrison D.A., Mattsson J.G., Hoglund J. Global patterns reveal strong population structure in Haemonchus contortus, a nematode parasite of domesticated ruminants. Int. J. Parasitol. 2006;36:1305–1316. doi: 10.1016/j.ijpara.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu G.H., Li J.Y., Xu M.J., Ye Y.G., Zhou D.H., Song H.Q., Lin R.Q., Zhu X.Q. Genetic variability among Trichuris ovis isolates from different hosts in Guangdong Province, China revealed by sequences of three mitochondrial genes. Mitochondrial DNA. 2013;24:50–54. doi: 10.3109/19401736.2012.710210. [DOI] [PubMed] [Google Scholar]
- Win S.Y., Win M., Thwin E.P., Htun L.L., Hmoon M.M., Chel H.M., Thaw Y.N., Nyein C.S., Phyo T.T., Thein S.S., Khaing Y., Than A.A., Bawm S. Occurrence of gastrointestinal parasites in small ruminants in the central part of Myanmar. J Parasitol. Res. 2020 doi: 10.1155/2020/8826327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A.K., Tandon V. Stereoscan studies of two species of the genus Oesophagostomum (Nematoda, Chabertiidae) Acta Parasitol. 1992;37(3):135–137. [Google Scholar]
- Yu S.K., Hu B., Deng Y., Li H.M., Ren W.X., Bian Q.Q., Gao M., Wang X.Y., Cong M.M., Song J.K., Lin Q., Xu M.J., Zhao G.H. Phylogenetic studies of Oesophagostomum asperum from goats based on sequences of internal transcribed spacers of ribosomal deoxyribonucleic acid (DNA) Afr. J. Microbiol. Res. 2012;6:3360–3365. [Google Scholar]
- Zhao G.H., Hu B., Cheng W.Y., Jia Y.Q., Li H.M., Yu S.K., Liu G.H. The complete mitochondrial genomes of Oesophagostomum asperum and Oesophagostomum columbianum in small ruminants. Infect. Genet. Evol. 2013;19:205–211. doi: 10.1016/j.meegid.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Zhao G.H., Hu B., Song J.K., Jia Y.Q., Li H.M., Wang C.R., Lin Q., Xu Q.X., Yu S.K., Deng Y. Characterization of Oesophagostomum asperum and O. columbianum by internal transcribed spacers of nuclear ribosomal DNA. J. Helminthol. 2014;88:74–81. doi: 10.1017/S0022149X12000764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.