Significance
Messenger RNA (mRNA) delivery is a powerful approach to treat rare genetic diseases via replacing or supplementing a protein of interest. One way to deliver mRNA into the cells is to envelop it in a delivery vector like a lipid nanoparticle. These nanoparticles tend to accumulate in the liver; this limitation may be overcome either by local administration to target organs or engineering materials that avoid the liver. Here, we present the identification of ionizable lipids that can achieve successful retinal and pulmonary delivery. Identification of potent lipids capable of extrahepatic delivery, such as the ones presented in this report, can broaden the potential of mRNA therapeutics for a larger spectrum of genetic diseases outside of the liver.
Keywords: mRNA delivery, lipid nanoparticle, ionizable lipid
Abstract
Lipid nanoparticles (LNPs) largely rely on ionizable lipids to yield successful nucleic acid delivery via electrostatic disruption of the endosomal membrane. Here, we report the identification and evaluation of ionizable lipids containing a thiophene moiety (Thio-lipids). The Thio-lipids can be readily synthesized via the Gewald reaction, allowing for modular lipid design with functional constituents at various positions of the thiophene ring. Through the rational design of ionizable lipid structure, we prepared 47 Thio-lipids and identified some structural criteria required in Thio-lipids for efficient mRNA (messenger RNA) encapsulation and delivery in vitro and in vivo. Notably, none of the tested lipids have a pH-response profile like traditional ionizable lipids, potentially due to the electron delocalization in the thiophene core. Placement of the tails and localization of the ionizable headgroup in the thiophene core can endow the nanoparticles with the capability to reach various tissues. Using high-throughput formulation and barcoding techniques, we optimized the formulations to select two top lipids—20b and 29d—and investigated their biodistribution in mice. Lipid 20b enabled LNPs to transfect the liver and spleen, and 29d LNP transfected the lung and spleen. Unexpectedly, LNP with lipid 20b was especially potent in mRNA delivery to the retina with no acute toxicity, leading to the successful delivery to the photoreceptors and retinal pigment epithelium in non-human primates.
The expansion of gene therapies relies on continuous improvement of delivery platforms, such as lipid nanoparticles (LNPs) (1). Among standard components of a LNP—ionizable lipid, cholesterol, structural lipid, and polyethylene glycol-conjugated lipid (PEG-lipid)—ionizable lipids hold the highest interest due to the impressive variability of chemical structures that can lead to successful nucleic acid delivery (2). The search for lipids that help further improve nucleic acid loading and delivery efficiency is ongoing.
Lipid-based materials generally have a natural propensity for accumulation in the liver from intravenous administration (1), limiting the scope of the diseases that can be targeted in this approach. A few examples exist suggesting that the modification of lipid materials or formulation may divert the nanoparticles from transfecting the liver. For example, it is possible to bypass the liver by modulating LNP formulation components (3), thereby changing the composition of the superficial protein corona and directing the LNPs to the lung and spleen (4). Incorporating cationic lipids as the fifth [selective organ-targeting or SORT (3)] LNP component may result in lung transfection whereas anionic lipids lead to splenic transfection although the surface of nanoparticles exhibited neutral charges. Lung transfection was also achieved in the case of N-series lipidoids, likely following the same mechanism (5). These observations indicate that there are some common molecular patterns that may lead to organ-specific delivery.
In this article, we report the identification and development of ionizable lipids containing a thiophene core (Thio-lipids). The 2-aminothiophenes are considered privileged structures in medicinal chemistry owing to their ease of synthesis and a broad spectrum of activity. Despite the popularity of this chemical motif in medicinal chemistry, its utility in ionizable lipids has not been explored. Thio-lipids can be prepared through the Gewald reaction, which offers a great degree of flexibility in terms of lipid modification. From a fundamental lipid design perspective, a successful ionizable lipid contains three parts: one or more hydrophobic tails, a linker group, and an ionizable head (2, 6). Hydrophobic tails determine the solubility of the lipids and their capability to form a conical shape, which helps to entrap hydrophilic macromolecules in an inverted micelle or inverted hexagonal channels (7) at the early stages of LNP formation. The ionizable headgroup determines apparent pKa of the lipids; secondary or tertiary amines are often used for this purpose due to ease of synthesis and modification and favorable pKa. It is postulated that the optimal apparent pKa of an LNP typically falls in the range of 6 to 7, which allows pH-triggered mRNA (messenger RNA) release during endocytosis of nanoparticles (2, 6). Lastly, linker, or backbone, is a chemical moiety connecting a hydrophilic headgroup to the hydrophobic tails resulting in an amphiphilic molecule. While the roles of lipid parts are relatively well understood, a correct combination of the chemical moieties usually can only be found empirically. Through the rational structural evolution of Thio-lipids, we prepared 47 different Thio-lipids and identified some structural criteria required to efficiently encapsulate and deliver mRNA in vitro and in vivo. Using high-throughput formulation and barcoding tools, we further dissected the biodistribution of these materials. Moreover, we found unexpected evidence that shows a potent delivery to the retina in mice and non-human primates (NHP) based on the specific structure of the Thio-lipids.
Results
The Gewald reaction is a multicomponent reaction between an α-cyanoester, an α-methylene carbonyl compound, and elemental sulfur, yielding 2-aminothiophene motif (8–10). This reaction allows to incorporate functional groups in all four positions of the thiophene core; here, we elected to include an amino group to serve as an ionizable head or an additional synthetic handle at 5-position of thiophene via either cyclical or linear derivatives. A variation of this reaction used in the present work is shown in Fig. 1. To evaluate the capabilities of Thio-lipids to produce robust and potent LNPs, we solubilized the lipids in ethanol and prepared LNPs containing firefly luciferase (FLuc) mRNA via microfluidic mixing (NanoAssemblr Benchtop) at lipid molar ratio of 50/10/38.5/1.5 (ionizable lipid, distearoylphosphatidylcholine(DSPC), cholesterol, and 1,2-dimyristoyl-rac-glycero-3-methoxypolyethylene glycol-2000(DMG-PEG2000), respectively), volumetric ratio of 3:1 (mRNA aqueous mix:ethanol lipid mix, respectively) and N/P 5.7 ratio (ratio of ionizable nitrogens, assuming 1 per molecule, to number of phosphates in mRNA).
Fig. 1.
