Abstract
Although the fundamental processes and chemical changes in metabolic programs have been elucidated in many cancers, the expression patterns of metabolism-related genes in head and neck squamous cell carcinoma (HNSCC) remain unclear. The mRNA expression profiles from the Cancer Genome Atlas included 502 tumour and 44 normal samples were extracted. We explored the biological functions and prognosis roles of metabolism-associated genes in patients with HNSCC. The results indicated that patients with HNSCC could be divided into three molecular subtypes (C1, C2 and C3) based on 249 metabolism-related genes. There were markedly different clinical characteristics, prognosis outcomes, and biological functions among the three subtypes. Different molecular subtypes also have different tumour microenvironments and immune infiltration levels. The established prognosis model with 17 signature genes could predict the prognosis of patients with HNSCC and was validated using an independent cohort dataset. An individual risk scoring tool was developed using the risk score and clinical parameters; the risk score was an independent prognostic factor for patients with HNSCC. Different risk stratifications have different clinical characteristics, biological features, tumour microenvironments and immune infiltration levels. Our study could be used for clinical risk management and to help conduct precision medicine for patients with HNSCC.
Keywords: Head and neck squamous cell carcinoma, Metabolism, Immune infiltration, Prognosis model
1. Introduction
According to the latest statistics, head and neck cancer ranks seventh among the cancer-related causes of death worldwide [1]. In 2018, approximately 700,000 new cases of head and neck cancer were reported globally, with approximately 350,000 deaths [2]. Head and neck cancers primarily refer to cancers that occur in the nasal cavity, sinuses, oral cavity, pharynx, and larynx, with more than 90% of cases being squamous cell carcinomas [3]. Despite advancements in comprehensive treatment techniques such as surgery, radiation, and chemotherapy in recent years, 30–40% of patients still experience distant metastasis within 5 years [4]. Research has found that approximately 58% of patients with head and neck squamous cell carcinoma (HNSCC) are diagnosed at advanced stages (stages III-IV) [5]. Therefore, conducting in-depth studies on the factors and mechanisms that affect the prognosis of HNSCC and identifying tumour markers that can be used for molecular diagnosis, prognosis prediction, and targeted therapy are highly significant.
Decomposition and synthesis are two crucial biological processes for tumour cells to maintain their energy supply and product synthesis. An important biological characteristic of tumour cells is the alteration or rewiring of genetically-regulated metabolic processes [6]. Research has shown that tumour cells can increase the synthesis of macromolecules and intermediate products to sustain abnormal cell proliferation, making them more prone than normal tissues to glucose uptake through the glycolysis pathway [7]. Moreover, abnormal metabolism and tumour development have a reciprocal cause-and-effect relationship because changes in the expression levels of certain proteases regulate alterations in metabolic products [8]. For example, some malignant tumours rely primarily on glycolysis for energy production, whereas others exhibit a metabolic phenotype that depends on oxidative phosphorylation [9]. Although the fundamental processes and chemical changes of metabolic programs have been elucidated in many cancers, the expression patterns of metabolism-related genes in HNSCC remain unclear. In this study, we explored the biological functions and prognostic roles of metabolism-associated genes in HNSCC. We explored the molecular subtypes of HNSCC based on metabolism-associated genes, and described their clinical and molecular features. We then developed a prognostic model based on metabolism-associated genes, built a risk evaluation tool, and compared clinical and molecular characteristics between different risk stratifications. This study provides novel insights into the treatment and management of HNSCC.
2. Materials and methods
2.1. Patients and samples
We obtained mRNA expression profiles from the Cancer Genome Atlas, which included 502 tumour and 44 normal samples. Clinical characteristics and follow-up data were also obtained. For tumour, samples without clinical or follow-up data were excluded. Copy number variation data were also downloaded. Raw counts were transformed into transcripts per kilobase million for further analysis. Additional processed microarray data from 270 patients with HNSCC from GSE65858 were used as the validation dataset. We identified the metabolism-related genes from a previously published dataset [10].
2.2. Identification of HNSCC subtypes
We first screened metabolism-related genes using clinical data and median absolute deviation (MAD), followed by univariate Cox regression. We overlapped the data, and the genes with MAD >0.5 and significant associations with prognosis were used for subtype identification. We identified the HNSCC subtypes using the non-negative matrix factorisation clustering [[11], [12]]. A class map and correlation rank helped us select the optimal values of K. Principal component analysis was used to validate the subtypes.
2.3. Gene set variation and enrichment analysis
Gene set variation analysis (GSVA) was performed using the GSVA R package. GSVA can be used to calculate the scores of metabolisms-related genes using nonparametric and unsupervised methods [13]. These pathways were divided into four types: amino acid, carbohydrate, lipid as well as other pathways. We explored the pathway differences among the subtypes. Gene Set enrichment analysis was performed using the GSEABase R package, which is a knowledge-based approach for interpreting genome-wide expression profiles [14]. We used GSEA to explore the pathway enrichment of certain gene sets.
2.4. GO and KEGG analysis
We first performed differential gene expression analysis and identified the differentially expressed genes (DEGs). The DEGs was then used for genes ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analyses, which were performed using the R “clusterProfiler” packages and org.Hs.eg.db dataset [[15], [16]].
