Abstract
The hepatitis B virus and the mammalian hepadnavirus genomes encode for a short open reading frame called x. Expression of the protein product (HBx) appears necessary for establishment of natural infection. However, in vitro studies have suggested a multifunctional role for HBx as an indirect transcriptional transactivator of a variety of different viral and cellular promoters. Indeed, HBx has no known direct DNA binding properties but may interact with transcription factors as well as activate intracellular signaling pathways associated with cell growth. To further address the possible functional role of HBx in the life cycle of hepatitis B virus, we performed an analysis using the yeast two-hybrid system to screen a cDNA library derived from a hepatocellular carcinoma cell line with a HBx fusion bait in an attempt to identify cellular partners that may bind to and alter the biologic properties of HBx. A HBx-interacting protein that specifically complexes with the carboxy terminus of wild-type HBx was identified and designated XIP. This 9.6-kDa protein is capable of binding to HBx in vitro, and transient and stable expression in hepatocellular carcinoma cells abolishes the transactivation properties of HBx on luciferase constructs driven by AP-1 and endogenous hepatitis B virus enhancer/promoter elements. Investigation of the role of XIP in hepatitis B virus replication in differentiated hepatocellular carcinoma cells revealed that XIP expression reduces wild-type hepatitis B virus replication to levels observed following transfection with an HBx-minus virus. In contrast, the replication levels of the duck hepatitis B virus, a hepadnavirus that lacks the x open reading frame, were unchanged in the context of XIP expression. We propose that one of the physiologic functions of the cellular protein XIP is to negatively regulate HBx activity and thus to alter the replication life cycle of the virus.
Chronic hepatitis B virus (HBV) infection is associated with a 100-fold-higher risk of developing hepatocellular carcinoma (HCC) (3). HCC also occurs in animals persistently infected with the mammalian hepadnaviruses. These viral species all contain a highly conserved small open reading frame (ORF) that encodes the HBV x protein (HBx). The HBx has been shown capable of transactivating many different viral and cellular promoters (25). Since this protein lacks a DNA binding motif or a nuclear localization signal, it has been proposed that HBx functions through protein-protein interactions. Several studies have shown that HBx possesses pleiotropic activity, serving both as a cytoplasmic activator of known mitogenic pathways (4, 5, 9, 16, 31) and as a nuclear transactivator (9, 12, 21, 28), and may contribute to the development of HCC by such cellular mechanisms. Regarding the viral life cycle, in the related woodchuck hepatitis virus (WHV), ablation of the WHx start codon or creation of C-terminal truncations has been found to decrease viral replication in vitro (34) and to inhibit the establishment of productive infection in vivo (7, 34). It is not known whether such cellular pathways influenced by HBx expression are needed or sufficient to establish an environment required for viral replication.
Several techniques have been applied to study possible protein-protein interaction with HBx. Using these approaches, HBx has been shown to interact with several nuclear proteins, including members of the general transcription complex machinery (8, 24), a component of proteasomes (15), and a probable DNA repair protein (17), as well as members of the CREB/ATF family of transcription factors (18, 32) where interaction with HBx appears to alter their specificity and DNA binding affinity. Nonetheless, the functional significance of these interactions has not yet been defined in the context of the HBV life cycle. Since HBx function is necessary to establish productive infection of the liver, we searched for proteins that physically interact with HBx and may alter its activity with respect to transactivation of cellular and endogenous viral promoters and enhancers and subsequently influence replication of HBV.
By means of the yeast two-hybrid screening with a LexA-HBx fusion protein (14) of a cDNA library derived from a hepatoblastoma cell line, we isolated cDNA clones encoding proteins that directly interact with HBx. A cDNA clone designated XIP (for HBx-interacting protein) was found to specifically bind to wild-type but not HBx mutant baits. The function of XIP was further investigated in the context of the HBx transactivation properties and effect on viral replication.
MATERIALS AND METHODS
Two-hybrid screen and cDNA isolation.
