Abstract
Objective
Millions of Americans are infected by influenza annually. A minority seek care in the emergency department (ED) and, of those, only a limited number experience severe disease or death. ED clinicians must distinguish those at risk for deterioration from those who can be safely discharged.
Methods
We developed random forest machine learning (ML) models to estimate needs for critical care within 24 h and inpatient care within 72 h in ED patients with influenza. Predictor data were limited to those recorded prior to ED disposition decision: demographics, ED complaint, medical problems, vital signs, supplemental oxygen use, and laboratory results. Our study population was comprised of adults diagnosed with influenza at one of five EDs in our university health system between January 1, 2017 and May 18, 2022; visits were divided into two cohorts to facilitate model development and validation. Prediction performance was assessed by the area under the receiver operating characteristic curve (AUC) and the Brier score.
Results
Among 8032 patients with laboratory‐confirmed influenza, incidence of critical care needs was 6.3% and incidence of inpatient care needs was 19.6%. The most common reasons for ED visit were symptoms of respiratory tract infection, fever, and shortness of breath. Model AUCs were 0.89 (95% CI 0.86–0.93) for prediction of critical care and 0.90 (95% CI 0.88–0.93) for inpatient care needs; Brier scores were 0.026 and 0.042, respectively. Importantpredictors included shortness of breath, increasing respiratory rate, and a high number of comorbid diseases.
Conclusions
ML methods can be used to accurately predict clinical deterioration in ED patients with influenza and have potential to support ED disposition decision‐making.
1. INTRODUCTION
1.1. Background and importance
The Centers for Disease Control and Prevention estimates that 9.3–45 million Americans experienced symptoms of influenza infection each year between 2010 and 2020. Most infections were mild and self‐limited, but a minority progressed to severe disease and death. Annual influenza hospitalizations ranged from 140,000 to 590,000 and 12,000–52,000 patients died from influenza‐related complications. 1 These numbers declined during the COVID‐19 pandemic, but are expected to return to similar or higher levels in coming years. 2
In the United States, emergency departments (EDs) are the primary gateway to hospital‐based care and treat up to a million patients with influenza each year. 3 , 4 , 5 Emergency clinicians must differentiate patients who can be safely discharged from those who require hospitalization, and those who require intensive monitoring in critical care settings. 3 , 4 These decisions are made within hours of presentation and can be challenging due to time constraints and limited information availability. Suboptimal ED disposition decision‐making has been linked to increased mortality as well as excess healthcare expenditures. 6
The electronic health record (EHR) is a valuable source for clinical insight generation. 7 Machine learning (ML) algorithms, trained on large EHR datasets, can identify subtle patterns and predict future outcomes. 8 , 9 , 10 Recent studies suggest ML‐based clinical decision support (CDS) may be useful for clinical decision‐making in the ED, including at the point of disposition. 11 , 12 , 13 , 14 Many ML models for COVID‐19 outcome prediction have been described. 14 , 15 Influenza has been a leading cause of past pandemics and remains a major threat for the future, yet has not received the same level of attention. 16 , 17 As an NIH Center of Excellence for Influenza Research and Response, we sought to determine whether ML methods could be leveraged to support management of severe influenza.
1.2. Goals of this investigation
Our primary objective was to derive and validate a series of ML models that reliably predict severe clinical outcomes in ED patients with influenza. We hypothesized that short‐term inpatient and critical care needs could be accurately predicted using routinely available EHR data, and designed ML models to support ED disposition decision‐making at the point‐of‐care. Because degradation of predictive performance is a potential barrier to the real‐world implementation of ML‐driven CDS, a key secondary objective of this study was to assess the stability of ML prediction models over time (multiple flu seasons) and location (across clinical sites). 18 This was achieved by employing and evaluating two distinct approaches to model derivation, out‐of‐sample testing and external validation.
2. METHODS
2.1. Setting and selection of participants
This retrospective cohort analysis was performed using data collected at five EDs within a university‐based health system between January 1, 2017 and May 18, 2022. Study sites included two urban academic EDs (Johns Hopkins Hospital [JHH] and Bayview Medical Center [BMC]) and three suburban community EDs (Howard County General Hospital [HCGH], Suburban Hospital [SH], and Sibley Memorial Hospital [SMH]) with a combined patient volume of 270,000 ED visits per year. All adult patients (≥18 years old) who tested positive for influenza during their index ED encounter were included in the study. Patients who tested negative for influenza or who were not tested were excluded from analysis.
2.2. Methods of measurement
Outcome and predictor data were extracted from the EHR (Epic, Verona, WI). Prediction time‐points were set as the time of disposition order entry (eg, discharge or hospitalization orders) for each patient. To be included in analysis, predictor data had to be recorded and available in the EHR prior to the time of prediction. Data elements included patient demographics (age, sex), chief complaint(s), active medical problems (identified based on ICD‐10 codes), vital signs, routine laboratory results, markers of inflammation (c‐reactive protein, d‐dimer, ferritin), and respiratory support requirements. Additional description of influenza testing and predictor processing procedures is included in a Supplementary Appendix. 19 , 24
2.3. Outcome definitions
The primary outcomes predicted were critical care needs and inpatient care needs within 24 and 72 h of ED disposition, respectively. Outcome definitions were developed by multidisciplinary (emergency medicine, internal medicine, and critical care) consensus as previously described and operationally defined. 14 In brief, outcome criteria were designed to capture events (eg, administration of supplemental oxygen) and physiologic states (eg, hypoxemia) reflective of a need for medical care provided in a hospital ward or critical care unit, as applicable. ED disposition decision‐making may be influenced by social factors (eg, unsafe living conditions) as well, but such considerations were not included in our models. 25 As described below, hospitalized patients were required to meet predefined physiologic parameters for cardiopulmonary dysfunction to meet inpatient care needs outcome criteria. Patients discharged without meeting outcome criteria before reaching 24 or 72 h were assumed to be outcome negative.
Criteria for critical care needs were met if a patient died, was admitted to an intermediate or intensive care unit (ICU), or developed cardiovascular or respiratory failure within 24 h of ED disposition. Cardiovascular failure was defined by hypotension requiring intravenous vasopressor support (dopamine, epinephrine, norepinephrine, phenylephrine, or vasopressin). Respiratory failure was defined by hypoxemia or hypercarbia requiring high‐flow oxygen (>10 L/min), high‐flow nasal canula, noninvasive positive pressure ventilation, or invasive mechanical ventilation. 26
Criteria for inpatient care needs were met if patients exhibited at least moderate cardiovascular dysfunction (systolic blood pressure < 80 mmHg, heart rate ≥ 125 for ≥ 30 min or any troponin measurement > 99th percentile), respiratory dysfunction (respiratory rate ≥ 24, hypoxemia with documented SpO2 < 88%, or administration of supplemental oxygen at a rate > 2 L/min sustained for ≥30 min) or were discharged at initial ED visit and had a return ED visit and hospitalization within 72 h.
