Abstract
Over the past decade, environmental metagenomics and polymerase chain reaction-based marker gene surveys have revealed that several lineages beyond just a few well-established groups within the Euryarchaeota superphylum harbor the genetic potential for methanogenesis. One of these groups are the Archaeoglobi, a class of thermophilic Euryarchaeota that have long been considered to live non-methanogenic lifestyles. Here, we enriched Candidatus Methanoglobus hypatiae, a methanogen affiliated with the family Archaeoglobaceae, from a hot spring in Yellowstone National Park. The enrichment is sediment-free, grows at 64–70°C and a pH of 7.8, and produces methane from mono-, di-, and tri-methylamine. Ca. M. hypatiae is represented by a 1.62 Mb metagenome-assembled genome with an estimated completeness of 100% and accounts for up to 67% of cells in the culture according to fluorescence in situ hybridization. Via genome-resolved metatranscriptomics and stable isotope tracing, we demonstrate that Ca. M. hypatiae expresses methylotrophic methanogenesis and energy-conserving pathways for reducing monomethylamine to methane. The detection of Archaeoglobi populations related to Ca. M. hypatiae in 36 geochemically diverse geothermal sites within Yellowstone National Park, as revealed through the examination of previously published gene amplicon datasets, implies a previously underestimated contribution to anaerobic carbon cycling in extreme ecosystems.
Keywords: archaea, MCR, methane, stable isotope tracing, thermophile, transcriptomics
Introduction
Methanogenesis is one of the most ancient metabolic pathways and plays a major role in the biogeochemical carbon cycle. Phylogenomic reconstructions and geological evidence suggest that methanogenesis was among the earliest metabolisms to evolve and that the last common ancestor of all extant archaea likely was a methanogen [1-9]. Therefore, the study of methanogens is essential for understanding the co-evolution of life and the biosphere. Methanogenic archaea are the primary producers of biogenic methane (CH4) and contribute ~60% to the estimated 576 Tg of annual global methane emissions to the atmosphere [10, 11]. Methanogenic pathways are classified by their carbon and electron sources [12-14]. All methanogenic pathways converge at the terminal methane-forming step catalyzed by the methyl-coenzyme M reductase (MCR) complex. MCR and its homologs also catalyze the reverse reaction in the anaerobic oxidation of alkanes in alkanotrophic archaea [15, 16]. MCR is uniquely present in all methanogens and is commonly used to identify potential methane and/or alkane cycling archaea in sequencing surveys [12, 17].
The physiology and biochemistry of methanogens have near-exclusively been investigated in axenic cultures of microorganisms belonging to the Euryarchaeota superphylum [12, 17-19]. These predominantly grow by acetoclastic or CO2-reducing hydrogenotrophic methanogenesis, with only rare observations of Euryarchaeotal methyl-reducing methanogens [12, 20, 21]. As a result, despite the dominance of methyl-based methanogenesis in anoxic environments with high salt and/or high sulfate concentration (e.g. anoxic marine sediments, coastal wetlands, hypersaline lakes), methylotrophic methanogenesis has in the past often been considered to be of comparatively limited environmental distribution. The extensive use of environmental metagenomics has led to the discovery of metagenome-assembled genomes (MAGs) encoding MCR from new lineages that are prevalent in anoxic environments, both within and outside the Euryarchaeota [2, 12, 22-26].
The majority of MAGs affiliated with archaeal phyla outside the Euryarchaeota are predicted to be methyl-reducing methanogens, with the exception of Candidatus (Ca.) Nezhaarchaeota [25, 27] and Ca. Methanomixophus affiliated with the order Archaeoglobales, which have been hypothesized to be CO2-reducing hydrogenotrophic methanogens [12, 25, 28]. This result is consistent with the observation that methylated methanogenic substrates, including methylamines and methanol, are prevalent in the environment, although their concentrations in hot springs is currently unknown. Furthermore, methyl-reducing methanogenesis is considered the predominant mode of methanogenesis in anoxic marine, freshwater, and hypersaline sediments (reviewed in Bueno de Mesquita et al. [20]).
Members of the class Archaeoglobi have long been considered non-methanogenic with isolates characterized as dissimilatory sulfate reducers brought into culture as early as 1987 [29]. To date, only nine species of the class Archaeoglobi have been obtained in axenic culture, and all were sourced from marine hydrothermal systems or off-shore oil reservoirs [30]. The discovery of both MCR [25, 31, 32] and methyl-H4M(S)PT:coenzyme M methyltransferase (MTR) complexes in genomes of the Archaeoglobaceae has suggested that members of this family may live by methanogenesis [28].
Very recently, important progress toward experimental verification of methanogenesis by members of this family has been made. Liu et al. reported the in situ expression of genes related to hydrogen-dependent methylotrophic methanogenesis and heterotrophic fermentation within populations of Archaeoglobi in a high-temperature oil reservoir [28]. Lynes et al. reported that Archaeoglobi can be enriched in hot spring mesocosms under methanogenic conditions [33]. Wang et al. reported that mcrABG and other methanogenesis marker genes encoded by two Archaeoglobales MAGs were highly expressed in hot spring microcosms incubated at 65°C and 75°C [34]. Importantly, one of these Archaeoglobales MAGs represented the only Mcr-encoding archaeon that expressed mcrABG genes in methanogenic microcosms performed without substrate addition or with the addition of 10 mM methanol at 75°C. This indirectly demonstrated the methanogenic nature of this archaeon [34]. Last, Buessecker et al. reported the establishment of a methanogenic enrichment culture of Ca. Methanoglobus nevadensis from Great Boiling Spring (GBS) (NV, USA) [35]. The culture yields up to 158 μM methane after 2 weeks of incubation at its optimal growth temperature of 75°C. Ca. M. nevadensis is represented by a 63% complete MAG obtained from the culture and a 98% complete MAG obtained a decade earlier [35].
Here, we report on the enrichment cultivation of Ca. Methanoglobus hypatiae LCB24, a methanogen affiliated with the family Archaeoglobaceae, from a hot spring in Yellowstone National Park (YNP). Using a combination of targeted cultivation, growth experiments, microscopy, stable isotope tracing, metagenomics, and metatranscriptomics, we demonstrate that Ca. M. hypatiae lives by methylotrophic methanogenesis and converts different methylamines to methane. By examining previously published datasets for the presence of Mcr-encoding Archaeoglobi, we demonstrate that these archaea are distributed in geothermal features of YNP, where they likely contribute to anaerobic carbon cycling. Our study presents direct evidence of methanogenesis within the Archaeoglobaceae and adds to the growing body of evidence demonstrating that methanogenesis is widely spread within the Euryarchaeota superphylum.
Materials and Methods
All chemicals used in this study were sourced from Sigma Aldrich unless otherwise specified.
Sample collection, enrichment, and cultivation
In November 2021, a slurry of sediment and water (1:9) was collected from an unnamed hot spring in the Lower Culex Basin of YNP, WY, USA. In our previous survey of Mcr-encoding archaea in YNP [33], this hot spring was given the identifier LCB024 (44.573294, −110.795388; pH 7.8, 67°C). A mixture of surface sediment (~1–2 cm deep) and hot spring water was collected into a glass bottle and sealed headspace-free with a thick butyl rubber stopper. Collected material was transported back to the lab and stored at room temperature. Using this material as inoculum, 30 ml enrichments were established in February 2022 in 60 ml serum bottles. Material was homogenized by mixing and was then diluted 1:10 (v/v) with anoxic medium in an anoxic glove box (N2/CO2/H2; 90/5/5%).
Medium was prepared anoxically as described previously [36]. Basal mineral medium contained a base of (per liter): KH2PO4, 0.5 g; MgSO4·7H2O, 0.4 g; NaCl, 0.5 g; NH4Cl, 0.4 g; CaCl2·2H2O, 0.05 g; HEPES, 2.38 g; yeast extract, 0.1 g; and 0.002% (w/v) (NH4)2Fe(SO4)2·6H2O. Medium was transferred to a Duran flask with a side opening and autoclaved for 20 m at 121°C. Medium was then further supplemented with 5 mM NaHCO3, 1 ml trace element solution SL-10, 1 ml Selenite-Tungstate solution, 1 ml CCM vitamins [37], 0.0005% (w/v) resazurin, 10 mg of coenzyme-M, 2 mg sodium dithionite, 1 mM dithiothreitol, 1 mM Na2S·9H2O, with pH adjusted to 7.8 using sodium hydroxide (NaOH, 12 N). Serum bottles were sealed with butyl rubber stoppers and aluminum crimps before the headspace were exchanged with N2 (99.999%) for 5 min and set to a 200 kPa N2 atmosphere. Monomethylamine (MMA) was added from a filter-sterilized methylamine-hydrochloride anoxic stock solution to a final concentration of 10 mM. The bacterial antibiotics streptomycin (50 mg/L; inhibitor of protein synthesis) and vancomycin (50 mg/L; inhibitor of peptidoglycan synthesis) were added from filter-sterilized anoxic stock solutions. The enrichments were incubated at 70°C in the dark without shaking. Cultures were maintained by regular transfer of 10% v/v into fresh media, which contained MMA and antibiotics. A sediment-free culture was obtained after the third transfer after which it was transferred at 10% v/v to 50 ml in 125 ml serum bottles.
