Significance
The enormous success of bacterial pathogens has been attributed to their ability to exchange genetic material between one another. Similarly, in eukaryotes, horizontal transfer of genetic material allowed the spread of virulence factors across species. The horizontal transfer of whole chromosomes could be an important pathway for such exchange of genetic material, but little is known about the origin of transferable chromosomes and how frequently they are exchanged. Here, we show that the transfer of accessory chromosomes—chromosomes that are nonessential but may provide fitness benefits—is common during fungal co-infections and is even possible between distant pathogenic species, highlighting the importance of horizontal gene transfer via chromosome transfer also for the evolution and function of eukaryotic pathogens.
Keywords: entomopathogen, Metarhizium, preferential horizontal chromosome transfer, accessory chromosomes, intra- and interspecies horizontal chromosome transfer
Abstract
Entire chromosomes are typically only transmitted vertically from one generation to the next. The horizontal transfer of such chromosomes has long been considered improbable, yet gained recent support in several pathogenic fungi where it may affect the fitness or host specificity. To date, it is unknown how these transfers occur, how common they are, and whether they can occur between different species. In this study, we show multiple independent instances of horizontal transfers of the same accessory chromosome between two distinct strains of the asexual entomopathogenic fungus Metarhizium robertsii during experimental co-infection of its insect host, the Argentine ant. Notably, only the one chromosome—but no other—was transferred from the donor to the recipient strain. The recipient strain, now harboring the accessory chromosome, exhibited a competitive advantage under certain host conditions. By phylogenetic analysis, we further demonstrate that the same accessory chromosome was horizontally transferred in a natural environment between M. robertsii and another congeneric insect pathogen, Metarhizium guizhouense. Hence, horizontal chromosome transfer is not limited to the observed frequent events within species during experimental infections but also occurs naturally across species. The accessory chromosome that was transferred contains genes that may be involved in its preferential horizontal transfer or support its establishment. These genes encode putative histones and histone-modifying enzymes, as well as putative virulence factors. Our study reveals that both intra- and interspecies horizontal transfer of entire chromosomes is more frequent than previously assumed, likely representing a not uncommon mechanism for gene exchange.
Eukaryotic genomes consist of essential core genomes and, in some species, may also contain accessory chromosomes that are not essential. Accessory chromosomes are defined by their presence/absence polymorphism within a species and are widely observed in fungi, plants, and animals. These unique chromosomes (also known as supernumerary, lineage-specific, conditionally dispensable, or B chromosomes) can provide additional functions such as genes encoding virulence factors and are particularly common in fungal plant pathogens (1–4). The fitness effects of fungal accessory chromosomes can vary from negative to positive and appear to depend on the plant host genotype, cultivar, and/or species (5–9). Many fungal pathogens that harbor accessory chromosomes can infect a range of host species, so the presence/absence polymorphism of the accessory chromosomes is currently thought to be mainly due to their varying fitness effects in different plant host species. Despite their importance, the origin of accessory chromosomes is unknown; they may have originated from within the own genome (2) or may have been horizontally acquired from another species. The latter is supported by genomic comparisons for a few species (8, 10). Experimentally, however, horizontal transfer of accessory chromosomes has only been observed in very few fungal species. In all these cases, the transfer was observed during growth in vitro and at very low frequencies, requiring the use of inserted selectable fungicide-resistance markers in these studies (10–13). Hence, to date, horizontal transfer of an entire chromosome has not been observed during the natural stages of the life cycle of a fungus, e.g., during infections of a host with a pathogenic fungus. As a result, neither the frequency nor the underlying mechanisms of horizontal chromosome transfer between fungal pathogens are well understood.
Horizontal transfer of genetic material between different species was first recognized among bacteria, but has since then been found to occur also in the genomes of many eukaryotes (14–17). Transferred genetic material can have an adaptive advantage (18–21), such as the ToxA gene, which encodes an effector that interferes with the host plant’s immune system, and was horizontally transferred between three fungal pathogens of wheat (22, 23). The processes and mechanisms by which such horizontal transfer of genetic material can occur in eukaryotes are less well established, but, by definition, should result in a small amount of a donor genome becoming part of a recipient genome (17). Anastomosis—the fusion between fungal hyphae, transfer by (viral) vectors, and parasexuality in fungi—could represent such processes that might result in horizontal transfer. For instance, in parasexuality, vegetative cells fuse, resulting in cells with multiple nuclei that could either exchange genetic material (24) or fuse through karyogamy (25, 26). Prominent examples of such parasexual cycles are the human pathogens Candida albicans and Aspergillus fumigatus (18, 27–29). Here, the fusion of the nuclei might be followed by mitotic recombination and random chromosome losses that will eventually re-establish a stable ploidy level (18, 19, 30). Therefore, the horizontal transfer of genetic material in eukaryotes is important, but observing such transfer events is very difficult because they seem to occur very rarely. Such horizontal transfer could create new advantageous variation, particularly in asexual organisms. Asexual reproduction has long-term disadvantages due to the accumulation of genotypes that are loaded with deleterious mutations, known as Muller’s ratchet (31, 32). In contrast, sexual reproduction offers the advantage of generating diversity in offspring by recombining beneficial alleles (33). However, the formation of recombinants between advantageous alleles is not possible in asexual populations unless new genetic information is acquired from a different individual, such as through horizontal gene transfer. The high prevalence of horizontal gene transfer in prokaryotes underscores its potential advantages in asexual populations. Since accessory chromosomes in pathogenic fungi frequently contain genes that impact virulence (1–4), the horizontal transfer of an accessory chromosome between asexual fungi could be especially beneficial for the recipient organism.
Here, we ask how often and to what extent genetic material can be transferred horizontally between asexual lineages of a fungal pathogen during co-infection of their insect host. To this end, we studied common insect pathogens of the genus Metarhizium (Ascomycota), which frequently infect and kill insects and are often used as biocontrol agents against insect pests (34). In addition to their parasitic lifestyle in insects, some are also associated with plants as rhizosphere colonizers and root endophytes (34). We used a set of fungal strains derived from a selection experiment, in which Argentine ants (Linepithema humile) were co-infected with a mixture of six strains of Metarhizium robertsii and Metarhizium brunneum and then kept under conditions either allowing only the ant’s individual immune defenses to act, or both their individual and cooperative social defenses (35). By analyzing the ancestral and the evolved fungal strains resulting from that experiment, we show that i) horizontal transfer of an accessory chromosome occurred frequently between two distinct strains of M. robertsii, and ii) only the accessory chromosome was transferred and spread through the population over 10 host infection cycles, which iii) depended on the presence or absence of the ant’s social immune defenses. iv) A phylogenetic analysis of 36 strains across the genus Metarhizium revealed that the same accessory chromosome, which easily transmits within species of M. robertsii, was horizontally transferred to a distantly related, congeneric Metarhizium guizhouense. Taken together, our results indicate that an accessory chromosome in Metarhizium is highly mobile and encodes factors that may influence its mobility as well as putative virulence factors.
Results
In a previous experiment we performed 10 serial infection passages of Argentine ants with six different strains of M. robertsii and M. brunneum in two treatment groups, consisting of either one individual ant (treatment: individual) or one ant in the presence of two nestmates performing social grooming (treatment: social), with 10 replicates for each treatment (see Fig. 1A for a depiction of the experimental procedure) (35). As a result, we had found that a co-infecting mix of M. robertsii and M. brunneum shows different phenotypic adaptations to the individual vs. the social immune defenses of their Argentine ant hosts (35). We here performed a detailed molecular analysis of all six ancestral and the 24 resulting evolved fungal strains to determine the genetic changes associated with this selection experiment.
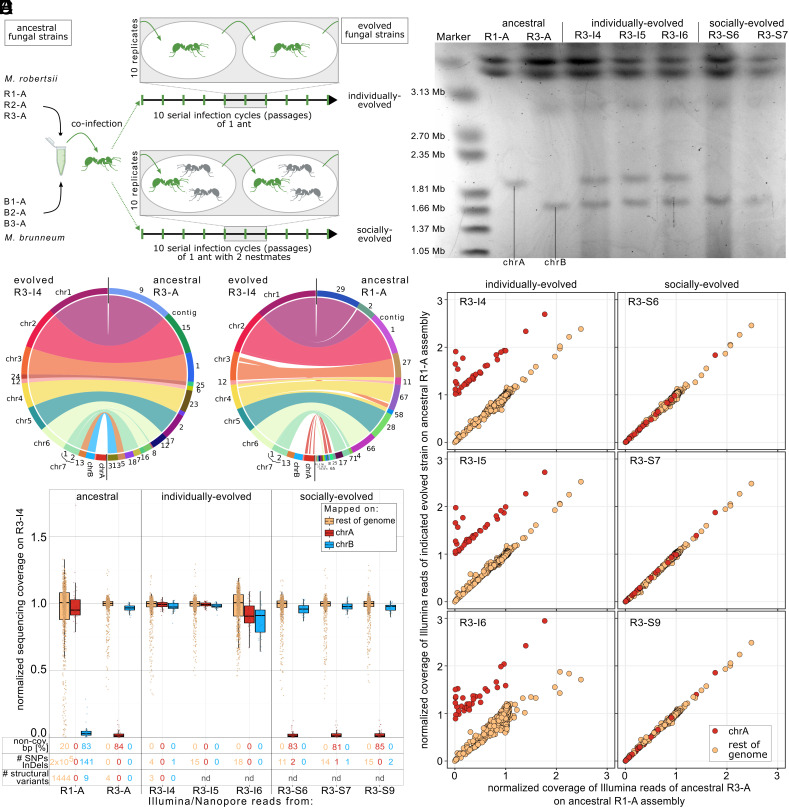
Fig. 1.
