Abstract
Background
Pyrazinamide is one of four first-line antibiotics used to treat tuberculosis; however, antibiotic susceptibility testing for pyrazinamide is challenging. Resistance to pyrazinamide is primarily driven by genetic variation in pncA, encoding an enzyme that converts pyrazinamide into its active form.
Methods
We curated a dataset of 664 non-redundant, missense amino acid mutations in PncA with associated high-confidence phenotypes from published studies and then trained three different machine-learning models to predict pyrazinamide resistance. All models had access to a range of protein structural-, chemical- and sequence-based features.
Results
The best model, a gradient-boosted decision tree, achieved a sensitivity of 80.2% and a specificity of 76.9% on the hold-out test dataset. The clinical performance of the models was then estimated by predicting the binary pyrazinamide resistance phenotype of 4027 samples harbouring 367 unique missense mutations in pncA derived from 24 231 clinical isolates.
Conclusions
This work demonstrates how machine learning can enhance the sensitivity/specificity of pyrazinamide resistance prediction in genetics-based clinical microbiology workflows, highlights novel mutations for future biochemical investigation, and is a proof of concept for using this approach in other drugs.
Introduction
Mycobacterium tuberculosis is an evolutionarily ancient human pathogen that is the leading cause of death by infectious disease worldwide, except during the SARS-CoV-2 pandemic. In 2021, TB was responsible for 1.6 million deaths and 10.6 million new infections.1 TB control efforts have been hampered by the evolution of resistance to antibiotics, threatening the efficacy of the standard four-drug antibiotic regimen consisting of rifampicin, isoniazid, ethambutol and pyrazinamide. Pyrazinamide plays a critical role in TB treatment through its specific action on slow-growing, ‘persister’ bacteria, which often tolerate other drugs due to their reduced metabolism.2–6 Due to its unique sterilizing effect and its synergy with new TB drugs such as bedaquiline, pyrazinamide is also included in new treatment regimens targeting drug-resistant TB.7–12 Therefore, accurately and rapidly determining whether a clinical isolate is resistant to pyrazinamide is critically important for the treatment of TB.
Most culture-based laboratory methods to determine pyrazinamide resistance are technically challenging, requiring highly trained technicians. Even then, results are often not reproducible, meaning these methods are rarely employed in low-resource and/or high-burden clinical settings.13 Even the current gold standard, the Mycobacteria Growth Indicator Tube (MGIT), which is relatively simple to use, can suffer from low precision, with false-resistance rates of 1%–68% reported.14–20 As the prevalence of MDR and XDR TB increases, this lack of precision will become more of a problem.
Resistance to rifampicin or isoniazid can be predicted in most isolates (90%–95% and 50%–97%, respectively) by the presence of a small number of highly penetrant genetic variants in short and well-delineated regions of one or two genes (rpoB and katG/fabG1, respectively).3 However, despite pyrazinamide being used to treat TB since 1952, comparatively less is known about which genetic variants confer resistance compared with other first-line drugs.4 In the recent catalogue of resistance-associated mutations of M. tuberculosis published by the WHO, the performance for pyrazinamide was markedly lower (72.3% sensitivity and 98.8% specificity) than either rifampicin or isoniazid (93.8% and 98.2% or 91.2% and 98.4% sensitivity and specificity, respectively).21,22 While some of this poor performance is likely due to inaccuracies in phenotypic testing, a comprehensive genetic catalogue for pyrazinamide resistance mutations remains elusive.
Pyrazinamide is a pro-drug that is converted to its active form of pyrazinoic acid by the action of PncA, a pyrazinamidase/nicotinamidase encoded by the pncA gene.23 While other genetic loci have been implicated in pyrazinamide resistance (notably rpsA, panD, clpC1, and the putative efflux pumps Rv0191, Rv3756c, Rv3008 and Rv1667c), the majority (70%–97%) of pyrazinamide-resistant clinical isolates harbour genetic variants in either the promoter region or coding sequence of pncA.21,24–34 In contrast to the well-delineated and relatively restricted ‘resistance-determining regions’ found in rpoB (rifampicin, 27 codons) and katG (isoniazid, single codon), pyrazinamide-resistant variants have been identified along the entire length of the pncA gene (Figure 1a) with no single variant predominating. Hence, while targeted genome sequencing or WGS approaches are capable of assaying the entire pncA gene, the number and diversity of resistance-conferring variants in pncA fundamentally limits the sensitivity and specificity of heuristic approaches that aim to predict the effectiveness of pyrazinamide based on a catalogue of previously observed genetic variants.3,13,24,29,30,35,36
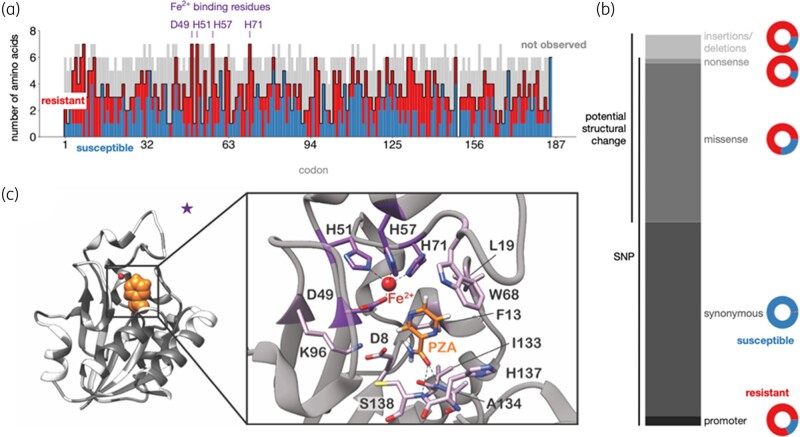
Figure 1.
Distribution of PncA mutations from published datasets. (a) Barplot of the impact of possible missense mutations in PncA by amino acid position. High-confidence resistant (red) and susceptible (blue) mutations are overlaid on the possible missense mutations whose effect on resistance is unknown or unclear (grey). (b) Distribution of the types of mutations reported by the CRyPTIC consortium et al. (c) Missense mutations from the dataset plotted onto the PncA structure (PDB ID: 3PL1) in dark grey. A pyrazinamide molecule (orange) has been modelled into the active site.