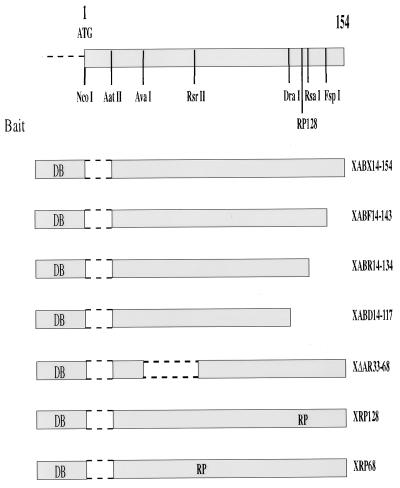
The yeast two-hybrid screening was performed with a HepG2-derived oligo(dT)-primed cDNA library (prepared by M. Melegari). This cDNA library was cloned into the yeast pJG4-5 shuttle vector (14) to prepare the prey constructs, and about 2 × 106 primary Escherichia coli colonies (Sure cells; Stratagene, San Diego, Calif.) were obtained. Maxipreparation of plasmids was followed by yeast transformation so that 200 × 106 transformants were plated for screening onto Ura− Leu− Thr− Leu− galactose-containing plates following 4 h of amplification. Three days later, single yeast-resistant colonies were identified, plated on 5-bromo-4-chloro-3-indolyl-β-d- galactopyranoside (X-Gal)-containing medium, and scored for blue color formation (33). The bait plasmid used in the screening, XABX (Fig. 1), was made of a fusion protein between the LexA DNA binding domain and an amino-terminally deleted HBx protein (amino acids [aa] 14 to 154). It should be noted that a fusion protein prepared with the full-length HBx proved to be unstable in yeast cells, leading to protein degradation at the hinge region formed with the LexA moiety, as assessed by Western blot analysis of LexA-HBx bait constructs with an anti-LexA polyclonal antibody (generous gift from R. Brent, Massachusetts General Hospital) (data not shown). To rule out nonspecific interacting clones, different nonrelevant baits including Max, Cdc2, cyclin D, cytoplasmic domain of transforming growth factor β type I receptor, and Myc-LexA fusion proteins (14, 33) as well as mutant constructs derived from the XABX vector were tested with all positive interacting cDNA clones. The XABX mutant baits were obtained by restriction enzymes digestion with FspI, RsaI, DraI, and AvaI-RsrII, followed by fill-in of the overhanging ends with Klenow DNA polymerase, and represent species with carboxy-terminal deletions at aa 143, 134, and 117 and an internally deleted mutant between aa 33 and 68. These constructs were named XABF, XABR, XABD, and XΔAR (Fig. 1). Moreover, by insertional mutagenesis (see below), 2 aa (arginine [R] and proline [P]) (26) were introduced at aa 68 and 128 in the HBx ORF to generate the XRP68 and XRP128 mutant baits, respectively.
FIG. 1.
Schematic diagram of the baits used in the study. To screen the HepG2 cell-derived cDNA library by the yeast two-hybrid screening, a stable bait was obtained by fusing the LexA DNA binding (DB) protein in frame with the HBx ORF deleted of its first 42 nt (XABX14-154) to create a 14-aa N-terminally truncated molecule. Several HBx mutants that include carboxy-terminal deletions (XABF14-143, XABR14-134, and XABD14-117) as well as an in-frame deletion (XΔAR33-68) and insertion of aa RP amino acid sequence (XRP128 and XRP68) were used to specifically identify and partially characterize, on a functional basis, the interacting XIP prey clone. A yeast vector carrying the amino acid substitution T36A in XIP failed to interact with the XABX14-154 bait, while a mutant with a T12A exchange did interact under the same experimental conditions.
The XIP-encoding cDNA clone was used to screen a normal human liver-derived oligo(dT)-primed cDNA phage library (generated by M. Melegari in the λZAP vector; Stratagene, San Diego, Calif.), and although numerous identical cDNAs were identified by sequence analysis, none had a larger coding sequence than the yeast-derived XIP cDNA clone. Sequencing of yeast- and human liver-derived cDNA clones as well as of mutant HBV vectors was performed with the Sequenase version 2.0 enzyme (U.S. Biochemicals, Cleveland, Ohio).
Plasmid constructs and mutagenesis.
The XABX bait construct was obtained by cloning into the EcoRI and XhoI sites of the vector pEG202 (33), an in-frame PCR-amplified x ORF starting at aa 14 in the HBx protein. The HBx-derived XRP68 and XRP128 baits were prepared by insertional mutagenesis using the pAlter system (Promega, Madison, Wis.). Using the same system, we also generated XIPT12A and XIPT36A mutant XIP constructs where the putative threonine (T) phosphorylation sites at aa 12 and 36 in the XIP ORF were changed to alanine (A). Constructs where bait and prey fragments were exchanged in the pJG4-5 and pEG202 vectors, respectively, were subsequently tested in the yeast two-hybrid system.
The EcoRI-XhoI-derived full-length XIP fragment obtained following excision from the λZAP clone was placed into a pGEX vector (Pharmacia Biotech Inc., Piscataway, N.J.), and the fusion protein glutathione S-transferase (GST)–XIP was purified from isopropythiogalactopyranoside-induced E. coli JM109 as instructed by the manufacturer. The full-length HBx and XIP ORFs were cloned into the EcoRI-XhoI and EcoRI-ApaI sites of the pcDNA3 vector (Invitrogen, San Diego, Calif.), respectively, to generate the pCMVX and pCMVXIP vectors. The FLAG epitope (Eastman Kodak Co., New Haven, Conn.) was inserted at the 3′ prime end of the pcDNA3 multiple cloning site by PCR to generate a construct expressing XIP fused with FLAG at its carboxy terminus (pCMVXIPF) for both in vitro transcription-translation (IVT) and eukaryotic expression experiments.
The replication-competent constructs that expressed wild-type HBV and duck hepatitis B virus (DHBV) under the control of their endogenous promoters have been described elsewhere and are designated payw1.2 (27) and pDHBV3 (30), respectively. Moreover, a payw1.2 mutant vector that contains an ochre termination signal (CAA to UAA) after codon 7 in the HBx ORF was generated by site-directed mutagenesis and named payw*7; no mutation was introduced in the overlapping HBV polymerase ORF. The vector expressing the HBV pregenome from the cytomegalovirus immediate-early promoter (pCMVHBV) has been described elsewhere (27).