Multiple independent transfers of accessory chrA from the ancestral M. robertsii R1-A to evolved M. robertsii R3 during experimental co-infection of an ant host. (A) Experimental procedure of the selection experiment performed by Stock et al. (35). Argentine ants were exposed (green) to a mix of six strains—three M. robertsii and three M. brunneum—and kept either alone (individual treatment, n = 10 replicate lines) or with two nestmates (gray; social treatment, n = 10 replicate lines). The produced infectious spores were used to expose ants in 10 serial infection cycles as described in ref. 35. We here performed whole genome sequencing of the six ancestral and the individually and socially evolved strains at the end of the experiment. (B) Synteny blot of the nanopore-based assemblies of the evolved R3-I4 compared to the two ancestral R1-A and R3-A. Accessory chrA is missing in R3-A but shows synteny to contigs in R1-A. (C) Normalized Illumina sequencing coverage mapped on the evolved R3-I4 nanopore-based assembly in 50 kb windows for chrA (red), chrB (blue), and the rest of the genome (yellow). The fraction of genomic compartments lacking a single sequencing read (non-cov. bp) and the number of SNPs/InDels absent in the ancestral R3-A strain are given. The number of structural variants (≥50 bp) determined by nanopore reads mapped onto the R3-I4 nanopore-based assembly is indicated (nd = nondetermined, as no nanopore reads were available for these samples). (D) PFGE image of small chromosomes from the ancestral R1-A and R3-A, as well as all three individually evolved and two socially evolved R3 strains. All three independent individually evolved R3 strains contained both chrA and chrB. (E) Normalized sequencing coverage comparison of Illumina reads mapped on the ancestral R1-A nanopore-based assembly in 50-kb windows. Only 50-kb windows that are syntenic to chrA of R3-I4 exhibited changes in sequencing coverage in the three individually evolved R3 strains, indicating that no other genetic material was transferred to them from the ancestral R1-A. Note: The plots exclude the rDNA cluster for clarity, due to its high coverage.
Frequent Horizontal Transfer of Solely Accessory Chromosome A in M. robertsii during Insect Infection.
Preliminary analysis using fragmented Illumina-based assemblies for M. robertsii and M. brunneum indicated that horizontal transfer may have occurred between two strains of M. robertsii (R1 and R3), but not between the other co-infecting strains. In order to further analyze this potential transfer, we generated nanopore-based assemblies for two ancestral strains (R1-A and R3-A) and one evolved strain (R3 from individual treatment replicate 4, i.e., R3-I4). These near-chromosome-level assemblies (SI Appendix, Fig. S1 A–C and Table S1) showed a high degree of synteny with the only published chromosome-level assembly for a Metarhizium species, the related M. brunneum (SI Appendix, Fig. S1 D–F) (36), confirming our assembly procedure. We obtained the best assembly, with the lowest number of contigs, for the evolved R3-I4 strain, and consequently, renamed our R3-I4 main contigs based on their synteny with M. brunneum’s chromosome-level assemblies. Interestingly, one complete chromosome with telomeres at both ends (1.81 Mb) was absent in the ancestral R3-A strain but showed synteny with eleven small contigs (size range: 30 kb to 437 kb) of the more fragmented ancestral R1-A assembly (Fig. 1B). Hence, this chromosome showed presence/absence polymorphism in M. robertsii and is therefore accessory, and we termed this accessory chromosome A (chrA). Based on the synteny, there were no single nucleotide polymorphism (SNPs) on chrA between R1-A and R3-I4 (SI Appendix, Fig. S2 A–C). Remarkably, the absence of SNPs for accessory chrA contrasted with the high SNP density observed in all other syntenic chromosomes between R1-A and R3-I4. Additionally, we found another accessory chromosome (1.64 Mb) in the evolved R3-I4, present in the R3-A ancestral strain but absent in the R1-A ancestral strain (Fig. 1B). We termed this chromosome accessory chromosome B (chrB).
To confirm that the absence of chrA in R3-A and of chrB in R1-A is not reflecting an assembly error, we mapped Illumina sequencing reads to the R3-I4 assembly, which contains both accessory chrA and chrB (Fig. 1C). Both accessory chromosomes showed minimal sequencing coverage when mapped with the reads from the ancestral R3-A and R1-A, respectively, with the majority of chrA and chrB lacking a mapped sequencing read (84% and 83% of bases without a single sequencing read, respectively) and low median normalized coverage in 50-kb windows (0.9% and 2.6%, respectively) (Fig. 1C). Therefore, we concluded that chrA was absent in R-3A and chrB was absent in R1-A. Thus, the two ancestral strains possess different accessory chromosomes. The presence/absence polymorphism for chrA and chrB in R1-A and R3-A was further confirmed by pulsed-field gel electrophoresis (PFGE), which showed chromosomal bands of corresponding size (1.83 Mb for chrA and 1.62 Mb for chrB) for R1-A and R3-A, respectively, and no additional bands for small chromosomes in either strain, indicating that the more fragmented assembly for R1-A did not prevent us from detecting any additional small accessory chromosomes.
Sequencing coverage analysis revealed that all three R3 strains that had independently evolved under the individual treatment (R3-I4, R3-I5, and R3-I6) acquired the accessory chrA from the ancestral R1-A strain, while chrA was not present at the end of the experiment in any of the three R3 strains that independently evolved under the social treatment (R3-S6, R3-S7, R3-S9; Fig. 1C), a pattern that was further confirmed by PFGE (Fig. 1D). Although the size of the chromosomal bands of the evolved strains on the PFGE gel matches the size of chrA and chrB in the R3-I4 assembly as well as the sizes of the chromosomal bands in the ancestral R1-A and R3-A strains on the PFGE gel, their identity can only be inferred. In order to verify the identity of these chromosomal bands for chrA and chrB on the PFGE gel we excised these bands from the gel and sequenced them using Illumina sequencing. The results confirm the identity of the chromosomal bands of the PFGE gel as chrA in R1-A, chrB in R3-A, as well as chrA and chrB in R3-I4 (SI Appendix, Fig. S3 A and B). In addition, we determined the number of SNPs and small InDels as well as larger structural variants (≥50 bp) by use of the Illumina and Nanopore reads, respectively, mapped on the R3-I4 assembly. Strikingly, when chrA is present in an evolved strain (R3-I4, R3-I5, or R3-I6), we could not detect any SNPs/InDels or structural variants (the latter analyzed only for R3-I4) between the chrA copy in the evolved strain and chrA in the ancestral R1-A (Fig. 1C). Therefore, we conclude that the horizontal transfer of chrA from R1-A to R3 was not accompanied by mutational processes or large-scale structural reorganization of chrA.
While all the above results clearly show the horizontal transfer of chrA, they do not yet exclude the possibility that other genetic material could have been transferred at the same time. To test for this, we, first, compared the normalized sequencing coverage of 50-kb windows between the six evolved R3 strains with their R3-A ancestor strain, when mapped to the ancestral R1-A assembly (Fig. 1E). We observed an increase in sequencing coverage by approximately one normalized sequencing coverage for those sequences syntenic to accessory chrA in all three chrA-recipient R3 strains. This suggests that only chrA was transferred and that the recipient strains now contained a single copy of chrA. Genomic regions of R1-A that were not syntenic to chrA showed no change in coverage between the ancestral and the evolved R3, indicating the absence of additional transferred genetic material. Second, we analyzed the presence of R1-A-specific SNPs in the individually evolved R3 strains. Despite the high number of 194,334 SNPs differentiating the ancestral strains R1-A and R3-A (SI Appendix, Table S2), we did not identify any R1-A-specific SNPs in the evolved R3 strains, ruling out chromosome replacement or mitotic recombination events (SI Appendix, Fig. S4). Hence, solely chrA was horizontally transferred independently multiple times with no other genetic material being transferred from R1-A to the individually evolved R3 strains.