Genetics-based clinical microbiology for TB currently depends on being able to infer the effect of any likely occurring pncA mutation on pyrazinamide susceptibility. Recent studies to identify pyrazinamide-resistance-determining mutations have focused on either classifying mutations from previously observed clinical isolates or discovering novel mutations through in vitro/in vivo screening approaches.21,22,30,37–39 However, these strategies are constrained, respectively, by the relative paucity of sequenced clinical isolates compared with the number of potential resistance-causing mutations and the lack of laboratory capacity to systematically generate and test mutants. Computational modelling approaches40 can potentially predict the effect of a significant number of missense mutations41–44 before they are observed in clinical isolates. Several studies have already trained machine-learning models on a number of anti-tuberculars,45–48 including pyrazinamide.49
As PncA is not essential and can be inactivated through defects in protein folding, reduced stability, distortion of active site geometry, abrogation of metal binding, or some combination of these, we expected a machine-learning approach to be ideally suited to simultaneously consider all these possible mechanisms of PncA inactivation, and hence more accurately predict pyrazinamide resistance/susceptibility. In this paper, we confirm using the largest Train/Test and Validation datasets used to date that machine-learning models that learn from a range of structural, chemical and evolutionary features can robustly and accurately predict the effect of missense amino acid mutations on pyrazinamide susceptibility.
Materials and methods
We first constructed independent Train, Test and Validation datasets (Table 1). The first two were built by combining a comprehensive in vitro/in vivo mutagenesis study38 with two published catalogues of M. tuberculosis genetic variants associated with resistance,21,39 resulting in a Train/Test dataset of 664 non-redundant missense mutations (349 associated with resistance) where there was no discrepancy in the predicted phenotype. This was then split 70:30 to produce independent Train and Test datasets containing 464 and 200 mutations, respectively (Table 1). The Validation dataset was constructed by aggregating 24 231 clinical samples from three collections,13,30,50 resulting in 4027 samples containing one of 367 non-redundant missense PncA mutations. Briefly, phenotypes for strains with single missense mutations in pncA were aggregated by mutation, tallying the results of the phenotypic testing. Mutations that were resistant or susceptible at least 75% of the time and that had been phenotyped at least four times were included. Additionally, mutations that had been phenotyped at least twice with no discrepancies were also added. Finally, a further independent Quantitative dataset was created by measuring the MIC on a small number of missense mutations to test if our models can predict the magnitude of the effect.
Table 1.
Description of datasets employed in this study
| Dataset | Phenotype | # Isolates | # Non-redundant missense mutations |
|---|---|---|---|
| Train | R/S | n/a | 464 |
| Test | R/S | n/a | 200 |
| Validation | R/U/S | 24 231 | 367 (199 with an R/S phenotype) |
| Quantitative | MIC | 71 | 57 |
R, resistant to antibiotic; S, susceptible; U, inconsistent results.
Pyrazinamide MIC determination
Isolates used for MIC determination came from the EXIT-RIF study and US CDC. Of the 366 M. tuberculosis clinical isolates, 333 were collected as part of a prospective cohort study (‘EXIT-RIF’) between November 2012 and December 2013 in three South African provinces (Free State, Eastern Cape and Gauteng). An M. tuberculosis databank housed at the SAMRC Centre for Tuberculosis Research, consisting of ∼45 000 drug-resistant isolates collected in the Western Cape province since 2001, was queried to identify isolates containing both pyrazinamide MIC data and pncA genotypic data; this produced the remaining 33 M. tuberculosis clinical isolates. Isolates that harboured single amino acid substitutions in PncA (39 out of 366 total) were selected for comparison to model predictions. An additional 32 clinical isolates (collected from 2000 to 2008) harbouring single missense mutations in pncA came from the culture collection at the Laboratory Branch, Division of Tuberculosis Elimination, US CDC.
All MICs were determined using the non-radiometric BACTEC MGIT 960 method (BD Diagnostic Systems, NJ, USA) with manufacturer-supplied pyrazinamide medium/supplement as previously described.51 This system makes use of modified test medium, which supports the growth of mycobacteria at a pH of 5.9. Isolates from the EXIT-RIF study were tested at 100, 75, 50 and 25 mg/L whilst the CDC used pyrazinamide concentrations of 50, 100, 200, 300, 400, 600 and 800 mg/L. A fully susceptible M. tuberculosis laboratory strain H37Rv (ATCC 27294) was included as a control for all isolates tested. The resulting 71 isolates contained one of 59 missense mutations; of the 10 mutations measured more than once, 2 had inconsistent phenotypes and were removed, leaving 57 missense mutations, of which 50 were resistant and 7 susceptible.
Adding pyrazinamide to the experimental structure
Since the M. tuberculosis structure of PncA52 (PDB: 3PL1) does not contain electron density for pyrazinamide, we first fitted it onto the Acinetobacter baumannii PncA structure53 and retained the coordinates of pyrazinamide. This was done using the MultiSeq analysis tool in VMD-1.9.4a57, which in turn used STAMP to perform the structural alignment. The root mean square deviation (RMSD) between the Cα atoms in 162 residues overlapped in the sequence alignment was 1.22 Å and the RMSD between the Cα atoms in the active site (Figure 1c) is 0.75 Å. Taken together these indicate that the structural core of both proteins is similar. The position and orientation of the pyrazinamide molecule is only required for one structural feature, the distance from the pyrazinamide molecule.