2.4. Model derivation and validation
For ML models designed to predict clinical outcomes, performance can be affected by evolution of disease pathology and changes in therapeutic approach over time, as well as by differences in local epidemiology, resource availability and clinical practice between sites. Two approaches to model derivation and external validation were used to measure the effects of time and practice location on out‐of‐sample performance separately. Under the first approach (temporal validation), the entire cohort was divided into two datasets by time. Models were trained and tested in a derivation cohort that comprised the earliest two out of three of ED encounters (January 1, 2017 to December 21, 2019) and validated in a cohort that comprised the latest one out of three of ED encounters (December 22, 2019 to May 18, 2022). Under the second approach (spatial validation), the cohort was divided into two datasets based on site of care. Models were trained and tested in a derivation cohort comprised of all ED visits to JHH, HCGH, and SMH during our study period and were validated using ED visit data from BMC and SH only.
The Bottom Line
A machine learning model was developed to estimate the need for critical care and inpatient care in patients presenting to the ED with influenza. The study model has the potential to support ED disposition decision making but may need further training and validation to be used outside the study setting.
Under each approach, separate ensemble‐based decision tree learning algorithms (random forest) were trained to predict each outcome (critical care needs within 24 h, inpatient care needs within 72 h). During training and testing, the random forest algorithm executed a randomized sampling process to train a set of individual decision trees and aggregated output to produce a single probabilistic prediction for each outcome. 27 To maximize opportunity for algorithmic learning, all encounters by influenza positive patients were included in training datasets, including those where patients met criteria for the outcome of interest prior to the prediction timepoint (i.e., time of disposition order entry by treating ED clinician). However, to prevent overestimation of model performance, we excluded patients who met outcome criteria at the time of prediction during model performance evaluation. This exclusion was applied to derivation cohort testing sets and external validation cohorts.
Model performance was measured using receiver operating characteristic (ROC) curve analysis, with confidence intervals and difference between performance determined using DeLong's method. 28 In derivation cohorts, performance was tested out‐of‐sample across all encounters using 10‐fold cross‐validation; training and testing was repeated 10 times to minimize the variance in prediction performance estimations. 29 Within each repetition, the cohort was randomly distributed into training (90% of data) and testing (10% of data). The models learned from the training data set, and their performance was assessed on the test data set, iteratively. During each iteration, isotonic regression was applied to achieve model calibration, thus optimizing the accuracy of individual model estimates across the full range of predicted probabilities. 30 For both derivation and validation cohorts, performance was reported for models that used all predictor data as well as for more parsimonious model versions. Specifically, we evaluated a model (model 1) that only included predictors available at ED triage (demographics, triage vitals, chief complaint, active medical problems); a second model (model 2) that included everything from model 1 as well as incorporated ED laboratory results; and finally, a third model (model 3) that included everything from model 2 as well as ED oxygen requirements, last vital signs measured prior to disposition decision and vital sign trends. Overall goodness of fit (Brier Score) and calibration curves (plots of observed versus predicted risk) were evaluated. 31 , 32 Model interpretation was performed using feature importance measures including SHapley Additive exPlanations (SHAP) values to assess predictor impact and directionality. 33 All model building and statistical analyses were performed using Python 3.6.
2.5. Ethical review
This research was performed under the approval of the Johns Hopkins Medicine Institutional Review Board (IRB00185078).
3. RESULTS
A total of 8032 patients with laboratory‐confirmed influenza infection were included in this study (Table 1). Females comprised 57.4% (n = 4614) of our cohort and most patients self‐identified as either non‐Latino Black or non‐Latino White (Table 1). The most common reasons for ED visit (e.g., chief complaint) were symptoms of upper or lower respiratory tract infection, fever, and shortness of breath (Table 1). Across the cohort, incidence of critical care needs within 24 h was 6.3% (n = 502) with 2.2% (n = 176) meeting criteria at the time of ED disposition order entry and 4.1% (n = 326) meeting criteria within 24 h of disposition. Incidence of inpatient care needs within 72 h was 19.6% (n = 1579), with 11.6% (n = 935) meeting criteria at the time of ED disposition and 8.0% (n = 644) meeting criteria within 72 h of disposition (Table 1).
TABLE 1.
Study cohort characteristics.