A retroviral vector (pBPXIP) that expresses the XIP ORF under the control of the Moloney mouse leukemia virus long terminal repeat was constructed in the EcoRI-SalI sites of the pBabepuro (pBP) vector (20). Recombinant and nonrecombinant retroviral stocks used to infect HCC cells were prepared by transfecting the cell line Bosc23 (22) with pBPXIP and pBP, recovery of the ecotropic retroviruses, and then infection of the amphotropic packaging cell line PA317 (American Type Culture Collection, Rockville, Md.) as previously described (27).
For transactivation assays, we used the vector -73colLuc, which contains one AP1 binding site (1) upstream from the luciferase gene, and a counterpart, -60colLuc, that has the AP-1 site deleted (a kind gift of M. Karin). Moreover, a vector, designated pSFLuc, that contains the HBV enhancers I and II (29) together with the core and precore promoter was generated by ligating the SpeI-FspI fragment (nucleotides [nt] 667 to 1798, where nt 1 [underlined] is by convention located at the GAATTC in the unique EcoRI site) into the pGL-basic vector (Promega) upstream from the luciferase gene. This vector is capable of expressing an HBx protein fused at aa 143 with few amino acids of the pGL-basic multiple cloning site. To design a construct with no HBx protein expression, plasmid pE was generated by filling in the pSFLuc AatII site with Klenow DNA polymerase to create a frameshift mutation in the x ORF.
In vitro binding assay.
The IVT XIP and HBx mRNAs were transcribed from 1 μg of the pCMVXIPF and pCMVX vectors by using T7 polymerase and translated with wheat germ extract (Promega) in the presence of [35S]methionine (NEN, Boston, Mass.). Immunoprecipitation was carried out at 4°C in 500 μl of NET-gelatin buffer in the presence of protease inhibitors (Boehringer Mannheim Co., Indianapolis, Ind.) with 3 μg of anti-x monoclonal antibody (MAb) (19a) or 5 μg of M2 anti-FLAG MAb (Eastman Kodak). Polyclonal rabbit anti-mouse antibodies were used to bind the immunoglobulin G1 MAbs, and protein A-Sepharose was added to pull down the complex. After five washes with NET-gelatin buffer at 4°C, the beads were boiled in sample loading buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis on a 15% gel (SDS-PAGE) followed by fluoroscopy.
Transfection of HCC cells.
The HuH-7 and HepG2 HCC cell lines as well as pooled puromycin-resistant BPHepG2 and BPXIPHepG2 cells were grown in Dulbecco’s modified essential medium supplemented with 10% fetal calf serum at 37°C in a 5% CO2-air mixture. Approximately 107 HCC cells were seeded on 10-cm-diameter dishes the day before transfection with plasmid DNA. Subsequently, 10 μg of payw1.2, pDHBV3, and payw*7 and 5 μg of pCMVHBV and pCMVXIP were transfected into HCC cells by the CaPO4 technique (CaPO4 transfection kit; 5′3′ Inc., Boulder, Colo.). In preliminary experiments, the vector pCMVX was used to restore the capacity of wild-type levels of viral replication to the payw*7 mutant vector. The final concentration of plasmid DNA was kept constant by adding pGEM-7Zf(+) DNA (Promega). The HepG2 cells were also infected with the retroviral BP and BPXIP stocks followed by puromycin selection to yield pools of transduced BPHepG2 and BPXIPHepG2 cells, respectively. The HBV, XIP, and DHBV probes were labeled with [α-32P]dCTP by using a Multiprime labeling kit as instructed by the manufacturer (Amersham International, Little Chalfont, England). Transactivation experiments were performed in HepG2 cells plated on 60-mm-diameter dishes by transfecting p-73colLuc or pE together with increasing amount of pCMVX and pCMVXIP; the cells were harvested 2 days after transfection, and the TNE–0.5% Nonidet P-40 cell lysate (23) was analyzed for luciferase activity.
HBV DNA from cytoplasm-derived nucleocapsid particles was prepared as described previously (23) and then analyzed by the Southern blot technique (27). The concentration of hepatitis B e antigen (HBeAg) secreted into the cell culture supernatant was measured by using an EBK kit (Incstar Co., Stillwater, Minn.), and the hepatitis B surface antigen (HBsAg) concentration was determined in the linear range (0.5 to 50 ng/ml) on serial dilutions of culture medium (19). Finally, transfection studies were carried out at least five times with each of the different plasmid preparations, and similar results were obtained in all instances. The transfection efficiency was controlled by cotransfection with a simian virus 40 promoter-driven β-galactosidase gene-containing plasmid. Subsequently, the β-galactosidase activity in cell lysates (Promega) and direct X-Gal staining of transfected cells were determined.
Nucleotide sequence.