Determination of structural-, chemical- and evolutionary features
A wide range of structural, chemical, thermodynamic and evolutionary features were added.54 Structural features included the distances from the Fe2+ ion and pyrazinamide molecule, solvent accessibility, backbone angles, secondary structure, temperature factor and depth from the protein surface. The effect on the chemistry was captured by the change in mass, volume, isoelectric point, hydrophobicity, chemistry55 and degree of hydrogen bonding. To assess a pncA mutation’s impact on the stability of PncA, we added scores from three meta-predictors (RaSP,56 mCSM57 and DeepDDG58). Finally we added MAPP scores, which aims to quantify the evolutionary constraints imposed on a given position in a protein,59 and SNAP2 scores. SNAP2 is a neural network trained to predict whether protein mutations are neutral or have a deleterious effect on function.60
Training and reproducibility
Logistic regression (LR), a multi-layer perceptron classifier (NN) and a gradient-boosted decision tree (XB) were all trained as described in our online code and data repository—this also contains saved states of the final models and Python3 Jupyter Notebooks, allowing one to reproduce in a web browser all results and figures.61
Results
Observed genetic variation in pncA
Since it includes the results of an in vitro mutagenesis study, the Train/Test dataset captures the most genetic variation in pncA. Mutations are observed at every codon bar one (Figure 1a) and all possible amino acids arising from a single nucleotide substitution are observed at several codons. Interestingly, there were a significant number of pncA codons where mutations associated with either resistance or susceptibility were seen, confirming that the change in local chemistry introduced by the mutant amino acid is an important factor in determining resistance (Figure 1a). The codons with the greatest mutational diversity in the dataset were all residues involved in active site formation or metal binding, suggesting that, consistent with our hypothesis, loss or alteration of these functions is a common mechanism for gaining pyrazinamide resistance. Indeed, previous studies have noted a negative correlation between a mutation’s distance from the active site and its tendency to cause resistance.30,38,52
Clinically observed association between genetic variation in pncA and pyrazinamide resistance
Overall, 3351 samples (14.7%) in the CRyPTIC dataset are resistant to pyrazinamide and 6851 samples have one or more genetic variants in either the promoter and/or ORF of pncA. The majority (6622 samples; 96.7%) have a single genetic variant with 93.9% (6221 samples) of these being substitutions. The remaining 401 samples (6.1%) contained insertions, deletions and frameshifts and these were strongly associated with resistance (343 samples; 85.5%),21,39 consistent with their likely disruption of the PncA enzyme. Most synonymous substitutions [present in 3288 (49.7%) of the single variant strains, Figure 1b] were not associated with resistance; however, seven variants were observed in resistant isolates. S65S (19 resistant isolates) is a phylogenetic SNP present in Lineage 1; however, it is susceptible in 3204 strains, suggesting that these 19 isolates are either phenotyping errors or that there is an alternative mechanism of pyrazinamide resistance at play in these strains. The remaining mutations—R2R, L19L, A46A, D63D, 131V and V155V—are each present a single time (twice for A46A), limiting our ability to associate these variants with resistance. Thus, non-synonymous substitution variants [present in 2766 (41.8%) of single variant strains] appear to be associated with most of the pyrazinamide resistance in M. tuberculosis.
Feature determination using Test/Train dataset
To understand the structural features that determine a mutation’s effect on pyrazinamide susceptibility, we mapped our combined Train/Test dataset onto the PncA structure. No obvious clustering was revealed, consistent with the previously observed distribution of resistant mutations across the gene sequence and protein structure (Figure 1a and c).13,24,29,30,38 Examining the PncA structure also suggested that resistant mutations were more likely to be buried in the hydrophobic core of the protein and therefore likely destabilizing, consistent with findings from previous in vitro and in vivo screens (Figure 2a).30,38 Indeed, some pyrazinamide-resistant mutations result in reduced production of functional PncA, perhaps due to impaired protein folding/stability.38,62 Despite having a similar learning objective there was only a moderate level of correlation between the different models that predicted the effect of a mutation on the protein stability (Figure 2b). Other more accurate methods exist, but these require several orders of magnitude of computational resource.63 Since SNAP2 uses evolutionary information derived from a multiple sequence alignment, one might expect some similarity to MAPP, but again there is only a moderate degree of correlation between the two scores (Figure 2b).
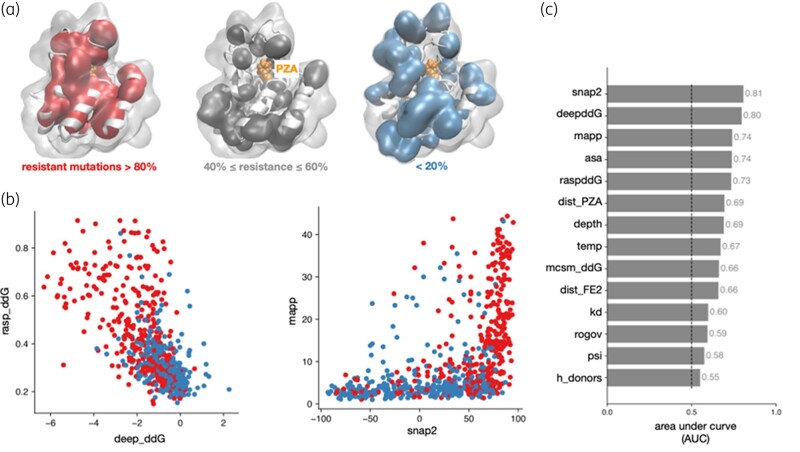
Figure 2.
Structural and evolutionary traits correlate with mutational impact on pyrazinamide susceptibility. (a) Amino acids where >80% of mutations confer resistance are more likely to be found in the core of PncA. (b) There is only a moderate correlation between RaSP and DeepDDG, which both predict the effect of a mutation on protein stability, and MAPP and SNAP2. Resistant and susceptible mutations are plotted in red and blue, respectively. (c) The performance of individual features, as measured by the AUC of a univariable logistic regression. The dashed line denotes random guessing.
Machine-learning models accurately predict pyrazinamide resistance
Univariable LR over the derivation dataset revealed that most of the individual predictors were associated with resistance [Figure 2c, Figure S1 (available as Supplementary data at JAC-AMR Online)]. The SNAP2 score and DeepDDG protein stability scores proved to be the most discriminatory individual features and six features (change in molecular weight, volume and isoelectric point, along with the secondary structure, ϕ backbone angle and number of hydrogen bond acceptors) were discarded at this point since their AUC lay below an arbitrary threshold of 0.55.