| Total | Spatial method | Temporal method | |||
|---|---|---|---|---|---|
| Derivation cohort | Validation cohort | Derivation cohort | Validation cohort | ||
| Total visits | 8032 | 5087 | 2945 | 5352 | 2680 |
| Age, N (%) | |||||
| 18–44 years | 3379 (42.1) | 2338 (46.0) | 1041 (35.3) | 1910 (35.7) | 1469 (54.8) |
| 45–64 years | 2311 (28.8) | 1441 (28.3) | 870 (29.5) | 1594 (29.8) | 717 (26.8) |
| 65–74 years | 978 (12.2) | 554 (10.9) | 424 (14.4) | 742 (13.9) | 236 (8.8) |
| >74 years | 1364 (17.0) | 754 (14.8) | 610 (20.7) | 1106 (20.7) | 258 (9.6) |
| Gender, N (%) | |||||
| Female | 4614 (57.4) | 2949 (58.0) | 1665 (56.5) | 3076 (57.5) | 1538 (57.4) |
| Race/ethnicity, N (%) | |||||
| Black non‐Latino | 3096 (38.5) | 2347 (46.1) | 749 (25.4) | 1950 (36.4) | 1146 (42.8) |
| White non‐Latino | 3154 (39.3) | 1726 (33.9) | 1428 (48.5) | 2306 (43.1) | 848 (31.6) |
| Latino | 910 (11.3) | 441 (8.7) | 469 (15.9) | 502 (9.4) | 408 (15.2) |
| Other | 872 (10.9) | 573 (11.3) | 299 (10.2) | 594 (11.1) | 278 (10.4) |
| Chief complaint, N (%) | |||||
| Upper respiratory infection symptoms | 2134 (26.6) | 1588 (31.2) | 546 (18.5) | 1270 (23.7) | 864 (32.2) |
| Fever | 1873 (23.3) | 1046 (20.6) | 827 (28.1) | 1288 (24.1) | 585 (21.8) |
| Lower respiratory infection symptoms | 1472 (18.3) | 828 (16.3) | 644 (21.9) | 1013 (18.9) | 459 (17.1) |
| Shortness of breath | 1332 (16.6) | 770 (15.1) | 562 (19.1) | 984 (18.4) | 348 (13.0) |
| Chest pain | 485 (6.0) | 311 (6.1) | 174 (5.9) | 332 (6.2) | 153 (5.7) |
| Comorbidities, N (%) | |||||
| Atrial fibrillation | 336 (4.2) | 203 (4.0) | 133 (4.5) | 264 (4.9) | 72 (2.7) |
| Coronary artery disease | 395 (4.9) | 219 (4.3) | 176 (6.0) | 305 (5.7) | 90 (3.4) |
| Cancer | 552 (6.9) | 366 (7.2) | 186 (6.3) | 418 (7.8) | 134 (5.0) |
| Cerebrovascular disease | 292 (3.6) | 219 (4.3) | 73 (2.5) | 216 (4.0) | 76 (2.8) |
| Diabetes | 653 (8.1) | 405 (8.0) | 248 (8.4) | 474 (8.9) | 179 (6.7) |
| Heart failure | 410 (5.1) | 242 (4.8) | 168 (5.7) | 318 (5.9) | 92 (3.4) |
| Hypertension | 1146 (14.3) | 688 (13.5) | 458 (15.6) | 835 (15.6) | 311 (11.6) |
| Immunosuppression | 550 (6.8) | 404 (7.9) | 146 (5.0) | 396 (7.4) | 154 (5.7) |
| Kidney disease | 554 (6.9) | 348 (6.8) | 206 (7.0) | 418 (7.8) | 136 (5.1) |
| Liver disease | 812 (10.1) | 493 (9.7) | 319 (10.8) | 570 (10.7) | 242 (9.0) |
| Pregnancy | 216 (2.7) | 149 (2.9) | 67 (2.3) | 107 (2.0) | 109 (4.1) |
| Prior respiratory failure | 220 (2.7) | 146 (2.9) | 74 (2.5) | 160 (3.0) | 60 (2.2) |
| Smoker | 318 (4.0) | 168 (3.3) | 150 (5.1) | 232 (4.3) | 86 (3.2) |
| Vital signs, mean (95% CI) | |||||
| Temperature | 99.2 (97.2–102.7) | 99.2 (97.2–102.7) | 99.2 (97.3–102.7) | 99.2 (97.3–102.7) | 99.2 (97.0–102.7) |
| Heart rate | 90.6 (60.0–125.0) | 91.2 (60.0–125.0) | 89.7 (59.0–126.0) | 90.1 (59.0–125.0) | 91.7 (61.0–126.0) |
| Respiratory rate | 18.6 (14.0–28.0) | 18.2 (14.0–26.0) | 19.3 (14.0–32.0) | 18.7 (14.0–29.0) | 18.3 (14.0–27.0) |
| Oxygen saturation | 97.0 (92.0–100.0) | 97.2 (92.0–100.0) | 96.7 (91.0–100.0) | 96.9 (91.0–100.0) | 97.3 (92.0–100.0) |
| Systolic blood pressure | 128.1 (95.0–177.0) | 127.1 (96.0–173.0) | 129.8 (94.0–180.0) | 128.8 (95.0–177.0) | 126.7 (94.5–175.0) |
| Labs, N tested, mean (95% CI) | |||||
| Absolute lymphocyte count, K/cu mm | 596, 2.2 (0.2–5.0) | 365, 1.7 (0.2–4.7) | 231, 3.0 (0.2–8.2) | 439, 2.4 (0.2–4.8) | 157, 1.7 (0.2–5.1) |
| Alanine aminotransferase, U/L | 5747, 31.6 (8.0–105.0) | 3385, 31.4 (8.0–101.0) | 2362, 31.8 (9.0–112.0) | 4054, 30.6 (8.0–98.7) | 1693, 34.1 (8.0–123.4) |
| Aspartate aminotransferase, U/L | 5166, 36.2 (13.0–113.0) | 3063, 36.6 (14.0–112.4) | 2103, 35.5 (12.0–118.0) | 3649, 35.0 (13.0–110.4) | 1517, 38.9 (13.0–118.1) |
| Bilirubin, mg/dL | 5853, 0.5 (0.0–1.6) | 3481, 0.5 (0.0–1.6) | 2372, 0.5 (0.0–1.6) | 4109, 0.5 (0.0–1.6) | 1744, 0.5 (0.0–1.6) |
| Blood urea nitrogen, mg/dL | 6098, 16.0 (5.0–49.0) | 3675, 15.8 (5.0–49.0) | 2423, 16.2 (5.0–49.4) | 4287, 16.6 (5.0–51.0) | 1811, 14.5 (5.0–43.0) |
| Creatinine, mg/dL | 6097, 1.2 (0.5–3.3) | 3675, 1.2 (0.5–3.4) | 2422, 1.1 (0.5–3.1) | 4286, 1.2 (0.5–3.6) | 1811, 1.1 (0.5–2.9) |
| D‐Dimer, mg/L | 203, 1.0 (0.0–4.0) | 119, 1.0 (0.0–3.1) | 84, 1.2 (0.0–4.0) | 117, 1.2 (0.0–7.0) | 86, 0.8 (0.0–2.3) |
| International normalized ratio | 1063, 1.3 (0.9–2.9) | 409, 1.3 (0.9–3.2) | 654, 1.2 (0.9–2.5) | 774, 1.3 (0.9–3.1) | 289, 1.2 (0.9–2.6) |
| Lactate, mmol/L | 2019, 1.6 (0.7–3.7) | 1507, 1.5 (0.7–3.4) | 512, 1.7 (0.7–4.2) | 1533, 1.6 (0.7–3.8) | 486, 1.5 (0.7–3.4) |
| Platelets, K/cu mm | 6411, 205.8 (93.0–364.0) | 3927, 207.1 (98.0–366.0) | 2484, 203.8 (89.0–360.9) | 4524, 202.3 (90.1–359.0) | 1887, 214.2 (103.2–375.0) |
| Partial thromboplastin time, s | 557, 11.8 (0.8–42.5) | 163, 25.8 (0.8–52.6) | 394, 6.0 (0.9–37.8) | 420, 10.8 (0.8–43.0) | 137, 14.8 (0.9–38.2) |
| Troponin, N tested, N positive (%) | 2593, 314 (3.9) | 1219, 136 (2.7) | 1374, 178 (6.0) | 1826, 240 (4.5) | 767, 74 (2.8) |
| White blood cell count, K/cu mm | 6477, 8.1 (0.0–20.1) | 3969, 7.8 (0.0–20.6) | 2508, 8.5 (0.0–19.6) | 4563, 8.4 (0.0–21.0) | 1914, 7.3 (0.0–18.9) |
| Oxygen requirements, N (%) | |||||
| Low‐flow oxygen, <2 L/min | 501 (6.2) | 278 (5.5) | 223 (7.6) | 373 (7.0) | 128 (4.8) |
| Mid‐flow oxygen, 2–9 L/min | 333 (4.1) | 174 (3.4) | 159 (5.4) | 253 (4.7) | 80 (3.0) |