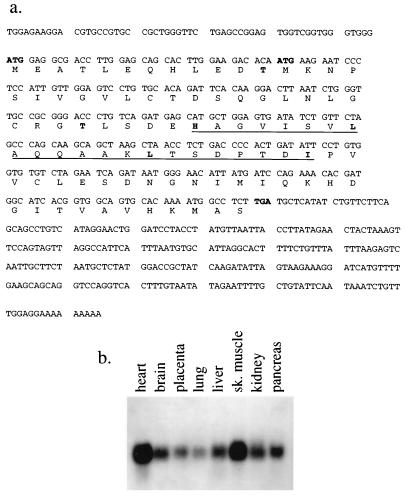
Sequencing of the full-length XIP cDNA revealed that it encodes a protein of 9.6 kDa with two in-frame ATGs (Fig. 2a). The XIP cDNA sequence has been submitted to GenBank (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Md.) and has accession no. AF029890.
FIG. 2.
(a) Nucleic and amino acid sequences of the XIP cDNA. The XIP cDNA encodes a protein of 9.6 kDa with two in-frame ATGs (boldface). There is a putative leucine zipper motif (underlined). There are two consensus phosphorylation sites at Thr12 and Thr36 for protein kinase C and casein kinase II, respectively (boldface). A partial expressed sequence tag (EST) sequence of the XIP cDNA has been reported (GenBank accession no. gb/W07170) (b) Northern blot analysis of poly(A)+ RNA (Clontech) derived from normal human tissues. The XIP cDNA recognizes a single 0.7-kb transcript in all tissues studied.
RESULTS
Cloning of the XIP cDNA by the yeast two-hybrid system.
In the screening of the HepG2-derived library, we used a stable HBx bait obtained by deleting the first 42 nt of the x ORF (XABX14-154) to create a 14-aa N-terminally truncated molecule. Pilot experiments had previously demonstrated that the LexA DNA binding protein fused in frame to the full-length HBx protein resulted in degradation of the bait, a phenomenon that can increase the selection of false-positive yeast colonies. In the screening of 2 × 103 positive colonies, the yeast-interacting clones were placed into several groups on the basis of their cDNA sequence, and the XIP proved to be the most numerous cDNA among the isolates (about 10% of the HBx-interacting clones) and exhibited the strongest reaction in terms of β-galactosidase expression (data not shown). To specifically identify and partially characterize on a functional basis the interacting prey clones, several mutants of the XABX bait were used (Fig. 1). Indeed, XIP was the only yeast-derived clone that failed to interact with carboxy-terminal mutant HBx baits such as XABD14-117 as well as the RP insertion mutant XRP128 (26). These experiments suggest that the B42 activation domain (14) fused to the XIP cDNA (XIP prey) functionally interacted with the XABX bait in the yeast cell and that this interaction was dependent on the C terminus of the HBx protein. Moreover, interchange of XABX14-154 with XIP inserts in the bait and prey plasmids also yielded a strong protein interaction pattern. This study suggests that the two yeast vectors did not contribute to a false-positive interaction due to a conformational change in the protein.
Sequencing data of the XIP cDNA revealed that it contains a putative leucine zipper motif and may form an amphipathic helix by helical wheel analysis (13). There are also two consensus phosphorylation sites at threonines 12 and 36 for protein kinase C and casein kinase II, respectively. A yeast vector carrying the amino acid substitution T36A in XIP failed to interact with the XABX14-154 bait, while a mutant with a T12A exchange did interact under the same experimental conditions (Fig. 1). Northern blot analysis of normal human tissues poly(A)+ RNA (Clontech Laboratories, Palo Alto, Calif.) showed that the XIP cDNA recognizes a single 0.7-kb transcript in all tissues studied and was particularly abundant in skeletal and cardiac muscle tissues (Fig. 2b).
In vitro interaction of HBx with XIP.
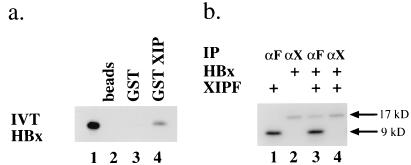
To rule out the possibility that the interaction between the XIP and HBx proteins did not occur through a yeast-derived bridging protein(s) and to provide evidence of direct binding in a different system, we purified a GST-XIP fusion protein and studied the interaction with an IVT [35S]methionine-labeled HBx. Indeed, IVT HBx was found to interact with GST-XIP but not with the GST protein or with the glutathione-linked beads (Fig. 3a). We then tested whether IVT HBx and a version of XIP engineered with a FLAG epitope directly interact in vitro. The anti-FLAG M2 MAb was capable of precipitating the complex only when the FLAG epitope was linked to the carboxy-terminal region of the XIP molecule (Fig. 3b), not at the amino terminus (data not shown). On the other hand, an anti-HBx MAb immunoprecipitated the IVT HBx (data not shown) but not the HBx-XIP complex. This finding suggests that the epitope recognized by the anti-HBx MAb was masked by the complex formation or that the FLAG extension on XIP disrupted detection of the complex. As controls, we performed identical reactions omitting the primary MAb or with [35S]methionine IVT luciferase in place of either HBx or XIPF, with negative results.
FIG. 3.
