Abstract
RNA helicases drive necessary rearrangements and ensure fidelity during the pre-mRNA splicing cycle. DEAD-box helicase DDX41 has been linked to human disease and has recently been shown to interact with DEAH-box helicase PRP22 in the spliceosomal C* complex, yet its function in splicing remains unknown. Depletion of DDX41 homolog SACY-1 from somatic cells has been previously shown to lead to changes in alternative 3′ splice site (3′ss) usage. Here, we show by transcriptomic analysis of published and novel data sets that SACY-1 perturbation causes a previously unreported pattern in alternative 3′ splicing in introns with pairs of 3′ splice sites ≤18 nt away from each other. We find that both SACY-1 depletion and the allele sacy-1(G533R) lead to a striking unidirectional increase in the usage of the proximal (upstream) 3′ss. We previously discovered a similar alternative splicing pattern between germline tissue and somatic tissue, in which there is a unidirectional increase in proximal 3′ss usage in the germline for ∼200 events; many of the somatic SACY-1 alternative 3′ splicing events overlap with these developmentally regulated events. We generated targeted mutant alleles of the Caenorhabditis elegans homolog of PRP22, mog-5, in the region of MOG-5 that is predicted to interact with SACY-1 based on the human C* structure. These viable alleles, and a mimic of the myelodysplastic syndrome-associated allele DDX41(R525H), all promote the usage of proximal alternative adjacent 3′ splice sites. We show that PRP22/MOG-5 and DDX41/SACY-1 have overlapping roles in proofreading the 3′ss.
Keywords: C* complex, DDX41, PRP22, RNA helicase, splicing
INTRODUCTION
Introns are removed from precursor messenger RNA (pre-mRNA) by the spliceosome, a dynamic multimegadalton ribonucleoprotein complex (Wilkinson et al. 2020). During the splicing cycle, five small nuclear RNAs (snRNAs) along with many protein splicing factors assemble onto the pre-mRNA and form spliceosomal complexes that transition through several rearrangements and composition changes to create the catalytically active spliceosome with a ribozyme core (Fica et al. 2013). The 5′ splice site (5′ss) and the branchpoint (BP) are joined in the first trans-esterification reaction to form a lariat-containing splicing intermediate and a free 5′ exon. Then the 5′ exon is joined to the 3′ exon in the second trans-esterification reaction, with the intron lariat released as a by-product. The spliceosome is then disassembled to start the cycle anew.
Many of the critical assembly, rearrangement, and disassembly steps of the spliceosome cycle require RNA helicases and ATP; eight helicases are conserved in eukaryotes, and five are found in metazoans but not in Saccharomyces cerevisiae (De Bortoli et al. 2021). In splicing, DEAH-box helicases translocate along single-stranded RNA in the 3′–5′ direction and enact conformation changes in the spliceosome, but, contrary to their name, do not necessarily unwind RNA helices during splicing (Semlow et al. 2016). The potential ATPase and helicase mechanisms of action in splicing of the DEAD-box helicases are less understood. Five spliceosomal RNA helicases are currently known to proofread against unfavorable mRNA substrates or sequence features (De Bortoli et al. 2021).
Many steps of the splicing cycle are functionally linked to the 3′ splice site (3′ss) choice. During early assembly, the approximate location of the 3′ end of the intron is initially bound by U2AF proteins which help recruit the U2snRNP. U2snRNA base pairs with the BP sequence (Parker et al. 1987). The candidate BP can then be proofread by the DEAH helicase Prp16 (Burgess and Guthrie 1993). When the first trans-esterification reaction happens, BP choice is cemented. For the second step of splicing, the spliceosome must load the 3′ss into its active site. A common model is that the spliceosome will scan for the first AG dinucleotide that occurs past a minimal distance downstream from the BP, and choose it for the 3′ss (Smith et al. 1989). However, AGs that are usable in mutant spliceosomes can be skipped by wild type (WT) spliceosomes (Chua and Reed 1999). Hundreds of human introns feature a NAGNAG sequence motif where either AG can be used as the 3′ss (Hiller et al. 2004). Scanning alone does not define 3′ss choice. DEAH-box helicase Prp22 can proofread the 3′ss before splicing is completed (Mayas et al. 2006).
New cryo-electron microscopy (cryo-EM) structures have furthered our understanding of the portion of the splicing cycle that happens after PRP16 remodeling but before the second trans-esterification reaction. Several proteins were recently modeled into the human C* complex for the first time, including DDX41 (Dybkov et al. 2023). siRNA-mediated knockdown of some of these C* proteins was found to alter the splicing of adjacent NAGNAG 3′ splice sites. DDX41 and PRP22 were shown to interact, with DDX41 modeled into the periphery of the spliceosome, on the opposite side of PRP22 from the 3′ exon (Dybkov et al. 2023). In another study, three populations of human C spliceosomes, termed pre-C*-I, pre-C*-2, and C* were modeled, providing new insight into the dynamic mechanism that prepares the spliceosome for the second trans-esterification reaction (Zhan et al. 2022). Between all three of these structures, PRP22 transitions through different conformations; these transitions may be involved in PRP22 proofreading activity. These structures have many features that suggest specific, novel functions for proteins and amino acids whose roles in splicing are unknown. Testing these novel functions will require applying novel splicing assays.
Disruption of factors involved in identifying 3′ss contributes to human disease, and because most eukaryotic transcripts require splicing for mRNA export and translation, all cellular processes are potentially vulnerable to disruption. For example, oncogenic mutation of SF3b1 alters BP choice and 3′ss usage (Darman et al. 2015). The pleiotropic effects of disrupting spliceosomal components often make it difficult to untangle the different pathogenic mechanisms; for example, SF3b1 also contributes to malignancy through R-loop formation and DNA damage (Singh et al. 2020). DEAD-box helicase DDX41 is another spliceosome-associated protein linked to cancers, and its disruption causes changes to both splicing and R-loop formation, leading to replicative stress (Shinriki et al. 2022). sacy-1, the Caenorhabditis elegans homolog of DDX41, also has multiple roles; it was initially identified in a screen for oocyte meiotic maturation factors (the name sacy-1 stands for “suppressor of acy-4”) (Kim et al. 2012), and was only later understood to be involved in splicing and to be associated with C complex proteins (Tsukamoto et al. 2020).