| High‐flow oxygen, >10 L/min | 171 (2.1) | 83 (1.6) | 88 (3.0) | 118 (2.2) | 53 (2.0) |
| Emergency department disposition, N (%) | |||||
| Discharged | 5686 (70.8) | 3676 (72.3) | 2010 (68.3) | 3621 (67.7) | 2065 (77.1) |
| Hospitalized | 2345 (29.2) | 1410 (27.7) | 935 (31.7) | 1731 (32.3) | 614 (22.9) |
| Intensive or intermediate care | 328 (4.1) | 226 (4.4) | 102 (3.5) | 234 (4.4) | 94 (3.5) |
| Critical care outcome | |||||
| Criteria met at time of disposition | 176 (2.2) | 85 (1.7) | 91 (3.1) | 119 (2.2) | 57 (2.1) |
| Criteria met after disposition | 326 (4.1) | 205 (4.0) | 121 (4.1) | 251 (4.7) | 75 (2.8) |
| Inpatient care outcome | |||||
| Criteria met at time of disposition | 935 (11.6) | 437 (8.6) | 498 (16.9) | 690 (12.9) | 245 (9.1) |
| Criteria met after disposition | 644 (8.0) | 360 (7.1) | 284 (9.6) | 502 (9.4) | 142 (5.3) |
Study population flow diagrams for both approaches to model derivation and validation are detailed in Figure 1. Under the spatial approach to external validation (Figure 1A), 5087 ED encounters from JHH, HCGH, and SMH that occurred across the entire study period were included in our derivation cohort. All encounter data were used for model training, while 85 (1.7%) and 437 (8.6%) visits were excluded from critical care and inpatient care testing sets due to meeting outcome criteria at the point of prediction (Figure 1B and Table 1). A cohort of 2945 ED visits to separate hospitals (BMC and SH) during the same period was utilized for external validation, with performance of critical and inpatient care needs prediction models measured in the 2854 (96.9%) and 2447 (83.1%) visits who had not met outcome criteria at the point of prediction, respectively. Under the temporal approach (Figure 1A), all ED encounters that occurred at any study site between January 1, 2017 and December 21, 2019 (n = 5352) were included in our model derivation cohort and ED encounters that occurred between January 22, 2019 and May 1, 2022 (n = 2680) were included in our external validation cohort. Two percent of encounters from each cohort were excluded from derivation testing set and validation cohort for meeting critical care outcome criteria prior to time of prediction, while 12.9 and 9.1% were excluded from the inpatient care testing set and validation cohort, respectively (Figure 1B and Table 1).
FIGURE 1.
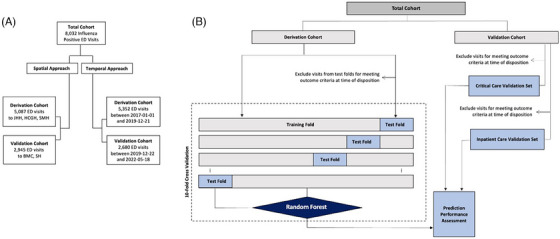
Study flow chart. (A) Two parallel approaches to model derivation and external validation were employed. Under the spatial approach, models were derived and cross‐validated (out of sample) using data from encounters at three study sites, then underwent external validation at two separate study sites. Under the temporal approach, models derived were derived and cross‐validated using data from encounters at all sites that occurred on or before December 21, 2019, then externally validated using data from encounters that occurred afterward. (B) Under both approaches, all encounters were used for model training, but patients who met outcome criteria prior to the prediction timepoint (disposition order entry) were excluded from model performance evaluation. These exclusions were applied during out‐of‐sample testing in the derivation cohorts and in the validation cohorts.
Predictive accuracy of all models, as measured by AUC, is shown in Table 2. Performance was high under all conditions, but overall accuracy during external validation was highest using a comprehensive model that included demographics, ED chief complaint, active medical problems, first and last vital signs with trends, ED oxygen requirements, and laboratory results (model 3). As shown in Table 2, no performance degradation for prediction of critical care needs was observed between derivation and validation cohorts regardless of whether a spatial or temporal approach to model derivation and validation was pursued, with comprehensive model 3 achieving similar AUCs during external validation in both scenarios (0.90, 95% CI 0.87–0.92 under the spatial approach and 0.89, 95% CI 0.86–0.93 under the temporal approach).
TABLE 2.
Prediction model performance.
| Critical care outcome | Inpatient care outcome | |||
|---|---|---|---|---|
| Derivation cohort | Validation cohort | Derivation cohort | Validation cohort | |
| Spatial method | ||||
| Model 1 | 0.86(0.84–0.88) | 0.89(0.86–0.91) | 0.86(0.84–0.88) | 0.83(0.81–0.86) |
| Model 2 | 0.89(0.87–0.91) | 0.89(0.87–0.92) | 0.88(0.87–0.90) | 0.86(0.83–0.88) |
| Model 3 | 0.90(0.89–0.92) | 0.9(0.87–0.92) | 0.90(0.88–0.91) | 0.86(0.84–0.89) |
| Temporal method | ||||
| Model 1 | 0.87(0.84–0.89) | 0.87(0.83–0.91) | 0.85(0.83–0.86) | 0.86(0.83–0.88) |
| Model 2 | 0.88(0.86–0.90) | 0.90(0.87–0.93) | 0.87(0.86–0.89) | 0.88(0.85–0.90) |
| Model 3 | 0.89(0.86–0.91) | 0.89(0.86–0.93) | 0.89(0.87–0.90) | 0.90(0.88–0.93) |
For each method, three models of increasing complexity were tested. Model 1 included predictors available at emergency department (ED) triage only (demographics, triage vitals, chief complaint, active medical problems). ED laboratory results were added to model 2. ED oxygen requirements, last vital signs measured prior to disposition decision and vital sign trends were added to model 3. Model prediction performance is shown as area under the receiver operating characteristic curve (AUC) with 95% confidence intervals in parentheses.