The Methylation Pattern of chrA Is Partially Retained in the Recipient Strain.
The intraspecific horizontal transfer of chrA from R1-A to R3 during our experiment represents the transfer of an entire chromosome from one genomic background to another. Here, we have the unique opportunity to study how epigenetic marks were affected by such a horizontal transfer, because we have access to both ancestral strains as well as the recipient strain that now harbors chrA. We focused on DNA methylation, which is known to vary between different Metarhizium strains (37–39). We hypothesized that chrA may possess a distinct DNA methylation pattern compared to the rest of the genome in the donor strain and that its pattern might be affected by the horizontal transfer to R3. Using Nanopore sequencing reads, we called methylated cytosines in CpG context (see also SI Appendix, Text S1). Overall, the ancestral R1-A strain exhibited significantly higher methylation levels than the ancestral R3-A strain (Fisher exact test, P < 4.6 × 10−15) (SI Appendix, Fig. S5A and Table S3). Furthermore, in both the ancestral R1-A strain and the evolved R3-I4 strain, chrA showed significantly higher methylation levels compared to the rest of the genome (Fisher exact test, P < 4.6 × 10−15 for both comparisons). Upon closer examination, we found that the subtelomeric regions of chrA in R3-I4 retained similar levels of methylation compared to R1-A, while the central portions displayed methylation levels similar to the genome-wide average (SI Appendix, Fig. S5B). Although the methylation level on chrA was substantially lower in the evolved R3-I4 strain compared to the ancestral R1-A, the majority (549 out of 741) of methylated sites in R3-I4 were also methylated in R1-A (SI Appendix, Fig. S5C). These results led us to conclude that the transferred chrA in the recipient R3-I4 strain retained some of the methylation pattern of its donor, the ancestral R1-A strain, although the absolute level of methylation decreased upon transfer to the recipient strains. This is therefore an example of how the genomic context can influence epigenetic marks, potentially also affecting gene expression.
chrA Provides a Competitive Advantage in Individual Ant Hosts.
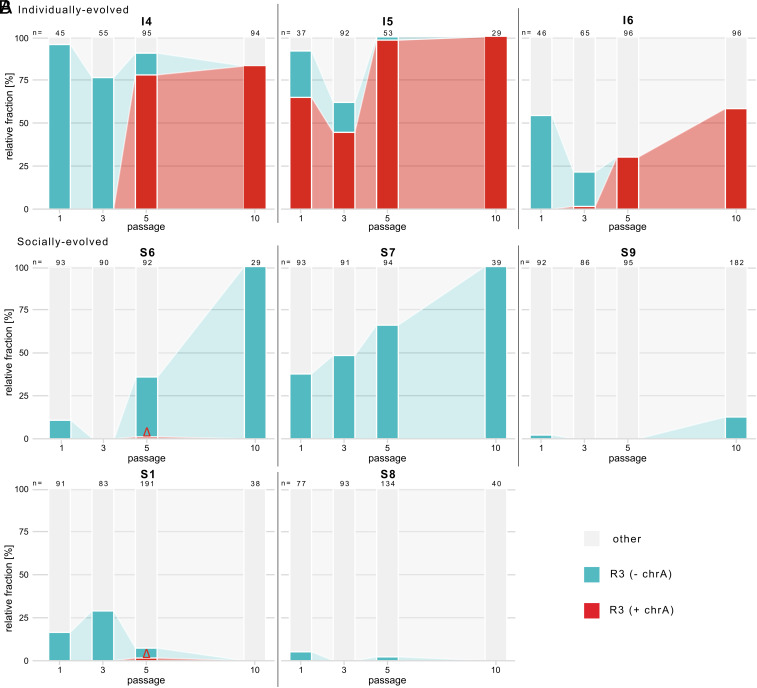
Next, we wanted to understand the temporal dynamics of the horizontal transfers and whether recipient strains had a competitive advantage or disadvantage compared to the same strain without chrA. To this end, we performed a detailed analysis of the clone diversity at passages 1, 3, 5, and 10 testing the proportion of R3 spores with present vs. absent chrA for the six above-mentioned R3-strains that had persisted until passage 10, plus another two that had persisted at least until passage 5, but had become outcompeted by passage 10 [R3-S1, R3-S8 (35); see SI Appendix, Text S1 for detailed information on how the presence/absence of chrA was determined]. We found that chrA was transferred from R1 to R3 in five out of the eight selection lines (in all three individually evolved, as well as in two of the five socially evolved) (Fig. 2 A and B). Hence, horizontal transfer occurred frequently. The main difference between the individual and social immunity treatments arose, however, in the persistence of the R3 strains that had acquired the chrA. When adapting to individual ants, chrA spread very quickly through the fungal population. After 10 passages, chrA was present in all R3 spores across all three replicate lines. This contrasts with the lack of the spread of chrA in R3 lines adapting to social ants. Despite being transferred from R1 to R3 in two of the five replicates (R3-S1 and R3-S6), R3 became completely extinct in the socially evolved replicate S1, while in replicate S6, R3 had outcompeted all other strains at the end of the experiment, but the winning R3 spores were the ones not containing chrA. This means that acquiring chrA improved the competitive ability of the evolved R3 against the R3 without chrA in individual ant hosts, yet when the exposed ants were with caretaking nestmates, the opposite was the case. This indicates that the establishment and long-term persistence of chrA in the pathogen population was influenced by the hosts’ social immunity, providing a further example of the modulatory power of social immunity on the competitive ability of coinfecting pathogens (40).
Fig. 2.
Comparative dynamics of R3 spores with either chrA present or absent over the course of the selection experiment. Proportion of R3 spores with chrA present (red) or absent (turquoise) in comparison to the remaining strains (gray) analyzed in passages 1, 3, 5, and 10 in (A) individual ant hosts (I4, I5, I6) or (B) ants with nestmates providing sanitary care [Upper row: replicate lines, in which R3 persisted until passage 10 (S6, S7, S9); Lower row: replicate lines, in which R3 still persisted in passage 5, but was outcompeted until passage 10 (S1, S8)]. The value of n represents the total number of genotyped single-spore derived colonies (n = total of 2,626 spores). Please note that the occurrence of chrA in R3 in two of the socially evolved replicate lines, where it later failed to establish, is highlighted by a red triangle.
ChrA Shows Presence/Absence Polymorphism in the Genus Metarhizium.
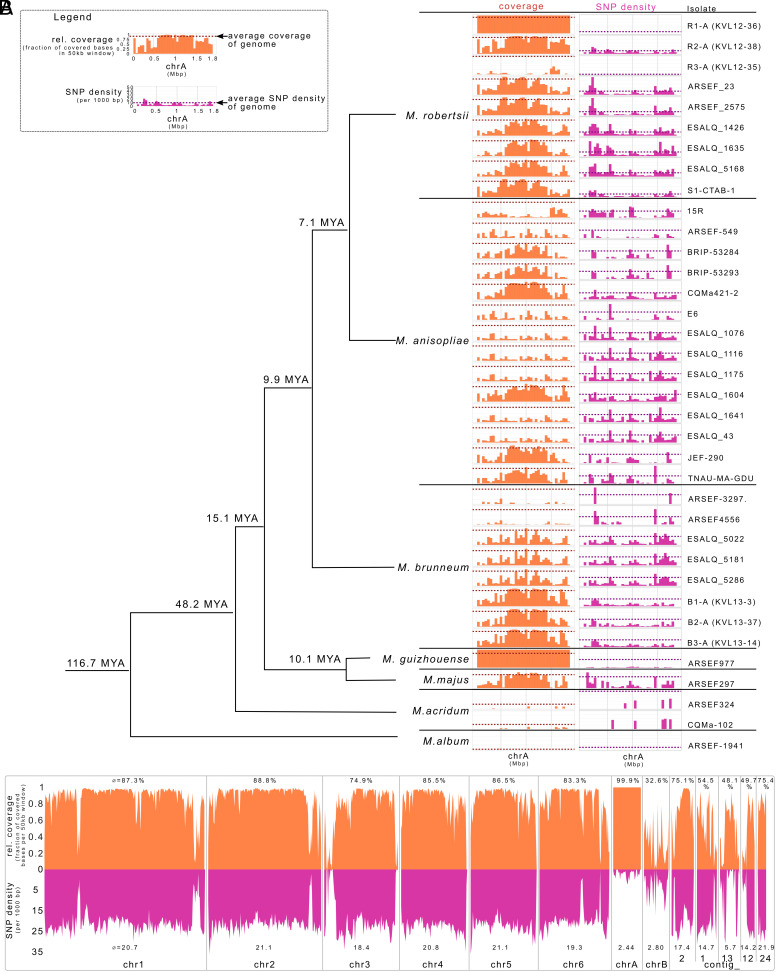
Our experimental data therefore revealed that chrA is easily horizontally transferred between different M. robertsii strains during co-infection in an insect host. We hypothesized that the accessory chromosome in M. robertsii might also be transferable between different species, since many species of Metarhizium have a broad host range (34), making congeneric co-infections a likely common scenario in the field. We therefore tested whether we could find evidence that chrA is present throughout the genus Metarhizium. To do this, we performed a phylogenetic analysis of the six ancestral strains of our experiment and all 30 published sequences of Metarhizium strains and species (M. robertsii, Metarhizium anisopliae, M. brunneum, M. guizhouense, Metarhizium majus, Metarhizium acridum, and Metarhizium album), all isolated from natural populations from the field (SI Appendix, Table S4).