Interaction of HBx and XIP in vitro. (a) SDS-PAGE analysis of IVT [35S]methionine-HBx binding to GST-XIP. Lane 1, input IVT [35S]methionine-labeled HBx; lanes 2 to 4, labeled HBx bound to Sepharose beads (lane 2), to Sepharose-GST beads (lane 3), and to GST-XIP fusion protein (lane 4). (b) SDS-PAGE analysis of immunoprecipitated IVT [35S]methionine-labeled HBx and XIPF. Lane 1, XIPF immunoprecipitated by anti-FLAG M2 MAb (αF); lane 2, HBx immunoprecipitated by an anti-HBx MAb (αX) (19a). In lane 3, the αF MAb immunoprecipitated the HBx-XIPF complex, whereas in lane 4, the αX MAb did not. The αF MAb does not immunoprecipitate IVT [35S]methionine-HBx, and positioning of the FLAG epitope at the amino terminus of XIP also did not permit immunoprecipitation of the HBx-FXIP complex (data not shown). We have been unable to prepare high-affinity antibodies against the small hydrophobic XIP.
XIP inhibits the transactivation properties of HBx.
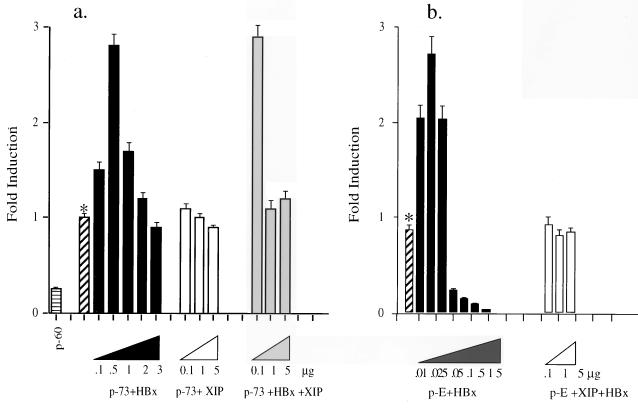
To test the possible functional consequences of an interaction between HBx and XIP, we determined if XIP alters the transcriptional transactivator properties of HBx. Since HBx has been reported to upregulate AP-1-responsive elements (5, 16, 18, 21, 28), cotransfection experiments were performed with the HBx-responsive HepG2 cell line with the p-73colLuc reporter plasmid containing an AP-1 binding site inserted upstream of the luciferase gene together with pCMVX. Expression of HBx induced transcription of the p-73colLuc construct in a dose-dependent manner up to threefold. However, cotransfection of increasing amounts of XIP expression plasmid with a previously determined optimal concentration of pCMVX (0.5 μg) decreased the luciferase activity to basal levels (Fig. 4a).
FIG. 4.
XIP inhibits the transactivation function of HBx. (a) HBx transactivates the AP-1-driven reporter construct -73colLuc, (p-73) severalfold at an optimal plasmid concentration of 0.5 μg, whereas XIP alone has no effect on p-73 expression. Addition of XIP reduces transactivation capacity of HBx on p-73 to basal levels (∗). p-60 represents the basal luciferase activity of the -60colLuc construct, the plasmid that does not have the AP-1 binding site. (b) The asterisk indicates the basal level of luciferase activity with the reporter construct p-E, where the luciferase gene is under the control of the HBV core promoter/enhancer I element. This construct contained a frameshift mutation in the HBx ORF to abolish the endogenous expression of HBx. Cotransfection with increasing amounts of pCMVX resulted in transactivation at low plasmid concentrations (0.01, 0.025, and 0.05 μg); at higher concentrations (0.1, 0.5, 1.0, and 5.0 μg), transactivation of the luciferase gene was inhibited. Cotransfection of pCMVX at an optimal concentration of 0.025 μg with increasing concentrations (0.1, 1, and 5 μg) of pCMVXIP resulted in a reduction of transactivation activity to basal levels. These experiments were repeated several times, and the data reported are averages of three experiments.
Studies were also performed with HepG2 cells to determine if HBx activated transcription of the luciferase gene when driven by the HBV enhancer and core promoter element (10). The HBV enhancers and core promoter region overlap with the HBx promoter and ORF such that coexpression of HBx will occur along with the luciferase protein. We observed that pSFLuc was not transactivated by HBx and reasoned that such a phenomenon might be due to the presence of HBx expression from the same pSFLuc construct. Indeed, we observed about a three- to fourfold increase in transcription of the luciferase gene in HepG2 cells cotransfected with pCMVX and the pE construct. The pE construct is identical to pSFLuc except for a frameshift mutation in the x ORF created to abolish the expression of HBx. Moreover, when XIP was coexpressed with the optimal amount of HBx (0.025 μg), the luciferase activity exhibited by pE was reduced to basal levels. These observations suggest that XIP modulates HBx transcriptional activity within the cell.
We observed that the optimal amount of HBx-expressing plasmid needed to promote transactivation of the pE vector containing the endogenous viral enhancers and core promoter was about 20-fold less than the amount necessary to detect a positive effect on the luciferase expression plasmid containing an AP-1 site. This difference in transactivation properties did not appear to be due to the cell type, since in both instances HepG2 cells were used. We also found by immunohistochemistry that HBx protein expression was appreciable and easily detectable in the cytoplasm of cells transfected with an amount of HBx expression plasmid that achieved activation of transcription through the AP-1 site. However, transactivation on the endogenous HBV enhancers was achieved at much lower levels of HBx protein expression, and such levels of expression were not detectable by immunohistochemistry (data not shown). These findings are important with respect to the proposed dual effect of HBx in the cytoplasm and in the nucleus (12) and may have physiologic relevance for the low level of HBx expression required to promote wild-type HBV replication, a function that would presumably occur in the nucleus.