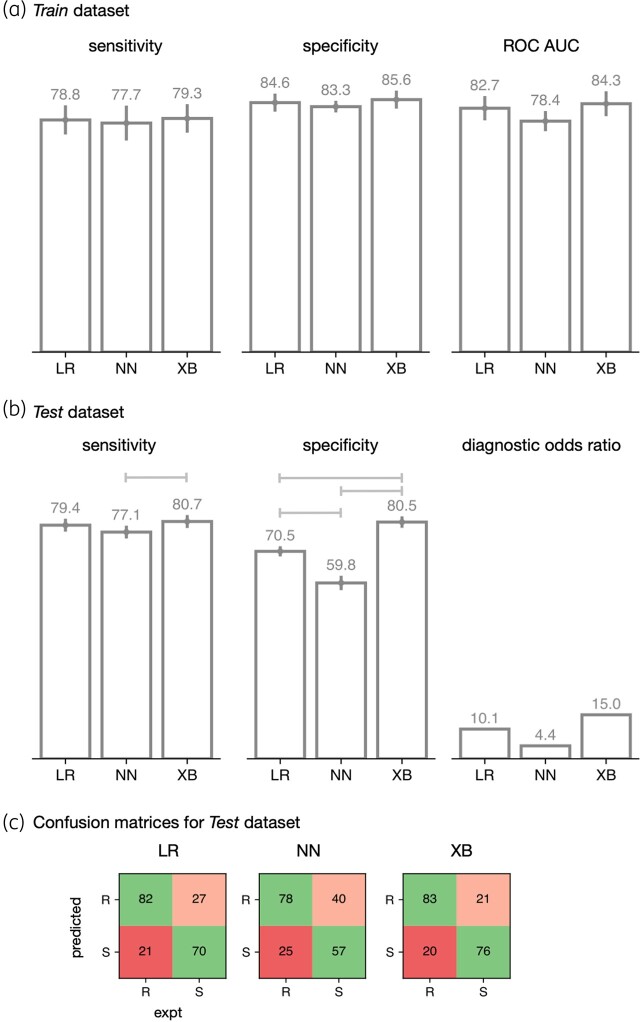
Following hyperparameter tuning, three different machine-learning models (LR; a gradient-boosted decision tree, XB; and a single layer neural network, NN) were trained on the Train dataset using 10-fold cross-validation. All three models performed similarly when applied to the Train dataset (Figure 3), with sensitivities of 78%–79% and specificities in the range 83%–86%. As expected, the models performed less well on the Test dataset and the XB model had a superior sensitivity (80.7%) to the NN model (77.1%) whilst was indistinguishable from the LR (79.4%) model. The XB model also had a improved specificity (80.5%) relative to the LR or NN models (70.5% and 59.8%, respectively). We conclude that the gradient-boosted decision tree (XB) model performed best since it resulted in the fewest number of resistant samples incorrectly classified as susceptible (so-called very major errors, VMEs) and had the highest diagnostic OR (Figure 3b and c).
Figure 3.
Machine-learning models predict pyrazinamide resistance from structural, chemical and evolutionary features. Performance of logistic regression (LR), a simple neural network (NN) and gradient-boosted decision tree (XB) models on the (a) Training and (b) Test sets. Error bars represent 95% CIs from bootstrapping (n = 10) and brackets indicate a significant difference (z-test, P < 0.05) (c) Confusion matrices are shown for the Test set. VMEs are considered worse than MEs and hence VMEs and MEs are shaded red and pink, respectively.
Most residues that were incorrectly predicted as susceptible are surface exposed
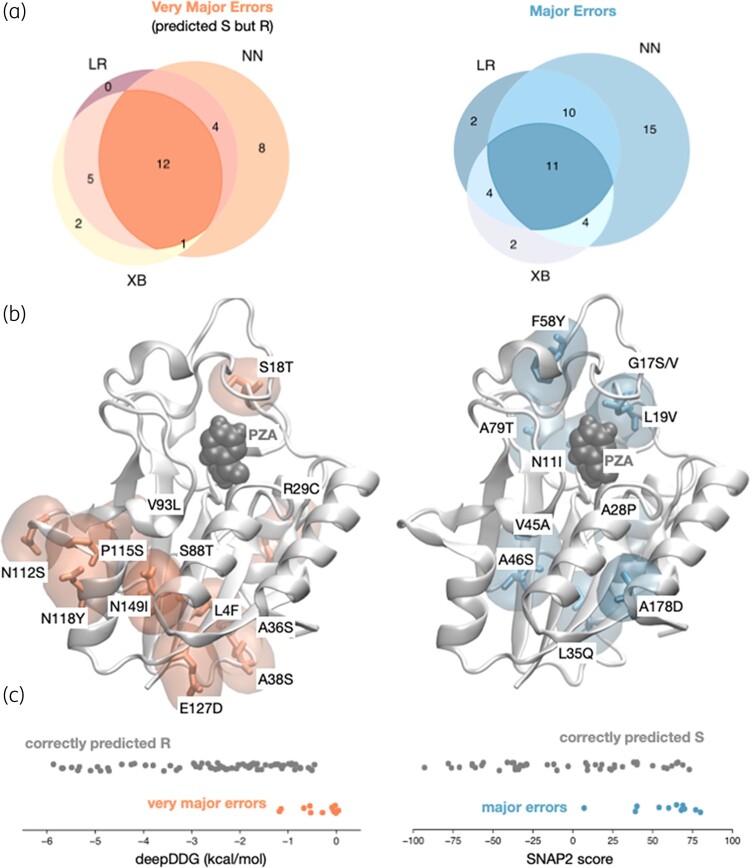
The models predicted 20–25 VMEs and misclassified a further 21–40 susceptible samples as resistant (major errors, MEs). Collectively 12 VMEs and 11 MEs were shared between all three models (Figure 4a). Although the mutations responsible for the shared VMEs were dispersed throughout the protein structure, most (11/12) were surface exposed (Figure 4b). All these mutations were predicted by DeepDDG, mCSM and RaSP to minimally decrease the stability of PncA compared with mutations correctly predicted to confer resistance, suggesting these errors may be partly due to inaccuracies in the predicted free energy change of unfolding (Figure 4c), although other features also contributed (Figure S2). Major errors were also dispersed throughout the protein and were more likely to be buried and to be predicted by SNAP2 to not have a functional effect compared with mutations correctly predicted to have no effect (Figure 4c), although again other features played a part (Figure S2).
Figure 4.
VMEs are concentrated on the surface of PncA. (a) The majority of VMEs and MEs are shared between the three models. (b) PncA with the corresponding residues highlighted where the shared VMEs (orange) and MEs (blue) are found. (c) The shared VMEs and MEs are predicted to have less and more effect, respectively, on the stability of the protein, as exemplified by DeepDDG and the function of the protein, according to SNAP2.
Examining the feature importances of the gradient-boosted decision tree (XB) models (Figure S3) shows that whilst all 16 features are incorporated to some extent, the first 4 are all scores from other machine-learning models (MAPP, DeepDDG, RaSP and SNAP2), with the next 4 all being derived from the protein structure (ψ backbone angle, residue depth and residue solvent accessible surface area) or describing the change in chemistry.55
Gradient-boosted decision tree model predictions generalize to a large clinical dataset
A Validation dataset was derived from 24 231 pncA gene sequences with MGIT antibiotic susceptibility results (Table 1). Most samples contained no mutations in pncA: only 4027 samples had 1 of 367 missense mutations. We assume this dataset is representative of the genetic diversity in PncA existing in clinical infections but it is likely biased due to oversampling of outbreak strains and other factors. Until very large unselected clinical datasets are collected and made publicly available, however, it is the best dataset available.