Stable isotope tracing
The conversion of 13C- or D3-MMA (13CH3-NH2, CD3-NH2) to 13CH4 or CD3H was tracked by incubating active enrichment cultures in the presence of 20% labeled substrate (98%; Cambridge Isotope Laboratories). Incubations were carried out in 30 ml culture volumes in 60 ml serum bottles with 8% v/v inoculum, 50 mg/L streptomycin, 50 mg/L vancomycin, 10 mM MMA, and N2 gas (99.999%) incubated in anoxic media (pH 7.8, 70°C) in six replicates (SI Appendix, Fig. S3). Duplicate control incubations included (i) 12C-MMA and (ii) inoculum without MMA. Triplicate control incubations were performed with (iii) 12C-MMA plus 10 mM bromoethanesulfonate (BES) added in mid-exponential phase (Day 33) to inhibit methanogenesis and (iv) 10 mM BES added at time of inoculation (Day 0) without substrate. Headspace samples were collected throughout the experiment as described above and analyzed using a Shimadzu QP2020 NX GCMS equipped with a GS-CarbonPLOT column (30 m × 0.35 mm; 1.5 μm film thickness; Agilent) and operated in Selected Ion Monitoring mode. The instrument was operated using the method described in Ai et al. [38] with helium as a carrier gas. All injections were performed by a Shimadzu AOC-6000 autosampler robot. Peak areas corresponding to m/z ratios of 16 for 12CH4, 17 for 13CH4, and 19 for CD3H were used for quantification.
Metagenomic sequencing, assembly, and annotation
Two metagenomes were obtained over the course of this study. A 42 ml aliquot of the fourth transfer of the enrichment (Fig. 1 T4-MG) was filtered onto a 0.22 μm filter. The filter was transferred to a lysing matrix E tube and DNA extracted immediately following filtration. Genomic DNA was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals, Irvine, CA) following the manufacturer’s guidelines.
Figure 1.
Community composition and methane production of the methanogenic enrichment culture containing Ca. M. hypatiae LCB24; (A) relative abundance of 16S rRNA gene amplicons in the initial sediment from hot spring LCB024, the slurry collected in November 2021, slurry material used to initiate enrichments in February 2022, the initial enrichment, and five subsequent transfers (T1–T5) are shown; for comparison, the estimated relative abundance of two metagenomic samples (T4-MG and SIT-MG) is included; the metagenome recovered from a replicate from the stable isotope tracing experiment incubated in the presence of deuterated methylamine (SIT-MG) revealed Ca. M. hypatiae grew to 92.8% relative abundance during the experiment; the two most abundant ASVs across enrichment transfers are shown with other taxa collapsed; other methanogenic archaea were not identified in the initial enrichment or in any subsequent transfer; relative sequence abundance for all ASVs is reported in SI Appendix, Table S1; (B) headspace methane produced over long-term cultivation; the time between transfers decreased while the average maximum concentration of methane increased over time; culture 1A represents the initial enrichment; a history of methane measurements can be found in SI Appendix, Table S2; (C) visualization of Ca. M. hypatiae cells at T6 labeled via CARD-FISH by the general archaea probe Arch915 (red). DAPI staining of cells is in blue; (D) cell morphologies in enrichment culture LCB24 at T7 as observed by SEM.
A second metagenome was recovered from one of the six culture replicates grown in the presence of CD3-NH2 and used for recruiting transcriptomic reads from the other replicates (Fig. 1 SIT-MG). A 60 ml syringe flushed with N2 gas was used to transfer 30 ml of culture to a sterilized oak ridge tube. Cells were harvested through centrifugation for 30 min at 10000 rpm at 4°C. The supernatant was removed, and DNA extracted from the pellet using the FastDNA Spin Kit for Soil (MP Biomedicals, Irvine, CA) following the manufacturer’s guidelines. Genomic DNA for both metagenomes was shipped to SeqCenter (Pittsburgh, PA), and sample libraries were prepared using the Illumina DNA Prep kit and 10 bp unique dual indices (UDIs). The first metagenome (T4-MG) was sequenced on a NextSeq 2000 System (Illumina) and the second (SIT-MG) sequenced on a NovaSeq 6000 System (Illumina), each producing 2 × 151 bp reads. Demultiplexing, quality control, and adapter trimming were performed with bcl-convert v3.9.3. The quality of the reads was evaluated with FastQC before quality, linker and adapter trimming, artifact and common contaminant removal, and error correction were performed with the rqcfilter2 pipeline (maxn = 3, maq = 10, trimq = 20) and bbcms (mincount = 2, hcf = 0.6). Resulting reads were assembled with SPAdes v3.15.13 (Nurk, 2017) (−k 33,55,77,99 127 —meta –only-assembler), and coverage was determined with bbmap v38.94 (ambiguous = random) (https://sourceforge.net/projects/bbmap) [39]. In addition to the initial assembly, co-assemblies using both T4-MG and SIT-MG metagenomes were also performed [1] with reads directly fed into SPAdes with the—only-assembler option excluded; and [2] with the trimmed and error corrected reads and the same SPAdes parameters as above. The statistics of MAGs generated through various assembly and quality control methods were evaluated, and the approach that produced the highest quality MAG was chosen for subsequent analysis (Dataset S1). The quality was determined by considering factors such as the number of resulting sequences, total length, completeness, and the minimum, maximum, and average sequence lengths. Annotation of the assembled sequences was performed with Prokka v1.14.16 [40]. Assembled scaffolds ≥2000 bp were binned using Maxbin v2.2.7 [41], Metabat v2.12.1 (with and without coverage) [42], Concoct v1.0.0 [43], Autometa v1 (bacterial and archaeal modes with the machine learning step) [44], followed by bin refinement with DAS_Tool v1.1.6 [45], as previously described [46]. Bin completeness and redundancy were assessed with CheckM v1.2.2 [47].
RNA extraction, sequencing, and transcriptomic processing
Total RNA was extracted for transcriptomics from four of the six replicates of Archaeoglobus cultivated in the presence of labeled substrate (13CH3-NH2 or CD3-NH2) for a total of eight replicates. Each replicate culture in the exponential growth phase (Day 32) was moved from the 70°C incubator to an ice bath placed at −20°C for 40 mins to stop cellular activity. A 60 ml syringe flushed with N2 gas was used to transfer 30 ml of culture to a sterilized oak ridge tube and kept on ice. Cells were harvested through centrifugation for 30 min at 10000 rpm at 4°C. The supernatant was removed, and the pellet transferred to a lysing matrix E tube (MP Biomedicals, Irvine, CA) to which 600 μL of RNA lysis buffer was added. Samples were homogenized in a MP Bioscience FastPrep instrument for 40 s at a speed setting of 6.0 m/s followed by centrifugation for 15 min at 14000 rpm. RNA was extracted using the Quick-RNA miniprep kit (Zymo Research, Irvine, CA) including a DNAse treatment step and eluted in 50 μL of RNAse free water. Centrifugation steps were performed at 15000 rpm and the final spin for elution at 10000 rpm. Of the eight replicates extracted, six measured >50 ng/μL (3× 13CH3-NH2 and 3× CD3-NH2) and were sent for transcriptomic sequencing at SeqCenter (Pittsburgh, PA). Samples were DNAse treated with Invitrogen DNAse (RNAse free). Library preparation was performed using Illumina’s Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit and 10 bp UDI. Sequencing was done on a NovaSeq 6000, producing paired end 151 bp reads. Demultiplexing, quality control, and adapter trimming were performed with bcl-convert (v4.1.5). Read quality was further evaluated with FastQC v0.11.9 [48] before quality trimming and artifact, rRNA, and common contaminant removal with the rqcfilter2 pipeline (trimq = 28, maxns = 3, maq = 20), and error correction with bbcms (mincount = 2, hcf = 0.6) from the BBTools suite v38.94 [39]. Additional rRNA gene reads were detected and removed with Ribodetector v0.2.7 [49], and any remaining rRNA gene reads were finally removed with bbmap, using rRNA genes recovered from the metagenomes (see below) as references. The resulting mRNA reads were mapped against annotated genes from the paired metagenomes with bbmap to calculate reads per kilobase of transcript per million mapped read (RPKM) (ambig = random).