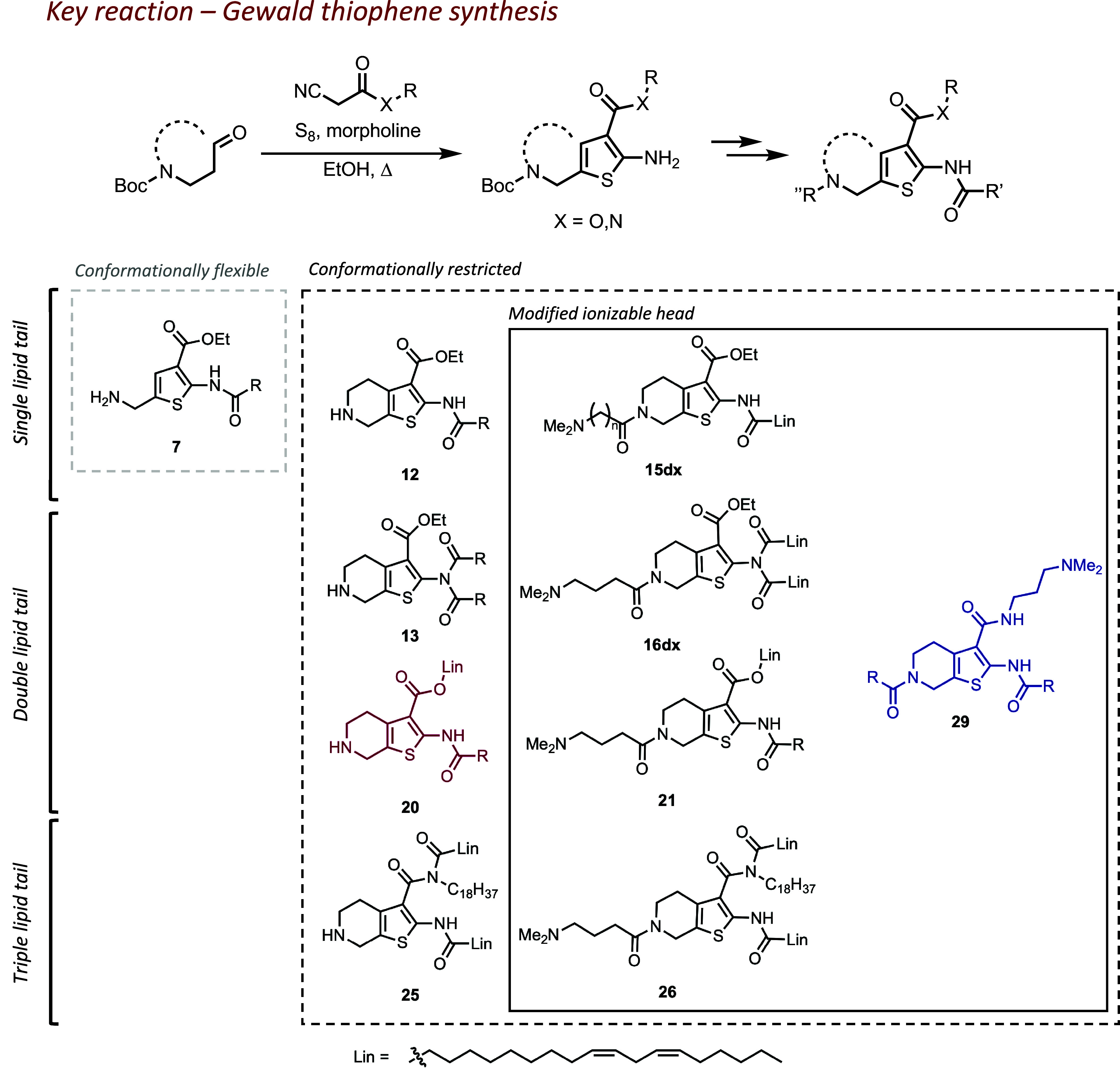
Synthetic evolution of thiophene-based lipids (Lin = linoleic lipid tail) and the scope of the structures discussed in this work. Lipids are grouped by structural criteria: 1) flexible or restricted conformation of the thiophene core; 2) modification of the ionizable head; and 3) number of lipid tails present in a molecule. Compound enumeration begins from precursors; full synthetic information is available in SI Appendix.
Initial Lipid Tail Screening.
During the initial development of thiophene core into ionizable lipids, we started off with a flexible thiophene core. Our first step was to evaluate whether attaching a single lipid tail to a 2-aminothiophene core 4 can yield lipids capable of forming LNPs (compound enumeration is available in SI Appendix). The resulting amides 5a-f and their Boc-deprotected counterparts 6a-f had both variable lengths of the lipid tail and degree of unsaturation. During the synthesis, we also isolated imide 7, an unexpected by-product in preparation of 6c. These amide derivatives produced stable nanoparticles with acceptable size albeit with a moderately heterogeneous population (ca. 100 nm, PDI > 0.2; see Fig. 2A). Most lipids in the series were able to quantitatively encapsulate mRNA; however, 6a and 6d failed to encapsulate to a satisfactory degree (<75%; Fig. 2B). Overall, we confirmed that thiophene can be a suitable starting group for lipidic materials. Unfortunately, none of these LNPs succeeded in delivering mRNA in vitro. Amides with lipid tails less than C16 (capric and lauric; 6a and 6b) also required significant solubilization efforts, so we limited the scope of reacting carboxylic acids to palmitic, stearic, oleic, and linoleic acids.
Fig. 2.
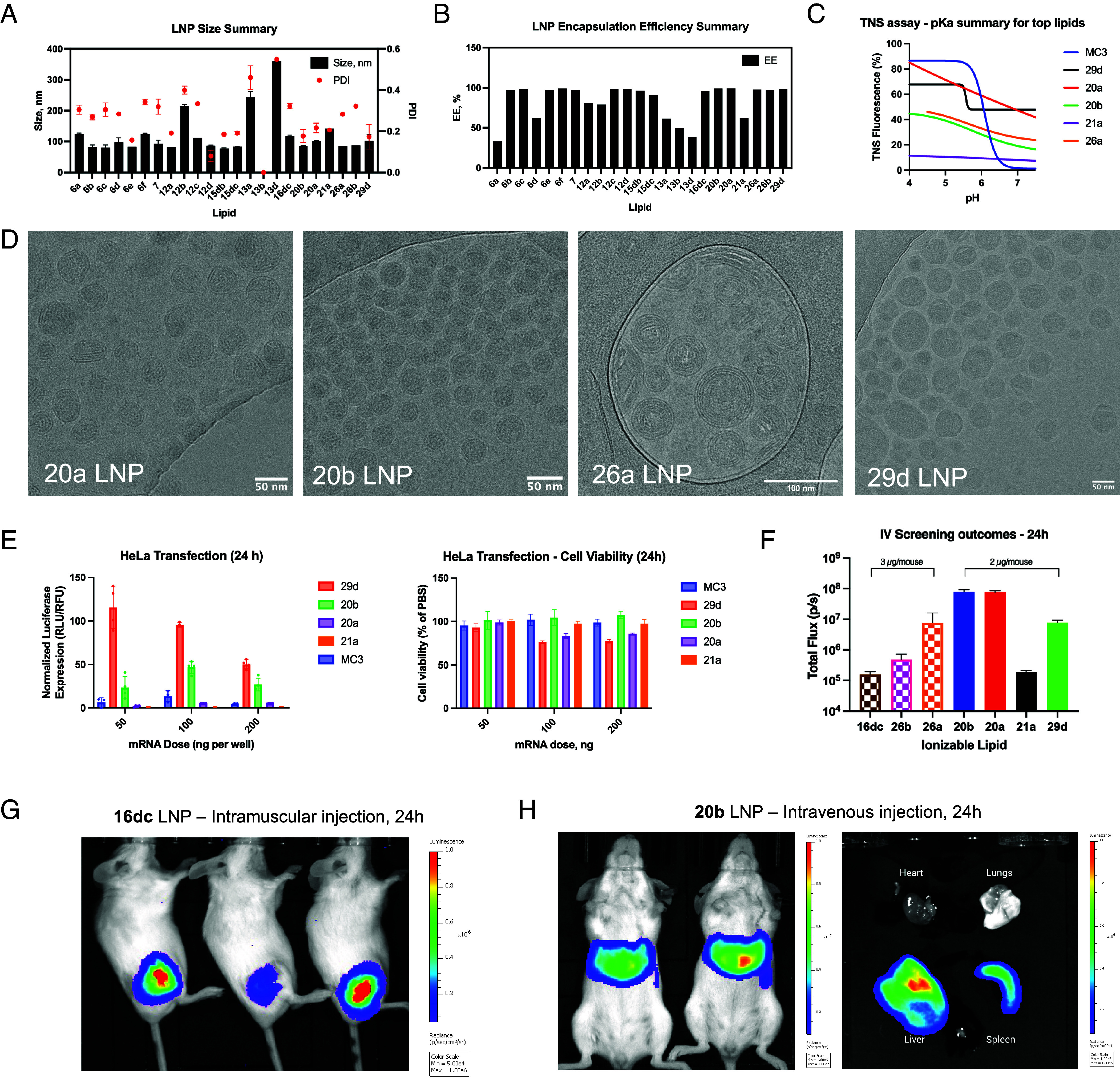
Physicochemical characterization of LNPs: (A) size and PDI of LNPs; (B) mRNA encapsulation efficiency as determined by the modified RiboGreen assay; (C) determination of pKa for select lipids in LNPs via 2-(p-toluidino) naphthalene-6-sulfonic acid (TNS) assay, typical pH-responsiveness of ionizable lipid represented by MC3 LNP; (D) cryoTEM micrographs illustrating LNP morphologies with select lipids; (E) comparison of 29d, 20b, 20a, and 21a LNP efficacy and cell viability in HeLa; (F) Bioluminescence imaging summary for intravenous administration of LNPs, note the dose difference. (G) Representative in vivo bioluminescence images after an IM administration of LNPs incorporating 16dc; (H) Representative in vivo and ex vivo bioluminescence images after an IV administration of LNPs incorporating lipid 20b, the most potent lipid in the tested series.