HCC-derived cell lines support different levels of HBV replication.
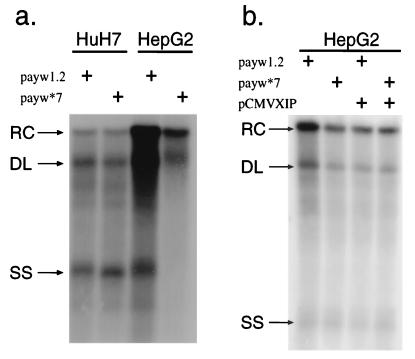
Analysis of HBV replication levels in HCC cells transfected with vectors expressing wild-type HBV (payw1.2) or with a mutant virus harboring a termination signal after codon 7 in the HBx ORF (payw*7) revealed that the presence or absence of HBx expression was important with respect to the level of HBV replication observed in the human hepatoblastoma HepG2 cell line (34); indeed, lack of HBx expression led to decreased HBV replication as well as reduced secretion of HBsAg and HBeAg proteins only in the HepG2, not in the HuH-7, cell line (10) (Fig. 5a and c). These findings confirm a previous observation of no effect of HBx on HBV replication when an HBx-minus vector was transfected into HuH-7 cells (6).
FIG. 5.
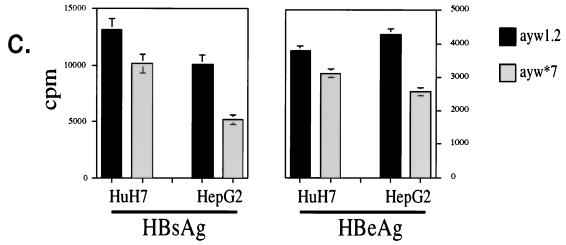
HBV replication is modulated by HBx and XIP expression, as shown by Southern blot analysis of HBV DNA derived from nucleocapsid particles purified from the cytoplasm of transfected HCC cells. (a) Higher levels of HBV replicative forms are found in HepG2 cells transiently transfected with the wild-type HBV, payw1.2, than with the payw*7 mutant virus carrying an ochre termination signal after aa 7 in the HBx ORF; in HuH-7 cells, the levels of HBV replicative forms were unaffected by the presence or absence of a functional HBx protein. (b) HepG2 cells transiently transfected with payw1.2 and pCMVXIP showed a reduction of viral replicative forms compared to payw1.2 alone, whereas cotransfection of payw*7 with pCMVXIP had no further effects. Transfection efficiency was controlled by β-galactosidase and luciferase assays as described in Materials and Methods. The experiments were repeated at least five times with comparable results. The relaxed circular (RC), double-stranded linear (DL), and single-strand (SS) DNA species are indicated by arrows on the left. (c) Concentrations of secreted viral antigens in the supernatant of transfected cells. While the levels of HBsAg, measured in its linear range, and HBeAg in the supernatants of HuH-7 cells transfected with payw1.2 or payw*7 were comparable, transfection of the wild-type plasmid payw1.2 in HepG2 cells yielded about twice the levels of HBsAg and HBeAg as in cells transfected with the HBx-minus plasmid payw*7.
XIP expression inhibited HBV replication.
Based on the foregoing observations, HepG2 cells were subsequently used to investigate the effect of XIP on replication of both wild-type and HBx-minus viral species. If HepG2 cells were cotransfected with the wild-type payw1.2 and pCMVXIP, the level of HBV replication was found to be substantially less than in cells transfected with payw1.2 alone. This level of replication of wild-type HBV was similar to that found in HepG2 cells transfected with the HBx-minus payw*7 mutant virus (Fig. 5b). Cotransfection of the payw*7 with pCMVXIP had no additional effects on viral replication.
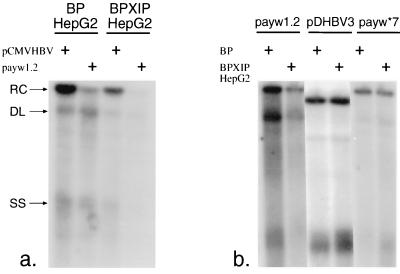
To further investigate the role of XIP on HBV replication, the HepG2 cell line was stably infected with a retrovirus that expresses the XIP ORF under the control of the Moloney mouse leukemia virus long terminal repeat. These BPXIPHepG2 cells express endogenous and retrovirus-encoded XIP with transcripts sizes of 0.7 and 3.2 kb, respectively, as assessed by Northern blot analysis with an XIP probe (data not shown). The BPXIPHepG2 cells demonstrated a reduced level of HBV replication following transient transfection with the wild-type payw1.2 vector compared to mock retrovirus-infected BPHepG2 control cells (Fig. 6a). This inhibitory effect was independent of the type of promoter driving the expression of the HBV pregenome. Indeed, BPXIPHepG2 cells supported a lower level of viral replication when the HBV pregenome was driven either by the cytomegalovirus immediate-early or the endogenous viral enhancer/core promoter. As a specificity control, both cell lines were transfected with a plasmid expressing a replication-competent DHBV genome (pDHBV3); the native DHBV lacks the x ORF. No difference in the level of DHBV replication was observed between the two retrovirus-transduced cell lines. Finally, when BPHepG2 and BPXIPHepG2 cells were transfected with the HBx-deficient payw*7, the level of replication was found to be unchanged and resembled the findings with wild-type HBV payw1.2 when transfected into the BPXIPHepG2 cells (Fig. 6b).