Results and Discussion
Cultivation
In our recent survey on the diversity of Mcr-encoding archaea in the geothermal features of YNP, mesocosms seeded with biomass from a hot spring located within the Lower Culex Basin (LCB024; pH 7–8, 56–74°C) had shown potential to enrich for methanogenic Archaeoglobi [33]. Using a sediment slurry collected from LCB024, we initiated incubations supplied with MMA and antibiotics incubated in anoxic media (pH 7.8, 70°C) under a N2 headspace. The relative abundance, as determined by 16S rRNA gene amplicon sequencing, of Archaeoglobi-affiliated organisms in LCB024 was 0.32% in the initial slurry and had fallen to 0.02% by the time incubations were initiated a few months after samples had been collected (Fig. 1A).
Methane was detected after 36 days in the initial enrichment, and the culture transferred to fresh media after reaching the late exponential phase of methane production following 70 days of incubation (447 μM; Fig. 1B). Five Archaeoglobi-related 16S rRNA gene amplicon ASVs were identified in the initial enrichment; however, one ASV grew to dominate the microbial community after the first transfer and reached 74.8% relative abundance after 62 days. In the transfers that followed, Archaeoglobi-related sequences became the only archaeal reads detected by 16S rRNA gene amplicon sequencing with the second most abundant organism a bacterium affiliated with the Pseudothermotoga at 6.8%. Although the CO2-reducing methanogen Methanothermobacter sp. was detected at 0.45% relative abundance in the slurry material used for inoculation, it was not detected in any subsequent transfers, nor were any known methanogens. Over subsequent transfers (238 days, T2–T5), the relative abundance of Archaeoglobi ASVs ranged from 46% to 69%, and the final methane yield steadily increased from 1844 to 2459 μM. A sediment-free enrichment was obtained by the third transfer. Starting with the fourth transfer, the culture volume was scaled from 30 ml to 50 ml. By the sixth transfer, the culture produced 3943 μM methane within 34 days. Metagenomic sequencing at two timepoints (Day 335 of the enrichment and Day 33 of the isotope tracing experiment described below) and 16S rRNA gene amplicon sequencing over recurring transfers (Fig. 1A) demonstrated that ASVs and MAGs affiliated with Archaeoglobi represented the only archaeon in culture LCB24. A single MCR complex (mcrAGCDB) belonging to the Archaeoglobi MAG was present, indicating this MAG represents the only methanogenic population.
Metagenomics and phylogenetics
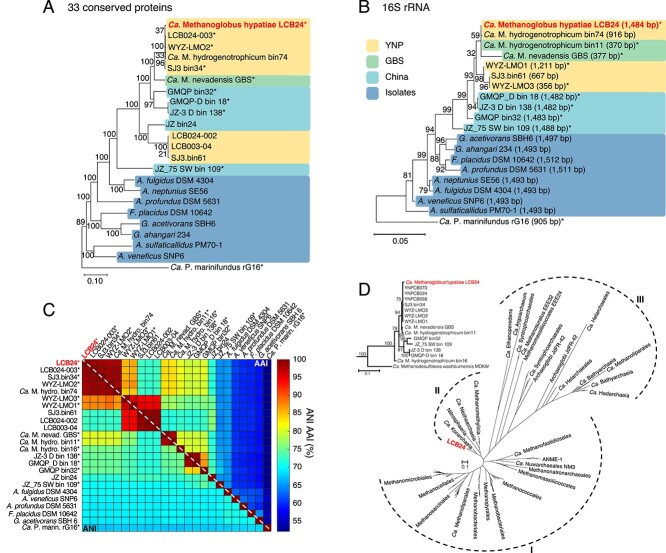
The reconstructed Mcr-encoding Archaeoglobi MAG from culture LCB24 was 1.62 Mbp in length with an estimated completeness of 100% according to checkM (SI Appendix, Table S3). This MAG was the result of a combined assembly of the T4-MG and SIT-MG metagenomes as this method yielded an improved assembly. Therefore, it was used for phylogenomic analysis against Archaeoglobi reference MAGs and genomes using 33 conserved single copy marker proteins and 16S rRNA genes (Fig. 2A and B, SI Appendix, Table S4). The phylogenomic analysis showed that MAGs encoding MCR complexes clustered separately from those lacking mcr gene sequences. Consistently, 16S rRNA gene phylogeny supported this clustering with a pronounced separation of hot spring reference genomes and MAGs from current known isolates of Archaeoglobi, resulting in three main clusters: (i) those retrieved from North American hot springs (YNP and GBS), (ii) those originating from hot springs in China, and (iii) isolates, all of which were obtained from deep-sea marine hydrothermal systems (Fig. 2B).
Figure 2.
Phylogenetic affiliation of Ca. M. hypatiae LCB24; (A) maximum-likelihood tree, inferred with fasttree and WAG model (midpoint root), using a concatenated alignment of 33 conserved single copy proteins (list provided in SI Appendix, Table S4); references are colored by the habitat or type from which sequences had been recovered: hot springs in YNP, yellow; GBS, green; hot springs in China, blue; isolates from marine hydrothermal vent systems, dark blue; (B) maximum-likelihood tree inferred with fasttree using 16S rRNA genes with length in base pairs (bp); (C) ANI and AAI analysis of reference Archaeoglobales MAGs and genomes; asterisks (*) indicate MAGs containing mcrA, apart from the MAG of Ca. M. hydrogenotrophicum bin74 which encodes a mcrA that is interrupted by a stop codon; AAI and ANI values are provided in SI Appendix, Fig. S1; (D) maximum-likelihood tree, inferred with IQtree2 and the LG + C60 + F + G model, from the amino acid alignment of McrA; dashed lines indicate McrA/AcrA groups: (I) McrA from methanogens and ANME (MCR-type), (II) McrA from TACK lineages (MCR-type), (III) McrA-like from proposed and experimentally confirmed alkane oxidizing archaea (ACR-type); insert shows MAGs closely related to Ca. M. hypatiae LCB24.
LCB24 and closely related reference MAGs and isolate genomes exhibited a range of amino acid identities (AAI, 52.6%–98.6%; Fig. 2C). Altogether, the LCB24 MAG was found to be highly related to previously obtained Archaeoglobi MAGs encoding the MCR complex and only distantly related to other Archaeoglobales sp. (AAI, 58.9%–65%; average nucleotide identity (ANI), 70.3%–70.6%; 16S rRNA ANI, 91.6%–93.8%; SI AppendixFigs S1 and S2). Based on AAI, MAG LCB24 was most closely related to Archaeoglobi LCB024-003 MAG (AAI, 98.6%), which we had obtained from the same hot spring in a previous study [33]. The ANI and AAI values to the closest cultured methanogen, Ca. Methanoglobus nevadensis GBS, are 80.2% and 83.3%, respectively. Based on these results, we designate this archaeon Ca. M. hypatiae strain LCB24, named after the philosopher Hypatia of Alexandria (for a protologue, see the SI Appendix, Results and Discussion). The estimated relative abundance of Ca. M. hypatiae based on the SIT-MG was 92.8%. Other community members in the LCB24 culture with >1% relative sequence abundance included members of the Pseudothermotoga (3.2%), Desulfovirgula (1.7%), and the family Moorellaceae (1.3%) (Fig. 1A, Dataset S1).
The only mcrAGCDB genes recovered from both metagenomes belong to the genome of Ca. M. hypatiae. Phylogenetic analysis of the single copy of McrA indicated its close relationship to McrA sequences found in members of the TACK superphylum (Fig. 2D). This contrasts with the placement of Ca. M. hypatiae within the Euryarchaeota based on phylogenomics (Fig. 2A), suggesting that Archaeoglobi could have obtained the MCR complex as a result of a horizontal gene transfer event from an archaeon in the TACK superphylum [7, 8]. Also, it could indicate that non-methanogenic Archaeoglobi lost the capacity for anaerobic methane cycling after they had diverged from a shared methanogenic ancestor.