2.5. Immune infiltration estimation
We used the CIBERSORT method to evaluate immune infiltration levels. CIBERSORT deconvolutes the expression matrix of human immune cell subtypes based on the linear support vector regression [17]. This algorithm uses microarray data to construct a feature matrix that describes the expression characteristics of 22 immune cell phenotypes, including various cell types and functional states of immune cells. In the CIBERSORT parameters setting, the relative modes were selected, quantile normalization was disabled, 22 immune cell phenotypes were used, and the permutations were 100.
We also used the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE; https://bioinformatics.mdanderson.org/estimate/index.html) tool to evaluate the tumour purity, levels of stromal cells, and infiltration levels of immune cells in tumour tissues based on expression data.
2.6. Establishment and validation of risk prediction model
We adopted the least absolute shrinkage and selection operator (LASSO) penalized Cox regression method to build a prognosis model. A penalty parameter (lambada) was used to construct an optimal prognostic gene set to prevent overfitting. When multiple lambadas appear, those with fewer variables are selected based on partial likelihood deviance. Using cox regression coefficient and gene expression levels, we can calculate the risk score using the following formula: risk score = β(1)*Gene(1)_expression+ … +β(n)*Gene(n)_expression, β is cox regression coefficient [18].Seventeen genes were included the final model, and risk score was calc −0.977*CACNA1A + 0.442*PIGP - 0.422*GLYCTK - 0.357*ABCB8 - 0.239*STAR+0.329*ELOVL6+0.421*HS3ST1+0.262*PNPLA4+0.235*PFKM+0.371*NAT10 + 0.501*ATP6V0E1-0.645*SLC25A45–0.275*CSGALNACT1+0.262*PNPLA4+0.309*HK1+0.297*DAD1. Patients were also divided into two groups based on the median risk score: high- and low-risk. The TCGA dataset was used as the training group, and the GEO dataset was used as the validation dataset (GSE65858; n = 270).
2.7. Clinical relevance and risk evaluation system
We further explored the clinical relevance of the risk score to the clinical characteristics. Univariate and multivariate cox regression were performed to investigate the independence of risk scores. Time-independent receiver operating characteristic curves were obtained to evaluate the predictive ability of the risk model. We also built a nomogram scoring system to evaluate the individual prognostic risk [19]. The nomogram included the risk score and clinical parameters (age, gender, grade, T: tumour N: node, M: metastasis). Calibration plots of the nomogram were used to depict the predictive values between the predicted and observed values, which can assess the stability of the scoring system.
2.8. Statistical analysis
Continuous variables with normal distributions were represented using the mean and standard deviation. Student's t-test was used for stromal, immune, ESTIMATE, between high- and low-risk groups, one-way analysis of variance (ANOVA) was used among multiple groups, and multiple testing was performed using the LSD method including comparisons of stromal, immune and ESTAMATE, risk score among C1, C2 and C3. The Mann-Whitney or Kruskal-Wallis H test was used for immune checkpoint genes expression among three components. Categorical variables were represented as counts and percentages, and chi-square tests were used to compare the two groups (gender, grade, stage, TNM). Kaplan-Meier survival curves were compared using log-rank tests. All analyses were performed using the R software version 4.0 [20]. Differences were considered statistically significant when P values were less than 0.05.
3. Results
3.1. Identification of metabolism-related subtypes in HNSCC
The flowchart of the study is presented in Fig. 1A. We obtained the expression profile data of 502 HNSCC and 44 normal control samples from the dataset, and 499 tumour samples were included in the analyses. We first performed DEGs analysis and identified 318 DEGs between HNSCC and normal samples (|log2 Fold change| (|Log2 FC|)>1, P < 0.05; Table S1 and Fig. S1A). The top 20 genes are presented in Fig. S1B. GO enrichment analysis indicated that these genes were enriched for amino acids, carbohydrate, lipid transport, biosynthetic processes, transporter and channel complex, and transmembrane transporter activities (Fig. S1C). KEGG pathway analyses indicated that DEGs were mainly enriched in glycerophospholipid, purine, carbon, amino, and o-glycan metabolism, and biosynthesis (Fig. S1D). Further details of the GO and KEGG analyses are presented in Tables S2 and S3.
Fig. 1.
Flow of study analyses and identification of HNSCC molecular subtypes using NMF consensus clustering. A: The flow chart of data analysis. B: NMF clustering using 249 metabolism-associated genes. Cophenetic correlation coefficient for K = 2–5 is shown. C: Consensus matrix of three HNSCC subclasses. D: PCA showed the distribution of three HNSCC subclasses. E: Individuals CA of three HNSCC subclasses. F: Kaplan-Meier curves of overall survival among three subclasses (C1, C2 and C3).
Finally, we identified 249 metabolism-related genes from 318 DEGs Using MAD>0.5and P < 0.05 in the univariate Cox regression (Table S4). Non-negative matrix factorisation clustering was performed, and the cophenetic correlation coefficients were calculated to determine the optimal k value. The cophenetic correlation coefficient is the maximum when k = 2. However, we want to explore potential subtypes based on current sample size as many as we can, but also keep subtypes some differences. In our study, k = 3 was eventually chosen as optimal number of clusters based on current sample size instead of k = 2, includingC1, C2, C3 (Fig. 1B and C). PCA and individual PCA confirmed the presence of these three components (Fig. 1D and E). Furthermore, we performed K-M survival analyses and found significant differences among C1, C2, and C3. Patients from C3 had favorable prognosis (Fig. 1F).