Several important splicing factors in C. elegans were initially discovered while studying germline development and were named after the masculinization of germline (Mog) phenotype (Graham and Kimble 1993); these were only later found to be homologs of splicing proteins. mog-1 (PRP16), mog-4 (PRP2), and mog-5 (PRP22) are all homologs of highly conserved DEAH-box helicases that proofread and then drive the splicing cycle forward. The sacy-1(P222L) allele also causes Mog phenotype (Tsukamoto et al. 2020), whereas a null allele causes sterility but not Mog for both sacy-1 (Kim et al. 2012) and mog-5 (C. elegans Deletion Mutant Consortium 2012). This highlights that hypomorphic alleles of these factors may prove more useful for studying direct effects on splicing than null mutations in essential genes.
Caenorhabditis elegans genetics provides an opportunity to study splice site choice at sites with unusual features. These features allow us to assay the splicing effects of homozygous mutations of conserved amino acids. In Ragle et al. (2015), we found a set of 203 regulated alternative adjacent (≤18 nt apart) 3′ss pairs that show tissue-specific splicing; in all cases, the proximal or upstream 3′ss (closer to the 5′ end of the intron) shows increased usage in germline tissue relative to somatic tissue (Ragle et al. 2015). Interestingly, the proximal splice sites generally do not have any sequence conservation besides an AG dinucleotide, whereas the distal or downstream (more toward the 3′ end of the pre-mRNA) splice sites closely match the C. elegans UUUCAG 3′ss consensus. We hypothesized that the distal 3′ss matching the C. elegans consensus sequence is a binding site for the U2AF homologs UAF-1/UAF-2 (Zorio and Blumenthal 1999), whereas the proximal site represents an AG dinucleotide that can enter the active site of the spliceosome at step 2 due to inefficient translocation from the BP to the 3′ss. These splice sites provide an opportunity to study how a metazoan spliceosome chooses between adjacent 3′ splice sites.
Although the function of DDX41 has been the focus of a growing intensity of research activity, and it is recognized as a C complex protein, its function in splicing is poorly understood. In this study, we report that the DDX41 homolog SACY-1 and a portion of the PRP22 homolog MOG-5 that is predicted to interact with SACY-1 both have overlapping phenotypes in 3′ss choice. Disruption of the putative interaction increases usage of proximal 3′ splice sites. These results provide direct evidence that conserved residues of PRP22/MOG-5 and DDX41/SACY-1 are required for a C*-linked proofreading mechanism.
RESULTS
SACY-1 depletion causes both directional and sequence preference changes in 3′ splice site choice
SACY-1 has been implicated in 3′ss choice (Tsukamoto et al. 2020), but the nature of its splicing phenotype has not been explored. RNA-seq analysis following SACY-1 protein depletion in somatic cells via the auxin-induced degron system predominantly detected changes in alternative (alt.) 3′ss choice. This data set provided an opportunity to study the 3′ss choice mechanism. We searched these data for additional patterns in alt. 3′ (A3) splicing. To do this, we downloaded the raw sequencing data (accession number GSE144003) from Tsukamoto et al. (2020) and analyzed alt. splicing using our custom workflow (Suzuki et al. 2022). We focused on the somatic cell 24 h auxin-treated depletion samples. We compared the control strain CA1200, which has somatic TIR1 expression, with DG4703, which has an auxin-induced degron tag on SACY-1 combined with somatic TIR1 expression. We expected to find fewer events than in Tsukamoto et al. (2020), because in our workflow we use highly stringent cutoffs. We require 15% ΔPSI (change in percent spliced in) for each of the six pairwise comparisons between replicates (three replicates for DG4703 against two for CA1200) to identify alt. splicing events, and then visually inspect each alt. event and the sequencing reads on the UCSC Genome Browser (Nassar et al. 2023) to remove miscalled events (see Materials and Methods).
We identified 122 A3 events between somatic SACY-1-depleted versus nondepleted samples that meet our stringent criteria (Table 1; Supplemental Table 1). The primary difference between our results compared to the original analysis is that we found many fewer alt. splicing events using our analysis pipeline. We only found three skipped exon events and one retained intron event. Additionally, 45 of the 122 A3 events that we identified were not called in the original analysis. We noticed that the interval between the two splice sites in a pair was usually a multiple of 3 (Fig. 1A). It is possible that some alt. splicing events were not captured because they created a frameshift, causing those isoforms to have premature stop codons and be targeted by nonsense-mediated decay (Losson and Lacroute 1979) and therefore not recovered for sequencing.
TABLE 1.
Alt. splicing events by category
| Event type | CA1200 vs. DG4703 (Tsukamoto et al. 2020) | N2 vs. DG3430 [sacy-1(G533R)] | N2 vs. SZ454 [mog-5(Δ17 + 9)] |
|---|---|---|---|
| Alt. 3′ss (A3) | 122 | 210 | 76 |
| Alt. 5′ss | |||
| Skipped exon | 3 | 1 | 1 |
| Retained intron | 1 | ||
| Multiskip exon | |||
| Mut. Excl. exon | |||
| Alt. first exon | 1 | ||
| Alt. last exon |
FIGURE 1.

SACY-1 depletion increases the usage of proximal splice sites that poorly match the C. elegans consensus sequence. (A) Histogram. X-axis: relative position of the splice site that increases usage upon SACY-1 depletion compared to the other site of the pair; a negative number indicates shift in the 5′ (upstream or proximal) direction. (B) Sequence logo. The height of each base represents significant enrichment of nucleotide identity at that position over random chance, created on WebLogo (https://weblogo.berkeley.edu/) (Crooks et al. 2004). T is used rather than U because these splice site sequences are inferred from alignments with the genome rather than directly sequenced.
Two interesting patterns emerged that were not previously reported. First, 121 of the events featured increased usage of a proximal 3′ss, and only 1 event featured increased usage of a distal 3′ss, upon depletion (hereafter called the directional effect) (Fig. 1A). Second, the distal site usually matched the C. elegans 3′ss consensus sequence closely, whereas the proximal site did not; the proximal consensus consists of an AG dinucleotide with no other consensus pattern (Fig. 1B) (hereafter called the sequence preference effect). Strikingly, both these effects were also seen in alt. 3′ss choice in germline tissue relative to somatic tissue (Ragle et al. 2015). In addition, 36% of the 122 A3 events in the SACY-1 somatic depletion matched events that we previously showed are developmentally regulated in the germline.