Methanogenic activity of Ca. M. hypatiae
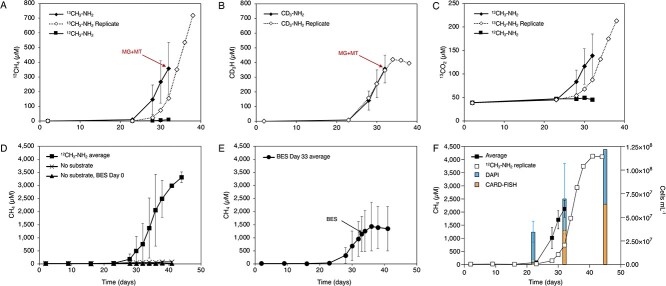
To gain insight into the activity of Ca. M. hypatiae under methanogenic and non-methanogenic conditions, a stable isotope tracing (SIT) experiment was conducted. Cultures were incubated in the presence of 10 mM of MMA; 8 mM of substrate were isotopically light, whereas the remaining 2 mM consisted of either 13C-MMA (13CH3-NH2) or D3-MMA (CD3-NH2). Addition of the methanogenesis inhibitor BES was used as a non-methanogenic control (Figs 1B and 3, SI Appendix, Fig. S3). On average across six replicates, the cultured converted 13CH3-NH2 to 356 μM 13CH4 (17.8%) and 138.71 μM 13CO2 (6.9%) by Day 32 (Fig. 3A and C, Dataset S2). The conversion of CD3-NH2 was nearly identical yielding 355 μM CD3H (Fig. 3B). In the exponential phase of methane production, five of the six replicates were harvested for metagenomic and metatranscriptomic sequencing, while the sixth replicate was allowed to grow to stationary phase. The replicate allowed to grow in each respective experiment converted the provided 13CH3-NH2 to 717.7 μM 13CH4 (35.9%) and 212.95 μM 13CO2 (10.65%) or CD3-NH2 to 394.76 μM CD3H (19.7%) by Day 38 (Fig. 3A–C). These results confirmed that MMA was converted to methane by the LCB24 culture. The production of 13CO2 may represent the dismutation of 13CH3-NH2 to generate the reducing power for methanogenesis via the methyl-branch of the Wood–Ljungdahl pathway (WLP) or may be explained by other organisms in the culture catabolizing MMA. Yet, no transcriptomic evidence for this activity was present in this experiment. No methane production was observed for cultures treated with BES or in cultures incubated without MMA (Fig. 3D). When BES was added to cultures in the exponential phase, methane production ceased indicating the generation of methane is reliant on the Archaeoglobi MCR (Fig. 3E).
Figure 3.
Conversion of stable isotope labeled MMA to methane by culture LCB24; (A) production of 13CH4 in cultures amended with 13CH3-NH2 vs. 12CH3-NH2 (six replicates); (B) production of CD3H in cultures amended with CD3-NH2 (six replicates); (C) production of 13CO2 in cultures amended with 13CH3-NH2 vs. 12CH3-NH2; for plots A and B, 10 total replicates across treatments were sacrificed during mid-exponential phase for metagenomic or metatranscriptomic sequencing indicated by red arrows; 13CH4, CD3H, or 13CO2 production for the replicate allowed to reach stationary phase is shown as a dashed line through open diamond symbols; (D) production of 12CH4 in cultures amended with 12C-MMA (12CH3-NH2; 2 replicates); cultures incubated without substrates (two replicates) and those to which the inhibitor BES was added on Day 0 (3 replicates) did not produce 12CH4 over the course of the experiment; (E) production of 12CH4 in cultures amended with 12CH3-NH2 to which BES was added on Day 33 of incubation (black arrow; three replicates); the average production of 12CH4 leveled off and ceased after the introduction of BES, indicating methane generation by Ca. M. hypatiae is MCR-dependent; error bars indicate standard deviation of biological replicates when applicable; measurements of 12CH4, 13CH4, CD3H, and 13CO2 for all replicates and controls are reported in Dataset S2; 12CH4 measurements for all controls and replicates are shown in SI Appendix, Fig. S4 and Dataset S3; (F) 12CH4 production and fraction of Ca. M. hypatiae cells in biological replicates incubated with 13CH3-NH2; relative abundance of cells was determined at three time points (Days 22, 32, 45) based on the fraction of Ca. M. hypatiae specific CARD-FISH counts (orange) versus total counts of DAPI-stained cells (blue); error bars indicate the standard deviation for four biological replicates on Days 22 and 32.
Visualization and cell enumeration
The growth of Ca. M. hypatiae was tracked in four replicates during the SIT experiment with catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) using a general archaea-targeted probe Arch915 [50] and DNA-staining (DAPI) (Fig. 1C). As the production of methane increased throughout the experiment, there was a concurrent rise in the relative cell abundance of Ca. M. hypatiae (Fig. 3F, SI Appendix, Table S5). The initial assessment on Day 22 across four replicates revealed the total cell density to be 3.45 × 107 ± 1.14 × 107 before substantial concentrations of methane had been detected in the headspace (<132 μM). By Day 32, methane concentrations reached 1777 ± 739 μM and the total cell density increased to 6.97 × 107 ± 3.73 × 107 cells ml−1 with 54% (±9.6%) of cells labeled as Ca. M. hypatiae (Fig. 3F). All but one of these replicates were then sacrificed for further analysis. Finally on Day 45, the remaining replicate reached a headspace methane concentration of 4109 μM and a total cell density of 1.22 × 108 with 53% of cells labeled as Ca. M. hypatiae.
Visualization of the enrichment culture via scanning electron microscopy (SEM) revealed that most cells exhibited a regular to irregular coccoid morphology, with a width ranging from 0.5 to 1 μm (Fig. 1D). This morphology has previously been described for other Archaeoglobi species [30, 51-53].
Alternative substrates and temperature optimum
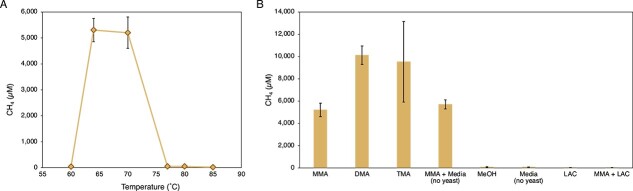
We determined the substrate and temperature range of Ca. M. hypatiae by growing the culture in the presence of several substrates at 70°C or with 10 mM MMA at 60–85°C (Fig. 4A and B). Conditions that lead to the production of methane included 10 mM trimethylamine (TMA), 10 mM dimethylamine (DMA), 10 mM MMA in media without yeast extract, and the control with 10 mM MMA and 0.01% yeast extract. Methane production of cultures grown with MMA in the presence or absence of yeast extract was indistinguishable (5202 ± 606 and 5703 ± 410 μM CH4, respectively) indicating that yeast extract is not essential for methanogenic growth. Observed methane concentrations were higher in incubations amended with DMA (10 115 ± 836 μM CH4) and TMA (9524 ± 3626 μM CH4, with a wide range of 5361–11 993 μM) on average more than the MMA controls, consistent with what has been observed for other methylotrophic methanogens [54]. Incubations amended with 10 mM methanol (MeOH) did not produce methane after 47 days of incubation at 70°C. Due to its use by sulfate-reducing organisms as an electron donor [55], 10 mM lactate (LAC) was tested, as well as 10 mM MMA with 10 mM LAC, but none of these incubations produced methane. Production of methane has not been observed in any attempted transfers where hydrogen (99.9999% purity) was present in the headspace, or hydrogen with MMA was added.
Figure 4.
Temperature and substrate range of culture LCB24; (A) methane production from MMA was observed between 64 and 70°C; (B) substrate range; methane production was observed for MMA, DMA, TMA, and in media prepared without yeast extract; MeOH, methanol; both experiments performed in triplicate; all measurements can be found in Dataset S4.
The enrichment grew optimally at both 64 and 70°C with relative amounts of methane produced at 5304 ± 451 μM and 5202 ± 606 μM, respectively. This deviates from the predicted optimal growth temperature of 74.4°C, which was derived from the translation of proteins in the Ca. M. hypatiae MAG using Tome [56]. This is lower than the observed range of growth and optimum temperatures for type strains of non-methanogenic Archaeoglobus which have been demonstrated to grow between 50 and 95°C with optimal temperatures between 75 and 83°C in organisms sourced predominantly from deep sea vent environments [30]. No methane production was detected at temperatures 77°C or above or lower than 64°C after 47 days of incubation (Dataset S4).