Available assemblies and short sequencing reads were mapped onto the assembly of our individually evolved M. robertsii strain R3-I4 that contains both, chrA and chrB. For each strain, we determined the coverage as a fraction of bases covered [excluding transposable elements (TEs)] for both, chrA (Fig. 3A) and chrB (SI Appendix, Fig. S6). Within M. robertsii, chrA shows a presence/absence polymorphism, with 100% coverage and an absence of SNPs in our ancestral R1-A donor strain and with very low coverage in the ancestral R3-A, indicating again its absence. Among other M. robertsii strains, the relative coverage of chrA varied from 39.9 to 70.3%. Similarly, in M. anisopliae, the relative coverage ranged from 12.7 to 51.5%, while in M. brunneum, it ranged from 2.8 to 61%. In M. majus, chrA had coverage of 47.3% of all bases, whereas it was poorly covered in M. acridum and M. album (1.4 to 1.9%). Interpreting intermediary coverage is difficult due to availability of only very fragmented assemblies for these strains. Hence, it is unclear whether the sequences are located on one chromosome (which would then be accessory) or distributed within the genome.
Fig. 3.
Distribution of chrA sequences across Metarhizium species. (A) Phylogeny of published Metarhizium strains [from Hu et al. (41)], for which assemblies or sequencing reads were available. These assemblies or sequencing reads were mapped onto the near-chromosome level assembly of M. robertsii R3-I4, which contains both accessory chrA and chrB. The fraction of covered bases of chrA (orange) and the density of SNPs (pink) were determined in 50 kb windows. Dotted lines represent the respective genome-wide averages of the covered sequences and the SNP density. TEs were excluded from the analysis. MYA: Million years ago. (B) Relative sequence coverage of Illumina reads from M. guizhouense ARSEF977 mapped to the evolved M. robertsii R3-I4 strain assembly. The fraction of bases covered per 50-kb window is shown in orange while SNP density per 1,000 bp in 50-kb windows is shown in pink. ChrA exhibits higher average sequence coverage and lower SNP density in M. guizhouense compared to the rest of the genome (see also SI Appendix, Fig. S6 for chrB).
Interestingly, M. guizhouense exhibited complete coverage (99.9%) of all bases on chrA as well as a very low number of SNPs. This clearly demonstrates that chrA is shared between M. robertsii and M. guizhouense, despite their separation 15.1 Mya. Performing the same analysis on chrB (SI Appendix, Fig. S6) did not detect the presence of this second accessory chromosome of M. robertsii in any of the 35 strains in its entirety other than our ancestral strain R3-A, but detected a high variability in the presence of part of its sequences. We could also not detect any relation with the presence/absence of the two accessory chromosomes. In conclusion, there is a high variation in the presence of sequences of the accessory chrA and chrB that does not relate to the phylogeny and therefore might be indicative of either frequent losses of these chromosomes or horizontal transfer.
Horizontal Transfer of chrA across Metarhizium Species Borders.
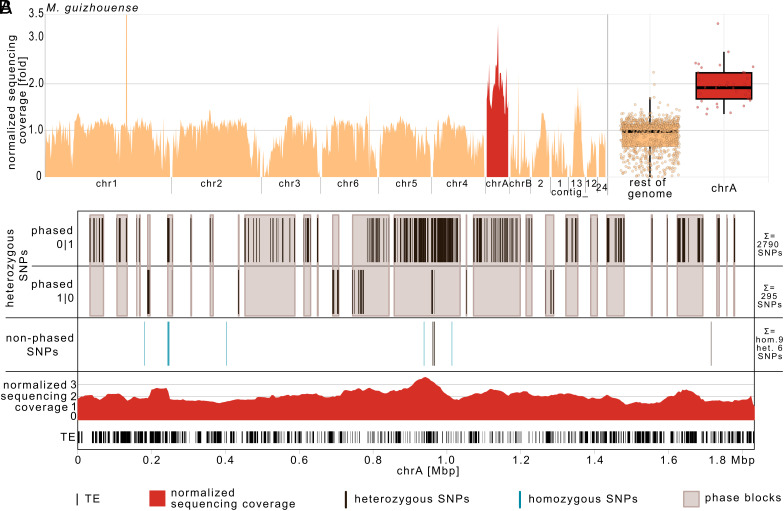
Accessory chrA showed a markedly reduced SNP density between M. robertsii and M. guizhouense compared to the rest of the genomes (2.44 compared to 19.2 SNPs per 1,000 bp, respectively) (Fig. 3B and SI Appendix, Table S5), which could be indicative of a horizontal transfer. We therefore analyzed chrA in M. guizhouense in more detail. Mapping the short sequencing reads of M. guizhouense revealed that chrA displayed approximately double the normalized sequencing coverage compared to the rest of the genome (Fig. 4A). These observations strongly suggest that the sequences present on chrA are duplicated in M. guizhouense, indicating the presence of a disomic state for chrA in this strain. PFGE confirmed the presence of at least two small chromosomes of approx. 1.9 Mb and 1.6 Mb in M. guizhouense (SI Appendix, Fig. S7). Illumina sequencing confirmed the presence of chrA sequences in both of these two chromosomal PFGE bands (SI Appendix, Fig. S3). In combination with the above phylogenetic analysis, we therefore conclude that chrA exists in two copies in M. guizhouense, and the relatively low SNP density separating chrA in M. robertsii from M. guizhouense, compared to the significantly higher SNP density in the rest of the genome, likely reflects a shorter separation time from their respective syntenic sequences for chrA than for the rest of the genome.
Fig. 4.
Sequencing coverage and SNP density indicate two copies of chrA in M. guizhouense that differ from each other in their SNP density. (A) Normalized Illumina sequencing coverage of M. guizhouense ARSEF977 on the M. robertsii R3-I4 assembly, presented separately for each contig and summarized for chrA and the rest of the genome in 50-kb windows. chrA shows approximate double the sequencing coverage compared to the rest of the genome. For visual clarity, the coverage of the rDNA cluster is not shown in full. (B) Phasing information of M. guizhouense SNPs located outside of TEs on the M. robertsii R3-I4, shown in individual phase blocks. The presence of separate phase blocks does not allow direct assignment of phase 0|1 or 1|0 to a copy of the disomic chrA in M. guizhouense—but most phase blocks contain SNPs exclusively from one phase (non-mixed phase blocks), so the relative frequency of non-mixed phase blocks was used to estimate the number of SNPs for each copy of chrA in M. guizhouense (see SI Appendix, Text S1 for details). The figure also depicts the normalized sequencing coverage in 50-kb (5-kb sliding) windows, along with the location of TEs (hom = homozygous, het = heterozygous).
Next, we investigated whether the two copies of chrA in M. guizhouense differ with respect to their separation time from chrA in M. robertsii (R1-A) by determining their SNP density separately for each copy. To do this, we phased the heterozygous SNPs of M. guizhouense, i.e., we determined on which of the two copies of chrA a SNP is located. Phasing SNPs is most efficient using long sequencing reads, so we used long PacBio reads. Phasing results in separate phase blocks, which are sections of the chromosome sequence for which the relative position of a heterozygous SNP on one of the two copies of the chromosome is known, relative to the other SNPs of the same phase block. Out of the 3,090 heterozygous SNPs on chrA, 2,790 were phased as 0|1 and 294 as 1|0. Due to the presence of multiple phase blocks, it was not possible to assign the phase 0|1 and 1|0 in different phase blocks directly to one of the chrA copies. We therefore used the distribution of SNPs within and between the different phase blocks to estimate the SNPs for each copy (see SI Appendix, Text S1 for more details). Based on this estimation, copy a of chrA contains an estimated 2,769 SNPs, while copy b contains approximately 345 SNPs. Therefore, the two copies of chrA in M. guizhouense differ in their SNP density from each other as well as from the rest of the genome compared to M. robertsii, indicating different separation times for the two copies of chrA.