3.2. Correlation of the HNSCC subtypes with metabolism-associated signatures
First, we performed DEGs analysis of the three subtypes. Using P < 0.05 and |log2 FC|>2, we identified 4744 DEGs in C1, 6613 DEGs in C2, and 7165 DEGs in C3 (Table S5). There were 1463 overlaps among the three subtypes (Fig. 2A and B). We detected the expression of several key metabolism-related genes among the three subtypes, and found that PCK2, ACLY, HK2, CPT1A, ACSS2, and IDH1 were highest expressed in C1. LDHA and GLS were the highest in C2, whereas IDH2 was the highest in C3 (Fig. 2C). GO functional enrichment and KEGG pathway analyses were performed. Cell -cell adhesion, junction, and binding were mainly enriched in C1. Signal transduction, immune response, ficolin-1-rich granule, and protease and phosphatase binding were enriched in C2. Immune-related signaling and function were mainly enriched in C3 (Fig. S2). KEGG pathway analyses indicated that focal adhesion, cell adhesion, and cell differentiation were enriched in C1, MAPK signaling pathway, cell cycle, apoptosis, p53 signaling pathways, and AGE-RAGE signaling pathways were enriched in C2, while the NF-kappa B, T cell receptor, and some immune-related signaling pathways were mainly enriched in C3 (Fig. S3).
Fig. 2.
Distributions of 249 metabolism-associated genes and key metabolic genes expressions among three HNSCC subclasses. A: Heatmap of correlation of metabolism-associated genes expression among three subclasses and different clinical features. B: Venn diagram showed the overlapped number of DEGs among three subtypes. C: Expression differences of several key metabolic genes among three subtypes (***: adjusted P values).
We performed GSVA for the three subtypes by dividing these pathways into four categories: amino acid, carbohydrate, lipid, as well as other metabolic pathways. The top five differential metabolism pathways in C1 were cholesterol biosynthesis, glyoxylate and dicarboxylate metabolism, ubiquinone and other terpenoid-quinone biosynthesis, propanoate metabolism, and ketone biosynthesis and metabolism. The top five differentially metabolism pathways in the C2 group were glycosaminoglycan biosynthesis, n-glycan biosynthesis, other types of o-glycan biosynthesis, glycan degradation, and ADP-ribosylation. There were no amino acid metabolism-related signaling pathways. Tryptophan metabolism valine, leucine and isoleucine degradation, kynurenine metabolism; taurine and hypotaurine metabolism; and selenocompound metabolism were the main differential signaling pathways (Fig. 3 and Table S6).
Fig. 3.
Heatmap of significantly differential metabolism-associated pathways among three HNSCC subclasses.
3.3. Correlation of HNSCC subtypes with immune infiltration
We then compared the immune infiltration levels among the three components (Fig. 4A and B). The results indicated that the C3 components had more B cells naïve, plasma cells, T cells CD8, T cells follicular helper, T cells regulatory (Tregs), mast cells resting than the C1 and C2 components. The C2 component had higher T cells CD4+memory resting, T cells CD4 memory activated, NK cells resting, macrophages M1 and M2 than C1 and C3. The C1 component had higher macrophages M0 than C2 and C3. The ESTIMATE results indicated that C2 and C3 had higher stromal score than C1 (Fig. 4C). The immune and ESTMATE scores increased with the C1, C2, and C3 (adjusted P values within groups<0.001, Fig. 4D and E). We also evaluated the expression levels of several key immune-related genes among the three components. Significant differential expression was observed among C1, C2 and C3 (Fig. 4F). C2 exhibited higher expression levels of CD276, IL6, CD274, PVR, and TGFB1 than C1 and C3. Expression levels of CCL2, CTLA4, LGALS9, CXCR4, LAG3, CD4, and PDCD1 were higher in C3 than C1 and C2.
Fig. 4.
Comparisons of immune status among three subclasses. A: Heatmap showed the immune cells infiltration levels among three subclasses and different clinical features. B: Box plot comparing the immune cells scale of fraction among three subclasses (***: adjusted P values). C-E: Violin plot shown stromal, immune, and ESTIMATE scores among three subtypes (***: adjusted P values). F: Comparisons of expression levels of several immune checkpoint genes among three subtypes (***: adjusted P values).
