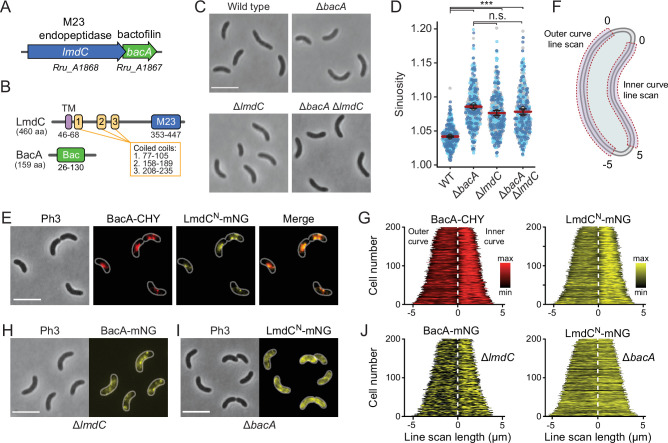
Figure 10. BacA and LmdC contribute to cell morphogenesis in R. rubrum.
(A) Schematic representation of the bacA-lmdC operon in R. rubrum. (B) Domain organization of LmdC and BacA from R. rubrum. LmdCRs consists of a transmembrane helix (TM) followed by a coiled-coil-rich region and a C-terminal M23 peptidase domain (M23). The bactofilin polymerization domain (green box) is flanked by non-structured N- and C-terminal regions. Numbers indicate the first and last amino acid of a domain. (C) Phenotypes of R. rubrum wild-type (S1), ΔbacARs (SP70), ΔlmdCRs (SP68), and ΔbacARs ΔlmdCRs (SP116) cells, imaged using phase-contrast microscopy. Bar: 5 µm. (D) Superplots showing the distribution of cell sinuosities in populations of the indicated R. rubrum strains. Small dots represent the data points, large dots represent the median values of the three independent experiments shown in the graphs (dark blue, light blue, gray). The mean of the three median values is indicated by a horizontal red line. Whiskers represent the standard deviation. ***p<0.005; ns, not significant; unpaired two-sided Welch's t-test. (E) Co-localization of BacARs-CHY and LmdCNRs-mNG in the ΔlmdCRs background (SP119). The Pearson’s Correlation Coefficient (PCC) of the two fluorescence signals in a random subpopulation of cells is 0.916. Scale bar: 5 µm. (F) Schematic showing the approach used to quantify the distribution of proteins along the outer and inner curve of a cell in panels G and J. (G) Demographs showing the enrichment of BacA-CYH and LmdCN-mNG at the inner curve the populations of strain SP119 analyzed in panel E (n=200 cells per strain). The white dashed line represents the point of transition from the outer to the inner curve. (H) Localization of BacARs-mNG in the ΔlmdCRs background (SP98). Bar: 5 µm. (I) Localization of LmdCRs-mNG in the ΔbacARs background (SP118). Bar: 5 µm. (J) Demographs showing the outer-versus-inner curve distribution of BacARs-mNG in the ΔlmdCRs background (SP98; n=200 cells) and LmdCNRs-mNG in the ΔbacARs background (SP118; n=200 cells). The white dashed line represents the point of transition from the outer to the inner curve.
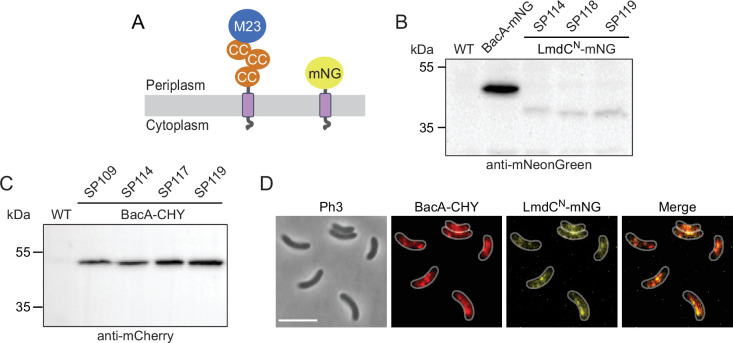
Figure 10—figure supplement 1. Analysis of the role of BacA and LmdC in R. rubrum morphogenesis.