When did the two copies of chrA in M. guizhouense separate from the chrA found in M. robertsii? The estimated divergence between M. guizhouense and M. robertsii is 15.1 Mya (41), which allows us to calibrate the molecular clock based on the rest of the genome for a comparison between M. robertsii and M. guizhouense. As TEs may be subject to different mutational processes and SNP calling is less reliable in TEs, we excluded all SNPs located in TEs (on core and accessory chromosomes) from this analysis. Using this calibration, we estimated that the separation of the two copies of chrA in M. guizhouense from chrA in M. robertsii took place approximately 1.72 Mya (copy a) and 0.214 Mya (copy b). Thus, the divergence of the two copies in M. guizhouense from chrA in M. robertsii occurred much more recently than the speciation event between the two fungal species 15.1 Mya. This suggests that the observed divergence is likely the result of a more recent horizontal transfer of chrA. Without additional sequence data on the population level for the accessory and the core chromosomes for both species—that is required to determine the ancestral state—the direction of this horizontal transfer remains currently unknown and could also include additional species. In conclusion, the presence of two copies of chrA in M. guizhouense, with different separation times from chrA in M. robertsii, implies either two horizontal chromosome transfers or a mixture of two biological mechanisms—thus, in either scenario, quite complex dynamics underlie this pattern.
chrA and chrB Differ in Their Composition from the Rest of the Genome.
Accessory chromosomes in fungi often differ in their sequence characteristics compared to the core chromosomes, with lower gene density and higher density of TEs (1). These differences may indicate other evolutionary constraints for accessory chromosomes than for the core genome, such as a smaller effective population size (1). We observed a comparable pattern for the accessory chromosomes chrA and chrB in M. robertsii. chrA and chrB were enriched in their proportion of TEs, which constituted approximately 31 to 32% of all sequences in the accessory chromosomes, with 9.1% in the rest of the genome (Fig. 5A). The proportion of genes on the accessory chromosomes was lower, with 22% compared to 29% in the rest of the genome. Taken together, TE and gene density resulted in a clear compositional separation of the two accessory chrA and chrB from the core chromosomes (Fig. 5B). In addition, the TE composition (SI Appendix, Fig. S8 A and B), and the codon usage of the genes (SI Appendix, Fig. S8C) located on the accessory chrA and chrB differed from that of the rest of the genome. The gene-wise relative synonymous codon usage (gRSCU), which measures the bias in the use of synonymous codons, was significantly lower for genes located on chrA and chrB compared to the rest of the genome of M. robertsii R3-I4 as well as compared to those genes located on the rest of the genome of the R1-A ancestral strain (all pairwise Wilcoxon rank-sum test with BH correction, all P < 2 × 10−16). Taken together, these differences in TE composition and codon usage suggest that the accessory chromosomes were horizontally acquired by M. robertsii. Furthermore, the codon usage of chrA also differed from that of all genes in the M. guizhouense genome, which suggests that chrA was also horizontally acquired by M. guizhouense. Therefore, chrA may have been horizontally acquired by both M. guizhouense and M. robertsii from a third, currently unknown species.
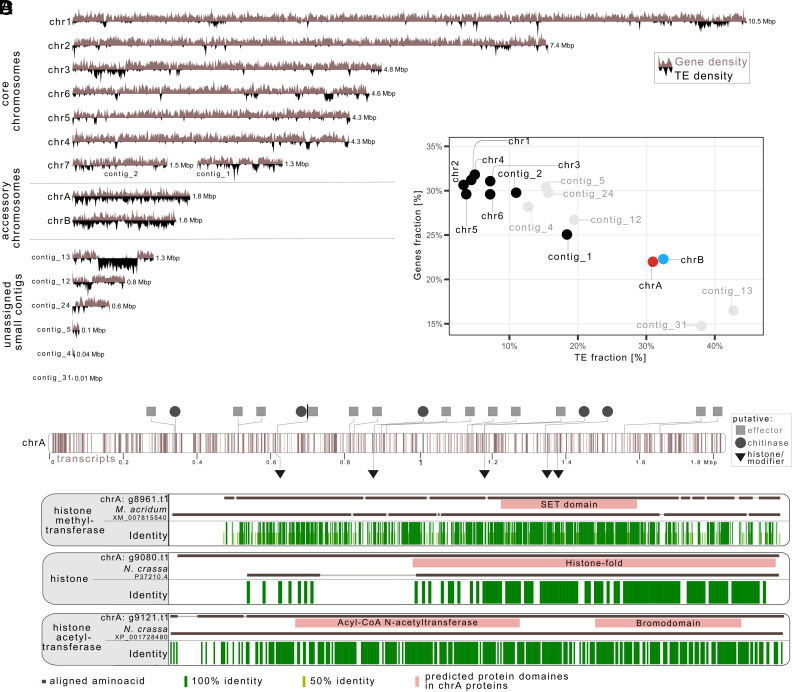
Fig. 5.
Overview of gene and TE annotation of the evolved M. robertsii R3-I4 strain. (A) Karyoplot illustrating the distribution of genes (light brown) and TEs (black) along the nanopore-based assembly of the evolved R3-I4 strain. (B) Distribution of gene density vs. TE density, highlighting the higher TE density and lower gene density observed in chrA (red) and chrB (blue) compared to other chromosomes (black) and unassigned small contigs (gray). (C) Gene transcript distribution along chrA, with putative candidates that might influence either the interaction with the host or the chromatin conformation being indicated. (D) Example alignments of proteins for one putative histone and one putative histone methyltransferase and one putative histone acetyltransferase with proteins of known function. Identical amino-acids are depicted in green. Although being similar to the proteins with known function, the putative histone and histone-modifying enzymes encoded on the accessory chromosomes show differences in their protein sequence.
chrA Encodes Putative Virulence Factors and May Influence Its Horizontal Transfer.
We were further interested to see if the proteins encoded by the genes on the accessory chromosomes could be potentially involved in the interaction with the insect host or the horizontal transfer of the chromosome and therefore functionally annotated the predicted transcripts. On accessory chrA, we could identify a total of 364 genes through ab initio annotation (SI Appendix, Table S6 and Datasets S1 and S2). Among them, 13 genes were predicted to encode putative effectors (i.e., small, secreted proteins that could potentially interfere with the host immune system) and 10 genes were identified as Carbohydrate-Active Enzymes (CAZymes). Notably, three of these CAZymes were putative chitinases, suggesting a possible role in host insect cuticle degradation, which is a crucial step during the infection process, since the fungal spores need to penetrate the host’s chitin cuticle to be able to infect and eventually kill the host (34). Hence, the accessory chromosomes might be involved in the interaction with the insect host, and by receiving the accessory chrA, the recipient strain may have gained a fitness benefit by increased virulence.
On both accessory chrA and chrB, we further found genes that might be involved in the horizontal transfer of the accessory chromosomes. A GO-term enrichment analysis revealed that several GO terms related to chromatin, nucleosome, and chromosome segregation are enriched on chrA and chrB (SI Appendix, Fig. S9A). A more detailed analysis showed that chrA contains two genes encoding putative histone H2B proteins (g9016.t1, g9080.t1) and three genes encoding putative enzymes involved in histone post-translational modifications (g8961.t1, g9112.t1, g9121.t1) (Fig. 5 C and D). Similarly, chrB contains a gene encoding a putative histone H3 protein and two genes encoding putative histone-modifying enzymes (g10208.t1, g10199.t1). A phylogenetic analysis showed that the putative histone proteins encoded by the genes located on the accessory chromosomes are not paralogs of those encoded by the genes located on the core chromosomes of the M. robertsii R3-I4 strain but group independently of these (SI Appendix, Fig. S9C). This suggests a different evolutionary origin and/or trajectory of the corresponding genes. These differences in origin, but more importantly the differences in protein sequence between histones and histone modifying enzymes encoded on the accessory chromosomes vs. those encoded on the core chromosomes, could indicate differences in the function of the corresponding proteins. In all our observed intraspecies horizontal transfers of accessory chrA, solely this chromosome and no other material was either transferred or retained. Hence, there must be a distinguishing characteristic that separates chrA from the rest of the donor genome and also from the genome of the recipient strain. Histone modifications are known to be involved in many programmed DNA elimination events in plants and animals (42). The presence of these putative histone proteins and histone-modifying enzymes on both chrA and chrB suggests their potential involvement in affecting the chromatin conformation and thereby potentially mechanistically influencing the horizontal transfer of the accessory chromosomes. Finally, we found that the genes located on the accessory chromosomes chrA and chrB have a significantly higher dN/dS ratio than the genes located on the core chromosomes when comparing SNPs between M. robertsii strain R3-I4 and M. guizhouense (SI Appendix, Fig. S9B). This suggests that genes on chrA and chrB are less affected by purifying selection and more affected by positive selection, but due to the lack of available population-level sequence data for chrA and chrB and the core chromosomes for both M. robertsii and M. guizhouense and the absence of an outgroup for the accessory chromosomes we cannot say whether and how these selective forces affected M. robertsii or M. guizhouense differently.