FIG. 6.
XIP expression reduced viral replication in stably transduced HepG2 cells. (a) Transient transfection of BPXIPHepG2 cells with wild-type pCMVHBV and payw1.2 produced lower levels of viral replicative forms than in BPHepG2 cells. The relaxed circular (RC), double-stranded linear (DL), and single-strand (SS) DNA species are indicated by arrows on the left. (b) There was no change in the levels of HBV replicative forms after transfection of the two cell lines with HBx-deficient payw*7, or in the levels of DHBV viral replication after transfection with plasmid pDHBV3, as measured by hybridization with a full-length DHBV probe.
DISCUSSION
The mammalian hepadnaviruses encode an HBx protein that may function as a weak transactivator of a variety of viral and cellular promoter and enhancer elements. In addition, the ability of HBx to alter signaling pathways involved in cellular growth and differentiation as well as apoptosis has led to the hypothesis that continuous low-level HBx expression due to persistent viral infection of the liver may contribute to the development of HCC. However, the biologic function(s) of HBx during natural HBV infection is not known. The HBx protein structure does not contain a defined DNA binding motif, nuclear localization signal, or any other protein domains that yield clues to its cellular localization or function in virus-infected hepatocytes. Studies of the molecular mechanism(s) by which HBx functions with respect to modulation of viral replication have been hindered by the fact that HBx expression occurs at very low levels in naturally HBV infected hepatocytes as well as in HCC cells transfected with replication-competent vectors expressing wild-type HBV. However, two groups independently provided evidence that the WHx protein was necessary for establishment of primary infection in the liver of the woodchucks (7, 34). Therefore, it is apparent that in the natural host, WHx expression plays a central role in the viral life cycle. In support of this concept, our findings and the observations of others (34) have established that transfection of HepG2 cells with HBx-minus constructs produce significantly lower levels of viral replicative forms.
Since little is known regarding how HBx functions within the cell, a yeast two-hybrid screening was performed with the principal goal to identify and characterize cellular proteins that specifically interact and influence the properties of the HBx protein. The identification of the HepG2 cell line as susceptible to the known biologic activity of HBx led us to believe that the screening of a cDNA library derived from this cell line would generate such candidate cDNAs. Here we reported the cloning and functional characterization of an HBx-interacting protein. In the yeast system, XIP failed to interact both with a carboxy-terminally truncated HBx and with an HBx protein carrying an RP insertion at aa 128, and these observations suggest that the interaction was authentic. It should also be noted that these HBx mutants failed to transactivate a reporter construct bearing an AP-1-responsive element in HepG2 cells (26). The interaction between HBx and XIP was found to occur not only in the yeast cell but also in vitro when a fusion protein of the yeast vector was exchanged with an E. coli-produced GST and the binding assays were performed with purified recombinant proteins. Moreover, mixing experiments with IVT HBx and XIP allowed us to establish the direct association between the two proteins.
We then determined if the binding of XIP to HBx produced an effect on the known transactivation properties of HBx or, more important, altered HBV replication levels. First, it was demonstrated that the transactivation effects of HBx on an AP-1 binding site as well as on the HBV enhancers were abolished by coexpression with the XIP protein. The HBx central and carboxy-terminal domains have been shown to be important for its transactivation properties (2, 25, 26). The results suggest that XIP may have interacted with a domain necessary for transactivation. Our results indicate that overexpression of the XIP protein prevented wild-type HBx activity on such promoters as well as reduced HBV replication to levels comparable to those observed with an HBx-minus variant strain. Due to the lack of XIP-specific antibodies, we cannot quantify XIP expression within the cell, and therefore our data do not allow us to determine natural levels of XIP expression. The fact that XIP overexpression did not affect the levels of DHBV replicative intermediates provided further indication that XIP synthesis does not alter cell viability or other pathways required for viral replication and clearly needs the presence of HBx to exert its biologic effect. Because XIP expression decreased HBV replication together with HBsAg and HBeAg synthesis, we are led to believe that the effect is mediated by inhibition of HBx action on the endogenous viral core promoter/enhancer elements.