These results show that SACY-1 has a specific role in 3′ss choice that was not previously reported. SACY-1 specifically decreases the usage of proximal splices that poorly match the consensus sequence, and the depletion of SACY-1 in somatic cells mimics to some extent the changes in A3 splicing that we previously identified in the germline.
sacy-1(G533R) increases proximal splice site usage of developmentally regulated alternative 3′ splice sites
Because of the confounding pleiotropic effects of a complete loss of SACY-1, we chose to study splicing changes caused by the sacy-1(G533R) mutation, which has a less severe phenotype than complete loss of function (Kim et al. 2012). In addition, the G533R mutation is in a region highly conserved with human DDX41 and adjacent to human disease allele R525H (Fig. 2A). We tested whether sacy-1(G533R) increases usage of proximal, developmentally regulated alt. 3′ splice sites with divergent sequences by performing reverse transcription-polymerase chain reactions (RT-PCR) followed by PAGE. We chose alt. splicing events from Ragle et al. (2015) in the genes icd-2 and lmd-1 to test if sacy-1(G533R) causes alternative splicing of at least a few of the same splice sites that are differentially spliced between germline and somatic tissues. The event in icd-2 included an especially divergent proximal splice site that lacks a canonical AG dinucleotide.
FIGURE 2.

sacy-1(G533R) increases proximal splice site usage of developmentally regulated alt. 3′ splice sites. (A) BLASTp alignment of C. elegans SACY-1 versus human DDX41 at the region around SACY-1(G533). (B) Flowchart of the experimental procedure. The hatched portion is the sequence for which inclusion requires proximal splice site choice. (C) Splicing assay of RNA from synchronized worms. RT-PCR products were run on 6% polyacrylamide denaturing gels. The strain and stage of RNA extraction are shown above each lane. The sequence of the alt. spliced 3′ss is shown to the right of each gel. Quantification and standard deviation are shown below (see Materials and Methods). Each sample shown is representative of three biological replicates; replicates are RNA extracted from independent worm samples grown under identical conditions.
We have previously developed a method to study the effects of a splicing mutation while minimizing germline-specific splicing changes (Suzuki et al. 2022) by studying only L3 worms. We collect synchronized embryos by bleaching gravid adults, allowing 34 h growth to reach the L3 larval stage, and then extracting RNA (Fig. 2B). The L3 stage is optimal for this assay as this stage is prior to germline expansion and these worms have minimal germline gene expression relative to somatic cells. This enables us to distinguish between expected normal developmental changes in alt. 3′ss usage in the germline and mutant-induced changes in somatic splicing. We also collect animals at 60 h postbleaching in order to obtain synchronized young adult RNA.
Comparing RNA samples extracted from WT animals at L3 versus adult, the WT samples show that the germline-specific splicing pattern is not detectable in L3s (Fig. 2C). For the event in icd-2, the adult WT sample shows increased proximal 3′ss usage. For the alt. splicing event in lmd-1, the adult WT does not show increased proximal splicing. A possible reason for this is that most of the lmd-1 mRNA in the samples may be from somatic tissue due to tissue-specific expression, so an increase in proximal splicing in this gene may only be visible using RNA from dissected gonads as in Ragle et al. (2015).
For both introns, sacy-1(G533R) increases usage of the proximal 3′ss relative to WT for the L3 samples (Fig. 2C). Thus, the sacy-1(G533R) reduction-of-function allele affects alt. 3′ss choice in the soma.
Increased proximal 3′ splice site usage is the predominant splicing effect of sacy-1 (G533R) on the transcriptome
To further study the effects of sacy-1(G533R) on splicing, we performed RNA-seq using synchronized mutant and WT L3 RNA. Three biological replicate libraries from each strain were analyzed to identify alt. splicing events. We called A3 and alt. 5′ (A5) splicing events de novo to identify previously unannotated alt. events (Suzuki et al. 2022). We also looked for all classes of alt. splicing events using existing annotations. Events that showed >15% ΔPSI in all nine pairwise comparisons between WT and sacy-1(G533R) libraries were flagged for further analysis. After verifying called events by hand, we identified 211 alt. splicing events between WT and sacy-1(G533R). All but one of these events are A3 events (Table 1; Fig. 3A), showing that the sacy-1(G533R) allele specifically affects 3′ss choice. This is consistent with the presence of its human homolog DDX41 in the C* complex, the complex in which the 3′ss is loaded into the spliceosome's active site after the first step of splicing.
FIGURE 3.
Overview of WT versus sacy-1(G533R) RNA-seq comparative splicing analysis. (A) Categories of alt. splicing events. Two hundred and eleven events are divided and subdivided into specific categories by the features described in the box (see Materials and Methods). (B) Sequence logo. The height of each base represents significant enrichment of nucleotide identity at that position over random chance, created on WebLogo (https://weblogo.berkeley.edu/) (Crooks et al. 2004). Note that for the sacy-1(G533R) proximal and distal sites, only the 206 events for which two adjacent sites were used could have their splice sites categorized as proximal or distal by definition.
Only 44 of the 122 A3 events we identified with SACY-1 depletion were also alt. spliced between WT and sacy-1(G533R). This likely reflects the functional difference between the full loss of SACY-1, which causes sterility, compared to the missense allele sacy-1(G533R), which is completely viable. However, some of the discrepancies may be due to differences in procedure between the two experiments, especially the difference in developmental staging at the time of RNA extraction, which would alter the relative expression of genes with alt. splicing and alter 3′ss choice patterns.
One hundred and eighty-seven of the A3 events featured a pair of splice sites in which sacy-1(G533R) increases usage of a proximal site that matches the C. elegans 3′ss consensus less closely than the pair's distal site (Fig. 3A,B). Most of the intervals are a multiple of 3 (Fig. 4A). To provide examples of the most common type of A3 events, two splice sites randomly chosen from this group of 187 events are shown (Fig. 4B). Both the directional effect and sequence preference effect are clearly associated with the sacy-1(G533R) mutation. We found 127 unused AG dinucleotides located ≤15 nt downstream from distal splice sites, so sacy-1(G533R) does not simply decrease sequence stringency to allow aberrant splicing in either direction (Fig. 3B). There are 14 instances of 2 AG dinucleotides being present ≤18 nt upstream of a distal site, and in only one case (hlh-11, discussed below) are both used. The 13 other AG dinucleotides are further upstream than a used proximal site; these are not used and have no sequence features to distinguish them. Two hundred and six of the events have ≤18 nt between a pair of used splice sites.