Discussion
In this study, we describe the horizontal transfer of an entire accessory chromosome (chrA) between asexual strains and species of common insect-pathogenic fungi. We show that: i) two accessory chromosomes are present in M. robertsii and that one of them is frequently transferred horizontally during experimental host co-infection between different M. robertsii strains. This transfer involved only the accessory chromosome, while no other genetic material was transferred. ii) Although horizontal transfers occurred frequently, the strains including the transferred accessory chromosome were only able to outcompete the same strains lacking it, and to spread in the pathogen population under one experimental condition—infection of single ant hosts, but not when infected ants were accompanied by nestmates that provided social immunity. Therefore, the accessory chromosome alters the competitive ability of the recipient strain, and whether or not this results in fitness benefits is condition-dependent. iii) Importantly, we show that this horizontal transfer is not restricted to exchange within the same species in experimental co-infections, but also occurs naturally across species under field conditions, as the same accessory chromosome has been transferred between different species of Metarhizium much more recently than their speciation time of 15.1 Mya. Such horizontal transfer between different species could therefore be a possible mechanism for the spread of accessory chromosomes to new species.
Our characterization of multiple field-isolated strains of M. robertsii showed that this entomopathogen contains two accessory chromosomes with presence/absence polymorphism. We therefore here describe accessory chromosomes in a pathogenic fungus of animals, where their presence and role is so far underexplored (43), in contrast to their common occurrence in plant–pathogenic fungi, where they have demonstrated effects on fitness and/or host range (1, 3). Notably, M. robertsii, like many other Metarhizium species, is—in addition to its parasitic lifestyle in insects—also closely associated with plants, in particular their roots. Therefore, the accessory chromosomes in M. robertsii may also be involved in the interaction with plants. Similar to accessory chromosomes in other fungal pathogens, the accessory chromosomes in M. robertsii (chrA and chrB) differed in TE and gene content as well as codon usage from the rest of the genome, which could indicate different evolutionary histories or constraints in accessory vs. core chromosomes.
In our experiment, chrA was transferred at least five times independently from M. robertsii R1 to the M. robertsii R3 strain, while no other genetic material was exchanged. Whereas horizontal transfer of accessory chromosomes has been reported in vitro for the plant pathogens Colletotrichum gloeosporioides (10, 12), members of the Fusarium oxysporum species complex (7, 8, 13, 44) and possibly in Alternaria alternata pathotypes (45), the low frequency of these transfers necessitated the use of selectable fungicide resistance markers in these studies to identify and isolate those fungal cells, in which a horizontal transfer had occurred. Consequently, to date, no horizontal transfer of chromosomes has been observed for these fungal pathogens during infection of their respective host (10, 12, 13, 44). In contrast, our study observed frequent horizontal transfer between M. robertsii strains during co-infection of ants, which represent a natural host of multiple Metarhizium species (34). This disparity suggests that the higher frequency of horizontal transfer observed in our study may in principle be attributable to parasexual-like processes described for species of the genus Metarhizium (46, 47), which is thought to lead to exchange of genetic material between co-infecting strains when both grow within the same host individual. However, the parasexual-like cycle in Metarhizium is thought to involve cell fusion resulting in cells with two different nuclei (heterokaryons), followed by karyogamy to produce unstable diploids that randomly lose chromosomes to regain haploidy (47). In our study, in contrast, we observed specific transfer of exactly one chromosome (chrA) without random genetic exchanges or loss of additional chromosomes. Thus, we propose that the observed horizontal transfer of chrA cannot be explained by the established parasexual-like processes alone. Instead, alternative mechanisms for accessory chromosome transfer must be in place. These could include either the transfer of accessory chromosomes between nuclei during the heterokaryon stage (12) or the selective degradation of all chromosomes except the one chromosome that was successfully transferred (12, 13). A process similar to the former has been described in Saccharomyces cerevisiae, where a mutant defective in nuclear fusion (kar1-1) generates transient heterokaryons, allowing chromosome transfer between nuclei before one of the nuclei is lost. This process, known as chromoduction, enables the transfer of whole chromosomes into a new genomic background (24, 48–50). A process similar to the latter (i.e., degradation of all but the transferred chromosomes) has been described as programmed DNA elimination in several plants and animals (reviewed in ref. 42), with the retained DNA sequences differing from the eliminated ones through DNA methylation, histones, or post-translational histone modifications, particularly in centromeric regions (42). We speculate that such a mechanism may also act during the horizontal transfer of chrA and possibly also chrB in M. robertsii. Since both accessory chromosomes contain genes encoding putative histones and histone-modifying enzymes, these may be involved in ensuring the transfer of only the accessory chromosomes. It is important to note that the putative histones encoded on the accessory chromosomes are not paralogs of the histones encoded on the core chromosomes and show differences in the protein sequence, which could indicate a functional difference. Therefore, we hypothesize that the putative histone and histone-modifying enzymes might provide a signal—e.g., a distinct chromatin conformation of the accessory chromosomes compared to the rest of the donor genome—that either allows their preferential transfer to the recipient nucleus (similar to chromoduction), or evasion of degradation [similar to the PSR chromosome in the parasitoid wasp, Nasonia vitripennis, where the presence of the PSR chromosome affects the post-translational methylation of at least three histones in the genome that later becomes eliminated (51, 52)]. An alternative, but not mutually exclusive, mechanism could be that the subtelomeric DNA methylation we observed on chrA, some of which was retained after transfer, could be the signal for preferential transfer or evasion of degradation of chrA. Since DNA methylation and histone modification are known to interact (53), possibly both the putative histones and histone-modifying enzymes and DNA methylation could be mechanistically involved in the horizontal transfer of chrA.
In addition to the horizontal transfer of chrA between strains of M. robertsii, we show phylogenetic support of a past horizontal chromosome transfer between two different species of the genus Metarhizium in the field. While interspecific chromosome transfer has been proposed as a potential mechanism for how species can gain new accessory chromosomes, no such transfer had been previously documented to our knowledge. Existing experimental evidence of horizontal transfers has focused on different pathovars (e.g., A. alternata), biotypes (e.g., C. gloeosporioides), or formae speciales within species complexes (e.g., F. oxysporum) (6–8, 10, 13, 44, 45). Although these studies provide insights into the distribution of accessory chromosomes within species, they fail to account for their spread between species. The scarcity of fungal accessory chromosomes that exhibit synteny with each other in different species was considered as evidence against the hypothesis that these chromosomes had been acquired through horizontal transfer (2) and evidence for their origin by endogenous processes [i.e., from within the genome, e.g., via degenerative breakage, duplication, missegregation, or Robertsonian chromosome fusion (2, 54, 55)]. Here, we report exactly such a syntenic accessory chromosome being present in two species (M. robertsii and M. guizhouense). Moreover, the codon usage pattern of the accessory chrA differed from the rest of the genome of both M. robertsii and M. guizhouense. This suggests that it might have originated from a third species and was then horizontally acquired. Taken together, we here show intra- and interspecific horizontal transfer of an accessory chromosome, supporting that horizontal chromosome transfers present a more common mechanism for a species to acquire novel accessory chromosomes than previously thought.
Our experimental study further gives details into the dynamics and constraints of the spread of an accessory chromosome after its horizontal transfer into a new pathogenic strain. In our experiment, chrA was horizontally transferred in five of the total 20 independent replicates. Strikingly, all three cases where the strain that horizontally acquired chrA outcompeted the same strain lacking chrA were observed during infection of single ants. The two instances where the strain that horizontally acquired chrA was outcompeted by the same strain that had not acquired it, leading to its extinction, was observed during infection of ants in the presence of nestmates that provide social immunity. Hence, even when transfer occurs frequently, whether or not the strain harboring the acquired accessory chromosome will be able to establish itself in the pathogen population likely is a condition-dependent evolutionary process. However, a positive fitness effect may also not be required in all cases, as the accessory chromosome may still spread even in the absence of a benefit, purely due to a preferential transfer mechanism similar to a meiotic chromosome drive (56). It is tempting to speculate that since chrA and chrB contain genes that could influence their preferential transmission—such as the putative histone and histone-modifying enzymes—these accessory chromosomes could also be selfish genetic elements that manipulate and propagate their horizontal transmission.