3.4. Development and validation of metabolism-related prognosis signature
Next, LASSO regression was performed to select the model genes, and 17 prognosis-related genes were included in the final model (Fig. 5A and B). Among these genes, CACNA1A, PIGP, GLYCTK, ABCB8, STAR, ELOVL6, HS3ST1, PNPLA4, PFKM, NAT10, and ATP6V0E1 were risk factors for patients with HNSCC, whereas SLC25A45, CSGALNACT1, PNPLA4, HK1, and DAD1 were favorable factors for patients with HNSCC (Table S7). We then calculated the risk score of each patient and divided the patients into high- and low-risk groups using the median risk score. The Kaplan-Meier curve indicated that the high-risk group had a poorer prognosis than the low-risk group in training (Fig. 5C) and validation cohorts (Fig. 5D). Finally, we performed the univariate and multivariate Cox regression analyses and identified the risk score was an independent prognostic factor for HNSCC (univariate: HR = 1.360, 95%CI: 1.277–1.448, P < 0.001, Fig. 5E; Multivariate: HR = 1.383, 95%CI: 1.289–1.484, P < 0.001, Fig. 5F). We also validated the prognostic signatures of three subtypes. The risk scores of C1 and C2 were significantly higher than those of C3 (P < 0.001), and there was no significant difference between C1 and C2 (Fig. 6A). Kaplan-Meier analysis also indicated that the high-risk group had a poorer prognosis than the low-risk group for the three different subtypes of patients (Fig. 6B–D).
Fig. 5.
Identification of metabolism-associated genes prognosis model for HNSCC. A: Elastic net solution path of LASSO regression. B: Determination of the number of prognosis-related genes via the LASSO regression. C and D: Overall survival Kaplan-Meier curves of high-risk group and low-risk group for HNSCC patients in TCGA training group and validation group; E and F: Forest plots of univariate and multivariate cox regression analysis exhibited the association between risk score and overall survival in HNSCC patients.
Fig. 6.
Association between risk score and three metabolism components. A: Comparisons of risk score among three subtypes (***: adjusted P values). B-D: Kaplan-Meier analysis curve of high- and low-risk groups in three subtypes.
3.5. Evaluation of prognostic model and individual risk scoring system
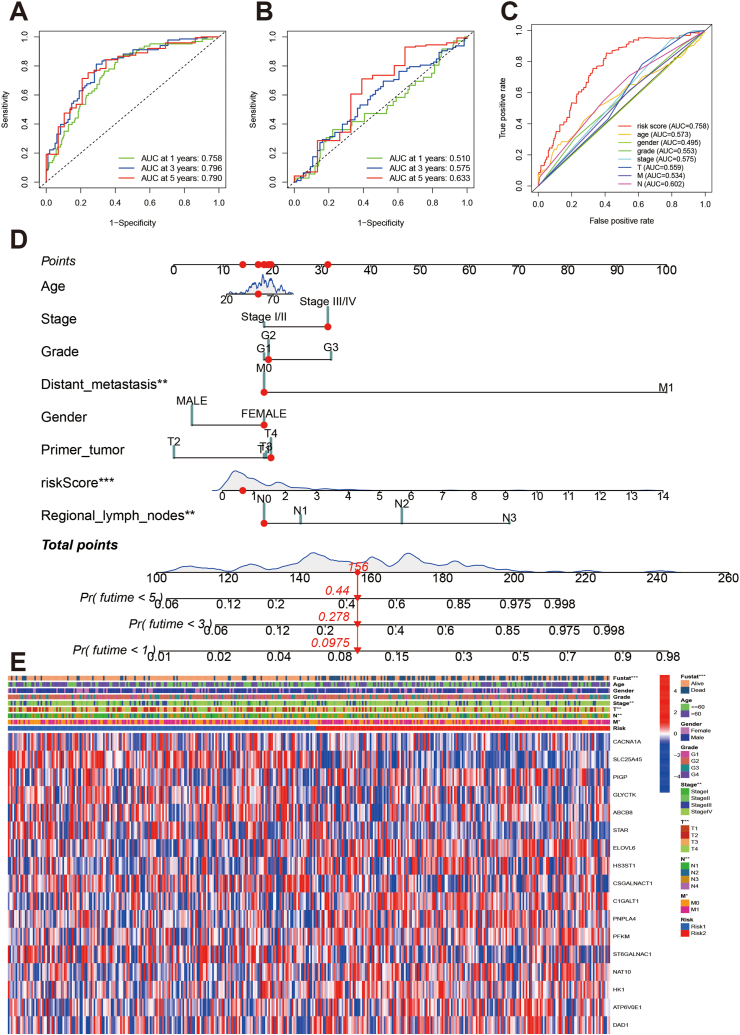
ROC curves were constructed for the training and validation datasets. The AUCs at 1-, 3-, and 5-year were 0.758, 0.796, and 0.790 in the training dataset (Fig. 7A), and 0.510, 0.675, and 0.633 in the validation dataset, respectively (Fig. 7B). We evaluated the predictive ability of the prognostic model using time-independent ROC curves (Fig. 7C). The results indicated that the risk score had the highest predictive ability compared to the other clinical parameters (AUC:0.758). Using the signature genes and clinical parameters, we established an individual risk scoring system (Fig. 7D). Furthermore, we evaluated the association among risk stratification, clinical parameters, and signature gene expression. We found that the high-risk group tended to have more advanced T and N stages, and higher expression of risk genes (Fig. 7E).
Fig. 7.
Evaluation of prognosis model and individual risk scoring system. A and B: ROC curves of prognostic signature in training dataset and validation dataset. C: ROC curves of risk score and other clinical parameters in HNSCC patients. D: Nomograph plot of predicted 1-,3-and 5-year overall survival probability based on metabolism-related genes signature. E: Heatmap showed prognosis signatures genes among clinical features.