FIGURE 4.

Features of WT versus sacy-1(G533R) A3 events. (A) Histogram. X-axis: relative position of the splice site that increases usage with sacy-1(G533R) compared to the other site of the pair; negative number indicates 5′ direction. (B) Sequences of two splice site pairs randomly chosen from the 187 pairs where the distal site has a closer match to the consensus sequence than the proximal site. (C) Sequences of the only two events found where sacy-1 (G533R) increases usage of a distal splice site ≥27 nt away from the proximal site. (D) Sequences of the only two events found where three adjacent splice sites are used.
We studied whether the directional effect or the sequence preference effect was stronger. For 208 of the 210 A3 events, sacy-1(G533R) increases proximal splice site usage. If we define adjacent splice sites as ≤18 nt between a pair, then 100% of the adjacent splice site pairs show the directional effect. For 187 of the 206 events with two splice sites, sacy-1(G533R) increases usage of the splice site with a weaker match to the C. elegans 3′ss consensus sequence (see Materials and Methods), so only 90.8% of the adjacent splice site pairs show the sequence preference effect. Interestingly, for all 18 adjacent alt. 3′ss pairs for which neither splice site was obviously closer to the consensus sequence, the directional effect still occurs. Even in the one instance where the proximal splice site has a closer match to the consensus site than the distal splice site, the directional effect was still seen. Furthermore, we found 106 pairs of used alt. 3′ss with one or more additional AGs in the vicinity that are not used, compared to only two instances of three adjacent splice sites being used, showing that sacy-1(G533R) does not increase usage of all nearby AG dinucleotides, but predominantly increases AGs directly upstream of a potential splice site.
We conclude that sacy-1(G533R) causes the directional effect in alt. 3′ss usage and does not act through sequence preference. The sequence difference may be due to evolutionary selection against ambiguous splice sites, limiting the number of introns with two adjacent 3′ splice sites that match the consensus.
The sacy-1(G533R) directional effect is local
Four sacy-1(G533R) A3 events are outliers in terms of the distance between the used splice sites or in their splicing pattern (Fig. 4C,D). Splice sites in nucb-1, F23H11.2, and F40A3.2 are separated by distances of 42, 40, and 27 nt, respectively, whereas the remaining 207 have ≤18 nt between 3′ splice sites. The A3 events in nucb-1 and hlh-11 are also outliers in that they have three used 3′ splice sites. The F23H11.2 and F40A3.2 events are outliers in that they are the only two events we found in which sacy-1(G533R) increases the usage of a distal splice site.
In nucb-1, the middle and downstream splice sites are close together, and if these two sites are looked at in isolation, their splicing pattern follows the directional effect in which sacy-1(G533R) increases usage of a nearby proximal site. Interestingly, there is another AG dinucleotide that is not used 24 nt upstream of the middle splice site, yet this AG cannot be too upstream to be usable, because an AG even further upstream is used. This indicates that the directional effect of sacy-1(G533R) does not increase usage of a theoretically usable AG 24 nt away. The same pattern occurs in the F40A3.2 event. In the F23H11.2 event, if the directionality effect of sacy-1(G533R) had no distance limit, we would see increased usage of the upstream splice site, yet the opposite occurs. The splicing patterns at these three A3 events imply that the directional effect of sacy-1(G533R) has a distance limit; that is, the effect is local.
A possible explanation, consistent with the splicing patterns at these four events, is that nucb-1, F23H11.2, and F40A3.2 events have alt. BP choice, and then the directional effect occurs after BP choice. If an upstream BP is used, it may cause the upstream 3′ss to be used. If a downstream BP is used, the most upstream site would be upstream of the BP chosen for that molecule, and therefore be unusable. The event in hlh-11 may feature a BP that is unusually far upstream, and the directionality effect of sacy-1(G533R) may increase the usage of the AG that is closest to the BP.
Disruption of the MOG-5 region predicted to interface with SACY-1 causes an overlapping splicing phenotype with that of sacy-1 alleles
Dybkov et al. (2023) reported a cryo-EM structural model of the human C* complex, which includes the first modeling of DDX41 into the spliceosome. In this model, DDX41 and PRP22 contact each other, and DDX41 only has contact with the spliceosome through PRP22 (Fig. 5A). Because PRP22 is linked to proofreading the 3′ss (Mayas et al. 2006), and because sacy-1 and mog-5 both have alleles with a masculinization of germline (Mog) phenotype, we hypothesized that SACY-1 and MOG-5 cooperate to proofread against proximal splice sites.
FIGURE 5.