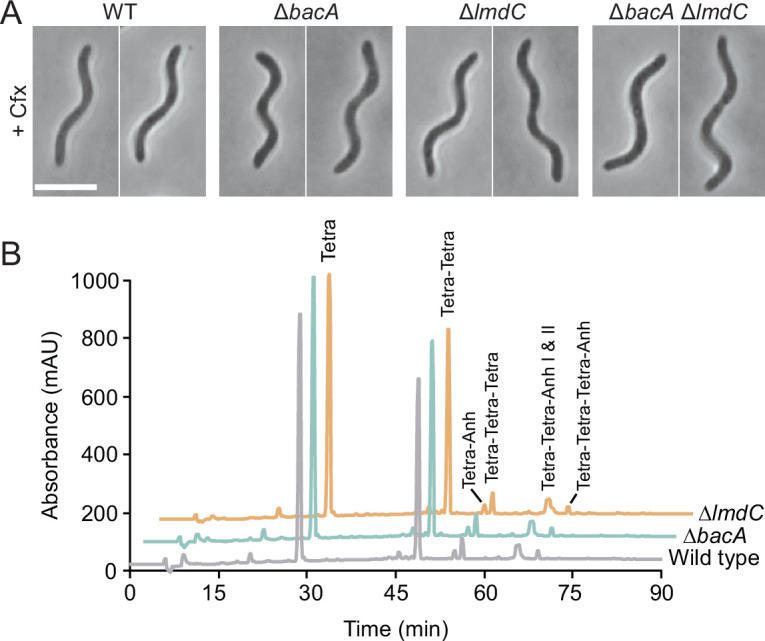
Figure 10—figure supplement 2. Cell lengths of R. rubrum strains.
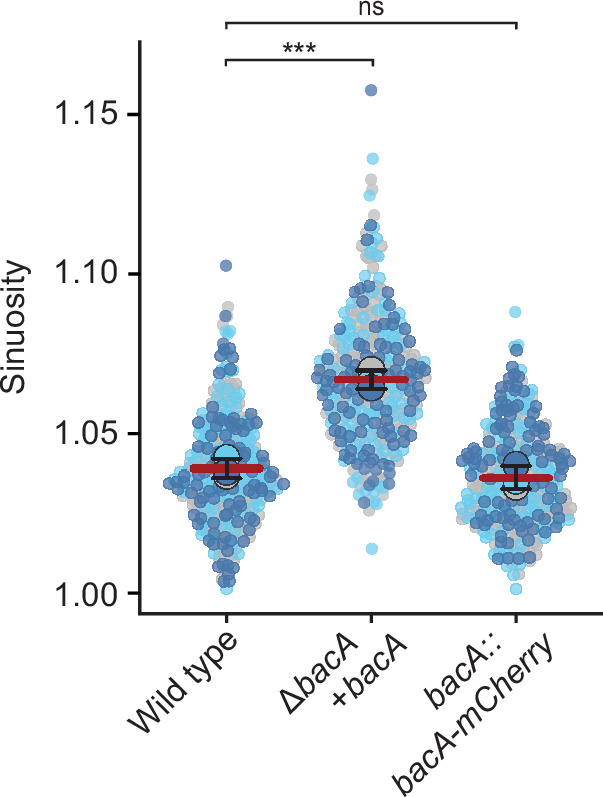
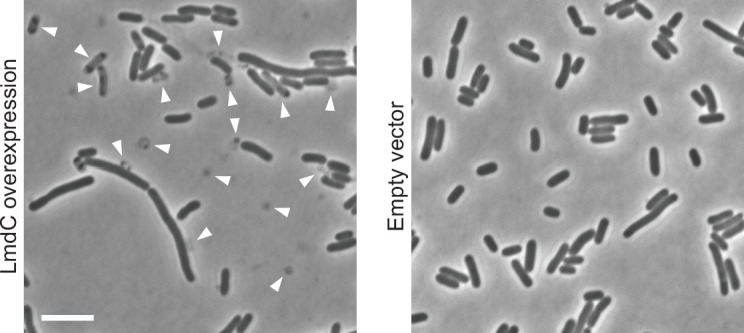
Figure 10—figure supplement 3. Lysis of E. coli upon heterologous expression of full-length R. rubrum lmdC.