HBx has been shown to be a weak transactivator of AP-1, NF-κB, or other cellular promoters as well as an activator of mitogenic pathways. However, it is not known if these effects are required for sustaining HBV replication in liver cells. Previous studies suggest that HBx displays dual specificity of function, serving as a cytoplasmic activator of the Ras–Raf–mitogen-activated protein kinase cascade and as a nuclear activator of AP-1 and NF-κB transcription factors (12) and the endogenous HBV enhancers. The WHx and HBx proteins in both WHV-infected woodchuck liver (11) and HCC cells overexpressing HBx (reference 12 and our unpublished results) have been found in the cytoplasm, as assessed by cell fractionation and immunohistochemistry with specific antibodies. However, because both the levels and the site of HBx expression during the natural course of viral infection are unknown, it is likely that multiple interactions with a cellular protein(s) may be required in the cytoplasm and the nucleus for HBx to exert its biologic function(s) within the cell. It is noteworthy that much lower levels of HBx expression were sufficient to activate transcription of the HBV enhancers/core promoter compared to activation of the luciferase gene by the AP-1-responsive element. Of the many factors identified by studies of protein-protein interactions, several have been reported to directly interact or to be functionally altered by HBx (17, 18, 24, 28, 32). Most of these putative partners have been tested in the context of HBx transactivation capacity (8, 15). Here we have shown for the first time that by binding to HBx, XIP is capable of altering the replicative life cycle of HBV. Determination of the cellular function(s) of XIP may provide further insights with respect to the characteristics of this HBV-host protein interaction and its regulatory role of HBV replication.
ACKNOWLEDGMENTS
M.M. and P.P.S. contributed equally to this work.
We thank Antonis S. Zervos for advice with the yeast two-hybrid system, Roger Brent for the gift of the anti-LexA MAb, and Michael Karin for plasmids -73colLuc and -60colLuc.
This work was supported by NIH grant CA-35711 and a grant from the American Cancer Society. M.M. was supported in part by a A. Castelnuovo fellowship (Cermenate, Italy).
REFERENCES
- 1.Angel P, Baumann I, Stein B, Delius H, Rahmsdorf H J, Herrlich P. 12-O-Tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol Cell Biol. 1987;7:2256–2266. doi: 10.1128/mcb.7.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arii M, Tanaka S, Koike K. Identification of three essential regions of hepatitis B virus X protein for transactivation function. Oncogene. 1992;7:397–403. [PubMed] [Google Scholar]
- 3.Beasley R P, Lin C C, Hwang L Y, Chien C S. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 4.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benn J, Su F, Doria M, Schneider R J. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J Virol. 1996;70:4978–4985. doi: 10.1128/jvi.70.8.4978-4985.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blum H E, Zhang Z-S, Galun E, von Weizsäcker F, Gardner B, Liang T J, Wands J R. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J Virol. 1992;66:1223–1227. doi: 10.1128/jvi.66.2.1223-1227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheong J-H, Yi M-K, Lin Y, Murakami S. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 1995;14:143–150. doi: 10.1002/j.1460-2075.1995.tb06984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chirillo P, Falco M, Puri P L, Artini M, Balsano C, Levrero M, Natoli G. Hepatitis B virus pX activates NF-κB-dependent transcription through a Raf-independent pathway. J Virol. 1996;70:641–646. doi: 10.1128/jvi.70.1.641-646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colgrove R, Simon G, Ganem D. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J Virol. 1989;63:4019–4026. doi: 10.1128/jvi.63.9.4019-4026.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dandri M, Schirmacher P, Rogler C E. Woodchuck hepatitis x protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J Virol. 1996;70:5246–5254. doi: 10.1128/jvi.70.8.5246-5254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;14:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferré-D’Amaré A R, Prendergast G C, Ziff E B, Burley S K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 14.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Kwong J, Sun E C-Y, Liang T J. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J Virol. 1996;70:5582–5591. doi: 10.1128/jvi.70.8.5582-5591.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kekulé A S, Lauer U, Weiss B L, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumor promoter signalling pathway. Nature (London) 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 17.Lee T-H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 19.Melegari M, Bruno S, Wands J R. Properties of hepatitis B virus pre-S1 deletion mutants. Virology. 1994;199:292–300. doi: 10.1006/viro.1994.1127. [DOI] [PubMed] [Google Scholar]
- 19a.Melegari, M., and J. R. Wands. Unpublished data.
- 20.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natoli G, Avantaggiati M L, Chirillo P, Costanzo A, Artini M, Balsano C, Levrero M. Induction of the DNA-binding activity of c-Jun/c-Fos heterodimers by the hepatitis B virus transactivator pX. Mol Cell Biol. 1994;14:989–998. doi: 10.1128/mcb.14.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high titer helper free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugh J C, Yaginuma K, Koike K, Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988;62:3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qadri I, Maguire H F, Siddiqui A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc Natl Acad Sci USA. 1995;92:1003–1007. doi: 10.1073/pnas.92.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossner M T. Hepatitis B virus X-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 26.Runkel L, Fischer M, Schaller H. Two-codon insertion mutations of the HBX define two separate regions necessary for its trans-activation function. Virology. 1993;197:529–536. doi: 10.1006/viro.1993.1626. [DOI] [PubMed] [Google Scholar]
- 27.Scaglioni P P, Melegari M, Wands J R. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seto E, Mitchell P J, Yen T S B. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcriptional factors. Nature (London) 1990;344:72–74. doi: 10.1038/344072a0. [DOI] [PubMed] [Google Scholar]
- 29.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 31.Su F, Schneider R J. Hepatitis B virus HBx protein activates transcription factor NF-κB by acting on multiple cytoplasmic inhibitors of Rel-related proteins. J Virol. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams J S, Andrisani O M. The hepatitis B virus X protein targets the basic region-leucine domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition site. Cell. 1993;72:525–524. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 34.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]