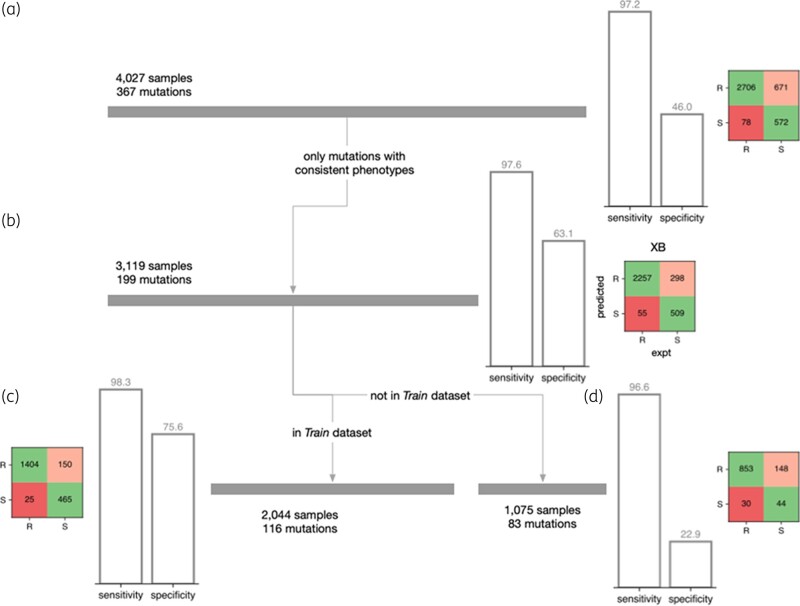
Applying the gradient-boosted decision tree (XB) model to this dataset (Figure 5a) resulted in a high sensitivity (97.2%) but a modest specificity (46.0%). The presence of a substantial number of samples in this dataset (908 samples; 22.5%) contained 1 of 168 (45.8%) mutations that either were only observed once, or whose phenotype varied between isolates was a key contributor to this reduction in performance. Whilst this dataset therefore captures the real-world variability of culture-based phenotypic methods for pyrazinamide susceptibility testing, it is not a good basis on which to assess performance and removing these samples improved the specificity to 63.1% (Figure 5b). Slightly over half (116; 58.3%) of remaining mutations were also present in the Train dataset and accounted for 2044 out of the remaining 3119 samples. The predictions for the samples in this group had a sensitivity of 98.3% and a specificity of 75.6%. As expected, the other 83 mutations found in 1075 samples had a lower performance, with the specificity notably being 22.9%. Examining the performance at the level of the mutations (rather than samples) yields a specificity of 48.0%; however, the size of the dataset is now small with only 25 out of 83 mutations having a susceptible phenotype. The XB model also outperforms a previously published model49 applied to this same dataset; SUSPECT-PZA achieved a sensitivity of 93.7% and a specificity of 44.3% on the original 4027 samples. Only considering the 199 mutations with a consistent phenotype improved the specificity to 47.7% with a slight fall in sensitivity (92.3%); however, this is less predictive than the performance of the gradient-boosted decision tree.
Figure 5.
Performance on a real set of clinical samples. (a) Whilst the sensitivity is high, the specificity of the gradient-boosted decision tree model on the Validation dataset is lower than observed on the Test dataset. (b) Removing samples containing a mutation that has an experimentally inconsistent phenotype increases the specificity. As expected, splitting into samples whose mutation either (c) belongs or (d) does not belong to the Train dataset further stratifies performance.
Comparison of model predictions with pyrazinamide MICs in vitro
Since it is difficult to assess how much of the discordance in the previous section can be attributed to either error in the measured clinical phenotype or deficiencies in our model, we compared its predictions to MIC data taken from a small but high-quality dataset of 71 M. tuberculosis isolates (59 unique missense mutations, quantitative dataset). This also enabled us to test the model’s capacity to predict the degree of pyrazinamide resistance conferred by a particular mutation, by comparing the calls and predicted probabilities of our model with the pyrazinamide MICs. Overall, our model correctly predicted the binary (resistant/susceptible) phenotype for 51 of 57 missense mutations in PncA (Figure S4) and, crucially, predicted the correct phenotype for six out of eight mutations that were not in either the Train or Test datasets. Ultimately, many more samples with a wide range of pyrazinamide MICs will be needed to accurately assess if quantitative prediction is possible for this drug.
Discussion
We have shown that machine-learning models trained on structural, chemical and evolutionary features can predict whether missense amino acid mutations in pncA confer resistance to pyrazinamide, adding to the growing body of work that is exploring different ways of predicting antibiotic resistance from genetics.41–49 While improvements to the model are necessary to achieve the sensitivity and specificity required for routine clinical use, this work increases our ability to classify rare resistance mutations, thereby potentially increasing the capability of WGS-based diagnostic susceptibility testing to respond to emerging and rare resistance patterns, as well as prioritizing rare resistance mutations for in vitro validation. Additionally, improving the classification of susceptible pncA mutations will allow us to begin to disentangle the involvement of other genes in pyrazinamide resistance, including determining the effect of mutations in other pyrazinamide resistance-associated genes such as panD and rpsA.
There are two principal limitations of our approach: (i) since the training set uses a binary resistant/susceptible phenotype, the models can only predict whether a mutation confers high-level resistance (>100 mg/L64) or not; and (ii) it can only make predictions for missense mutations in the coding sequence of pncA. It is known that genetic variation can lead to small changes in MIC of pyrazinamide and other first-line antitubercular compounds and that, whilst these may not change the binary phenotype, they do affect clinical outcome.65,66 In addition, while we have shown that missense mutations represent most of the possible resistant genetic variants in pncA, insertions/deletions and non-sense mutations must also be considered, as they are generally associated with resistance. Likewise, promoter mutations that result in reduced transcription of pncA will likely also lead to resistance.
Our predictive capabilities will improve with time: the largest potential improvement is likely to come from the availability of larger datasets, preferably with pyrazinamide MICs. Quantitative labels would help delineate mutations that result in an MIC similar to the 100 mg/L breakpoint as one suspects that this effect is the reason why many mutations test inconsistently in the laboratory, which has complicated both our training and validation. New machine-learning approaches and better general-purpose predictors, especially those that aim to predict the effect of a mutation on protein stability, will no doubt come.