Accuracy of prediction for inpatient care needs was also preserved during external validation under the temporal approach (AUC 0.90, 95% CI 0.88–0.93). However, some performance degradation was observed for prediction of inpatient care needs under the spatial approach, when a model derived and tested out‐of‐sample using data from JHH, HCGH, and SMH hospitals (AUC 0.90, 95% CI 0.88–0.91) was evaluated in a contemporaneous cohort from BMC and SH hospitals (AUC 0.86, 95% CI 0.84–0.89).
Based on superior prediction performance and consistent stability during external validation, comprehensive models (“model 3”) developed under the temporal approach were selected for further analysis. Prediction performance of these models during external validation is visualized in Figure 2. ROC curves that correspond to data included in Table 22 and demonstrate overall prediction accuracy are shown in panels 2A and 2B and calibration curves in panels 2C and 2D. Critical and inpatient care prediction models were both well calibrated, with Brier scores of 0.026 (95% CI 0.021–0.031) and 0.042 (95% CI 0.036–0.048), respectively. As demonstrated in kernel density estimation plots, predicted probabilities of critical care and inpatient care outcomes were low for most of the population (Figures 2E and F). Binary performance metrics including sensitivity, specificity, predictive values, and likelihood ratios across a wide range of operating points are shown for both models in Figure S1. If optimized toward specificity, with a single cut‐off set to achieve 95% specificity while maintaining 53% sensitivity, patients labeled “at‐risk” by our critical care model would have a positive likelihood ratio of 11.3 for manifestation of critical care needs within 24 h. Similar operating point selection for our inpatient care needs model would generate a positive likelihood ratio of 9.7. While Figure S1 provides insight into model performance using a single cut‐off, it is more likely that real‐world decision support would function best as a scale with risk levels (e.g., 1–10) assigned across a range of prediction probabilities. Development of decision support software for these models under a user‐centered design framework is an important future aim.
FIGURE 2.
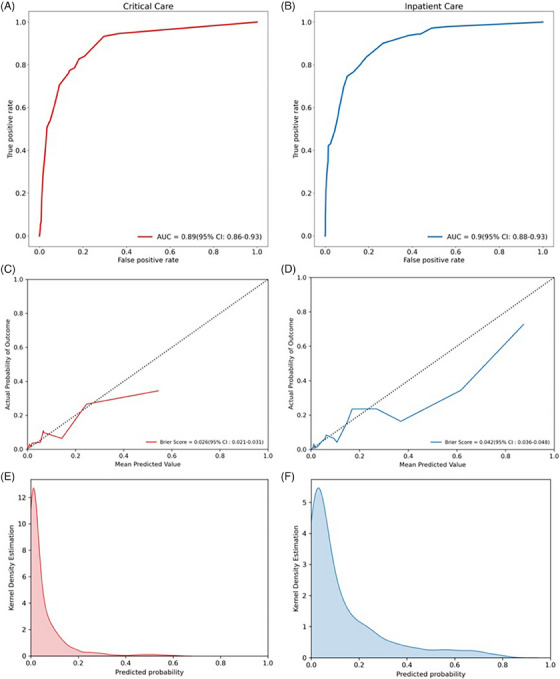
Final model performance. Receiver operating characteristics curves with area under the curve (AUC) for critical care (red) and inpatient care (blue) prediction models are shown in panels A and B, respectively; model calibration curves with Brier scores are shown in panels C and D; distribution of predicted probabilities across the cohort is shown using kernel density estimation plots in panels E and F.
Individual predictor importance, determined by SHAP analysis and demonstrated using bee swarm plots, is shown in Figure 3. Markers of respiratory dysfunction were important predictors of adverse outcomes in both models. Interestingly, patients who complained of upper respiratory symptoms at arrival were less likely to develop either outcome. Respiratory rate trend was the most important predictor for each model, with rising respiratory rates portending high likelihood for the development of severe illness and declining respiratory rates predictive of good outcome. Several laboratory derangements, including elevated lactate, leukocytosis, and coagulopathy, were predictive of both critical and inpatient care needs. Only blood urea nitrogen exhibited bimodal predictive properties, with high values predictive of adverse outcome and normal values predictive of their absence. Elevated troponin was highly predictive of critical outcome but was not a leading predictor of inpatient care needs. The only comorbidity with high predictive value in isolation was liver disease, but the presence of numerous comorbid diseases were highly predictive of both severe disease outcomes.
FIGURE 3.
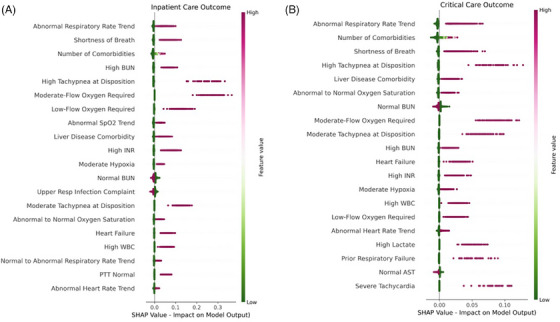
Shapley additive explanations (SHAP) values for (A) inpatient care and (B) critical care prediction models. The 20 most important features for each predictive model are shown using bee swarm plots, with color representing original feature value, kernel density representing relative frequency of feature values in the cohort, and location on x‐axis representing impact on model output. AST, aspartate aminotransferase; BUN, blood urea nitrogen; INR, international normalized ration; PTT, activated partial thromboplastin time; SpO2, peripheral capillary oxygen saturation; WBC, white blood cell count.
4. LIMITATIONS
This study has limitations. Most importantly, models were developed and evaluated using data from a single health system. While we sought to mitigate this limitation through inclusion of five diverse sites across the region, its potential impacts on our conclusions should be considered. Events that occurred outside our health system, including return ED visits by discharged patients, are not captured in our dataset. Differences in local population characteristics and clinical practice patterns, including influenza diagnostic testing procedures, could also affect performance and utility. For example, our institution is an NIH Center of Excellence for Influenza Research and Response; we frequently test for influenza infection (see Table S1). Our models were developed for patients with confirmed influenza and may not be useful at sites where influenza testing is not commonly performed. Prediction performance should be evaluated in local populations prior to use of these models at any external site.