Genomic and transcriptomic basis for methanogenesis
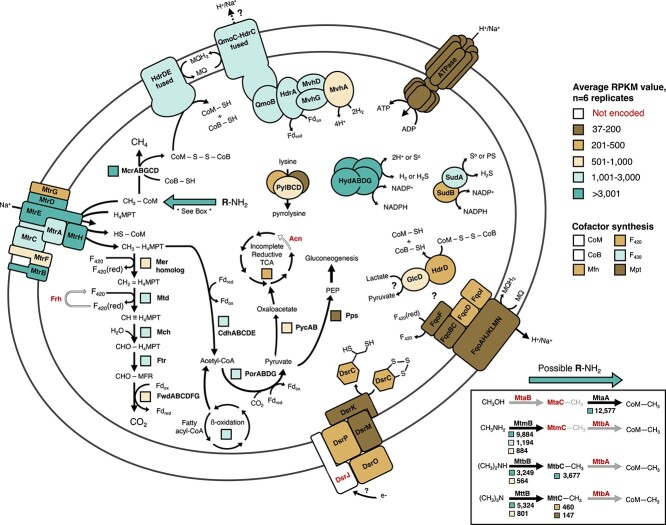
The assembled metagenome obtained at the end of the SIT experiment was used to align a total of 23 376 154 metatranscriptome mRNA reads obtained from six replicates harvested in the exponential growth phase and to create a detailed reconstruction of the metabolism of Ca. M. hypatiae (Figs 3A and B and 5, Dataset S5). A total of 22 891 651 reads, i.e. 97.8% of all recovered reads, were recruited to the Ca. M. hypatiae MAG. Only 2.1% of the total mRNA reads (484503) were aligned with other co-enriched organisms. Among these, only 13 genes across four MAGs were expressed above 200 RPKMs and just five genes exceeded >1000 RPKM. Genes required for the conversion of methylamine to methane were among the top 2% of highest expressed genes transcribed by Ca. M. hypatiae, including genes encoding the MCR complex (mcrAGCDB; 13 046–18 098 RPKM), one of three MMA methyltransferase copies (mtmB; 9884 RPKM), DMA corrinoid (mtbC; 3677 RPKM), and methanol:coenzyme M methyltransferase (mtaA; 12 577 RPKM) (Fig. 5). Seven copies of substrate-specific methyltransferases for MMA (mtmB; 3 copies), DMA (mtbB; 2 copies), and TMA (mttB; 2 copies) were present in the genome, but methanol methyltransferase (mtaB) was not identified. These genes were differentially expressed with one copy for each type of methylamine expressed above 3200 RPKM. In addition to mtbC, two gene copies of the TMA corrinoid protein (mttC) were found in the genome but their expression was relatively low (<460 RPKM average). MMA corrinoid (mtmC) or methanol corrinoid (mtaC) proteins were not identified in Ca. M. hypatiae. Additionally, genes were expressed for pyrrolysine synthesis (pylBCD; 819, 343, 37 RPKM) and the methyltransferase corrinoid activation protein (ramA; 1076 RPKM), both of which support methylamine methyltransferases in methylotrophic methanogenesis [57, 58]. The absence of mtmC and the high expression levels of mtbC (3677 RPKM) and mtaA (12 577 RPKM) suggest that they are responsible for the transfer of a methyl group from MMA to coenzyme M (CoM) after it has been transferred by a substrate-specific methyltransferase (mtmB). Consistent with the observed methane production from DMA and TMA, Ca. M. hypatiae can use these methylamines and expressed the corresponding genes (mtbB, mttB) at comparatively high levels (JOOIALLP_01813 mtbB 3249 RPKM; JOOIALLP_01787 mttB 5324 RPKM; Figs 4B and 5). It is worth noting that the expression of mtbB/mttB was detected despite the culture not having been previously exposed to DMA or TMA at the time of the transcriptomics experiment. We hypothesize that Ca. M. hypatiae could employ one of two strategies: it either (i) constitutively expresses all substrate-specific methyltransferases and corrinoid proteins as a precautionary measure to accommodate substrates potentially encountered in situ, or (ii) Ca. M. hypatiae transcriptionally co-regulates the genes responsible for these functions.
Figure 5.
Transcriptional activity in Ca. M. hypatiae grown under methanogenic conditions (N2 headspace, 10 mM MMA, and 0.01% yeast extract); transcriptionally active proteins are shown in bold black font; proteins not encoded in the MAG are colored in white and denoted in bold red font; average RPKMs values of six biological replicates are depicted; RPKM values are represented by boxes or colored subunits close to each protein and are colored according to their expression level with the RPKM value of the lowest expressed gene depicted, 37 RPKM; for enzymes comprising multiple subunits, the beta-oxidation pathway, and the TCA cycle, an average RPKM value representing the transcribed enzymes is used. Ca. M. hypatiae is transcriptionally active under methanogenic conditions and encodes the ability to convert methyl-groups from mono-, di-, and TMA to methane; this ability is enabled by several copies of substrate-specific methyltransferases and corrinoid proteins highlighted in the box to the bottom right; a complete list of genes described in this figure, their transcription levels, and their abbreviations is provided in Dataset S5.
Ca. M. hypatiae expressed the methyl-branch of the WLP and the acetyl-CoA decarbonylase/synthase complex (Cdh, cdhABCDE), which is consistent with genes observed and shown to be expressed in sulfate-reducing Archaeoglobi genomes [55]. This includes two paralogous copies of 5,10-methylenetetrahydromethanopterin reductase (mer) which might function as a traditional Mer, considering that these genes are also members of the large luciferase-like monooxygenase family (pfam00296) [35]. The expression of genes in the WLP varied. Methylenetetrahydromethanopterin dehydrogenase (mtd), methenyltetrahydromethanopterin cyclohydrolase (mch), formylmethanofuran-tetrahydromethanopterin N-formyltransferase (ftr), formylmethanofuran dehydrogenase (fwdABC), and one copy of the mer homologs were expressed at comparatively high levels (456–2763 RPKM), whereas FwdDEFG and the other mer copy were only minimally expressed (<180 RPKM) (Dataset S5). The high expression of the Cdh complex (cdhACDE; 3063 ± 362, cdhB 677 RPKM average across subunits) suggests that Ca. M. hypatiae is capable of autotrophically fixing CO2 to acetyl-CoA as has been shown for other Archaeoglobus species [59]. Acetyl-CoA could also be derived from the degradation of fatty acids present in yeast extract through the process of beta-oxidation. Enzymes involved in this pathway were expressed at moderate to high levels during growth (Dataset S6). Pyruvate synthase (Por) was highly expressed providing a way for acetyl-CoA to be converted to pyruvate and subsequently be fed into major biosynthetic pathways. Specifically, Ca. M. hypatiae encodes pyruvate carboxylase (PycAB), an incomplete reductive tricarboxylic acid cycle (rTCA), phosphoenolpyruvate synthase (Pps), most enzymes needed for gluconeogenesis, and several enzymes associated with the pentose phosphate pathway in archaea, which were all expressed at varying levels (Dataset S5). Together, these pathways provide Ca. M. hypatiae the capacity to synthesize amino acids, carbohydrates, integral components of the cell wall, and vital sugars for nucleic acids.
Several complexes related to energy conservation and electron transport were moderately to highly expressed. Ca. M. hypatiae encodes a fused heterodisulfide reductase (hdrDE) that was highly expressed (1106 ± 120 RPKM) in addition to a fused hdrD/mvhD and four copies of hdrD that were all expressed at much lower levels (<500 RPKM). The differing levels of transcription suggest that the membrane-bound HdrDE is responsible for the regeneration of coenzymes M and B through the reduction of heterodisulfide (CoM-S-S-CoB). Additionally, the absence of HdrB, which contains the active site for disulfide reduction, eliminates the possibility that disulfide reduction could occur via a HdrABC complex [60]. As reported for Ca. M. nevadensis [35], a unique gene cluster was identified containing F420-non-reducing hydrogenase (MvhAGD), two HdrA copies and a QmoC fused to a HdrC. One HdrA copy (JOOIALLP_01710) was predicted by DiSCo analysis as a quinone-modifying oxidoreductase (QmoB), a protein related to the HdrA of methanogens [61, 62]. This cluster was expressed at high levels (995–2431 RPKM average), suggesting its importance for electron transfer in Ca. M. hypatiae. We hypothesize that these subunits are associating together in vivo to bifurcate electrons from hydrogen (H2) to reduce both menaquinone (MQ) and ferredoxin (Fdox), as proposed recently [35, 63]. Lastly, Ca. M. hypatiae moderately expressed a membrane-bound F420H2:quinone oxidoreductase (Fqo) complex (88–280 RPKM across subunits) and a V-type ATP synthase (24–442 RPKM across subunits).