Conformation Restriction and pKa of the Headgroup.
In the second stage, we chose to evaluate whether restricting the conformation of the 5-amino group to a piperidine ring brings any advantage. Resulting lipids 12a-d also produced heterogeneous LNPs (Fig. 2A). LNPs containing lipids with saturated tails (12a,b) were worse at encapsulating mRNA than those carrying unsaturated tails (12c,d; see Fig. 2B), a trend in line with literature reports (11). We also isolated imides 13a,b,d; however, they failed to form stable nanoparticles, likely due to the increased steric hindrance and crystallinity. Although conformationally restricted lipids did not offer a delivery advantage compared to their conformationally flexible counterparts, we found that the Gewald reaction producing conformationally restricted aminothiophenes was more efficient in terms of yield and scale-up. Therefore, we proceeded to utilize this synthetic advantage in the next stages of lipid evolution.
Since lipids 6 and 12 yielded robust LNPs but failed to deliver mRNA, we hypothesized that their ionizable headgroup (or resulting pKa) may be suboptimal. Indeed, the calculated pKa (cpKa) of these lipids was ca. 7.8 and 8.0, respectively, which is lower than presumptively optimal cpKa of 9 to 10 (6). To adjust pKa, we found via a cheminformatic analysis that it was necessary to add a spacer between the piperidine ring and an ionizable headgroup such as a tertiary amine. We selected lipid 12d as a parent structure for further modifications due to the good nanoparticle properties, yielding amides 15da-c. We expected that the transfection efficiency may be proportional to the spacer length since the cpKa became progressively more basic (from 8.1 to 9.8). Indeed, we found that 15dc produced the greatest in vitro mRNA transfection among these series although cytotoxicity was also observed (SI Appendix, Fig. S1A), proportional to the treated dose. We were also curious to see if adding this potent ionizable head to a disubstituted amide 13d would improve nanoparticle stability and mRNA delivery. Resulting lipid 16dc not only produced stable LNPs but also yielded greater levels of mRNA transfection in HeLa even compared to the positive control (DLin-MC3-DMA LNPs) by ca. 2.5-fold (SI Appendix, Fig. S1). We posit that an additional spacer group may have improved the capability of these lipids to form an inverted micelle, one of the key sterical requirements to produce LNPs (11). Intravenous administration of 16dc LNPs containing FLuc mRNA produced poor bioluminescence signal (Fig. 2F); however, administering 16dc LNPs via intramuscular injection resulted in robust bioluminescence at the site of injection (Fig. 2G), suggesting that lipid design could be further improved upon to produce LNPs capable of navigating in vivo environment.
Double Lipid Tails.
Thinking that the proximity of lipid tails in 16dc may have made the LNPs too unstable for intravenous administration, we sought to separate the lipid tails into different positions of the thiophene ring. This way, we could also investigate the potential of lipidic cyanoesters to participate in the Gewald reaction. Indeed, the reactions proceeded with high yield (SI Appendix). Resulting lipids 20a,b formed LNPs with good size and encapsulation efficiency (Fig. 2 A and B). However, LNPs with lipid 21a (carrying the ionizable head found in the previous step) presented reduced potency and mRNA encapsulation efficiency compared to its unmodified counterpart 20a (Fig. 2 E and F). We observed an inconsistent dose response to LNPs upon in vitro HeLa transfection (Fig. 2E); we attribute this phenomenon to the saturation of the translation machinery due to relatively low cell density or excessive dose (12, 13). Suspecting that the loss of efficacy was related to the changes in pKa, we performed the TNS assay to compare the ionizability profile of these lipids (Fig. 2C). None of the Thio-lipid LNPs displayed a sigmoidal curve characteristic for LNPs with traditional ionizable lipids like DLin-MC3-DMA (MC3) and indicating distinct ionized/unionized states. Thio-lipids may have a buffering property, as suggested by the slow decline in fluorescence over pH. This may indicate an alternative, unknown mechanism of action in nucleic acid delivery for Thio-lipid LNPs. Despite the suboptimal cpKa of ionizable headgroup (ca. 8), LNPs with lipids 20a and 20b successfully transfected the liver and spleen from an intravenous administration (Fig. 2 F and H). Cryogenic transmission electron microscopy (cryoTEM) imaging of LNPs with lipid 20a revealed slightly elongated and more hydrophobic particles (the latter indicated by preferential deposition on the carbon support material; Fig. 2D) than LNPs with lipid 20b, likely due to the saturated lipid tail.
Triple Lipid Tails.
We then explored the option of adding a third lipid tail to the thiophene linker. Instead of pursuing modifications of 11d, a minor acylation byproduct that led to lipid 16dc, we opted out to incorporate an amide at 3-position of the thiophene ring for a more convenient tail conjugation. Comparing the characteristics of 26a,b LNPs against 16dc revealed that LNPs containing triple-tail lipids still could form small but relatively heterogeneous nanoparticles with great mRNA encapsulation efficiency (Fig. 2 A and B). In vitro mRNA transfection data revealed that 16dc LNPs were the most potent in HeLa, 26a LNPs in the HepG2, and 26b LNPs in HEK293 (SI Appendix, Fig. S1). Notably, only 26a LNPs could transfect the liver from intravenous administration but to a lesser extent than 20a,b LNPs (Fig. 2F). It was also not as potent as lipid 16dc in intramuscular delivery (SI Appendix, Fig. S2). Therefore, there may not be a significant delivery benefit from adding a third lipid tail.
Rearrangement of the Building Blocks.