In conclusion, we here describe accessory chromosomes with the potential to spread both within and across species of fungal pathogen species of the genus Metarhizium, which comprises many important insect-pathogenic species. By describing the characteristics of this accessory chromosome and its transfer and spread dynamics, we find horizontal transfer to be a very important mechanism in shaping the evolution of accessory chromosomes and whole genomes. Based on its preferential horizontal transfer, chrA might be able to spread through the recipient pathogen populations in a process highly reminiscent of the spread of selfish genetic elements.
Methods
Fungal Strains.
We used the six ancestral strains and the evolved lines of the fungal pathogens M. robertsii and M. brunneum obtained during the selection experiment performed by Stock et al. (35). In this experiment, the six ancestral strains were mixed in equal amounts and used to experimentally co-infect Argentine ants, which subsequently were either kept alone [individual treatment (abbreviation: I)] or in the presence of two nestmates [social treatment (abbreviation: S)] over 10 consecutive host infection cycles (passages). An outline of the experiment is shown in Fig. 1A, and details described in Stock et al. (35) and SI Appendix, Text S1. The six ancestral strains included three M. robertsii strains [R1-A: KVL 12-36 (C17), R2-A: KVL 12-35 (E81) and R3-A: KVL 12-38 (F19)] and three M. brunneum strains [B1-A: KVL 13-13 (G39), B2-A: KVL 12-37 (J65), and B3-A: KVL 13-14 (L105); all obtained from the University of Copenhagen, Denmark], which had all been isolated from the same field population and characterized by Steinwender et al. (57). For the evolved lines (35), we focused particularly on M. robertsii R3, for which we determined overall strain proportion and integration of chrA over the course of the experiment (at passages P1, P3, P5, and P10). M. guizhouense ARSEF977 was obtained from the ARS Collection of Entomopathogenic Fungal Cultures, Ithaca NY. An overview of previously published reads and assemblies that were included in this study is given in SI Appendix, Table S4.
Molecular Analysis of Fungal Strains.
We performed both long-read sequencing (Nanopore and PacBio) as well as short-read sequencing (Illumina) for ancestral and evolved strains, as well as microsatellite analysis for spore strain identification and chrA presence/absence determination, as detailed in SI Appendix, Text S1.
Nanopore sequencing was performed by the Next Generation Sequencing Facility at Vienna BioCenter Core Facilities, member of the Vienna BioCenter (VBC). PacBio sequencing of M. guizhouense was performed at the Max Planck Genome Centre Cologne, Germany using Sequel IIe (Pacific Biosciences). Illumina Sequencing was performed at Eurofins Genomics GmbH (Ebersberg) and the Max Planck Genome Centre Cologne, Germany. An overview of the sequencing reads generated in this study is given in SI Appendix, Table S7.
Bioinformatic Analysis.
For the generation of the Nanopore-based assemblies, we used the pipeline described in ref. 36. TE and gene annotations were generated using the REPET pipeline (58, 59) following the pipeline as described in ref. 60 and Braker 2.0 (version 2.1.6) (61), respectively. Comparison between published assemblies and the R3-I4 assembly was determined by nucmer (version 4.0.0rc1) (62). Comparison between published short reads as well as the 150 PE Illumina reads generated in this study and the R3-I4 and the R1-A and R3-A assembly was determined by mapping and SNP calling using bowtie2 (version 2.4.4) (63) and bcftools mpileup (version = 1.14) (64). Phasing of SNPs and small InDels for M. guizhouense was determined using PacBio Hifi reads generated in this study and WhatsHap (version 1.6) (65). gRSCU was estimated using BioKIT (version 0.1.3). Methylated cytosines in a CpG context were identified using nanopore reads by mod_kit (version 0.1.4). Structural variants were called using Sniffles2 (66). For a detailed description of the bioinformatical steps, see SI Appendix, Text S1.
Statistical Data Analysis.
All statistical analyses were conducted in R (version R3.6.0) using the suite R Studio (1.2.1335), as described in more detail in SI Appendix, Text S1. A summary of all statistical test results is given in SI Appendix, Tables S10–S12.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
Dataset S14 (XLSX)
Acknowledgments
We thank Bernhardt Steinwender, Jorgen Eilenberg, and Nicolai V. Meyling for the fungal strains. We further thank Chengshu Wang for providing the short sequencing reads for M. guizhouense ARESF977 he used for his published genome assembly, and Kristian Ullrich for help in the bioinformatics analysis for methylation pattern in Nanopore reads, and the VBC and the Max Planck Society for the use of their sequencing centers. We thank Barbara Milutinović and Hinrich Schulenburg for discussion, and Tal Dagan and Jens Rolff for comments on a previous version of the manuscript. Fig. 1A was created with BioRender.com. This study received funding by the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (No. 771402; EPIDEMICSonCHIP) to S.C. and by the German Research Foundation (DFG grant HA9263/1-1) to M.H.
Author contributions
M.H., A.V.G., and S.C. designed research; M.H., A.V.G., J.M., E.H.S., and H.L. performed research; M.H. contributed new reagents/analytic tools; M.H. and A.V.G. analyzed data; M.H. and S.C. provided funding; E.H.S. provided funding, revised paper; and M.H. and S.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Michael Habig, Email: mhabig@bot.uni-kiel.de.
Sylvia Cremer, Email: sylvia.cremer@ist.ac.at.
Data, Materials, and Software Availability
Sequencing reads have been deposited in the Sequence Read Archive and are available under the BioProject PRJNA1017668 (67). The Nanopore-based assemblies and Gene annotations were deposited at NCBI under the BioProjects PRJNA1015426 (68), PRJNA1015429 (69), and PRJNA1015431 (70). All source data for the figures are given in Datasets S3–S14. Note that of the total of 2,626 spore-clones shown in Fig. 2, 872 had been produced, analyzed for strain identity and previously published in Stock et al. (35). The production and strain-identification of the remaining 1,754 spore-derived clones, as well as the presence/absence characterization of chrA within all 883 R3 spore-clones were performed in this study.
Supporting Information
References
- 1.Bertazzoni S., et al. , Accessories make the outfit: Accessory chromosomes and other dispensable DNA regions in plant-pathogenic fungi. Mol. Plant Microbe Interact. 31, 779–788 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Galazka J. M., Freitag M., Variability of chromosome structure in pathogenic fungi–of “ends and odds”. Curr. Opin. Microbiol. 20, 19–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komluski J., Stukenbrock E. H., Habig M., Non-Mendelian transmission of accessory chromosomes in fungi. Chromosome Res. 30, 241–253 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehrabi R., Mirzadi Gohari A., Kema G. H. J., Karyotype variability in plant-pathogenic fungi. Ann. Rev. Phytopathol. 55, 483–503 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Han Y., Liu X., Benny U., Kistler H. C., VanEtten H. D., Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 25, 305–314 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Hatta R., et al. , A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161, 59–70 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li J., Fokkens L., Conneely L. J., Rep M., Partial pathogenicity chromosomes in Fusarium oxysporum are sufficient to cause disease and can be horizontally transferred. Environ. Microbiol. 22, 4985–5004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L.-J., et al. , Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Habig M., Quade J., Stukenbrock E. H., Forward genetics approach reveals host genotype-dependent importance of accessory chromosomes in the fungal wheat pathogen Zymoseptoria tritici. mBio 8, e01919-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masel A. M., He C., Poplawski A. M., Irwin J. A. G., Manners J. M., Molecular evidence for chromosome transfer between biotypes of Colletotrichum gloeosporioides. Mol. Plant Microbe Interact. 9, 339–348 (1996). [Google Scholar]
- 11.Akagi Y., Akamatsu H., Otani H., Kodama M., Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant-pathogenic fungus. Eukaryotic Cell 8, 1732–1738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C., Rusu A. G., Poplawski A. M., Irwin J. A. G., Manners J. M., Transfer of a supernumerary chromosome between vegetatively incompatible biotypes of the fungus colletotrichum gloeosporioides. Genetics 150, 1459–1466 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlaardingerbroek I., et al. , Exchange of core chromosomes and horizontal transfer of lineage-specific chromosomes in Fusarium oxysporum. Environ. Microbiol. 18, 3702–3713 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Thomas C. M., Nielsen K. M., Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Etten J. V., Bhattacharya D., Horizontal gene transfer in eukaryotes: Not if, but how much? Trends Genet. 36, 915–925 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Keeling P. J., Palmer J. D., Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Rosewich U. L., Kistler H. C., Role of horizontal gene transfer in the evolution of fungi. Ann. Rev. Phytopathol. 38, 325–363 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Bennett R. J., The parasexual lifestyle of Candida albicans. Curr. Opin. Microbiol. 28, 10–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steensels J., Gallone B., Verstrepen K. J., Interspecific hybridization as a driver of fungal evolution and adaptation. Nat. Rev. Microbiol. 19, 485–500 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Hessenauer P., et al. , Hybridization and introgression drive genome evolution of Dutch elm disease pathogens. Nat. Ecol. Evol. 4, 626–638 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Menardo F., et al. , Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat. Genet. 48, 201–205 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Friesen T. L., et al. , Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38, 953–956 (2006). [DOI] [PubMed] [Google Scholar]