The electrons required for reducing the CoM-S-S-CoB heterodisulfide could originate from two possible routes. The first possibility would rely on sourcing electrons from hydrogen, which could be oxidized by the Mvh-Qmo-Hdr complex coupled to MQ reduction. H2 may be produced through the activity of a group 3b [NiFe]-sulfhydrogenase (HydABDG), which was the highest expressed hydrogenase complex with an average RPKM of 4421 across subunits [64, 65]. To evolve hydrogen via HydABDG, reducing power, via NADPH, could be supplied by sulfide dehydrogenase (SudAB; SudA, 1088 RPKM; SudB, 495 RPKM). Alternatively, NADPH could instead be provided to biosynthesis pathways and therefore be decoupled from methanogenic metabolism. H2 could also potentially be sourced from fermentative bacteria in the enrichment culture; however, the low number of hydrogenases encoded by co-enriched organisms was only very lowly expressed at the time of sampling for metatranscriptomics (<51 RPKM). At this point, the source of H2Ca. M. hypatiae uses remains uncertain, as no H2 was added to the headspace. The second option for reducing the CoM-S-S-CoB heterodisulfide involves a hydrogen-independent electron transport system, where reduced F420 and ferredoxin are generated through the dismutation of methylated substrate to CO2 via the WLP. Reduced F420 could be oxidized by the Fqo complex and contribute to a reduced MQ pool that could be used by the fused HdrDE complex to reduce CoM-S-S-CoB. Reduced ferredoxin could be oxidized at a soluble FqoF to reduce F420 or at an Fqo complex lacking FqoF to reduce MQ [66, 67]. Based on the low expression levels of the Fqo complex (171 ± 67 RPKM) and the absence of F420-reducing hydrogenase (frh) from the genome, it is not likely the WLP runs in the reductive direction as a source of reduced F420 would be required. Resolving the exact configuration of the electron transport system encoded by Ca. M. hypatiae will require biochemical confirmation in future investigations.
Genes necessary for dissimilatory sulfate reduction typically observed in sulfate-reducing members of the Archaeoglobi, including dissimilatory sulfite reductase (dsrAB), sulfate adenylyltransferase (sat), and adenylylsulfate reductase (aprAB), were neither identified in the genome of Ca. M. hypatiae nor in the unbinned fraction of the metagenome. They were also absent from the comparatively incomplete MAG of Ca. M. nevadensis GBS [35]. However, Ca. M. hypatiae encodes subunits dsrMK and dsrOP of the Dsr complex in addition to dsrC. This complex is strictly conserved in sulfate-reducing organisms [68] where it mediates electron transfer from the periplasm to the cytoplasm reducing the disulfide bond found in DsrC cysteines [69]. The expression of the Dsr complex and dsrC was low (450 ± 63 RPKM) during growth on MMA suggesting it is not vital to the metabolism of Ca. M. hypatiae. The presence of the Dsr complex, DsrC, and subunits QmoC and QmoB in the genome may be explained as evolutionary remnants from ancestral Archaeoglobi, growing initially as sulfate-reducing organisms but later transitioning to a methanogenic lifestyle [7, 8]. This raises the question whether intermediate of this process, Archaeoglobi capable of both methanogenesis and sulfate-reduction (and possible anaerobic oxidation of methane), still exist today [25, 28].
Collectively, the metagenomic and transcriptomic data confirmed that Ca. M. hypatiae is not only the sole archaeon but the sole methanogen in our culture. The metabolic reconstruction and metatranscriptomic results are consistent with methylotrophic methanogenesis from methylamines. The absence of genes required for sulfate reduction eliminates the possibility for this metabolism in Ca. M. hypatiae. A unique gene cluster (Mvh-Qmo-Hdr) potentially involved in energy conservation was expressed; however, future studies will be required to test how Ca. M. hypatiae internally cycles electrons for methanogenesis and if it sources H2, or other reductants, from the medium or co-enriched bacteria.
Distribution of Ca. methanoglobus across geothermal features in YNP
16S rRNA and mcrA gene amplicon sequence data generated in a recent microbial diversity survey of 100 geothermal features in YNP [33] were used to analyze the distribution of Archaeoglobi related to Ca. M. hypatiae (SI Appendix, Fig. S5). 16S rRNA gene amplicons closely related to Ca. M. hypatiae (96.7%–100% sequence identity) were found in seven DNA samples from six hot springs (pH 5.1–9.35, 31–78°C) in addition to hot spring LCB024 (the source of this culture) at relative abundances ranging from 0.02% to 0.22%. In addition, mcrA gene ASVs affiliated with Archaeoglobi were PCR-amplified from 53 DNA samples, out of 201 total samples that had been screened by PCR. These 53 samples had been collected from microbial mats or sediments originating from 36 geothermal features distributed across various thermal regions within YNP by Lynes et al. [33]. Archaeoglobi-related mcrA genes were found in geothermal features with a pH range of 2.61 to 9.32 and a temperature range of 18.4–93.8°C. Collectively, our results and the studies by Wang et al. and Buessecker et al., who reported that Mcr-encoding Archaeoglobi are present [35] and transcriptionally active in hot spring mesocosms [34], demonstrate the previously overlooked role that Archaeoglobi might play in the anaerobic carbon cycle of geothermal environments.
Conclusion
The cultivation of Ca. M. hypatiae LCB24 provides direct experimental evidence that members of the Archaeoglobi are methanogens. Ca. M. hypatiae can use MMA, DMA, and TMA as methanogenic substrates and grows optimally at 64–70°C, as evidenced by metagenomics, metatranscriptomics, and isotope tracing experiments. Metagenomic sequencing and phylogenomic analysis confirmed the close relationship of Ca. M. hypatiae to other Mcr-encoding Archaeoglobi and the relatedness of its mcrA to MAGs of the TACK superphylum, some of which have recently been shown to also be methanogens [70, 71]. Together, this supports the idea that the capacity for methanogenesis is deeply rooted in the archaea and possibly dates to the last common ancestor of archaea [1, 3, 7, 8, 72]. The wide distribution of Archaeoglobi-affiliated mcrA gene sequences and Ca. M. hypatiae-related 16S rRNA gene sequences in geothermal features across YNP suggests that members of this lineage play a hitherto unaccounted-for role in anaerobic carbon cycling in these extreme ecosystems. Future studies of Ca. M. hypatiae and other methanogens will provide valuable insights into the evolution of methane metabolism and the significance of these archaea in biogeochemical cycles across geothermal and other environments.
Supplementary Material
Acknowledgements
We thank the US National Park Service for permitting work in YNP under permit number YELL-SCI-8010. We thank George Schaible (MSU) for the help with SEM imaging, Dr Viola Krukenberg (MSU) for initial FISH methodology development, Sylvia Nupp, Dr Andrew Montgomery, and Paige Schlegel (all MSU) for the assistance with field sampling, Dr Christopher Lemon (MSU) for allowing use of his cooling centrifuge, and Dr Marike Palmer (UN Las Vegas) for discussing naming of this archaeon.
Contributor Information
Mackenzie M Lynes, Department of Chemistry and Biochemistry, Center for Biofilm Engineering, Thermal Biology Institute, Montana State University, Bozeman, MT 59717, United States.
Zackary J Jay, Department of Chemistry and Biochemistry, Center for Biofilm Engineering, Thermal Biology Institute, Montana State University, Bozeman, MT 59717, United States.
Anthony J Kohtz, Department of Chemistry and Biochemistry, Center for Biofilm Engineering, Thermal Biology Institute, Montana State University, Bozeman, MT 59717, United States.
Roland Hatzenpichler, Department of Chemistry and Biochemistry, Center for Biofilm Engineering, Thermal Biology Institute, Montana State University, Bozeman, MT 59717, United States; Department of Microbiology and Cell Biology, Montana State University, Bozeman, MT 59717, United States.
Author contributions
Mackenzie M. Lynes and Roland Hatzenpichler developed the research project. Mackenzie M. Lynes, Zackary J. Jay, Anthony J. Kohtz, and Roland Hatzenpichler designed experiments. Mackenzie M. Lynes and Anthony J. Kohtz conducted field sampling. Mackenzie M. Lynes performed cultivation, extracted DNA for amplicon and metagenomic sequencing, extracted RNA for transcriptomic sequencing, and conducted physiology and stable isotope experiments. Anthony J. Kohtz developed GC/GCMS protocols and processed GCMS samples. Zackary J. Jay processed and annotated metagenomic and transcriptomic data, assembled MAGs, mapped transcripts, assigned taxonomy, constructed 16S rRNA gene phylogeny, and performed phylogenetic analysis of MAGs. Mackenzie M. Lynes conducted phylogenetic analysis of amplicon data, refined gene annotations, reconstructed, and interpreted the metabolic potential of Ca. M. hypatiae with insight from Zackary J. Jay and Anthony J. Kohtz. Roland Hatzenpichler was responsible for funding and supervision of the project. Mackenzie M. Lynes and Roland Hatzenpichler wrote the manuscript, which was then edited by all authors.