Next, we investigated the arrangement of the building blocks. We separated the lipid tails to the opposing sides of the thiophene core and modified the ester to carry the ionizable headgroup. The resulting lipid 29d maintained the capability to form robust LNPs, although somewhat heterogeneous in shape and internal structure (Fig. 2D). Unexpectedly, ex vivo imaging of the mouse organs 24 h after an intravenous injection indicated that 29d LNPs transfected lungs and spleen (Fig. 3B). The tropism to these specific organs was peculiar, since delivery to the lungs and spleen was previously achieved by incorporating cationic and anionic lipids, respectively (3, 14). Similar to 20a,b, the TNS assay of 29d LNPs did not show a distinct sigmoidal curve (Fig. 2C). We are investigating the mechanism behind this organ tropism.
Fig. 3.
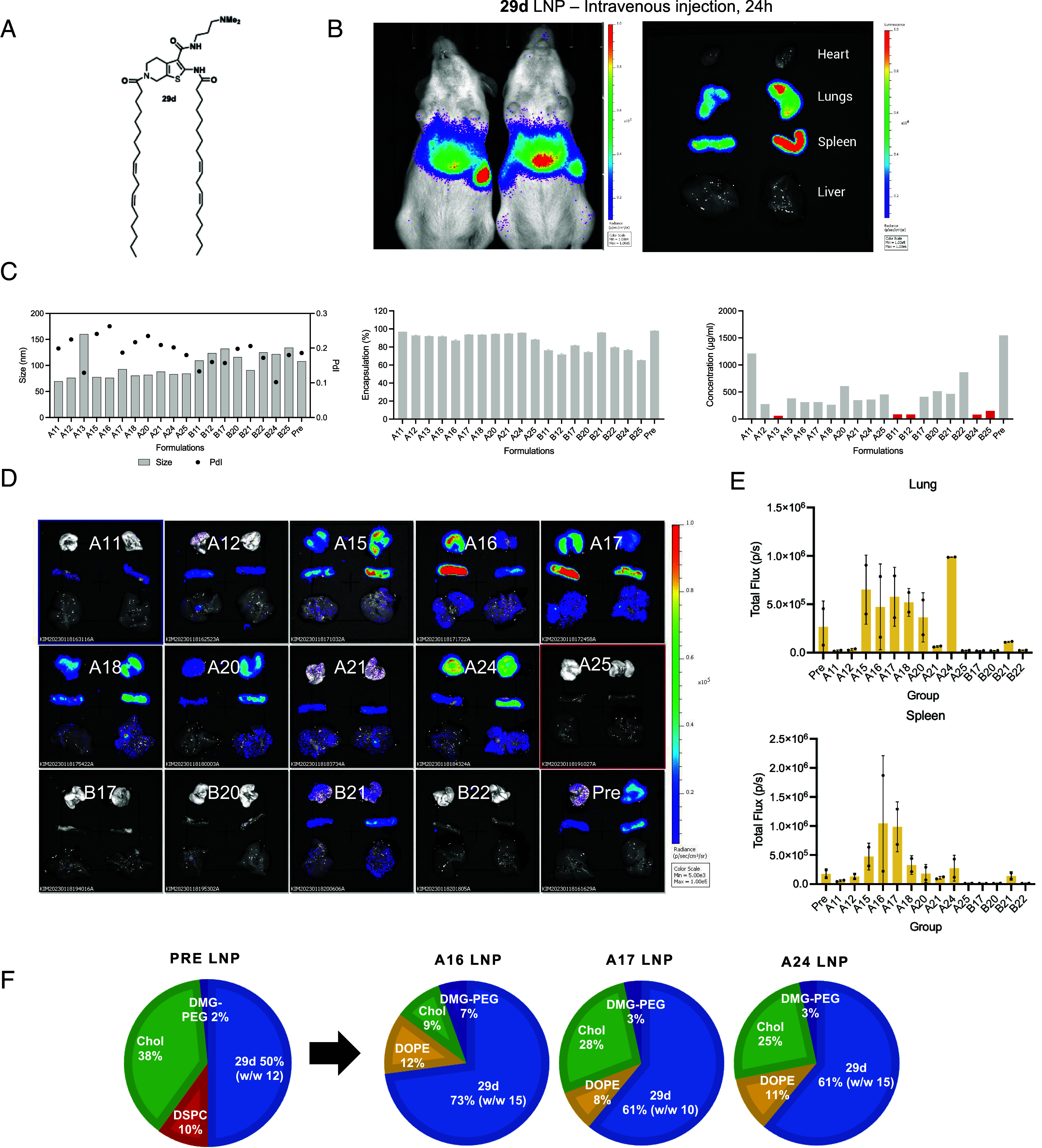
29d LNP formulation optimization for improved lung delivery. (A) Structure of lipid 29d; (B) Representative in vivo and ex vivo bioluminescence images after an IV administration of LNPs incorporating lipid 29d, (C) Summary of LNP formulation prescreening includes analysis of nanoparticle size, polydispersity, and mRNA encapsulation and loading efficiency; (D) results of intravenous screening of most robust formulations. Most LNPs showed preferential tropism toward the lung and spleen; the quantification of luminescence in those organs 24 h after IV administration is shown in panel (E). The top formulations A16, A17, and A24 are illustrated in panel (F).
Seeking further improvement to both the ease of synthesis and the transfection efficiency of 29d, we explored an alternative synthetic pathway—incorporate the ionizable headgroup directly into the cyanoester precursor prior to the Gewald reaction. This pathway allowed us to prepare compounds 29a-w in fewer steps since the same intermediates could be modified with various lipid tails. The scope of modifications included variable length, unsaturation, and branching of the lipid tails, and the addition of biodegradable ester spacer (SI Appendix). Once incorporated into LNPs, these lipids showed comparable encapsulation efficiency, size, and polydispersity to 29d LNPs; however, no significant improvement was achieved in terms of IV mRNA delivery (SI Appendix, Fig. S3). Although we were not able to improve on the structure of 29d, we confirmed the feasibility of an alternative synthetic pathway allowing us to reduce the number of reaction steps.
Formulation Optimization.
Next, we sought to optimize the LNP formulations of lipids 20b and 29d due to the high transfection efficiency and peculiar organ tropism. The design of the formulation library is presented in SI Appendix, Fig. S4A and included screening of lipid molar ratios, compatibility with 2 commonly used phospholipids [1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE; group A) or DSPC (group B)], and weight-to-weight ratio of ionizable lipid to mRNA. A previously used “baseline” formulation of 50/10/38.5/1.5 (ionizable lipid/DSPC/cholesterol/DMG-PEG2000, respectively) is labeled hereafter as Pre. We prepared the formulations via high-throughput robotic mixing with a small quantity of nucleic acid (5 to 30 µg) in 50 mM citrate buffer (pH 4) and first evaluated their size and encapsulation efficiency (SI Appendix, Fig. S4 B and C). To classify a formulation as a “success”, we deployed the quality assurance criteria of diameter <200 nm, PDI < 0.2, and encapsulation efficiency of >75%. Interestingly, a clear bias emerged toward preferred structural lipids. 20b LNPs showed a strong preference toward DSPC as a structural lipid (group B), while 29d LNPs were more compatible with DOPE (group A), suggesting that these lipids may have different spatial requirements due to their molecular geometry despite both carrying linoleic tails. The most robust formulations for lipid 29d incorporated either a higher percentage of ionizable lipid (>60 mol%), PEG-lipid (>3 mol%), or less cholesterol (<28 mol%). However, the resulting formulation space for lipid 20b had no clear preference for specific molar ratios, suggesting that lipid 20b can be incorporated in variable formulations depending on the application.