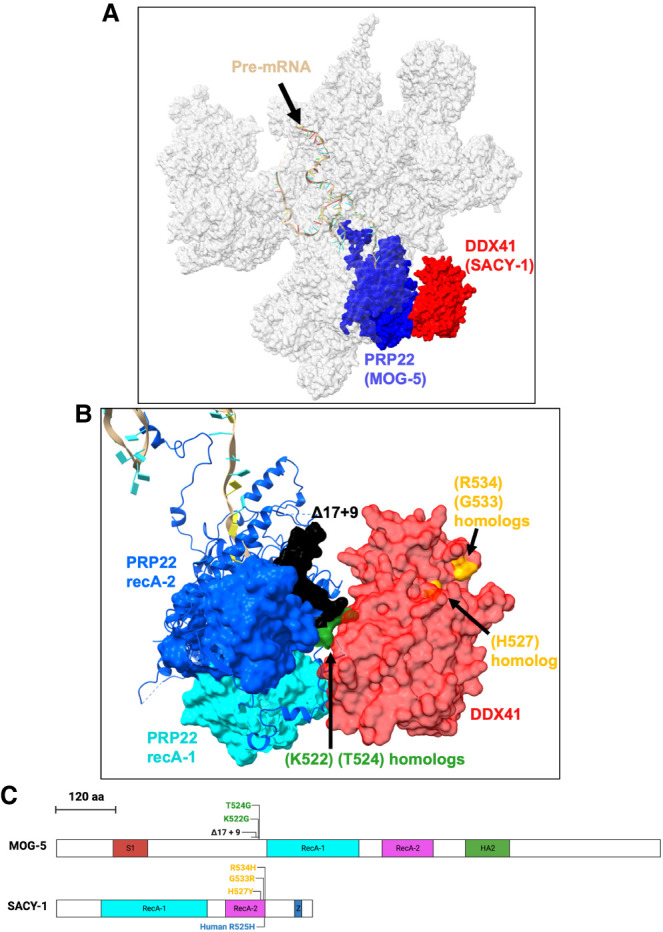
Diagram of sacy-1 and mog-5 mutations and the locations of their homologs in C*. (A) Model of C* structure based on coordinates from Dybkov et al. (2023), PDB ID 8C6J, image generated with ChimeraX (Pettersen et al. 2021). Shown are the pre-mRNA 3′ lariat intermediate (tan), PRP22 (blue), and DDX41 (red), and all other spliceosomal components are in transparent gray. (B) Closer view of PRP22/DDX41 interface showing the positions in human that correspond to C. elegans mutations, the same orientation as Figure 5A. Shown are homologs of SACY-1 (H527), (G533), and (R534) (yellow), MOG-5(K522, T524) (green), the region of amino acids that are altered in mog-5(Δ17 + 9) (black), the RecA-1 domain of PRP22 (light teal), and the RecA-2 domain of PRP22 (purple). (C) Protein structure diagram of SACY-1 and MOG-5. Scale bar shows the distance measured in amino acids for both proteins. Portions of amino acid sequences corresponding to domains are colored. Domain boundaries were taken from SMART (Letunic et al. 2021). Z stands for ZnF C2HC. Mutations in C. elegans are shown above the protein diagram, whereas a mutation in human is shown below.
There are amino acids in PRP22 that appear to interact with DDX41 and are very near both of the PRP22 recA domains (Fig. 5B). These regions are well conserved between PRP22 and MOG-5. To test if this interaction is involved 3′ss choice, we performed CRISPR/Cas9 genome editing to disrupt this interface in the C. elegans PRP22 homolog, MOG-5.
We obtained one mog-5 allele, az194, that matched our repair template; this allele has two missense mutations, K522G and T524G. We also obtained an allele resulting from nonhomologous end joining rather than the programmed homology-directed repair; this allele replaces the amino acid sequence between K508-T524 in mog-5, which is KEMPEWLKHVTAGGKAT, with the nine amino acids NIMEEIGSS (referred to as mog-5(Δ17 + 9)) creating a much stronger disruption of the interface. The homologous human residues for mog-5(K522, T524) and sacy-1(G533) and the 17 amino acids deleted in mog-5(Δ17 + 9) are shown in Figure 5B,C. Both of these mog-5 strains are viable and fertile as homozygotes. This contrasts with the two mog-5 alleles available from the Caenorhabditis Genetics Center that require maintenance over a balancer; mog-5(E608K) that is sterile due to the Mog phenotype (Graham et al. 1993), or the mog-5(ok1101) knockout allele (C. elegans Deletion Mutant Consortium 2012) that we confirmed causes developmental arrest before the worms reach adulthood for the homozygous mutant progeny of the balancer strain. Thus, mog-5(Δ17 + 9) and mog-5(K522G, T524G) alleles are hypomorphic and not full loss of function alleles. The large change in amino acid sequence caused by mog-5(Δ17 + 9) raises the possibility that it would cause misfolding and decrease stability. To test this possibility, we compared the existing AlphaFold prediction of SACY-1 (Jumper et al. 2021) to a newly generated AlphaFold prediction made using ColabFold (Mirdita et al. 2022) that includes the (Δ17 + 9) mutation. Regions of MOG-5 that are predicted with high confidence do not change their structure between the two predictions (Supplemental Fig. 1).
We tested whether these two new targeted mog-5 alleles cause changes to alt. 3′ss usage. We tested them by RT-PCR on 3 alt. splicing events chosen because they show a high ΔPSI in sacy-1(G533R) in the RNA-seq data. The alt. splicing event in dcp-66 was also alt. spliced upon SACY-1 depletion in our analysis of the data from Tsukamoto et al. (2020). For all three introns tested, mog-5 (Δ17 + 9) showed a strong increase in proximal splice site usage in L3 animals, but not quite to the same extent as sacy-1(G533R) (Fig. 6). The mog-5(K522G, T524G) mutant increased proximal splice site usage, but to a much lower degree. These results demonstrate that mog-5 and sacy-1 have an overlapping proofreading phenotype. Given the targeted nature of the new mog-5 alleles at the interaction site with sacy-1, we hypothesize that DDX41/SACY-1 functions in 3′ss choice by acting through PRP22/MOG-5.
FIGURE 6.

Disruption of the MOG-5 region predicted to interface with SACY-1 and the mimic of human (R525H) show overlapping proofreading phenotypes. Splicing assay of RNA from synchronized L3 populations. RT-PCR products were run on 6% polyacrylamide gels. The strains are indicated above each lane. The sequence of the alt. spliced 3′ss is shown to the right of each gel. Quantification and standard deviation from three independent experiments for each condition are shown below the gels.
A mimic of myelodysplastic syndrome-associated allele DDX41(R525H) increases proximal splice site usage
To study the human disease allele DDX41 (R525H), the C. elegans sacy-1(R534H) mimic allele was previously generated, but it was not studied for possible splicing phenotypes (Tsukamoto et al. 2020). Another sacy-1 allele in close proximity to G533 and R534 was previously isolated, sacy-1(H527Y). Human homologs of these three amino acids are shown in Figure 5B,C. These three sacy-1 alleles are hypomorphic and viable as homozygotes, whereas the full loss of function phenotype of sacy-1 is sterile with gamete degeneration (Tsukamoto et al. 2020).