3.6. Correlation of HNSCC risk stratification with immune infiltration and mutation
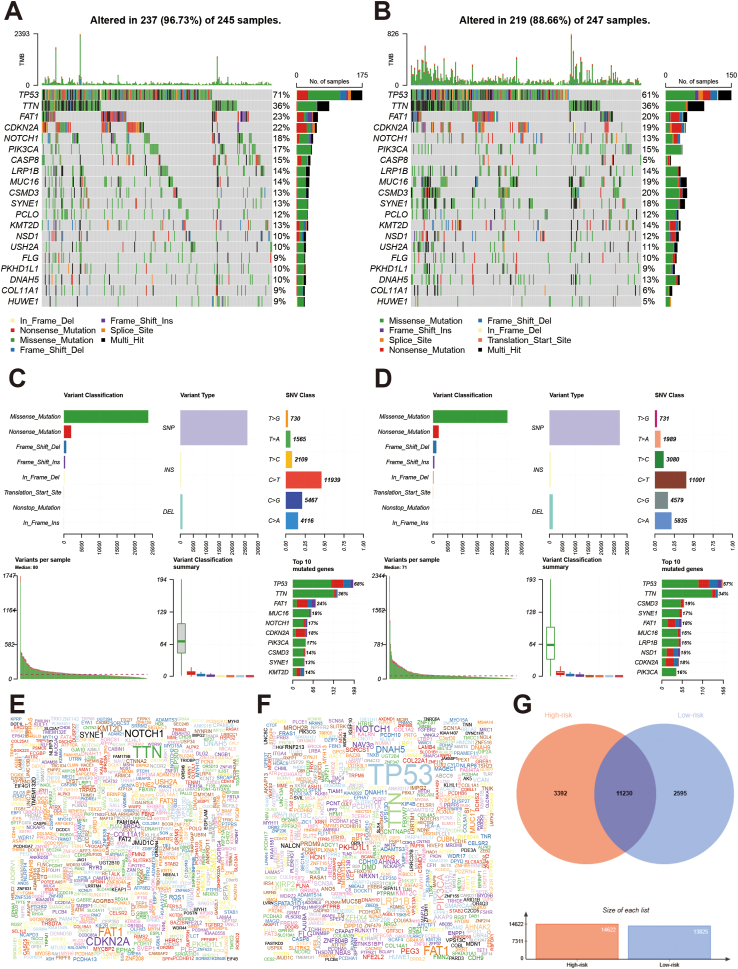
The high-risk group had lower stromal (Fig. 8A), immune (Fig. 8B), and ESTMATE scores (Fig. 8C). Next, we evaluated the immune infiltration levels between the high-risk and low-risk groups (Fig. 8D). The high-risk group had fewer naïve B cells, plasma cells, follicular helper, and T cells regulatory, and more T cells CD4 memory resting, and macrophages M1. Other immune cells showed no significant differences (Fig. 8E). We also compared the gene alterations between the high-risk group and low-risk groups (Fig. 9). Gene alteration levels in the high-risk group were higher than those in the low-risk group (96.73% vs. 88.66%; Fig. 9A and B). Variant classification, variant type, and SNV class showed similar trends. Among the top 10 mutated genes in both the high and low-risk groups, 8 of them are shared, including TP53, TTN, NOTCH1, FAT1, CDKN2A, LRP1B, MUC16, PIK3CA, and CSMD3 (Fig. 9C and D). The top five mutation genes in the high-risk group were TP53, TTN, NOTCH1, CDKN2A, and SYNE1, and those in the low-risk group were TP53, FAT1, NOTCH1, TTN, and PIK3CA (Fig. 9E and F). The Venn plot showed that there 11230 overlapped genes between the high- and low-risk groups (Fig. 9G).
Fig. 8.
Comparisons of immune status between high- and low-risk groups. A-C: Violin plot comparing the stromal, immune, and ESTIMATE scores between high- and low-risk groups. D: Heatmap showed the immune cells distributions between high- and low-risk groups. E: Violin plot comparing the immune cells infiltration levels between high- and low-risk groups.
Fig. 9.
Landscape of mutation profiles between high-and low-risk group in HNSCC patients. A and B: Waterfall plots of mutation information in each sample of high-and low-risk HNSCC patients; C and D: The variant classification and type, SNV class summary of in high-and low-risk groups of HNSCC patients; E and F: Gene cloud plots of mutations frequencies in high-and low-risk groups of HNSCC patients. G: Venn plot of overlapped mutation genes between high-risk and low-risk groups.
4. Discussion
HNSCC is among the most common human malignancies, ranking sixth in the global incidence, with approximately 600,000 new cases and 380,000 deaths annually [1]. At present, the main treatment method for most head and neck cancers is surgical resection combined with radiotherapy and chemotherapy, however, its curative effect is not ideal, many patients eventually experience relapse or distant metastasis, the median survival time of patients with incurable or metastatic recurrence is low, and the long-term survival and quality of life of patients are unsatisfactory [21]. The survival rate of the first 5 years is only about 60%, and only 20–30% for patients at advanced stages [22]. Therefore, it is urgent to improve the cure rate, optimise the treatment, and find a practical prognostic evaluation method for HNSCC.