Conflicts of interest
None declared.
Funding
This study was funded through a NASA Exobiology program award (80NSSC19K1633) to R.H.
Data availability
All metagenomic, metatranscriptomic, and amplicon data discussed in this manuscript are available under NCBI BioProject ID PRJNA1014417. McrA gene amplicon data from YNP hot springs discussed in this manuscript have been previously published (Lynes et al.) and are available under NCBI under BioProject PRJNA859922.
References
- 1. Wolfe JM, Fournier GP. Horizontal gene transfer constrains the timing of methanogen evolution. Nat Ecol Evol 2018;2:897–903. 10.1038/s41559-018-0513-7 [DOI] [PubMed] [Google Scholar]
- 2. Sorokin DY, Makarova KS, Abbas Bet al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol 2017;2:17081. 10.1038/nmicrobiol.2017.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Martin WF, Sousa FL. Early microbial evolution: the age of anaerobes. CSH Perspect Biol 2016;8:a018127. 10.1101/cshperspect.a018127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sauterey B, Charnay B, Affholder Aet al. Co-evolution of primitive methane-cycling ecosystems and early Earth’s atmosphere and climate. Nat Commun 2020;11:2705. 10.1038/s41467-020-16374-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ueno Y, Yamada K, Yoshida Net al. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 2006;440:516–9. 10.1038/nature04584 [DOI] [PubMed] [Google Scholar]
- 6. Spang A, Ettema TJG. Archaeal evolution: the methanogenic roots of Archaea. Nat Microbiol 2017;2:17109. 10.1038/nmicrobiol.2017.109 [DOI] [PubMed] [Google Scholar]
- 7. Adam PS, Kolyfetis GE, Bornemann TLVet al. Genomic remnants of ancestral methanogenesis and hydrogenotrophy in Archaea drive anaerobic carbon cycling. Sci Adv 2022;8:eabm9651. 10.1126/sciadv.abm9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Y, Wegener G, Williams TAet al. A methylotrophic origin of methanogenesis and early divergence of anaerobic multicarbon alkane metabolism. Sci Adv 2021;7:eabj1453. 10.1126/sciadv.abj1453 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Hinrichs K-U. Microbial fixation of methane carbon at 2.7 Ga: was an anaerobic mechanism possible? Geochem Geophy Geosy 2002;3:1–10. 10.1029/2001GC000286 [DOI] [Google Scholar]
- 10. Saunois M, Stavert AR, Poulter Bet al. The global methane budget 2000–2017. Earth Syst Sci Data 2020;12:1561–623. 10.5194/essd-12-1561-2020 [DOI] [Google Scholar]
- 11. Rosentreter JA, Borges AV, Deemer BRet al. Half of global methane emissions come from highly variable aquatic ecosystem sources. Nat Geosci 2021;14:225–30. 10.1038/s41561-021-00715-2 [DOI] [Google Scholar]
- 12. Garcia PS, Gribaldo S, Borrel G. Diversity and evolution of methane-related pathways in archaea. Annu Rev Microbiol 2022;76:727–55. 10.1146/annurev-micro-041020-024935 [DOI] [PubMed] [Google Scholar]
- 13. Ferry JG, Kastead KA. Methanogenesis. In: Cavicchioli R. (ed.), Archaea: Molecular and Cellular Biology. ASM Press, 2007, 288–314. [Google Scholar]
- 14. Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Env Microbiol Rep 2009;1:285–92. 10.1111/j.1758-2229.2009.00038.x [DOI] [PubMed] [Google Scholar]
- 15. Scheller S, Goenrich M, Boecher Ret al. The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 2010;465:606–8. 10.1038/nature09015 [DOI] [PubMed] [Google Scholar]
- 16. Thauer RK. Methyl (alkyl)-coenzyme M reductases: nickel F-430-containing enzymes involved in anaerobic methane formation and in anaerobic oxidation of methane or of short chain alkanes. Biochemistry 2019;58:5198–220. 10.1021/acs.biochem.9b00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evans PN, Boyd JA, Leu AOet al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol 2019;17:219–32. 10.1038/s41579-018-0136-7 [DOI] [PubMed] [Google Scholar]
- 18. Thauer RK, Kaster AK, Seedorf Het al. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 2008;6:579–91. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- 19. Baker BJ, De Anda V, Seitz KWet al. Diversity, ecology and evolution of Archaea. Nat Microbiol 2020;5:887–900. 10.1038/s41564-020-0715-z [DOI] [PubMed] [Google Scholar]
- 20. Bueno de Mesquita CP, Wu D, Tringe SG. Methyl-based methanogenesis: an ecological and genomic review. Microbiol Mol Biol Rev 2023;87:e00024–2. 10.1128/mmbr.00024-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Söllinger A, Urich T. Methylotrophic methanogens everywhere — physiology and ecology of novel players in global methane cycling. Biochem Soc Trans 2019;47:1895–907. 10.1042/BST20180565 [DOI] [PubMed] [Google Scholar]
- 22. Nobu MK, Narihiro T, Kuroda Ket al. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J 2016;10:2478–87. 10.1038/ismej.2016.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vanwonterghem I, Evans PN, Parks DHet al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol 2016;1:16170. 10.1038/nmicrobiol.2016.170 [DOI] [PubMed] [Google Scholar]
- 24. McKay LJ, Dlakic M, Fields MWet al. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat Microbiol 2019;4:614–22. 10.1038/s41564-019-0362-4 [DOI] [PubMed] [Google Scholar]
- 25. Wang Y, Wegener G, Hou Jet al. Expanding anaerobic alkane metabolism in the domain of Archaea. Nat Microbiol 2019;4:595–602. 10.1038/s41564-019-0364-2 [DOI] [PubMed] [Google Scholar]
- 26. Evans PN, Parks DH, Chadwick GLet al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015;350:434–8. 10.1126/science.aac7745 [DOI] [PubMed] [Google Scholar]
- 27. Berghuis BA, Yu FB, Schulz Fet al. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc Natl Acad Sci U S A 2019;116:5037–44. 10.1073/pnas.1815631116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu YF, Chen J, Zaramela LSet al. Genomic and transcriptomic evidence supports methane metabolism in Archaeoglobi. mSystems 2020;5:e00651–19. 10.1128/mSystems.00651-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stetter KO, Lauerer G, Thomm Met al. Isolation of extremely thermophilic sulfate reducers: evidence for a novel branch of archaebacteria. Science 1987;236:822–4. 10.1126/science.236.4803.822 [DOI] [PubMed] [Google Scholar]
- 30. Slobodkina G, Allioux M, Merkel Aet al. Physiological and genomic characterization of a hyperthermophilic archaeon Archaeoglobus neptunius sp. nov. isolated from a deep-sea hydrothermal vent warrants the reclassification of the genus Archaeoglobus. Front Microbiol 2021;12:679245. 10.3389/fmicb.2021.679245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boyd JA, Jungbluth SP, Leu AOet al. Divergent methyl-coenzyme M reductase genes in a deep-subseafloor Archaeoglobi. ISME J 2019;13:1269–79. 10.1038/s41396-018-0343-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hua ZS, Wang YL, Evans PNet al. Insights into the ecological roles and evolution of methyl-coenzyme M reductase-containing hot spring Archaea. Nat Commun 2019;10:4574. 10.1038/s41467-019-12574-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lynes MM, Krukenberg V, Jay ZJet al. Diversity and function of methyl-coenzyme M reductase-encoding archaea in Yellowstone hot springs revealed by metagenomics and mesocosm experiments. ISME Commun 2023;3:22. 10.1038/s43705-023-00225-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J, Qu Y-N, Evans PNet al. Evidence for nontraditional mcr-containing archaea contributing to biological methanogenesis in geothermal springs. Sci Adv 2023;9:eadg6004. 10.1126/sciadv.adg6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buessecker S, Chadwick GL, Quan MEet al. Mcr-dependent methanogenesis in Archaeoglobaceae enriched from a terrestrial hot spring. ISME J 2023;17:1649–59. 10.1038/s41396-023-01472-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laso-Perez R, Krukenberg V, Musat Fet al. Establishing anaerobic hydrocarbon-degrading enrichment cultures of microorganisms under strictly anoxic conditions. Nat Protoc 2018;13:1310–30. 10.1038/nprot.2018.030 [DOI] [PubMed] [Google Scholar]
- 37. Brandis A, Thauer RK. Growth of Desulfovibrio species on hydrogen and sulphate as sole energy source. Microbiology 1981;126:249–52. 10.1099/00221287-126-1-249 [DOI] [Google Scholar]