There are additional limitations related to our inclusion and outcome criteria. First, the cohort used to develop our models was heterogeneous and included adults of varying age, race, ethnicity, and baseline health status, but did not include children. Future studies to determine whether similar data and methods could be used to predict severe influenza outcomes in pediatric populations are needed. Second, we required confirmation of influenza infection, but made no differentiation based on genetic strain. It is possible that inclusion of strain as a predictor would improve model performance, but was excluded because this information is not routinely available in real time. Third, our models were designed to predict clinical outcomes and would not identify patients who require hospitalization for social reasons.
Finally, although we have shown that we can make early and reliable predictions of clinically meaningful events, we have not shown that these predictions out‐perform clinician gestalt or would lead to any change in patient management or outcomes. Studying these questions under a prospective interventional framework is an important future aim of our team.
5. DISCUSSION
We leveraged artificial intelligence to estimate short‐term risk for severe disease in ED patients with influenza. ML predictions were made using information routinely available in the EHR, populated within hours of hospital arrival. Our final models were well calibrated and accurately predicted the development of disease states that would necessitate inpatient care within 72 h and ICU‐level care within 24 h. Output from these models could be a useful adjunct to ED clinicians and staff, allowing them to anticipate the clinical course of individual patients more accurately and to make more informed treatment and healthcare resource allocation decisions. Optimal use requires incorporation into a system capable of real‐time EHR data processing and provision of specific, actionable decision support at the point of care. 34 This is an important future aim of our research group, and one we have achieved for other conditions. 14 , 35
Artificial intelligence has been used in the study of influenza previously. Its most popular application has been forecasting of seasonal influenza at the population level. 36 , 37 , 38 , 39 , 40 Many have used artificial intelligence‐based methods to forecast the arrival and intensity of seasonal influenza using social media and widely used search engines such as Google or Baidu, 39 , 40 while others have utilized prior patterns of healthcare utilization and external variables such as weather patterns to predict future outbreaks. 36 , 38 Artificial intelligence has also been used to predict influenza vaccine uptake and response. 41 , 42 These applications have shown great promise and may soon provide useful guidance to policy makers and healthcare administrators, but they are less useful to front‐line clinicians making time‐sensitive decisions for individual patients.
Hundreds of studies have been published describing ML models developed to predict clinical outcomes for patients with COVID‐19, 15 but notably few have pursued a similar objective for patients with influenza, which remains a significant source of morbidity and mortality each year. Small studies (each with less than 350 patients) have been performed in the ICU setting, with ML models designed to predict 30‐day mortality in adults and prolonged (≥7 days) hypoxemic respiratory failure in children. 43 , 44 These models exhibited good predictive performance, but were trained in more homogenous critically ill populations than we have studied here and focused on longer‐term outcomes less relevant to ED decision‐making. Two groups in Taiwan have published ED‐focused studies on the topic. 44 , 45 Hu et al. 44 employed a two‐stage logistic regression modeling approach at a single large academic medical center to predict critical illness (ICU admission or death) and in‐hospital mortality among 1680 adult ED patients with influenza. Tan et al 45 evaluated multiple ML modeling approaches to predict several clinical outcomes in the ED including hospitalization, ICU admission, and death in 5508 elderly patients with influenza. Performance of leading models was comparable to what we have reported here using random forests. Probability estimates of leading models also were made available to treating ED clinicians, though no specific decision support was provided and impact was not measured. To our knowledge, the ML models reported here are the first designed to guide disposition decision‐making for all ED patients with influenza and have important clinical and administrative utility. Our models predict clinically important composite outcomes and can be rapidly embedded into an existing CDS system with proven effectiveness. 14
Potential challenges to the clinical utility of ML‐driven CDS are degradation of predictive performance over time and limited transportability of ML models between clinical settings. ML relies on past events to predict the future. Changes in disease prevalence, innate and acquired host immunity, pathogen evolution and therapeutic intervention availability may render these predictions less accurate over time. Similarly, ML models derived and validated in one clinical or geographic setting may not perform as expected in another due to differences in the patient population or practice patterns of clinicians. 18 Transportability also can be affected by local health information technology practices including timestamping and labeling of predictor and outcome data used by ML models.
In this study, we employed a novel approach to model development and validation that allowed us to test the durability of our models under both conditions directly. Retrospective datasets were divided into two cohorts: one for model development (derivation and out‐of‐sample validation) and the other for external validation. This process was performed twice, with development and validation cohorts separated either by space or time; distinct models were generated and evaluated for each iteration. Under the first scenario, models derived, tested, and validated out‐of‐sample at two hospitals underwent external validation at three others. Under the second, models were derived, tested, and validated out‐of‐sample during the 2017–2019 flu seasons and externally validated using data from future flu seasons. We found that greater performance degradation occurred when models were transported from one location to another than when models developed during one time period were used to make outcome predictions in another (Table 2). These results reinforce the concept that the performance of models developed and validated in one setting should not be assumed in another. 18 , 46 Under real‐world conditions, ML models may require retraining at each destination site. At the very least, verification of function and validation of performance using locally derived datasets must be a prerequisite to clinical implementation. Our finding that prediction performance remained stable across influenza seasons is reassuring, as real‐world implementation of model‐driven CDS cross‐seasonal model training and prediction.
In conclusion, we developed artificial intelligence‐based ML models which accurately estimate the need for critical and inpatient care in patients presenting with influenza, using data that were routinely gathered during the ED encounter. Our results reinforce the concept that predictive models derived in one setting may need to be retrained and validated at each new target site but suggest model fidelity can be maintained over time and over multiple influenza seasons. The results of this study serve as a basis for the implementation and evaluation of a CDS system to optimize clinical care and resource utilization for patients with influenza.
AUTHOR CONTRIBUTIONS
J. H. and S. L. conceived and designed the study. J. H., S. L., and R. R. obtained research funding. J. H., S. L., and E. K. supervised data collection and the conduct of the study. S. L., M. T., A. S., and X. Z. performed data analysis. R. R. and K. F. provided content‐specific expertise and M. C. provided methodologic expertise. J. H. drafted the manuscript, and all authors contributed substantially to its revision. J. H. takes responsibility for the paper as a whole.
CONFLICT OF INTEREST STATEMENT
A. S., M. T., and S. L. participated in this work while employed by Johns Hopkins University but subsequently became employees of Beckman Coulter Diagnostics, where they contribute to the development of CDS tools. J. H. is a paid scientific consultant for Beckman Coulter Diagnostics. Beckman Coulter played no role in this study and no technology owned or licensed by Beckman Coulter was used. The remaining authors declare no competing financial or non‐financial interests.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
This study was supported by the National Institute of Allergies and Infectious Diseases (NIAID) Centers of Excellence for Influenza Research and Response (NIAID N272201400007C). Data infrastructure used to facilitate this work was built with the support of grant number R18 HS026640 from the Agency for Healthcare Research and Quality (AHRQ), U.S. Department of Health and Human Services (HHS). The funders had no role in the design and conduct of the study and the authors are solely responsible for this document's contents, findings, and conclusions, which do not necessarily represent the views of NIAID or AHRQ.