DDX41 (R525H) is the most common somatic mutation in myeloid neoplasms that have DDX41 mutation (Makishima et al. 2023). R525H decreases, but does not abolish ATPase activity (Kadono et al. 2016) and unwinding activity (Singh et al. 2022), but it has not been linked specifically to 3′ss choice or proofreading. We tested if a homologous mutation to R525H, sacy-1(R534H), affects 3′ss choice by RT-PCR of L3 worm RNA. We also tested sacy-1(H527Y) to determine if various disruptions of this region of SACY-1 all cause increased proximal splice site choice. Both sacy-1(R534H) and sacy-1(H527Y) substantially increase proximal splice site choice relative to WT, with H527Y showing a stronger effect (Fig. 6).
A global phenotypic overlap between mog-5(Δ17 + 9) and sacy-1(G533R)
If mog-5(Δ17 + 9) is specifically linked to the SACY-1 function, there would be a strong overlap in the set of A3 events between mog-5(Δ17 + 9) and sacy-1(G533R). To find the degree of phenotypic overlap between the two alleles, we performed high-throughput RNA sequencing on WT and mog-5(Δ17 + 9) RNA extracted from L3 animals. We identified 76 A3 events that meet our criteria for alt. splicing (Table 1; Supplemental Table 1). In every event, mog-5(Δ17 + 9) increased usage of the proximal splice site. The splice site sequences were similar to those seen in both sacy-1 RNA-seq experiments (Fig. 7A). Fifty-three (70%) of these A3 events were also identified with sacy-1(G533R) (Fig. 7B). For 51 of the 53 overlapping A3 events, WT versus mog-5(Δ17 + 9) showed a smaller ΔPSI compared to WT versus sacy-1(G533R) (Fig. 7C). On average, mog-5(Δ17 + 9) showed a ΔPSI that was 21 percentage points lower than sacy-1(G533R). We show that the mog-5(Δ17 + 9) has a weaker splicing effect than sacy-1(G533R). We conclude that there is a substantial, transcriptome-wide phenotypic overlap between mog-5 and sacy-1.
FIGURE 7.

Global analysis of splicing phenotypic overlap between sacy-1(G533R) and mog-5(Δ17 + 9). (A) Sequence logo. The height of each base represents significant enrichment of nucleotide identity at that position over random chance, created on WebLogo (https://weblogo.berkeley.edu/) (Crooks et al. 2004). (B) Venn diagram. Numbers correspond to the number of overlapping A3 events between RNA-seq experiments. Image created on WebTools (https://bioinformatics.psb.ugent.be/webtools/Venn/). (C) Histogram. Difference in the strength of splicing change by sacy-1(G533R) and mog-5(Δ17 + 9). For every A3 event that was alt. spliced in both analyses, the average ΔPSI of all the pairwise comparisons within that event for mog-5(Δ17 + 9) was subtracted from the corresponding number for sacy-1(G533R).
The overlap between sacy-1(G533R) and mog-5(Δ17 + 9) A3 events was much stronger than the overlaps with the germline events from Ragle et al. (2015) and the SACY-1 depletion events from Tsukamoto et al. (2020) (Fig. 7B). Because many differences exist between these experiments, we cannot draw conclusions from the absence of overlap. The depletion data are from adult animals on auxin plates, and the germline-specific data are from gonads isolated by dissection. In the two new sequencing libraries prepared for this study, sacy-1(G533R) and mog-5(Δ17 + 9), the RNA was prepared from L3 staged worms under the exact same conditions, and these data showed a substantially overlapping splicing phenotype.
If sacy-1(G533R) completely ablated a proofreading mechanism, and if mog-5(Δ17 + 9) disrupted the exact same mechanism but to a lesser degree, the double mutant would have the same phenotype as sacy-1(G533R). To test this, we performed a cross, and found that worms carrying the sacy-1(G533R) I; mog-5(Δ17 + 9) II double mutation are sterile, falsifying that hypothesis. We then hypothesize that the Δ17 + 9 mutation in PRP22/MOG-5 leads to inefficient recruitment of DDX41/SACY-1. When inefficient recruitment is combined with a hypomorphic allele of DDX41/SACY-1 with reduced proofreading activity, the weak recruitment of a factor with reduced activity might lead to the additive synthetic sterile phenotype that appears similar to the sacy-1(null) phenotype.
DISCUSSION
Here, we show that mog-5 and sacy-1 have a phenotypic overlap in 3′ss choice. Both genes proofread against a large and substantially overlapping set of proximal 3′ splice sites, in favor of adjacent distal splice sites.
The precise location within the human C* spliceosome of an intron's path between the BP and a candidate 3′ss has recently been observed (Dybkov et al. 2023). Intriguingly, this intron loops around the carboxyl terminus of PRP22, and the BP-to-3′ss-distance could change the tension of this interaction, with a longer distance potentially causing a looser interaction. This suggests that PRP22 could sense whether the candidate 3′ss in the catalytic core is a short distance from the BP. We hypothesize that PRP22/MOG-5 can sense the short distance and then cooperate with DDX41/SACY- to proofread against that 3′ss. The protein NKAP interacts with the same portion of the intron as the PRP22 carboxyl terminus, and NKAP also interacts with Slu7. Slu7 prevents splicing of a 3′ss that is a short distance from the BP (Chua and Reed 1999), suggesting that these proteins may all be involved in a BP-to-3′ss-distance proofreading mechanism.
Because DDX41/SACY-1 does not have a homolog in S. cerevisiae, our results highlight a difference between S. cerevisiae and metazoan 3′ss choice mechanisms. It is possible that in metazoans, DDX41/SACY-1 assists or regulates PRP22/MOG-5 function in splice site choice, whereas in S. cerevisiae, Prp22 performs the analogous function by itself. Metazoans likely have additional splice site choice mechanisms to create more flexibility in alt. splicing, and to enable 3′ss choice with less conserved BP and 3′ss sequences than found in S. cerevisiae. In addition to DDX41/SACY-1, metazoans have four more helicases involved in splicing that are not conserved in S. cerevisiae (De Bortoli et al. 2021). In Schizosaccharomyces pombe, helicases Prp2 and Aquarius act sequentially to activate the spliceosome, whereas in S. cerevisiae, Prp2 performs activation without an Aquarius homolog (Schmitzová et al. 2023). The spliceosome may have divided helicase duties among an increasing number of helicases as it evolved, and PRP22 and DDX41 may likewise share a role in activating the second step of splicing.