Considering that 20b lipid was the most potent in the studied series, we also wanted to evaluate the compatibility of this lipid with cargo other than mRNA and compare the delivery efficiency to a benchmark lipid DLin-MC3-DMA. To do so, we deployed the “standard” (50/10/38.5/1.5, DSPC) 20b formulation to encapsulate DNA barcode with or without mRNA. DNA barcode is an attractive choice of cargo to evaluate the LNP delivery efficiency since it can easily accumulate in various organs at early timepoint upon successful delivery and does not degrade as rapidly as mRNA (15); however, co-encapsulating barcode DNA with mRNA may be a better approach to predict formulation potency across different kinds of nucleic acids and even different species (16, 17). Therefore, the experiment scope included a naked barcode and 2 different types of LNPs (20b and MC3 LNPs at the same molar ratios) encapsulating either barcode with FLuc mRNA mix [1:10 w/w ratio (16)] or only the barcode. The major organs (heart, liver, lung, spleen, and kidneys) were harvested 6 h after an intravenous injection in BALB/c mice and processed for NGS sequencing. The results for barcode abundance in different organs after normalization to the injection solution are shown in SI Appendix, Fig. S4D. Overall, MC3 and 20b LNPs co-loaded with DNA barcode and mRNA showed higher abundance in DNA barcode counts compared to their barcode-only LNP counterparts. Lungs, heart, and kidney analyses showed marginally higher barcode abundance for LNP groups than the naked barcode, suggesting poor delivery efficiency to those organs in agreement with luminescence data. In the liver, 20b LNPs were comparable to MC3 in terms of barcode delivery efficiency regardless of the cargo; yet, Thio-lipids overall had a higher effective mRNA dose for delivery to the liver than MC3. In the spleen, co-loaded 20b LNPs yielded almost 4-fold higher barcode counts than co-loaded MC3 LNPs, and almost 100-fold higher than barcode-only 20b LNPs. It may be caused by an inherent affinity of 20b toward the spleen in addition to DSPC (18). The overall discrepancy between barcode delivery for barcode-only vs barcode co-encapsulated with mRNA LNPs suggests that the nanoscale differences in particle morphology caused by the cargo may significantly affect delivery efficiency.
Lastly, we investigated the lung transfection efficiency of the different formulations of lipid 29d. Out of 50 formulations described above, only 20 formulations of lipid 29d fully met quality criteria as described above (SI Appendix, Fig. S4B). These remaining 20 formulations were then prepared via microfluidic mixing at a larger scale (75 µg mRNA). The size of all 20 LNPs maintained at 150 nm or less; however, 5 more formulations were disqualified due to the low mRNA recovery and encapsulation (Fig. 3C). The LNPs were then administered intravenously at 5 µg mRNA dose to CD1 mice, and their organs were subject to ex vivo bioluminescence analysis at 24 h post-administration. Transfection tropism to the lungs and spleen persisted for all formulations although some signal was also detected in the liver (Fig. 3D). 29d LNPs with DOPE (group A) led to the greater mRNA transfection than DSPC (group B), and formulations A15-18 and A24 were more potent than the initial formulation (Pre). Formulation A24 was the most potent in lung transfection (ca. fourfold improvement as compared to Pre; Fig. 3 D and E) while formulations A16 and A17 were most potent in the spleen transfection (ca. sixfold improvement). Although no clear formulation trends emerged for delivery specifically to the lung or spleen (Fig. 3F), we confirmed that incorporation of DOPE and reducing the cholesterol content can help to divert 29d LNPs from the liver.
Subretinal Delivery.
Encouraged by the performance of the Thio-lipids in intravenous delivery, we sought to evaluate the translational potential of lipid 20b via subretinal delivery. Out of 11 formulations identified in the barcoding study, we chose formulation B06 as base for evaluation (60/20/19/0.5 mol%, DSPC as structural lipid; see Fig. 4B) due to its reduced PEG and cholesterol content in favor of phospholipid. These factors were previously identified as beneficial for subretinal delivery (19). We encapsulated Cre mRNA in 20b LNPs and administered them to Ai9 mice at 300 ng mRNA dose. The eyes were harvested after 96h, frozen, and processed for immunofluorescence. Confocal imaging of retinal sections revealed that 20b LNPs transfected retinal pigment epithelium (RPE), photoreceptors, and Müller glia (SI Appendix, Fig. S7). In comparison, 29d LNPs only transfected RPE, equivalent to conventional LNPs (20).
Fig. 4.
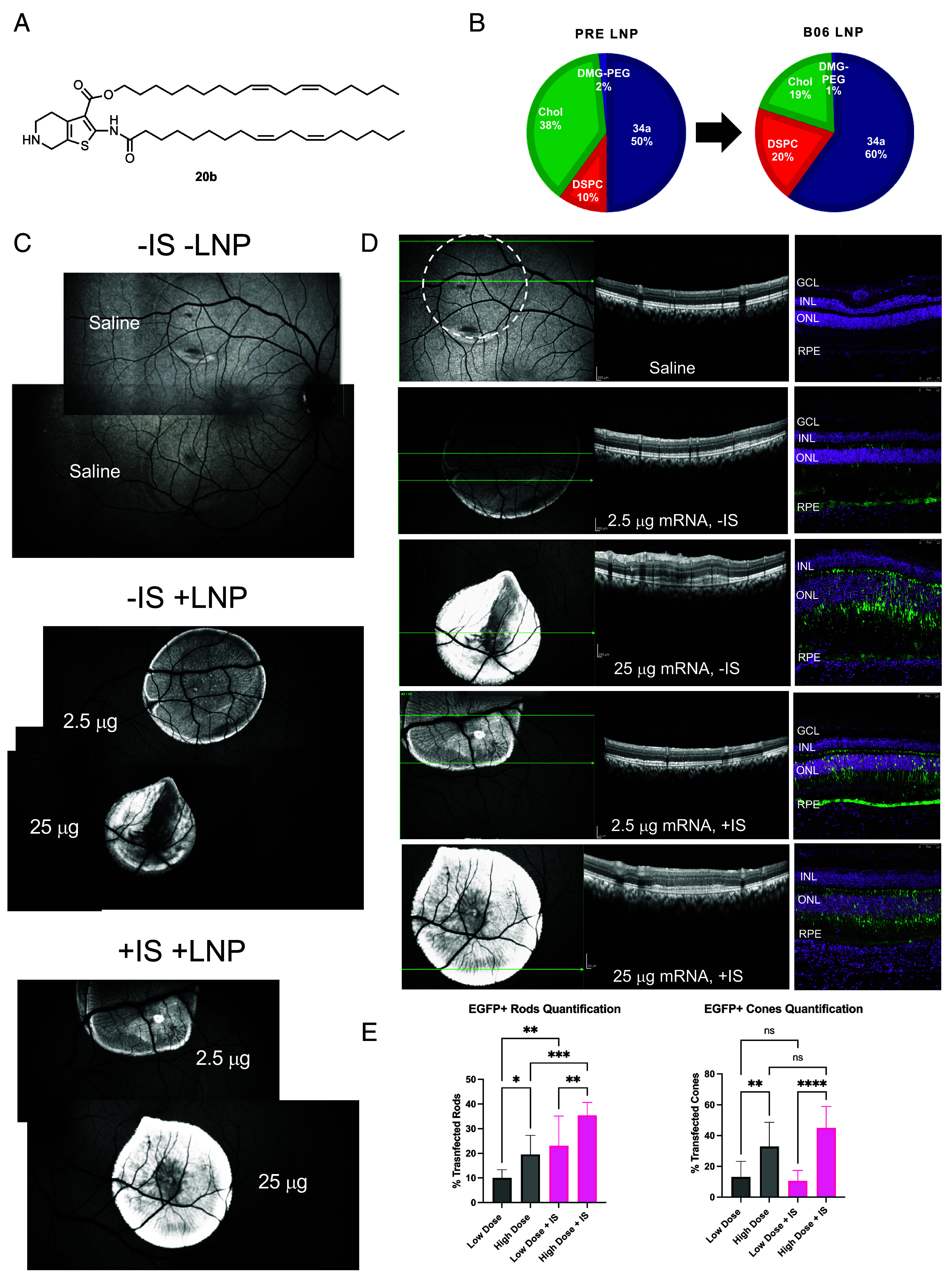
Evaluation of the top ionizable lipid 20b in the NHP eye after a subretinal administration of EGFP mRNA 20b LNPs. (A) structure of lipid 20b and the formulation used (B). (C) FAF image montage illustrating the relative EGFP intensity in the back of the eye changing in response to the dose. Saline was included as negative control. −IS animal received standard treatment, while +IS animal also received an immunosuppressive treatment (SI Appendix). (C) Representative OCT suggests negligible retinal toxicity for low-dose treatment (the dashed circle indicates the location of the saline bleb; green arrows indicate the scan plane). Significant perturbation was observed in high-dose −IS treatment compared to saline control, less so in high-dose +IS. (D) Representative IF images confirm disruption of retinal morphology in the 25 μg −IS group (DAPI—magenta, EGFP—green; orientation preserved between OCT and IF). (E) image-based quantification of photoreceptor transfection. Stronger EGFP signal correlated with higher dose or IS treatment (N = 1 eye; the statistical analysis is shown for the quantification in different areas of the injection blebs (n = 10). Results are reported as mean and SD after 1-way ANOVA with the Tukey multiple comparison test; *: P 0.0332; **: P 0.0021; ***: P 0.0002; ****: P < 0.0001).