Biography
Jeremiah Hinson, MD, PhD, is an Associate Director of Research in the Department of Emergency Medicine and Co‐Director of the Center for Data Science in Emergency Medicine at the Johns Hopkins University School of Medicine in Baltimore, Maryland. His research interests include emergency department operations, acute kidney injury, and infectious disease. He is particularly focused on the improvement of patient outcomes using data‐driven methods.

Hinson JS, Zhao X, Klein E, et al. Multisite development and validation of machine learning models to predict severe outcomes and guide decision‐making for emergency department patients with influenza. JACEP Open. 2024;5:e13117. 10.1002/emp2.13117
Meetings: An early version of this work was presented in abstract format at the NIH‐Sponsored National Meeting of the Centers for Excellence in Influenza Research and Response, held in Memphis, TN from August 16–18, 2022.
Supervising Editor: Adam Landman, MD, MS
DATA AVAILABILITY STATEMENT
The clinical data used in this study are from the Johns Hopkins Health System (JHHS). These individual‐level patient data are protected for privacy. Qualified researchers affiliated with Johns Hopkins University (JHU) may apply for access through the Johns Hopkins Institutional Review Board (IRB) (https://www.hopkinsmedicine.org/institutional_review_board/). Those not affiliated with JHU seeking to collaborate may contact the corresponding author. Access to these data for research collaboration with JHU must ultimately comply with IRB and data sharing protocols (https://ictrweb.johnshopkins.edu/ictr/dmig/Best_Practice/c8058e22‐0a7e‐4888‐aecc‐16e06aabc052.pdf).
REFERENCES
- 1. CDC . Burden of Influenza. Disease Burden of Influenza. Published January 10, 2020. Accessed February 15, 2020. https://www.cdc.gov/flu/about/burden/index.html
- 2. Ali ST, Lau YC, Shan S, et al. Prediction of upcoming global infection burden of influenza seasons after relaxation of public health and social measures during the COVID‐19 pandemic: a modelling study. Lancet Glob Health. 2022;10(11):e1612‐e1622. doi: 10.1016/S2214-109X(22)00358-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention . National Ambulatory Medical Care Survey: 2016 National Summary Tables. Published online January 25, 2020. Accessed January 25, 2020. Published 2016. https://www.cdc.gov/nchs/data/ahcd/namcs_summary/2016_namcs_web_tables.pdf
- 4. Augustine J. Latest data reveal the ED's role as hospital admission gatekeeper. ACEP Now. 2019;38(12):26. [Google Scholar]
- 5. Fingar KR, Liang L, Stocks C. Inpatient Hospital Stays and Emergency Department Visits Involving Influenza, 2006–2016. Agency for Health Research and Quality; 2019:1‐24. [PubMed] [Google Scholar]
- 6. Fernando SM, Rochwerg B, Reardon PM, et al. Emergency Department disposition decisions and associated mortality and costs in ICU patients with suspected infection. Crit Care. 2018;22(1):172. doi: 10.1186/s13054-018-2096-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet. 2012;13(6):395‐405. [DOI] [PubMed] [Google Scholar]
- 8. Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319(13):1317‐1318. doi: 10.1001/jama.2017.18391 [DOI] [PubMed] [Google Scholar]
- 9. Janke AT, Overbeek DL, Kocher KE, Levy PD. Exploring the potential of predictive analytics and big data in emergency care. Ann Emerg Med. 2016;67(2):227‐236. doi: 10.1016/j.annemergmed.2015.06.024 [DOI] [PubMed] [Google Scholar]
- 10. Kruse CS, Goswamy R, Raval Y, Marawi S. Challenges and opportunities of big data in health care: a systematic review. JMIR Med Inform. 2016;4(4):e38. doi: 10.2196/medinform.5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Levin S, Toerper M, Hamrock E, et al. Machine‐learning‐based electronic triage more accurately differentiates patients with respect to clinical outcomes compared with the emergency severity index. Ann Emerg Med. 2018;71(5):565‐574. [DOI] [PubMed] [Google Scholar]
- 12. Martinez DA, Levin SR, Klein EY, et al. Early prediction of acute kidney injury in the emergency department with machine‐learning methods applied to electronic health record data. Ann Emerg Med. 2020;76(4):501‐514. doi: 10.1016/j.annemergmed.2020.05.026 [DOI] [PubMed] [Google Scholar]
- 13. Delahanty RJ, Alvarez J, Flynn LM, Sherwin RL, Jones SS. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann Emerg Med. 2019;73(4):334‐344. doi: 10.1016/j.annemergmed.2018.11.036 [DOI] [PubMed] [Google Scholar]
- 14. Hinson JS, Klein E, Smith A, et al. Multisite implementation of a workflow‐integrated machine learning system to optimize COVID‐19 hospital admission decisions. NPJ Digit Med. 2022;5(1):94. doi: 10.1038/s41746-022-00646-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wynants L, Calster BV, Collins GS, et al. Prediction models for diagnosis and prognosis of covid‐19: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Past Pandemics | Pandemic Influenza (Flu) | CDC. Published June 11, 2019. Accessed January 7, 2022. https://www.cdc.gov/flu/pandemic‐resources/basics/past‐pandemics.html
- 17. Sampath S, Khedr A, Qamar S, et al. Pandemics throughout the history. Cureus. 2021;13(9):e18136. doi: 10.7759/cureus.18136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song X, Yu ASL, Kellum JA, et al. Cross‐site transportability of an explainable artificial intelligence model for acute kidney injury prediction. Nat Commun. 2020;11(1):5668. doi: 10.1038/s41467-020-19551-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dugas AF, Hsieh YH, LoVecchio F, et al. Derivation and validation of a clinical decision guideline for influenza testing in 4 US emergency departments. Clin Infect Dis. 2020;70(1):49‐58. doi: 10.1093/cid/ciz171 [DOI] [PubMed] [Google Scholar]
- 20. Pinto D, Lunet N, Azevedo A. Sensitivity and specificity of the Manchester Triage System for patients with acute coronary syndrome. Rev Port Cardiol. 2010;29(6):961‐987. [PubMed] [Google Scholar]
- 21. Dugas AF, Kirsch TD, Toerper M, et al. An electronic emergency triage system to improve patient distribution by critical outcomes. J Emerg Med. 2016;50(6):910‐918. doi: 10.1016/j.jemermed.2016.02.026 [DOI] [PubMed] [Google Scholar]
- 22. Schneider D, Appleton R, McLemorem T. A Reason for Visit Classification for Ambulatory Care. US Department of Health, Education and Welfare; 1979:72. Accessed October 22, 2019 https://www.cdc.gov/nchs/data/series/sr_02/sr02_078.pdf [Google Scholar]