DDX41/SACY-1 splicing function and its location in the C* complex have only recently been uncovered, so its mechanism of action is still unknown. Recent advances in understanding PRP22's dynamics between pre-C*-I, pre-C*-2, and C* (Zhan et al. 2022) suggest models of how DDX41/SACY-1 could work in conjunction with PRP22/MOG-5. It appears highly likely that the DDX41/PRP22 interaction is not compatible with all the conformations of PRP22 in these structures, so DDX41 binding might change the kinetics of the transitions between the conformations. DDX41/SACY-1 may drive the spliceosome toward a proofreading state, or it may hinder the transition to a catalytic state when an unfavored potential splice site is in the catalytic core. DDX41 binding may alter PRP22 ATPase kinetics. It is possible that DDX41/SACY-1 plays no role in splicing when the first potential 3′ss loaded into the catalytic core is favorable and proofreading is not needed. Because DDX41(R525H) decreases ATPase activity (Kadono et al. 2016) and unwinding activity (Singh et al. 2022), ATPase activity might be involved in the proofreading effect demonstrated here. To our knowledge, there are currently no data providing hints of any potential RNA substrate of DDX41 helicase activity during splicing.
Although human DDX41 proofreading function has not yet been found, this function is likely to be conserved with C. elegans SACY-1. There is a high level of sequence conservation between SACY-1 and DDX41, and the human structural interaction between DDX41 and PRP22 was recapitulated by the C. elegans phenotypic overlap between sacy-1 and mog-5. However, the siRNA-mediated knockdown in HeLa cells of many C* complex proteins, including DDX41, to study adjacent 3′ss NAGNAG splicing resulted primarily in skipped exon events, and DDX41 knockdown resulted in only a low percentage of A3 events (Dybkov et al. 2023). It may be that these NAGNAGs, alternative 3′ splice sites separated by only 3 nt, are too close together to be recognized by a BP-to-3′ss-distance proofreading mechanism. Also, it is likely that DDX41 knockdown in HeLa cells disrupted additional mechanisms besides the proofreading mechanism, which may have overshadowed the loss of a proofreading function.
Should the activation of proximal alternative 3′ splice sites in our data be defined as alternative splicing or aberrant splicing, or perhaps both? Given the unidirectional change in splicing in the germline and the mutants, proximal splicing usage could be considered the product of a compromised spliceosome. However, many of the proximal splice sites are biologically relevant to C. elegans because they receive significant usage in the germline, and for several events this alternative splicing is conserved between C. elegans and Caenorhabditis briggsae (Ragle et al. 2015). In addition, many of these proximal sites are used even by WT, somatic spliceosomes. We found 23 events in which the average usage of the proximal AG was >40% in the somatic tissue WT replicates. However, some of the proximal splice sites should be considered aberrant sites. For example, the gene ptb-1 has a proximal splice site that is not used in a single read in this work's novel WT samples despite ample coverage of the locus, and the event was not found to be developmentally regulated (Ragle et al. 2015). Many of the sacy-1(G533R) alt. 3′ splice sites have very low proximal usage in WT somatic tissue, and eight proximal sites are highly unusual in that they are preceded by something other than an AG dinucleotide. We did not find a clear way to demarcate the splice sites into alternative versus aberrant categories. However, defining these events based on a functional understanding provides a much clearer picture. All structural and genetic evidence implicates MOG-5 and SACY-1 functioning in the C* complex. This mechanism is clearly very distinct from the SR protein/hnRNP-based paradigm of regulated alternative splicing (Howard and Sanford 2015). We consider this a proofreading phenotype that is likely related to but potentially distinct from the currently understood proofreading functions of PRP22.
Our work demonstrates that C. elegans genetic studies can complement human biochemical and structural studies to aid the understanding of the fidelity of loading the 3′ss into the active site of the spliceosome. The rich collection of alt. adjacent 3′ splice sites that our laboratory has uncovered provides a useful variety of intriguing alt. splicing substrates. These substrates differ from human NAGNAG splicing substrates yet are now functionally linked to residues conserved in the human spliceosome. Given that mutations to sacy-1 and mog-5 in C. elegans lead to splicing changes in somatic cells that overlap with the changes we previously identified as specific to the germline, we hypothesize that the C. elegans germline naturally has an altered, C*-linked 3′ss proofreading mechanism compared to somatic tissue. These results naturally lead to the hypothesis that SACY-1 or MOG-5 have less expression in the germline, yet both genes and many other mRNA processing factors actually show increased mRNA expression in the germline relative to somatic tissue (Ragle et al. 2015; see their Supplemental Table S6). Studying this difference may provide insight into conserved proofreading mechanisms.
DDX41 plays multiple roles in human cells, and despite recent progress in studying DDX41 oncogenic perturbation, uncovering the pathogenic mechanisms of mutant DDX41 remains an important goal. Our results implicate a mimic of myelodysplastic syndrome-associated DDX41(R525H) in C*-linked 3′ss choice. We note that a subtle direct effect on a small number of 3′ splice sites could lead to many downstream phenotypes. This study draws attention to the hypothesis that mutations in DDX41 might contribute to pathogenesis by altering 3′ss choice.
MATERIALS AND METHODS
Strains used in this study
The genotypes and mutations found in C. elegans strains used in this study are listed in Table 2.
TABLE 2.
Strains used
| Strain | Genotype | Description |
|---|---|---|
| N2 | - - | Wild type |
| DG3430 | sacy-1(tn1385) I | G533R - - Viable, synthetic phenotypes (Kim et al. 2012) |
| DG3738 | sacy-1(tn1480) I | H527Y - - Viable, synthetic phenotypes (Tsukamoto et al. 2020) |
| DG4724 | sacy-1(tn1887) I | R534H - - Viable, synthetic phenotypes (Tsukamoto et al. 2020). Designed to mimic DDX41 human myeloid cancer-associated allele (R525H) |
| SZ454 | mog-5(az192) II | (17Δ9) - - Viable, this work |
| SZ456 | mog-5(az194) II | (K522G, T524G) - - Viable, this work |
| VC724 | mog-5 (ok1101) / mIn1 [mIs14 dpy-10 (e128)] II | Balancer strain, mIn1 Homozygotes Dumpy pharyngeal GFP signal, heterozygotes WT with GFP, mog-5 (ok1101) homozygotes late-larval arrest and sterile adult arrest - - Partial deletion of mog-5 - - Used as confirmation that mog-5 null is not viable (C. elegans Deletion Mutant Consortium 2012) |
Caenorhabditis elegans staging
Mixed staged worms were treated with bleach to isolate embryos in their egg shells for a rough synchronization of larval stages. See “Protocol 4. Egg prep” from Wormbook: Maintenance of C. elegans (http://www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html) (Stiernagle 2006). Worm embryos were plated on NGM agar with Escherichia coli as food and grown at 20°C. For L3 samples, we extracted RNA 34 h postbleaching, and for adult samples, we extracted RNA 60 h postbleaching.