Since we observed the capability of lipid 20b to deliver mRNA to the photoreceptors in the Ai9 mouse model, we decided to formulate 20b LNPs with enhanced green fluorescent protein (EGFP) mRNA and administer them subretinally to NHP. Due to the larger size of the NHP retina compared to the mouse, we were able to evaluate two doses—2.5 µg (“low dose”, 50 µL bleb at 50 ng/µL) and 25 µg (“high dose”, 50 µL bleb at 500 ng/µL)—in the same retina to estimate minimum effective dose. Saline treatment was included as negative control. Another important part of the study was incorporation of immunosuppressive treatment (IS) since our group previously observed immune cell recruitment after a subretinal injection in NHP (21). The EGFP signal in the fundus autofluorescence (FAF) images of the NHP retina clearly indicated dose response 48 h after administration (Fig. 4C). For both low-dose blebs, the EGFP fluorescence signal was relatively consistent throughout the bleb. In the animal treated with IS (+IS), the high-dose bleb was consistently bright and had higher signal than the low-dose bleb. In the animal without IS (−IS), the high-dose bleb had higher signal than the low-dose bleb, but it also had pronounced dark areas. Anticipating that high-dose treatment may have contributed to toxicity and/or immune response, we evaluated retina health via optical coherence tomography (OCT). Indeed, high-dose treatment led to subretinal accumulation of debris, especially in the −IS animal, while retinal morphology after low-dose treatment was unremarkable and similar to that after the saline treatment (Fig. 4D). Therefore, darker areas in the high-dose −IS bleb likely correspond to the subretinal accumulation of debris. Immunofluorescence images of retinal sections revealed that the EGFP was localized to the RPE and photoreceptors (Fig. 4D). We then counted the total number of EGFP+ rods and EGFP+ cones in the outer nuclear layer, providing a transfection efficiency value for treatments (Fig. 4E). We found that high-dose treatments led to higher transfection of both rods and cones, while immunosuppressive treatment only had a significant impact on the rod transfection. The highest number of transfected rod photoreceptors were in the high-dose +IS treated retina (35%), followed by the low-dose +IS treated retina (23%), the high-dose treated retina (20%), and the low-dose treated retina (10%). The highest number of transfected cone photoreceptors were in the high-dose +IS treated retina (45%), followed by the high-dose treated retina (33%), the low-dose treated retina (13%) and the low-dose +IS treated retina (11%). We also assessed the immune cell infiltration by staining the retina sections with antibodies specific to microglia (IBA1), macrophages (CD68), and T-cells (CD3). The choroidal space had some microglia infiltration, correlating with LNP dose, but not IS treatment (SI Appendix, Fig. S8). We did not observe any other immune cell infiltration or activation in the neural retina, subretinal space, or choroid (SI Appendix, Fig. S9). In conclusion, these data indicate the translational potential of lipid 20b across different species and provide photoreceptor transfection efficiency values that can be used when considering therapeutic interventions with LNP-mediated mRNA delivery to the retina.
Discussion
The delivery efficiency of LNPs is often determined by the potency of ionizable lipids. Ionizable lipids are a broad class of molecules that, as the name suggests, contain an ionizable or pH-responsive functional group such as tertiary amine, linkers, and lipid tails. Theoretically, these groups may be attached via a wide variety of chemistries, but most ionizable lipids share the same design strategies—highly flexible structure, ester or amide linker bonds, and tertiary amines as ionizable heads (2). Even though heterocycles are extensively used in medicinal chemistry—more than 80% of small molecules approved by the FDA in 2022 contain at least one heterocycle—their utility in the design of ionizable lipids is very limited (2). Here, we present heterocyclic ionizable lipids containing a thiophene core (Thio-lipids) that is readily accessible by the Gewald reaction. The thiophene intermediate presents a highly customizable core for diverse lipid library generation.
We were able to identify some key structural criteria of Thio-lipids leading to stable LNPs with good efficacy and tolerability. First, incorporating at least one lipid tail with >16 carbon atoms can result in a sufficiently robust LNP, likely due to optimal hydrophilic-lipophilic balance. We found that incorporation of two unsaturated linoleic tails was especially beneficial for potency, in line with previous reports (11). Second, the localization of the lipid tails and ionizable group may direct organ-specific transfection. A case in point is lipid 29d, with linoleic tails separated to the opposing sides of the molecule and capable of reaching the lung and spleen, while lipid 20b with linoleic tails attached on the same side of the thiophene backbone directed LNPs to the liver and spleen. Third, regardless of the nature of the amine introduced (primary, secondary, or tertiary), these lipids were able to efficiently encapsulate mRNA. Thio-lipids may rely on more than electrostatic interactions between the positively charged ionizable headgroup and negatively charged mRNA phosphates to form LNPs with high encapsulation efficiency—π–π and sigma hole interactions with mRNA may also be implicated in the encapsulation process (22–25). The lipids that failed to quantitatively encapsulate mRNA had either steric or solubility constraints. However, not all these lipids could deliver mRNA into cytoplasm. Investigating the cpKa of the most potent lipids and experimental pKa of the LNP formulations revealed no clear correlation between lipid efficacy and ionizability. For example, lipid 20b contained only a secondary amine incorporated into the thiophene backbone structure yet was more potent than its modified counterpart. Moreover, the resulting LNPs had an unusual pH response profile, suggesting a partial positive charge across a broad pH range. We acknowledge that more work is needed to fully understand the structure–activity relationship of these lipids.