The present study had the following findings: (1) Patients with HNSCC can be divided into three molecular subtypes (C1, C2, and C3) based on 249 metabolism-related genes. (2) There were markedly different clinical characteristics, prognosis outcomes and biological function among the three subtypes. (3) Different molecular subtypes also have different tumour microenvironments and immune infiltration levels. (4) The established prognosis model with 17 signature genes could predict the prognosis of patients with HNSCC, and this prognosis model was validated using an independent prognosis model. (5) An individual risk-scoring tool was built with the risk score and clinical parameters; the risk score was an independent prognostic factor for HNSCC. (6) Different risk stratification had different clinical characteristics, biological features, tumor microenvironment and immune infiltration. The present study may provide novel insights for the prognosic risk evaluation and clinical treatment of patients with HNSCC.
Currently, classification, grading, and staging are the three most important indicators for evaluating the biological behaviour and diagnosis of tumours, among the latter two are mainly used for evaluating the biological behaviour and prognosis of malignant tumours [23]. In recent decades, owing to breakthroughs in life science and medical technology, the detection of targets related to the clinical application of precision and personalized medicine, including targeted therapy, has not only greatly improved the detection rate of early tumours, but also significantly improved the prognosis of many tumours [24]. The clinical value and significance of traditional tumor classification, grading and staging have also changed to varying degrees. Owing to the great heterogeneity of tumours, with the development of personzlised medicine, traditional clinical classification can no longer meet these needs [[25], [26]]. The molecular classification of HNSCC is helpful for better clinical management.
The established prognosis model with 17 signature genes could predict the prognosis of patients with HNSCC, and this prognosis model was validated using an independent prognosis model. Patients with HNSCC patients were divided into high- and low-risk groups, and the high-risk group had a poorer prognosis. Multiple Cox regression analysis indicated that a high-risk score was an independent prognostic factor for HNSCC. The time-independent curve indicates that the predictive ability of the model is moderate. Previous studies also built some other models of gene sets such as pyroptosis‐related (AUC: 0.676) [27], stemness-related (AUC:0.732) [28], and immune-related gene signatures in HNSCC(AUC:0.626) [29]. Two studies built prognostic models for HNSCC using metabolism-related genes. Du et al. and Qiang et al. developed 7 and 12 metabolism-related gene models for HNSCC, respectively [[30], [31]]. There are some remarkable differences between our study and the previous studies. First, the number used metabolism-related genes differed. The two previous studies included 669 and 1454 metabolism-related genes, whereas our study included 2752 genes. Second, this study provided additional results. Previous studies have developed prognostic model, and analyzed the characteristics of high- and low-risk group. Our study not only developed a prognostic model but also established molecular subtypes based on metabolism-related genes, including amino acid, carbohydrate, and lipid metabolism as well as other metabolic pathways. Third, the developed prognostic model was completely different between our study (17 genes) and previous studies (7 and 12 genes). No overlapped genes were found among the three prognostic models. We also constructed a risk scoring-system for personal survival based on metabolism-related genes, which has not been used in previous studies. Compared with the previous prognostic model, our predictive ability was the highest (0.758 for 5-year survival) based on training set. The AUCs in previous studies based on metabolism-related genes were 0.670 and 0.660 using training set; However, this finding still has some significance. However, there are some similar points that the results showed the high-risk group had low stromal, immune and ESTAMTE scores. The number of immune cells was lower in the high-risk groups.
We also found that different molecular subtypes and risk stratifications had different tumour microenvironment and immune infiltration levels, which may help in carrying out immunotherapy. In fact, for immunotherapy of HNSCC, researchers have further developed compound system therapy based on single cytokine therapy and have achieved good efficacy [32]. Various tumor vaccines, including protein/polypeptide-dendritic cell vaccines, have entered clinical trials [33]. In addition, immunotherapy targeting immune checkpoints such as PD-1 has received considerable attention, and related inhibitory antibodies have been approved for treating recurrent or metastatic HNSCC [[34], [35]]. Immunotherapy is possible for HNSCC.
This study had several limitations. First, although a prognostic model was developed, the validation cohort had a small sample size and the statistical significance level is very close to 0.05. Therefore, validation datasets with larger sample size are required for this model. Second, its predictive ability for survival prognosis of HNSCC is moderate, and its clinical application is restricted. However, the present results provide some clues for evaluating prognosis. Third, in our study, k = 3 was eventually chosen as optimal number of clusters based on current sample size instead of k = 2 to explore potential subtypes based on current sample size as many as we can. However, some noticeable small mini clusters can be found when k = 3 was chosen and, will also need to be tested and re-evaluated in a future larger study. Finally, the biological functions and molecular mechanisms need to be validated in vivo and in vitro. Considering the rise of immunotherapy and its application in HNSCC, future studies should focus on the effect of metabolism on the immune response.
5. Conclusions
In conclusion, metabolism-related genes can be used to identify the molecular subtypes of HNSCC characterized by different clinical and prognostic outcomes, biological functions, and immune infiltration levels. A prognostic model established using metabolism-related genes could predict the prognosis of patients with HNSCC. Multiple functions and signaling pathways are involved in the development of HNSCC. Risk stratification could be associated with immune infiltration, tumor microenvironment, and gene alterations. Our study could be used for clinical risk management and to help conduct precision medicine for patients with HNSCC.