- 23. Chobanian AV, Bakris GL, Black HR, et al. The seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289(19):2560‐2571. doi: 10.1001/jama.289.19.2560 [DOI] [PubMed] [Google Scholar]
- 24. Barfod C, Lauritzen MMP, Danker JK, et al. Abnormal vital signs are strong predictors for intensive care unit admission and in‐hospital mortality in adults triaged in the emergency department—a prospective cohort study. Scand J Trauma Resusc Emerg Med. 2012;20:28. doi: 10.1186/1757-7241-20-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Andrew MK, Powell C. An approach to ‘the social admission’. Can J Gen Int Med. 2016;10(4). doi: 10.22374/cjgim.v10i4.80 [DOI] [Google Scholar]
- 26. Haimovich A, Ravindra NG, Stoytchev S, et al. Development and validation of the quick COVID‐19 severity index (qCSI): a prognostic tool for early clinical decompensation. Ann Emerg Med. 2020;76(4):442‐453. doi: 10.1016/j.annemergmed.2020.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Breiman L. Classification and Regression Trees. 1st ed. Routledge; 2017. [Google Scholar]
- 28. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837‐845. doi: 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 29. Krstajic D, Buturovic LJ, Leahy DE, Thomas S. Cross‐validation pitfalls when selecting and assessing regression and classification models. J Cheminform. 2014;6(1):10. doi: 10.1186/1758-2946-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zadrozny B, Elkan C, Transforming classifier scores into accurate multiclass probability estimates. Proceedings of the eighth ACM SIGKDD international conference on Knowledge discovery and data mining . Published online July 23, 2002:694‐699. doi: 10.1145/775047.775151 [DOI]
- 31. Steyerberg EW, Vickers AJ, Cook NR, et al. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology. 2010;21(1):128‐138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DiCiccio TJ, Efron B. Bootstrap confidence intervals. Stat Sci. 1996;11(3):189‐212. [Google Scholar]
- 33. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Guyon I, Luxburg UV, Bengio S, eds. Advances in Neural Information Processing Systems. Curran Associates, Inc; 2017. https://proceedings.neurips.cc/paper/2017/file/8a20a8621978632d76c43dfd28b67767‐Paper.pdf [Google Scholar]
- 34. Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ. 2005;330(7494):765. doi: 10.1136/bmj.38398.500764.8F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levin S, Toerper M, Hinson J, et al. 294 Machine‐learning‐based electronic triage: a prospective evaluation. Ann Emerg Med. 2018;72(4):S116. doi: 10.1016/j.annemergmed.2018.08.299 [DOI] [PubMed] [Google Scholar]
- 36. Cheng HY, Wu YC, Lin MH, et al. Applying machine learning models with an ensemble approach for accurate real‐time influenza forecasting in taiwan: development and validation study. J Med Internet Res. 2020;22(8):e15394. doi: 10.2196/15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miliou I, Xiong X, Rinzivillo S, et al. Predicting seasonal influenza using supermarket retail records. PLoS Comput Biol. 2021;17(7):e1009087. doi: 10.1371/journal.pcbi.1009087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Poirier C, Hswen Y, Bouzillé G, et al. Influenza forecasting for French regions combining EHR, web and climatic data sources with a machine learning ensemble approach. PLoS One. 2021;16(5):e0250890. doi: 10.1371/journal.pone.0250890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dai S, Han L. Influenza surveillance with Baidu index and attention‐based long short‐term memory model. PLoS One. 2023;18(1):e0280834. doi: 10.1371/journal.pone.0280834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Q, Gel YR, Ramirez Ramirez LL, Nezafati K, Zhang Q, Tsui KL. Forecasting influenza in Hong Kong with Google search queries and statistical model fusion. PLoS One. 2017;12(5):e0176690. doi: 10.1371/journal.pone.0176690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benis A, Chatsubi A, Levner E, Ashkenazi S. Change in threads on twitter regarding influenza, vaccines, and vaccination during the COVID‐19 pandemic: artificial intelligence‐based infodemiology study. JMIR Infodemiol. 2021;1(1):e31983. doi: 10.2196/31983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tomic A, Tomic I, Dekker CL, Maecker HT, Davis MM. The FluPRINT dataset, a multidimensional analysis of the influenza vaccine imprint on the immune system. Sci Data. 2019;6(1):214. doi: 10.1038/s41597-019-0213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sauthier MS, Jouvet PA, Newhams MM, Randolph AG. Machine learning predicts prolonged acute hypoxemic respiratory failure in pediatric severe influenza. Crit Care Explor. 2020;2(8):e0175. doi: 10.1097/CCE.0000000000000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu CA, Chen CM, Fang YC, et al. Using a machine learning approach to predict mortality in critically ill influenza patients: a cross‐sectional retrospective multicentre study in Taiwan. BMJ Open. 2020;10(2):e033898. doi: 10.1136/bmjopen-2019-033898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan TH, Hsu CC, Chen CJ, et al. Predicting outcomes in older ED patients with influenza in real time using a big data‐driven and machine learning approach to the hospital information system. BMC Geriatr. 2021;21(1):280. doi: 10.1186/s12877-021-02229-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wong A, Otles E, Donnelly JP, et al. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern Med. 2021;181(8):1065‐1070. doi: 10.1001/jamainternmed.2021.2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The clinical data used in this study are from the Johns Hopkins Health System (JHHS). These individual‐level patient data are protected for privacy. Qualified researchers affiliated with Johns Hopkins University (JHU) may apply for access through the Johns Hopkins Institutional Review Board (IRB) (https://www.hopkinsmedicine.org/institutional_review_board/). Those not affiliated with JHU seeking to collaborate may contact the corresponding author. Access to these data for research collaboration with JHU must ultimately comply with IRB and data sharing protocols (https://ictrweb.johnshopkins.edu/ictr/dmig/Best_Practice/c8058e22‐0a7e‐4888‐aecc‐16e06aabc052.pdf).