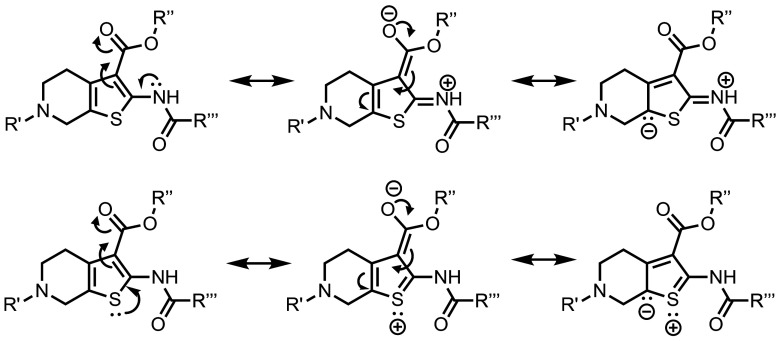
Organ-specific delivery via LNPs and nanomedicines can be achieved via either active targeting [i.e., targeting ligands (21, 26, 27)] or passive targeting (i.e., specific properties of the nanoparticle such as size or charge). Little is understood about the adjustments required to achieve passive targeting for specific organs; however, strong evidence suggests that the nature of the biomacromolecular corona formed on the nanoparticle surface may direct the delivery to specific organs (28–31). For example, ionizable but not cationic LNPs can bind to apolipoprotein E and utilize low-density lipoprotein receptor to enter hepatic tissues (32). The LNP ionization seems to be a major determinant in the corona composition, although LNP reorganization upon exposure to serum proteins may also contribute to this phenomenon (31). In the case of Thio-lipids, we found that the tested lipids did not have a defined ionizability profile even when ionizable headgroups with desired cpKa were introduced. This may be the case in three different scenarios: 1) A permanent cationic charge is present in these molecules after the formulation process, 2) the charge is offset by other lipid components or trace impurities, or 3) their ionization mechanism is different from ionizable lipids like MC3. Scenarios 1 and 2 seem unlikely because a) there was no significant toxicity typically attributed to cationic lipids and b) although incorporating permanently cationic lipids into an ionizable formulation would direct LNPs to the lungs (3, 4, 33), only 29d LNPs could reach lung tissues. Scenario 3, on the other hand, may occur due to the extended delocalization of the electron density through the thiophene ring and its substituents (Fig. 5). The electron delocalization might be a limitation for the TNS assay, presenting in a non-traditional ionizability profile. While Thio-lipids may not be ionizable in the traditional sense, we speculate that Thio-lipids still rely on the electrostatic binding to the endosomal membrane to deliver mRNA.
Fig. 5.
Example resonance structures of Thio-lipids demonstrating extended electron delocalization. The contribution of resonance forms may explain the broad ionization profile of Thio-lipids since heteroatoms (e.g., nitrogen and sulfur) may carry a partial positive charge.
Thio-lipids have slightly different formulation requirements from a frequently used 50/10/38.5/1.5 formulation for successful nucleic acid delivery—specifically, an increased percentage of ionizable lipid at the expense of cholesterol (>60 mol% IL, <30 mol% Chol). The phospholipid needs for intravenous administration of LNPs containing lipids 20b and 29d were different (DSPC vs. DOPE, respectively). Cholesterol and phospholipids stabilize the lipid membrane and facilitate the formation of an inverted hexagonal phase (34), and it is hardly a surprise that different phospholipids were preferred to produce a robust formulation. The lesser extent of cholesterol, however, indicates that Thio-lipids may be capable of producing an inverted hexagonal phase on their own. We are expanding the lipid library and conducting relevant studies to fully understand the phase behavior of these lipids.
Despite the unusual ionizability profile, we achieved successful delivery in vivo for several different cargos (FLuc, Cre recombinase, and EGFP mRNA) in mouse and non-human primate models. The majority of the Thio-lipid LNPs reached the liver and spleen from the intravenous administration, similar to most lipid-based nanomaterials (1). We identified lipid 20b as the most robust and potent lipid in the series. Lipid 29d was less potent; however, it was able to reach the lungs and spleen. Previously, tropism to these specific organs in case of SORT-lipids was attributed to cationic and anionic charge, respectively (3, 14). A zwitterionic nature of 29d would be a logical explanation to this observation; however, zwitterionic ionizable lipids may still possess a defined ionizability profile (14). Nonetheless, the scope of organs transfected by Thio-lipid LNPs is unusual, and we are currently investigating the biological causes for the lung tropism of the 29d LNPs.
When Thio-lipid LNPs were administered to the NHP retina, they mediated robust expression across the entire bleb and transfected not only RPE but also photoreceptors, a highly sought-after target in the retina. To determine if immune suppression would modulate mRNA expression in the retina, we evaluated a combined local and systemic immunosuppressive regimen, following routine practices in ophthalmology (35). Photoreceptor transfection generally increased with LNP dose; however, low LNP dose correlated with better retinal health outcomes. Interestingly, immune suppression was able to increase rod but not cone transfection efficiency. This suggests that immune suppression may enhance mRNA expression. In general, immune cell infiltration was minimal and limited to the choroidal space; however, the outcomes were captured only at 48 h post-injection. While these results are highly encouraging, more studies are needed to evaluate the biological variability in EGFP expression and the long-term impact of Thio-lipids on the retinal health. We believe that these data serve as a proof-of-concept for translation in higher animals and will continue to investigate the utility of Thio-lipids in potential treatments of pulmonary and retinal genetic diseases.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This project was supported through funding from the National Eye Institute 1R01EY033423-01A1 (G.S.), Oregon National Primate Research Center Pilot Grant (R.C.R.), Oregon National Primate Research Center Core Grant P51 OD011092 and S10RR024585, Casey Eye Institute Core Grant P30 EY010572 from the NIH, and unrestricted departmental funding from Research to Prevent Blindness. Electron microscopy was performed at the Multiscale Microscopy Core at Oregon Health and Science University.
Author contributions
Y.E., M. Gupta, J.K., J.R., A.T., M.N., and R.C.R. designed research; Y.E., M. Gupta, J.K., A.J., M. Gautam, J.R., D.N., E.B., A.T., J.S., R.R., A.K.L., and R.C.R. performed research; Y.E., M. Gupta, J.K., A.J., J.R., D.N., A.T., and J.S. contributed new reagents/analytic tools; Y.E., M. Gupta, J.K., A.J., M. Gautam, J.R., A.T., J.S., R.R., and R.C.R. analyzed data; and Y.E., M. Gupta, J.K., R.C.R., and G.S. wrote the paper.