- 23.McDonald M. C., et al. , Transposon-mediated horizontal transfer of the host-specific virulence protein ToxA between three fungal wheat pathogens. mBio 10, e01515-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutcher S. K., Internuclear transfer of genetic information in kar1-1/KAR1 heterokaryons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 245–253 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manners J. M., He C., Slow-growing heterokaryons as potential intermediates in supernumerary chromosome transfer between biotypes of Colletotrichum gloeosporioides. Mycol. Progress 10, 383–388 (2011). [Google Scholar]
- 26.Strom N. B., Bushley K. E., Two genomes are better than one: History, genetics, and biotechnological applications of fungal heterokaryons. Fungal Biol. Biotechnol. 3, 4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engel T., et al. , Parasexual recombination enables Aspergillus fumigatus to persist in cystic fibrosis. ERJ Open Res. 6, 00020–02020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherwood R. K., Bennett R. J., Fungal meiosis and parasexual reproduction—Lessons from pathogenic yeast. Curr. Opin. Microbiol. 12, 599–607 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varga J., et al. , Aspergillus: Sex and recombination. Mycopathologia 178, 349–362 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Moore D., Robson G. D., Trinci A. P. J., 21st Century Guidebook to Fungi (Cambridge University Press, 2020). [Google Scholar]
- 31.Hörandl E., et al. , “Genome evolution of asexual organisms and the paradox of sex in eukaryotes” in Evolutionary Biology—A Transdisciplinary Approach, Pontarotti P., Ed. (Springer International Publishing, 2020), pp. 133–167. [Google Scholar]
- 32.Muller H. J., The relation of recombination to mutational advance. Mutat. Res. 106, 2–9 (1964). [DOI] [PubMed] [Google Scholar]
- 33.Maynard Smith J., The Evolution of Sex (Cambridge University Press, Cambridge [England]; New York, 1978), (20 December 2023). [Google Scholar]
- 34.St Leger R. J., Wang J. B., Metarhizium: Jack of all trades, master of many. Open Biol. 10, 200307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stock M., et al. , Pathogen evasion of social immunity. Nat. Ecol. Evol. 7, 450–460 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saud Z., Kortsinoglou A. M., Kouvelis V. N., Butt T. M., Telomere length de novo assembly of all 7 chromosomes and mitogenome sequencing of the model entomopathogenic fungus, Metarhizium brunneum, by means of a novel assembly pipeline. BMC Genomics 22, 87 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., et al. , DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 101, 4215–4226 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Sbaraini N., et al. , Genome-wide DNA methylation analysis of Metarhizium anisopliae during tick mimicked infection condition. BMC Genomics 20, 836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu S., Bidochka M. J., DNA methyltransferase implicated in the recovery of conidiation, through successive plant passages, in phenotypically degenerated Metarhizium. Appl. Microbiol. Biotechnol. 104, 5371–5383 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Milutinović B., et al. , Social immunity modulates competition between coinfecting pathogens. Ecol. Lett. 23, 565–574 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Hu X., et al. , Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, 16796–16801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dedukh D., Krasikova A., Delete and survive: Strategies of programmed genetic material elimination in eukaryotes. Biol. Rev. 97, 195–216 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C., Skrobek A., Butt T. M., Concurrence of losing a chromosome and the ability to produce destruxins in a mutant of Metarhizium anisopliae. FEMS Microbiol. Lett. 226, 373–378 (2003). [DOI] [PubMed] [Google Scholar]
- 44.van Dam P., et al. , A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 7, 9042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akagi Y., et al. , Chromosome constitution of hybrid strains constructed by protoplast fusion between the tomato and strawberry pathotypes of Alternaria alternata. J. General Plant Pathol. 75, 101–109 (2009). [Google Scholar]
- 46.Li S., Yi W., Chen S., Wang C., Empirical support for the pattern of competitive exclusion between insect parasitic fungi. J. Fungi 7, 385 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang S., O’Brien T. R., Pava-Ripoll M., St Leger R. J., Local adaptation of an introduced transgenic insect fungal pathogen due to new beneficial mutations. Proc. Natl. Acad. Sci. U.S.A. 108, 20449–20454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji H., et al. , Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73, 1007–1018 (1993). [DOI] [PubMed] [Google Scholar]
- 49.Conde J., Fink G. R., A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. U.S.A. 73, 3651–3655 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y., et al. , Debugging and consolidating multiple synthetic chromosomes reveals combinatorial genetic interactions. Cell 186, 5220–5236.e16 (2022), 10.1016/j.cell.2023.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Aldrich J. C., Leibholz A., Cheema M. S., Ausiό J., Ferree P. M., A ‘selfish’ B chromosome induces genome elimination by disrupting the histone code in the jewel wasp Nasonia vitripennis. Sci. Rep. 7, 42551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nur U., Werren J. H., Eickbush D. G., Burke W. D., Eickbush T. H., A “selfish” B chromosome that enhances its transmission by eliminating the paternal genome. Science 240, 512–514 (1988). [DOI] [PubMed] [Google Scholar]
- 53.Freitag M., Histone methylation by SET domain proteins in fungi. Ann. Rev. Microbiol. 71, 413–439 (2017). [DOI] [PubMed] [Google Scholar]
- 54.McClintock B., The stability of broken ends of chromosomes in Zea mays. Genetics 26, 234–282 (1941). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croll D., Zala M., McDonald B. A., Breakage-fusion-bridge cycles and large insertions contribute to the rapid evolution of accessory chromosomes in a fungal pathogen. PLoS Genet. 9, e1003567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Habig M., Kema G. H., Stukenbrock E. H., Meiotic drive of female-inherited supernumerary chromosomes in a pathogenic fungus. Elife 7, e40251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinwender B. M., et al. , Molecular diversity of the entomopathogenic fungal Metarhizium community within an agroecosystem. J. Invertebr. Pathol. 123, 6–12 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Flutre T., Duprat E., Feuillet C., Quesneville H., Considering transposable element diversification in de novo annotation approaches. PLoS One 6, e16526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simin K., et al. , pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2, e22 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorrain C., Feurtey A., Möller M., Haueisen J., Stukenbrock E., Dynamics of transposable elements in recently diverged fungal pathogens: Lineage-specific transposable element content and efficiency of genome defenses. G3 (Bethesda) 11, jkab068 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brůna T., Hoff K. J., Lomsadze A., Stanke M., Borodovsky M., BRAKER2: Automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 3, lqaa108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marçais G., et al. , MUMmer4: A fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danecek P., et al. , Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin M., et al. , WhatsHap: Fast and accurate read-based phasing. bioRxiv [Preprint] (2016). 10.1101/085050 (Accessed 16 May 2023). [DOI]
- 66.Sedlazeck F. J., et al. , Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Habig M., et al. , Metarhizium robertsii and Metarhizium brunneum experimental evolution during co-infection of insect (ant) host for 10 serial infections. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1017668. Deposited 18 September 2023.
- 68.Habig M., et al. , Metarhizium robertsii strain:R1-A | breed:KVL12-36 | cultivar:C17 Genome sequencing and assembly. BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1015426. Deposited 26 September 2023.
- 69.Habig M., et al. , Metarhizium robertsii Genome sequencing and assembly (R3-A). BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1015429. Deposited 26 September 2023.
- 70.Habig M., et al. , Metarhizium robertsii Genome sequencing and assembly (R3-I4). BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1015431. Deposited 26 September 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Dataset S12 (XLSX)
Dataset S13 (XLSX)
Dataset S14 (XLSX)
Data Availability Statement
Sequencing reads have been deposited in the Sequence Read Archive and are available under the BioProject PRJNA1017668 (67). The Nanopore-based assemblies and Gene annotations were deposited at NCBI under the BioProjects PRJNA1015426 (68), PRJNA1015429 (69), and PRJNA1015431 (70). All source data for the figures are given in Datasets S3–S14. Note that of the total of 2,626 spore-clones shown in Fig. 2, 872 had been produced, analyzed for strain identity and previously published in Stock et al. (35). The production and strain-identification of the remaining 1,754 spore-derived clones, as well as the presence/absence characterization of chrA within all 883 R3 spore-clones were performed in this study.