Ethics declarations
The ethics are not applicable because this study is based on publicly available data.
Funding
None.
Data availability statement
All data can be available without any restriction, and these data can be download from TCGA database (https://portal.gdc.cancer.gov/). We also would like to share all data and code details if they had any questions.
CRediT authorship contribution statement
Mengxian Jiang: Writing – original draft. Xiang Gu: Software, Investigation, Formal analysis, Data curation. Yexing Xu: Writing – review & editing, Project administration, Formal analysis, Conceptualization. Jing Wang: Writing – review & editing, Supervision, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27587.
Contributor Information
Yexing Xu, Email: xuyexing1004@163.com.
Jing Wang, Email: 280827451@qq.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
figs3.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. https://10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. https://10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Pai S.I., Westra W.H. Molecular pathology of head and neck cancer: implications for diagnosis, prognosis, and treatment. Annu. Rev. Pathol. 2009;4:49–70. doi: 10.1146/annurev.pathol.4.110807.092158. https://10.1146/annurev.pathol.4.110807.092158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shui C.Y., Li C., Liu W., Cai Y.C., Jiang J., Sun R.H., Zhou Y.Q., Qin G. [Research progress in pathogenesis, treatment and prognosis of HPV positive head and neck squamous cell carcinoma] Zhonghua er bi yan hou tou jing wai ke za zhi. 2018;53:392–396. doi: 10.3760/cma.j.issn.1673-0860.2018.05.014. https://10.3760/cma.j.issn.1673-0860.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 5.Li T., Lu J., Yin Y., Qiu Q. Editorial: prognosis prediction and risk stratification in head and neck cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.1037001. https://10.3389/fonc.2022.1037001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan Y., Li H., Pu W., Chen L., Guo D., Jiang H., He B., Qin S., Wang K., Li N., Feng J., Wen J., Cheng S., Zhang Y., Yang W., Ye D., Lu Z., Huang C., Mei J., Zhang H.F., Gao P., Jiang P., Su S., Sun B., Zhao S.M. Cancer metabolism and tumor microenvironment: fostering each other? Sci. China Life Sci. 2022;65:236–279. doi: 10.1007/s11427-021-1999-2. https://10.1007/s11427-021-1999-2 [DOI] [PubMed] [Google Scholar]
- 7.Vitale I., Manic G., Coussens L.M., Kroemer G., Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metabol. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. https://10.1016/j.cmet.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 8.Bader J.E., Voss K., Rathmell J.C. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol. Cell. 2020;78:1019–1033. doi: 10.1016/j.molcel.2020.05.034. https://10.1016/j.molcel.2020.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Yu J., Huber A., Kryczek I., Wang Z., Jiang L., Li X., Du W., Li G., Wei S., Vatan L., Szeliga W., Chinnaiyan A.M., Green M.D., Cieslik M., Zou W. Metabolism drives macrophage heterogeneity in the tumor microenvironment. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110609. https://10.1016/j.celrep.2022.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Possemato R., Marks K.M., Shaul Y.D., Pacold M.E., Kim D., Birsoy K., Sethumadhavan S., Woo H.K., Jang H.G., Jha A.K., Chen W.W., Barrett F.G., Stransky N., Tsun Z.Y., Cowley G.S., Barretina J., Kalaany N.Y., Hsu P.P., Ottina K., Chan A.M., Yuan B., Garraway L.A., Root D.E., Mino-Kenudson M., Brachtel E.F., Driggers E.M., Sabatini D.M. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. https://10.1038/nature10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J.J., Wang X., Gao X. Non-negative matrix factorization by maximizing correntropy for cancer clustering. BMC Bioinf. 2013;14:107. doi: 10.1186/1471-2105-14-107. https://10.1186/1471-2105-14-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P., Li Y., Li N., Shen L., Li Z. Comprehensive analysis of pyroptosis-associated in molecular classification, immunity and prognostic of glioma. Front. Genet. 2022;12 doi: 10.3389/fgene.2021.781538. https://10.3389/fgene.2021.781538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. https://10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ai H. GSEA-SDBE: a gene selection method for breast cancer classification based on GSEA and analyzing differences in performance metrics. PLoS One. 2022;17 doi: 10.1371/journal.pone.0263171. https://10.1371/journal.pone.0263171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Combes F., Loux V., Vandenbrouck Y. GO enrichment analysis for differential proteomics using ProteoRE. Methods Mol. Biol. 2021;2361:179–196. doi: 10.1007/978-1-0716-1641-3_11. https://10.1007/978-1-0716-1641-3_11 [DOI] [PubMed] [Google Scholar]