Competing interests
Y.E., M. Gupta, J.K., M. Gautam, J.R., and G.S. are inventors on a patent application pertinent to this work filed by the Oregon State University. Y.E., D.N., and A.T. have stock options and advisory roles with EnterX Bio. G.S. is a cofounder of EnterX Bio. EnterX Bio has a scientific research agreement with Oregon State University. G.S. has a conflict management plan at Oregon State University. The other authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Materials can be made available upon request through a Materials Transfer Agreement (MTA) from Oregon State University. Raw NGS barcoding data are publicly available in the NCBI Sequence Read Archive (SRA). SRAs for each sample are available under NCBI Bioproject Accession # PRJNA1024541 (https://www.ncbi.nlm.nih.gov/bioproject/) (36). Python script used to interpret NGS data is available on GitHub (https://github.com/antonyjozic/lnp_barcode_script) (37). mRNA was sourced from TriLink Biotechnologies (San Diego, CA; substituted with 5-methoxy-U); the ORF sequences are available on the manufacturer's website (38–40).
Supporting Information
References
- 1.Hou X., Zaks T., Langer R., Dong Y., Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Sun C., Wang C., Jankovic K. E., Dong Y., Lipids and lipid derivatives for RNA delivery. Chem. Rev. 121, 12181–12277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Q., et al. , Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dilliard S. A., Cheng Q., Siegwart D. J., On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 118, e2109256118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu M., et al. , Lung-selective mRNA delivery of synthetic lipid nanoparticles for the treatment of pulmonary lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. U.S.A. 119, e2116271119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eygeris Y., Gupta M., Kim J., Sahay G., Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 55, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Yanez Arteta M., et al. , Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 115, E3351–E3360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabnis R. W., Rangnekar D. W., Sonawane N. D., 2-aminothiophenes by the Gewald reaction. J. Heterocyclic Chem. 36, 333–345 (1999). [Google Scholar]
- 9.Huang Y., Dömling A., The Gewald multicomponent reaction. Mol. Divers. 15, 3–33 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Nguyen T. B., Recent advances in the synthesis of heterocycles via reactions involving elemental sulfur. Adv. Synth. Catal. 362, 3448–3484 (2020). [Google Scholar]
- 11.Rietwyk S., Peer D., Next-generation lipids in RNA interference therapeutics. ACS Nano 11, 7572–7586 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Patel S., et al. , Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 17, 5711–5718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eygeris Y., Patel S., Jozic A., Sahay G., Deconvoluting lipid nanoparticle structure for messenger RNA delivery. Nano Lett. 20, 4543–4549 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S., et al. , Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 20, 701–710 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahlman J. E., et al. , Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. U.S.A. 114, 2060–2065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sago C. D., et al. , High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. U.S.A. 115, E9944–E9952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatit M. Z. C., et al. , Species-dependent in vivo mRNA delivery and cellular responses to nanoparticles. Nat. Nanotechnol. 17, 310–318 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R., et al. , Helper lipid structure influences protein adsorption and delivery of lipid nanoparticles to spleen and liver. Biomater. Sci. 9, 1449–1463 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryals R. C., et al. , The effects of PEGylation on LNP based mRNA delivery to the eye. PLoS One 15, e0241006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon W., Freeman S. A., Phagocytosis by the retinal pigment epithelium: Recognition, resolution, recycling. Front. Immunol. 11, 604205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrera-Barrera M., et al. , Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci. Adv. 9, eadd4623 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan P. F., et al. , Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 114, E4492–E4500 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foister S., Marques M. A., Doss R. M., Dervan P. B., Shape selective recognition of T·A base pairs by hairpin polyamides containing N-terminal 3-methoxy (and 3-chloro) thiophene residues. Bioorg. Med. Chem. 11, 4333–4340 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Stolić I., et al. , Effect of 3,4-ethylenedioxy-extension of thiophene core on the DNA/RNA binding properties and biological activity of bisbenzimidazole amidines. Bioorg. Med. Chem. 17, 2544–2554 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Padroni G., Patwardhan N. N., Schapira M., Hargrove A. E., Systematic analysis of the interactions driving small molecule–RNA recognition. RSC Med. Chem. 11, 802–813 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., et al. , Identification of a peptide that crosses the blood-cerebrospinal fluid barrier by phage display technology. Amino Acids 53, 1181–1186 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Leal J., et al. , Peptides as surface coatings of nanoparticles that penetrate human cystic fibrosis sputum and uniformly distribute in vivo following pulmonary delivery. J. Controlled Release 105, 72–80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amici A., et al. , In vivo protein corona patterns of lipid nanoparticles. RSC Adv. 7, 1137–1145 (2017). [Google Scholar]
- 29.Francia V., Schiffelers R. M., Cullis P. R., Witzigmann D., The biomolecular corona of lipid nanoparticles for gene therapy. Bioconjug. Chem. 31, 2046–2059 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Francia V., et al. , Corona composition can affect the mechanisms cells use to internalize nanoparticles. ACS Nano 13, 11107–11121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebastiani F., et al. , Apolipoprotein E binding drives structural and compositional rearrangement of mRNA-containing lipid nanoparticles. ACS Nano 15, 6709–6722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akinc A., et al. , Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Therapy 18, 1357–1364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LoPresti S. T., Arral M. L., Chaudhary N., Whitehead K. A., The replacement of helper lipids with charged alternatives in lipid nanoparticles facilitates targeted mRNA delivery to the spleen and lungs. J. Controlled Release 345, 819–831 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hafez I., Maurer N., Cullis P., On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8, 1188–1196 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Fung A. T., et al. , Local delivery of corticosteroids in clinical ophthalmology: A review. Clin. Exp. Ophthalmol. 48, 366–401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuttle A., affiliation at EnterX Biosciences, DNA Barcoding reads from in vivo mouse delivery. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1024541. Deposited 6 October 2023.
- 37.Jozic A., affiliation at Oregon State University, LNP Barcode script. GitHub. https://github.com/antonyjozic/lnp_barcode_script. Deposited 6 October 2023.
- 38.Trilink Biotechnologies, mRNA sequences, Firefly Luciferase mRNA. https://www.trilinkbiotech.com/media/folio3/productattachments/product_insert/fluc_orf_catno_l-7202_l-7602_l-7702__1.txt. Accessed 9 October 2023.
- 39.Trilink Biotechnologies, mRNA sequences, Enhanced Green Fluorescent Protein mRNA. https://www.trilinkbiotech.com/media/folio3/productattachments/product_insert/egfp__orf_catno_l-7201_l-7601_l-7701_.txt. Accessed 9 October 2023.
- 40.TTrilink Biotechnologies, mRNA sequences, Cre mRNA. https://www.trilinkbiotech.com/media/folio3/productattachments/product_insert/cre_orf_catno_l-7211_.txt. Accessed 9 October 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. Materials can be made available upon request through a Materials Transfer Agreement (MTA) from Oregon State University. Raw NGS barcoding data are publicly available in the NCBI Sequence Read Archive (SRA). SRAs for each sample are available under NCBI Bioproject Accession # PRJNA1024541 (https://www.ncbi.nlm.nih.gov/bioproject/) (36). Python script used to interpret NGS data is available on GitHub (https://github.com/antonyjozic/lnp_barcode_script) (37). mRNA was sourced from TriLink Biotechnologies (San Diego, CA; substituted with 5-methoxy-U); the ORF sequences are available on the manufacturer's website (38–40).