Even before that, predictions made by this or similar models could potentially provide clinicians with an initial estimate of pyrazinamide susceptibility after a novel mutation is observed but before traditional phenotypic testing has been completed. Given the latter can take weeks or even months, this could help guide initial therapy and further antibiotic susceptibility testing. In addition, the putative classification of additional pncA mutations potentially enables genetic variants conferring pyrazinamide resistance that do not involve the pncA gene to be discovered. The identification of pyrazinamide-susceptible mutations is also crucial, as it has been suggested that any non-synonymous mutation in pncA that is not catalogued as susceptible confers resistance, an incorrect assumption that would lead to overprediction of pyrazinamide resistance.67
The approach used here should be extensible to any pro-drug system where the enzyme is non-essential, such as delamanid, pretomanid or ethionamide, as well as to pro-drug systems in other pathogens. One promising area for future work is in the anti-tubercular bedaquiline, where resistance is caused in part by mutations in a transcriptional repressor (Rv0678) that causes loss of DNA binding and up-regulation of efflux pumps.68,69 Predictive methods, as shown here, will help accelerate the rate at which WGS approaches move to the forefront of global TB control efforts.
Supplementary Material
Contributor Information
Joshua J Carter, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
Timothy M Walker, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
A Sarah Walker, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; National Institute of Health Research Oxford Biomedical Research Centre, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; NIHR Health Protection Research Unit in Healthcare Associated Infection and Antimicrobial Resistance, University of Oxford, Oxford, UK.
Michael G Whitfield, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, SAMRC Centre for Tuberculosis Research, DST/NRF Centre of Excellence for Biomedical Tuberculosis Research, Stellenbosch University, Tygerberg, South Africa.
Glenn P Morlock, Division of Tuberculosis Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Charlotte I Lynch, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
Dylan Adlard, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
Timothy E A Peto, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; National Institute of Health Research Oxford Biomedical Research Centre, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
James E Posey, Division of Tuberculosis Elimination, National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Derrick W Crook, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; National Institute of Health Research Oxford Biomedical Research Centre, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; NIHR Health Protection Research Unit in Healthcare Associated Infection and Antimicrobial Resistance, University of Oxford, Oxford, UK.
Philip W Fowler, Nuffield Department of Medicine, University of Oxford, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK; National Institute of Health Research Oxford Biomedical Research Centre, John Radcliffe Hospital, Headley Way, Oxford OX3 9DU, UK.
Funding
The study was funded by the National Institute for Health Research (NIHR) Health Protection Research Unit (HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at Oxford University in partnership with Public Health England (PHE) [HPRU-2012-10041]; the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC); the CRyPTIC consortium, which is funded by a Wellcome Trust Collaborative Award [200205/Z/15/Z] and the Bill & Melinda Gates Foundation Trust [OPP1133541]; the European Commission for EU H2020 CompBioMed2 Centre of Excellence (grant no. 823712) and the South African Medical Research Council. Funded by Wellcome. This research was funded in whole, or in part, by the Wellcome Trust [200205/Z/15/Z]. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The EXIT-RIF project (Prof. Annelies Van; Prof. Rob Warren, Prof. Lesley Scott, Prof. Wendy Stevens, Dr Michael Whitfield) is funded by National Institutes of Health grant [#R01 AI099026]. J.J.C. is supported by a fellowship from the Rhodes Trust. T.M.W is a Wellcome Trust Clinical Career Development Fellow [214560/Z/18/Z]. D.A. is supported by the EPSRC Sustainable Approaches to Biomedical Science: Responsible & Reproducible Research CDT. T.E.A.P. and D.W.C. are NIHR Senior Investigators. T.M.W. was an NIHR Academic Clinical Lecturer. J.J.C would like to thank Spencer Dunleavy for thoughtful discussions on statistical analysis and modelling. The content is the solely the responsibility of the authors and does not necessarily represent the official views of the South African Medical Research Council. M.G.W. is supported by the TORCH project which is funded through the Research Foundation Flanders (FWO G0F8316N) and a fellowship from the Claude Leon Foundation. The findings and conclusions in this report are solely the responsibility of the authors and do not necessarily represent the official views of the NHS, the NIHR, the Department of Health, the Centers for Disease Control and Prevention (CDC), or the US Department of Health and Human Services. References in this manuscript to any specific commercial products, process, service, manufacturer, or company do not constitute its endorsement or recommendation by the US government or CDC.
Transparency declarations
The authors have no interests to declare.
Author contributions
J.J.C., P.W.F. and T.M.W. designed experiments, C.I.L., D.A. and P.W.F. wrote computer code used by the experiments, J.J.C. and P.W.F. carried out the experiments, J.J.C., P.W.F. and A.S.W. performed statistical analyses, J.J.C., P.W.F., A.S.W. and T.E.A.P. wrote the manuscript. P.W.F. and D.W.C. supervised the work. M.G.W., J.E.P. and G.P.M. contributed samples and edited the manuscript.
Supplementary data
Figures S1 to S4 are available as Supplementary data at JAC-AMR Online.
References
- 1. WHO . Global Tuberculosis Report 2022. 2022. https://www.who.int/publications/i/item/9789240061729.