- 16.Manyam G., Birerdinc A., Baranova A. KPP: KEGG pathway painter. BMC Syst. Biol. 2015;9(Suppl 2) doi: 10.1186/1752-0509-9-S2-S3. https://10.1186/1752-0509-9-S2-S3 S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B., Khodadoust M.S., Liu C.L., Newman A.M., Alizadeh A.A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. https://10.1007/978-1-4939-7493-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanzhu G., Li N., Li Z., Zhou R., Shen L. Molecular subtypes and prognostic signature of pyroptosis-related lncRNAs in glioma patients. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.779168. https://10.3389/fonc.2022.779168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Li Y., Shen L., Shen L., Li N. Molecular characterization, clinical relevance and immune feature of m7G regulator genes across 33 cancer types. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.981567. https://10.3389/fgene.2022.981567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sipos B., Massingham T., Jordan G.E., Goldman N. PhyloSim - Monte Carlo simulation of sequence evolution in the R statistical computing environment. BMC Bioinf. 2011;12:104. doi: 10.1186/1471-2105-12-104. https://10.1186/1471-2105-12-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel B., Tiwari A.K., Pandey R.K., Singh A.P., Kumar S., Sinha A., Jain S.K., Khattri A. Therapeutic approaches for the treatment of head and neck squamous cell carcinoma-An update on clinical trials. Transl. Oncol. 2022;21 doi: 10.1016/j.tranon.2022.101426. https://10.1016/j.tranon.2022.101426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang E., Lahmi L., Meillan N., Pietta G., Albert S., Maingon P. Treatment strategy for distant synchronous metastatic head and neck squamous cell carcinoma. Curr. Oncol. Rep. 2019;21:102. doi: 10.1007/s11912-019-0856-5. https://10.1007/s11912-019-0856-5 [DOI] [PubMed] [Google Scholar]
- 23.Belbin T.J., Singh B., Barber I., Socci N., Wenig B., Smith R., Prystowsky M.B., Childs G. Molecular classification of head and neck squamous cell carcinoma using cDNA microarrays. Cancer Res. 2002;62:1184–1190. [PubMed] [Google Scholar]
- 24.Li L., Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett. 2016;379:191–197. doi: 10.1016/j.canlet.2015.07.018. https://10.1016/j.canlet.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 25.Oramas D.M., Moran C.A. Thymoma: histologically a heterogenous group of tumors. Semin. Diagn. Pathol. 2022;39:99–104. doi: 10.1053/j.semdp.2021.06.002. https://10.1053/j.semdp.2021.06.002 [DOI] [PubMed] [Google Scholar]
- 26.Tuasha N., Petros B. Heterogeneity of tumors in breast cancer: implications and prospects for prognosis and therapeutics. Sci. Tech. Rep. 2020;2020 doi: 10.1155/2020/4736091. https://10.1155/2020/4736091 (Cairo) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Shen L., Li Y., Shen L., Li N. Identification of pyroptosis‐related gene prognostic signature in head and neck squamous cell carcinoma. Cancer Med.-Us. 2022;11:5129–5144. doi: 10.1002/cam4.4825. https://10.1002/cam4.4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Y., Xu W.B., Ma B., Wang Y. Novel stemness-related gene signature predicting prognosis and indicating a different immune microenvironment in HNSCC. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.822115. https://10.3389/fgene.2022.822115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang P., Li Y., Xu Z., He S. A signature of 17 immune-related gene pairs predicts prognosis and immune status in HNSCC patients. Transl. Oncol. 2021;14 doi: 10.1016/j.tranon.2020.100924. https://10.1016/j.tranon.2020.100924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du K., Zou J., Wang B., Liu C., Khan M., Xie T., Huang X., Shen P., Tian Y., Yuan Y. A metabolism-related gene prognostic index bridging metabolic signatures and antitumor immune cycling in head and neck squamous cell carcinoma. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.857934. https://10.3389/fimmu.2022.857934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiang W., Dai Y., Xing X., Sun X. Identification of a metabolic reprogramming-related signature associated with prognosis and immune microenvironment of head and neck squamous cell carcinoma by in silico analysis. Cancer Med. 2022;11:3168–3181. doi: 10.1002/cam4.4670. https://10.1002/cam4.4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Witzleben A., Wang C., Laban S., Savelyeva N., Ottensmeier C.H. HNSCC: tumour antigens and their targeting by immunotherapy. Cells-Basel. 2020;9 doi: 10.3390/cells9092103. https://10.3390/cells9092103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Przybylski K., Majchrzak E., Weselik L., Golusinski W. Immunotherapy of head and neck squamous cell carcinoma (HNSCC). Immune checkpoint blockade. Otolaryngol. Pol. 2018;72:10–16. doi: 10.5604/01.3001.0012.4367. https://10.5604/01.3001.0012.4367 [DOI] [PubMed] [Google Scholar]
- 34.Economopoulou P., Perisanidis C., Giotakis E.I., Psyrri A. The emerging role of immunotherapy in head and neck squamous cell carcinoma (HNSCC): anti-tumor immunity and clinical applications. Ann. Transl. Med. 2016;4:173. doi: 10.21037/atm.2016.03.34. https://10.21037/atm.2016.03.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puntigam L.K., Jeske S.S., Gotz M., Greiner J., Laban S., Theodoraki M.N., Doescher J., Weissinger S.E., Brunner C., Hoffmann T.K., Schuler P.J. Immune checkpoint expression on immune cells of HNSCC patients and modulation by chemo- and immunotherapy. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21155181. https://10.3390/ijms21155181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data can be available without any restriction, and these data can be download from TCGA database (https://portal.gdc.cancer.gov/). We also would like to share all data and code details if they had any questions.