- 38. Ai G, Zhu J, Dong Xet al. Simultaneous characterization of methane and carbon dioxide produced by cultured methanogens using gas chromatography/isotope ratio mass spectrometry and gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom 2013;27:1935–44. 10.1002/rcm.6651 [DOI] [PubMed] [Google Scholar]
- 39. Bushnell B. BBMap: A Fast, Accurate, Splice-Aware Aligner. Berkeley, CA: Lawrence Berkeley National Lab (LBNL), 2014 [Google Scholar]
- 40. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 41. Wu Y-W, Tang Y-H, Tringe SGet al. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2014;2:26. 10.1186/2049-2618-2-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kang DD, Froula J, Egan Ret al. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015;3:e1165. 10.7717/peerj.1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alneberg J, Bjarnason BS, de Bruijn Iet al. Binning metagenomic contigs by coverage and composition. Nat Methods 2014;11:1144–6. 10.1038/nmeth.3103 [DOI] [PubMed] [Google Scholar]
- 44. Miller IJ, Rees ER, Ross Jet al. Autometa: automated extraction of microbial genomes from individual shotgun metagenomes. Nucleic Acids Res 2019;47:e57. 10.1093/nar/gkz148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sieber CMK, Probst AJ, Sharrar Aet al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol 2018;3:836–43. 10.1038/s41564-018-0171-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kohtz AJ, Jay ZJ, Lynes MMet al. Culexarchaeia, a novel archaeal class of anaerobic generalists inhabiting geothermal environments. ISME Commun 2022;2:86. 10.1038/s43705-022-00175-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parks DH, Imelfort M, Skennerton CTet al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 2015;25:1043–55. 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Andrews S, Krueger F, Seconds-Pichon Aet al. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics. Babraham Institute, 2015. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 49. Deng Z-L, Münch PC, Mreches Ret al. Rapid and accurate identification of ribosomal RNA sequences via deep learning. Nucleic Acids Res 2022;50:e60. 10.1093/nar/gkac112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stahl DA. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E., Goodfellow M. (eds.), Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: John Wiley & Sons Ltd, 1991, 205–49 [Google Scholar]
- 51. Stetter KO. Archaeoglobus fulgidus gen. nov., sp. nov.: a new taxon of extremely thermophilic archaebacteria. Syst Appl Microbiol 1988;10:172–3. 10.1016/S0723-2020(88)80032-8 [DOI] [Google Scholar]
- 52. Huber H, Jannasch H, Rachel Ret al. Archaeoglobus veneficus sp. nov., a novel facultative chemolithoautotrophic hyperthermophilic sulfite reducer, isolated from abyssal black smokers. Syst Appl Microbiol 1997;20:374–80. 10.1016/S0723-2020(97)80005-7 [DOI] [Google Scholar]
- 53. Mori K, Maruyama A, Urabe Tet al. Archaeoglobus infectus sp. nov., a novel thermophilic, chemolithoheterotrophic archaeon isolated from a deep-sea rock collected at Suiyo Seamount, Izu-Bonin Arc, western Pacific Ocean. Int J Syst Evol Micr 2008;58:810–6. 10.1099/ijs.0.65422-0 [DOI] [PubMed] [Google Scholar]
- 54. Watkins AJ, Roussel EG, Webster Get al. Choline andN,N-dimethylethanolamine as direct substrates for methanogens. Appl Environ Microbiol 2012;78:8298–303. 10.1128/AEM.01941-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hocking WP, Stokke R, Roalkvam Iet al. Identification of key components in the energy metabolism of the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus by transcriptome analyses. Front Microbiol 2014;5:1–20. 10.3389/fmicb.2014.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li G, Rabe KS, Nielsen Jet al. Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth Biol 2019;8:1411–20. 10.1021/acssynbio.9b00099 [DOI] [PubMed] [Google Scholar]
- 57. Mahapatra A, Patel A, Soares JAet al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol Microbiol 2006;59:56–66. 10.1111/j.1365-2958.2005.04927.x [DOI] [PubMed] [Google Scholar]
- 58. Ferguson T, Soares JA, Lienard Tet al. RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea. J Biol Chem 2009;284:2285–95. 10.1074/jbc.M807392200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Estelmann S, Ramos-Vera WH, Gad'on Net al. Carbon dioxide fixation in ‘Archaeoglobus lithotrophicus’: are there multiple autotrophic pathways? FEMS Microbiol Lett 2011;319:65–72. 10.1111/j.1574-6968.2011.02268.x [DOI] [PubMed] [Google Scholar]
- 60. Ferry JG. How to make a living by exhaling methane. Annu Rev Microbiol 2010;64:453–73. 10.1146/annurev.micro.112408.134051 [DOI] [PubMed] [Google Scholar]
- 61. Chernyh NA, Neukirchen S, Frolov ENet al. Dissimilatory sulfate reduction in the archaeon 'Candidatus Vulcanisaeta moutnovskia' sheds light on the evolution of sulfur metabolism. Nat Microbiol 2020;5:1428–38. 10.1038/s41564-020-0776-z [DOI] [PubMed] [Google Scholar]
- 62. Neukirchen S, Sousa FL. DiSCo: a sequence-based type-specific predictor of Dsr-dependent dissimilatory sulphur metabolism in microbial data. Microb Genom 2021;7:000603. 10.1099/mgen.0.000603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramos AR, Keller KL, Wall JDet al. The membrane QmoABC complex interacts directly with the dissimilatory adenosine 5′-phosphosulfate reductase in sulfate reducing bacteria. Front Microbiol 2012;3:137. 10.3389/fmicb.2012.00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Adams MWW. The metabolism of hydrogen by extremely thermophilic, sulfur-dependent bacteria. FEMS Microbiol Rev 1990;75:219–37. 10.1111/j.1574-6968.1990.tb04096.x [DOI] [Google Scholar]
- 65. Ma K, Schicho RN, Kelly RMet al. Hydrogenase of the hyperthermophile Pyrococcus furiosus is an elemental sulfur reductase or sulfhydrogenase: evidence for a sulfur-reducing hydrogenase ancestor. Proc Natl Acad Sci U S A 1993;90:5341–4. 10.1073/pnas.90.11.5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hocking WP, Roalkvam I, Magnussen Cet al. Assessment of the carbon monoxide metabolism of the hyperthermophilic sulfate-reducing archaeon Archaeoglobus fulgidus VC-16 by comparative transcriptome analyses. Archaea 2015;2015:235384. 10.1155/2015/235384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Welte C, Deppenmeier U. Membrane-bound electron transport in Methanosaeta thermophila. J Bacteriol 2011;193:2868–70. 10.1128/JB.00162-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pereira IA, Haveman SA, Voordouw G. Biochemical, genetic and genomic characterization of anaerobic electron transport pathways in sulphate-reducing Delta-proteobacteria. In: Barton L.L., Hamilton W.A. (eds.), Sulphate-Reducing Bacteria: Environmental and Engineered Systems. Cambridge, UK: Cambridge University Press, 2007, 215–40 [Google Scholar]
- 69. Grein F, Pereira IA, Dahl C. Biochemical characterization of individual components of the Allochromatium vinosum DsrMKJOP transmembrane complex aids understanding of complex function in vivo. J Bacteriol 2010;192:6369–77. 10.1128/JB.00849-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu K, Zhou L, Tahon Get al. Isolation of a methyl-reducing methanogen outside the Euryarchaeota. 2023. 10.21203/rs.3.rs-2501667/v1 [DOI] [PubMed]
- 71. Kohtz A, Krukenberg V, Petrosian Net al. Cultivation and visualization of a methanogen of the phylum Thermoproteota. 2023. 10.21203/rs.3.rs-2500102/v1 [DOI] [PubMed]
- 72. Spang A, Caceres EF, Ettema TJG. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 2017;357:eaaf3883. 10.1126/science.aaf3883 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All metagenomic, metatranscriptomic, and amplicon data discussed in this manuscript are available under NCBI BioProject ID PRJNA1014417. McrA gene amplicon data from YNP hot springs discussed in this manuscript have been previously published (Lynes et al.) and are available under NCBI under BioProject PRJNA859922.