- 2. Njire M, Tan Y, Mugweru J et al. Pyrazinamide resistance in Mycobacterium tuberculosis: review and update. Adv Med Sci 2016; 61: 63–71. 10.1016/j.advms.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis 2015; 19: 1276–89. 10.5588/ijtld.15.0389 [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis 2003; 7: 6–21. [PubMed] [Google Scholar]
- 5. Mitchison DA. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 1985; 66: 219–25. 10.1016/0041-3879(85)90040-6 [DOI] [PubMed] [Google Scholar]
- 6. WHO . WHO Consolidated Guidelines on Tuberculosis: Module 4: Treatment: Drug-Susceptible Tuberculosis Treatment. 2022. https://www.who.int/publications-detail-redirect/9789240048126. [PubMed]
- 7. Dawson R, Diacon AH, Everitt D et al. Efficiency and safety of the combination of moxifloxacin, pretomanid (PA-824), and pyrazinamide during the first 8 weeks of antituberculosis treatment: a phase 2b, open-label, partly randomised trial in patients with drug-susceptible or drug-resistant pulmonary tuberculosis. Lancet 2015; 385: 1738–47. 10.1016/S0140-6736(14)62002-X [DOI] [PubMed] [Google Scholar]
- 8. Chang KC, Leung CC, Yew WW et al. Pyrazinamide may improve fluoroquinolone-based treatment of multidrug-resistant tuberculosis. Antimicrob Agents Chemother 2012; 56: 5465–75. 10.1128/AAC.01300-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zumla AI, Gillespie SH, Hoelscher M et al. New antituberculosis drugs, regimens, and adjunct therapies: needs, advances, and future prospects. Lancet Infect Dis 2014; 14: 327–40. 10.1016/S1473-3099(13)70328-1 [DOI] [PubMed] [Google Scholar]
- 10. Nuermberger E, Tyagi S, Tasneen R et al. Powerful bactericidal and sterilizing activity of a regimen containing PA-824, moxifloxacin, and pyrazinamide in a murine model of tuberculosis. Antimicrob Agents Chemother 2008; 52: 1522–4. 10.1128/AAC.00074-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenthal IM, Zhang M, Williams KN et al. Daily dosing of rifapentine cures tuberculosis in three months or less in the murine model. PLoS Med 2007; 4: 1931–9. 10.1371/journal.pmed.0040344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veziris N, Ibrahim M, Lounis N et al. A once-weekly R207910-containing regimen exceeds activity of the standard daily regimen in murine tuberculosis. Am J Respir Crit Care Med 2009; 179: 75–9. 10.1164/rccm.200711-1736OC [DOI] [PubMed] [Google Scholar]
- 13. Whitfield MG, Soeters HM, Warren RM et al. A global perspective on pyrazinamide resistance: systematic review and meta-analysis. PLoS One 2015; 10: e0133869. 10.1371/journal.pone.0133869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang KC, Yew WW, Zhang Y. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis: a systematic review with meta-analyses. Antimicrob Agents Chemother 2011; 55: 4499–505. 10.1128/AAC.00630-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hewlett D, Horn DL, Alfalla C. Drug-resistant tuberculosis: inconsistent results of pyrazinamide susceptibility testing. JAMA 1995; 273: 916–7. 10.1001/jama.1995.03520360030022 [DOI] [PubMed] [Google Scholar]
- 16. Miller MA, Thibert L, Desjardins F et al. Testing of susceptibility of Mycobacterium tuberculosis to pyrazinamide: comparison of Bactec method with pyrazinamidase assay. J Clin Microbiol 1995; 33: 2468–70. 10.1128/jcm.33.9.2468-2470.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffner S, Angeby K, Sturegard E et al. Proficiency of drug susceptibility testing of Mycobacterium tuberculosis against pyrazinamide: the Swedish experience. Int J Tuberc Lung Dis 2013; 17: 1486–90. 10.5588/ijtld.13.0195 [DOI] [PubMed] [Google Scholar]
- 18. Pandey S, Newton S, Upton A et al. Characterisation of pncA mutations in clinical Mycobacterium tuberculosis isolates in New Zealand. Pathology 2009; 41: 582–4. 10.1080/00313020903071587 [DOI] [PubMed] [Google Scholar]
- 19. Simons SO, Van Ingen J, Van Der Laan T et al. Validation of pncA gene sequencing in combination with the mycobacterial growth indicator tube method to test susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol 2012; 50: 428–34. 10.1128/JCM.05435-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chedore P, Bertucci L, Wolfe J et al. Potential for erroneous results indicating resistance when using the Bactec MGIT 960 system for testing susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Clin Microbiol 2010; 48: 300–1. 10.1128/JCM.01775-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. WHO . Catalogue of Mutations in Mycobacterium tuberculosis Complex and their Association With Drug Resistance. 2021. https://www.who.int/publications/i/item/9789240028173.
- 22. Walker TM, Miotto P, Köser CU et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe 2022; 3: e265–73. 10.1016/S2666-5247(21)00301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yüksel P, Tansel Ö. Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Turkey. New Microbiol 2009; 32: 153–8. [PubMed] [Google Scholar]
- 24. Ramirez-Busby SM, Rodwell TC, Fink L et al. A multinational analysis of mutations and heterogeneity in PZase, RpsA, and PanD associated with pyrazinamide resistance in M/XDR Mycobacterium tuberculosis. Sci Rep 2017; 7: 3790. 10.1038/s41598-017-03452-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheen P, Requena D, Gushiken E et al. A multiple genome analysis of Mycobacterium tuberculosis reveals specific novel genes and mutations associated with pyrazinamide resistance. BMC Genomics 2017; 18: 769. 10.1186/s12864-017-4146-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gopal P, Yee M, Sarathy J et al. Pyrazinamide resistance is caused by two distinct mechanisms: prevention of coenzyme a depletion and loss of virulence factor synthesis. ACS Infect Dis 2016; 2: 616–26. 10.1021/acsinfecdis.6b00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y, Zhang J, Cui P et al. Identification of novel efflux proteins Rv0191, Rv3756c, Rv3008, and Rv1667c involved in pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agent Chemo 2017; 61: e00940-17. 10.1128/AAC.00940-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirano K, Takahashi M, Kazumi Y et al. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung Dis 1997; 78: 117–22. 10.1016/S0962-8479(98)80004-X [DOI] [PubMed] [Google Scholar]
- 29. Stoffels K, Mathys V, Fauville-Dufaux M et al. Systematic analysis of pyrazinamide-resistant spontaneous mutants and clinical isolates of Mycobacterium tuberculosis. Antimicrob Agent Chemo 2012; 56: 5186–93. 10.1128/AAC.05385-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miotto P, Cabibbe AM, Feuerriegel S et al. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 2014; 5: e01819-14. 10.1128/mBio.01819-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med 1996; 2: 662–7. 10.1038/nm0696-662 [DOI] [PubMed] [Google Scholar]
- 32. Kim N, Petingi L, Schlick T. Network theory tools for RNA modeling. WSEAS Trans Math 2013; 12: 941–55. [PMC free article] [PubMed] [Google Scholar]
- 33. Yee M, Gopal P, Dick T. Missense mutations in the unfoldase ClpC1 of the caseinolytic protease complex are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2017; 61: e02342-16. 10.1128/AAC.02342-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang S, Chen J, Shi W et al. Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect 2017; 6: e8. 10.1038/emi.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Driesen M, Kondo Y, de Jong BC et al. Evaluation of a novel line probe assay to detect resistance to pyrazinamide, a key drug used for tuberculosis treatment. Clin Microbiol Infect 2018; 24: 60–4. 10.1016/j.cmi.2017.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kalokhe AS, Shafiq M, Lee JC et al. Multidrug-resistant tuberculosis drug susceptibility and molecular diagnostic testing: a review of the literature. Am J Med Sci 2013; 345: 143–8. 10.1097/MAJ.0b013e31825d32c6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitfield MG, Warren RM, Streicher EM et al. Mycobacterium tuberculosis pncA polymorphisms that do not confer pyrazinamide resistance at a breakpoint concentration of 100 micrograms per milliliter in MGIT. J Clin Microbiol 2015; 53: 3633–5. 10.1128/JCM.01001-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yadon AN, Maharaj K, Adamson JH et al. A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nat Commun 2017; 8: 588. 10.1038/s41467-017-00721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. The CRyPTIC Consortium, 100000 Genomes Project . Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. New Eng J Med 2018; 379: 1403–15. 10.1056/NEJMoa1800474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tunstall T, Portelli S, Phelan J et al. Combining structure and genomics to understand antimicrobial resistance. Comput Struct Biotechnol J 2020; 18: 3377–94. 10.1016/j.csbj.2020.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davis JJ, Boisvert S, Brettin T et al. Antimicrobial resistance prediction in PATRIC and RAST. Sci Rep 2016; 6: 27930. 10.1038/srep27930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deelder W, Christakoudi S, Phelan J et al. Machine learning predicts accurately Mycobacterium tuberculosis drug resistance from whole genome sequencing data. Front Genet 2019; 10: 922. 10.3389/fgene.2019.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kouchaki S, Yang Y, Walker TM et al. Application of machine learning techniques to tuberculosis drug resistance analysis. Bioinformatics 2019; 35: 2276–82. 10.1093/bioinformatics/bty949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brankin AE, Fowler PW. Predicting antibiotic resistance in complex protein targets using alchemical free energy methods. J Comput Chem 2022; 43: 1771–82. 10.1002/jcc.26979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Portelli S, Myung Y, Furnham N et al. Prediction of rifampicin resistance beyond the RRDR using structure-based machine learning approaches. Sci Rep 2020; 10: 18120. 10.1038/s41598-020-74648-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karmakar M, Rodrigues CHM, Holt KE et al. Empirical ways to identify novel bedaquiline resistance mutations in AtpE. PLoS One 2019; 14: e0217169. 10.1371/journal.pone.0217169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang Y, Walker TM, Walker AS et al. DeepAMR for predicting co-occurrent resistance of Mycobacterium tuberculosis. Bioinformatics 2019; 35: 3240–9. 10.1093/bioinformatics/btz067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. The CRyPTIC Consortium, Lachapelle AS. Quantitative drug susceptibility testing for M. tuberculosis using unassembled sequencing data and machine learning. bioRxiv 2022; 10.1101/2021.09.14.458035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Karmakar M, Rodrigues CHM, Horan K et al. Structure guided prediction of pyrazinamide resistance mutations in pncA. Sci Rep 2020; 10: 1875. 10.1038/s41598-020-58635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. The CRyPTIC Consortium . A data compendium associating the genomes of 12,289 Mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics. PLoS Biol 2022; 20: e3001721. 10.1371/journal.pbio.3001721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Piersimoni C, Mustazzolu A, Giannoni F et al. Prevention of false resistance results obtained in testing the susceptibility of Mycobacterium tuberculosis to pyrazinamide with the Bactec MGIT 960 system using a reduced inoculum. J Clin Microbiol 2013; 51: 291–4. 10.1128/JCM.01838-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petrella S, Gelus-Ziental N, Maudry A et al. Crystal structure of the pyrazinamidase of Mycobacterium tuberculosis: insights into natural and acquired resistance to pyrazinamide. PLoS One 2011; 6: e15785. 10.1371/journal.pone.0015785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fyfe PK, Rao VA, Zemla A et al. Specificity and mechanism of Acinetobacter baumanii nicotinamidase: implications for activation of the front-line tuberculosis drug pyrazinamide. Angew Chem Int Ed 2009; 48: 9176–9. 10.1002/anie.200903407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fowler PW, Lynch CI, Adlard D et al. sbmlcore: v0.2.5. 2023. https://github.com/fowler-lab/sbmlcore.
- 55. Rogov SI, Nekrasov AN. A numerical measure of amino acid residues similarity based on the analysis of their surroundings in natural protein sequences. Protein Eng 2001; 14: 459–63. 10.1093/protein/14.7.459 [DOI] [PubMed] [Google Scholar]
- 56. Blaabjerg LM, Kassem MM, Good LL et al. Rapid protein stability prediction using deep learning representations. Elife 2023; 12: e82593. 10.7554/eLife.82593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pires DEV, Ascher DB, Blundell TL. Structural bioinformatics mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2014; 30: 335–42. 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cao H, Wang J, He L et al. DeepDDG: predicting the stability change of protein point mutations using neural networks. J Chem Inf Model 2019; 59: 1508–14. 10.1021/acs.jcim.8b00697 [DOI] [PubMed] [Google Scholar]
- 59. Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res 2005; 15: 978–86. 10.1101/gr.3804205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics 2015; 16: S1. 10.1186/1471-2164-16-S8-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fowler PW. predict-pyrazinamide-resistance. 2023. https://github.com/fowler-lab/predict-pyrazinamide-resistance.
- 62. Yoon JH, Nam JS, Kim KJ et al. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Korea and analysis of the correlation between the mutations and pyrazinamidase activity. World J Microbiol Biotechnol 2014; 30: 2821–8. 10.1007/s11274-014-1706-0 [DOI] [PubMed] [Google Scholar]
- 63. Fowler PW, Cole K, Gordon NC et al. Robust prediction of resistance to trimethoprim in Staphylococcus aureus. Cell Chem Biol 2018; 25: 339–349.e4. 10.1016/j.chembiol.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 64. WHO . Technical Report on Critical Concentrations for Drug Susceptibility Testing of Medicines Used in the Treatment of Drug-Resistant Tuberculosis. 2018. https://www.who.int/publications/i/item/WHO-CDS-TB-2018.5.
- 65. Gumbo T, Chigutsa E, Pasipanodya J et al. The pyrazinamide susceptibility breakpoint above which combination therapy fails. J Antimicrob Chem 2014; 69: 2420–5. 10.1093/jac/dku136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Colangeli R, Jedrey H, Kim S et al. Bacterial factors that predict relapse after tuberculosis therapy. New Eng J Med 2018; 379: 823–33. 10.1056/NEJMoa1715849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zignol M, Dean AS, Alikhanova N et al. Population-based resistance of Mycobacterium tuberculosis isolates to pyrazinamide and fluoroquinolones: results from a multicountry surveillance project. Lancet Infect Dis 2016; 16: 1185–92. 10.1016/S1473-3099(16)30190-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Milano A, Pasca MR, Provvedi R et al. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 2009; 89: 84–90. 10.1016/j.tube.2008.08.003 [DOI] [PubMed] [Google Scholar]
- 69. Nguyen TVA, Anthony RM, Bañuls AL et al. Bedaquiline resistance: its emergence, mechanism, and prevention. Clin Infect Dis 2018; 66: 1625–30. 10.1093/cid/cix992 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.