RNA extraction
Staged worms were washed three times in 0.1 M NaCl to remove E. coli. Worms were gently pelleted, and the supernatant was removed to create a pellet of ∼50 µL. Worm pellets were flash frozen in liquid N2. Five hundred microliters of TRIzol were added to samples, vortexed, and incubated for 5 min at room temperature. One hundred microliters of CHCl3 were added to samples, vortexed, and incubated for 3 min at room temperature. Phases were separated by 15-min centrifugation at 13,000 rpm in a microcentrifuge. The aqueous phase was spin-column purified and DNase treated using Zymo RNA Clean and Concentrator Kit, following the manufacturer's instructions.
Library preparation and RNA sequencing
Azenta performed rRNA depletion followed by library preparation to create 150 × 150 paired-end reads. The WT versus sacy-1(G533R) libraries have strand-specific reads, whereas the SACY-1 somatic depletion (Tsukamoto et al. 2020) and WT versus mog-5(Δ17 + 9) libraries are not strand-specific.
RNA-seq analysis
Data were downloaded from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (accession number GSE144003) (Tsukamoto et al. 2020) or new high-throughput sequencing runs were performed by Azenta. Duplicates were removed, quality control analyses were performed, and reads were two-pass aligned to the UCSC Genome Browser (Nassar et al. 2023) C. elegans reference assembly (WS220/ce10) using STAR (Dobin et al. 2013).
For the data downloaded from GEO, we trimmed the 150 × 150 nt reads to 75 × 75 due to quality falloff. All five libraries had a least 11 million unique reads mapped and at least 16.5× average coverage. PCA analysis of this data set is shown in Tsukamoto et al. (2020).
For the 150 × 150 nt paired-end reads sequenced for this work, the average quality was high throughout the entire length of the reads, so no trimming was required. All 12 libraries prepared for this manuscript had at least 10 million uniquely mapped 150 × 150 paired-end reads and at least 30× average coverage. The reads had no 5′–3′ bias. PCA analysis was performed to check for replicate clustering (Supplemental Fig. 2). One WT replicate for the sacy-1(G533R) experiment was an outlier in PC2 but was not an outlier in PC1, and PC1 explained an unusually high amount of variation, so we decided to use that replicate after inspecting by eye that it was not a splicing outlier. One mog-5(Δ17 + 9) replicate (Supplemental Fig. 2) was an outlier and was excluded from further analysis.
We examined alt. first exon (AF), alt. last exon (AL), skipped exon (SE), retained intron (RI), mutually exclusive exon (MX) and multiple skipped exon (MS) events annotated in the Ensembl gene predictions Archive 65 of WS220/ce10 (EnsArch65), using the junctionCounts “infer pairwise events” function (https://github.com/ajw2329/junctionCounts).
We identified A3 and A5 alternative splicing events de novo from the sacy-1(G533R) and N2 libraries. To make our search for alternative events very open to events that have not been annotated, we made de novo lists of splice sites with any potential for A5 and A3 splicing. To do this, we combined all libraries from this experiment and identified exon junctions with at least five reads of support (total across all samples). By combining libraries in this process, it allowed us to look in the broadest way for splicing junctions, and we then applied stronger filters for A3 and A5 events described in the next paragraph. We then identified A3 and A5 events by looking for splice junctions that shared one end in common with an alt. 5′ss or 3′ss at the other end, with a maximum of 50 nt between the alt. splice sites (Suzuki et al. 2022). To identify A3 and A5 events in the SACY-1 depletion data from Tsukamoto et al. (2020), we used a list of potential splice sites created de novo previously (Suzuki et al. 2022). We then made a new list of potential splice sites using star mappings from the sacy-1(G533R) experiment. The SACY-1 depletion data and the WT versus mog-5(Δ17 + 9) data did not have strand-specific reads, so they were not used to generate lists of potential splice sites. The new list created for this work was used for both the WT versus sacy-1(G533R) and WT versus mog-5(Δ17 + 9) alt. splicing analysis.
For all events in all libraries, PSI corresponding to the proximal splice site usage was measured for each alternative event with at least 10 spanning reads in total for the isoforms in a library, and a ΔPSI comparing control and test sample was calculated. For the SACY-1 depletion data, we did six pairwise comparisons, two replicates of CA1200 (one replicate had RNA degradation before sequencing) versus three replicates of DG4703 with auxin treatment. For the sacy-1(G533R) data, we then did nine pairwise comparisons, three replicates of N2 versus three replicates of DG3430. For N2 versus mog-5(Δ17 + 9), we did six pairwise replicates, three N2 replicates versus two mog-5(Δ17 + 9) samples (one replicate was an outlier in PCA analysis, which may have indicated it was not staged properly, and so was excluded from splicing analysis). Those events with a >15% ΔPSI for all of the pairwise comparisons (pairSum = 6 or pairSum = 9 in the different experiments) were then analyzed by eye by viewing .bam tracks on the UCSC Genome Browser to verify all alt. splicing events reported in this manuscript. The events with three adjacent splice sites were found by eye, but the event calling pipeline and PSI calculations only assumed two splice sites. Supplemental Table 1 lists all confirmed A3 events for the three experiments described in the manuscript, their chromosomal location, information about splice site sequences, and the interval between the splice sites.
Splicing event subclassification
The subclassifications shown in Figure 2A were done by hand. The number of used splice sites in an event was determined by viewing aligned .bam reads at the locus of the alt. events. When a pair of adjacent AG splice sites was used, if one splice site had more bases at the -5 and -3 positions (TTTCAG) that matched the C. elegans 3′ss consensus sequence, it was binned as stronger. If one site of the pair was not an AG dinucleotide, it was considered weaker. If neither site of the pair was binned stronger by these checks, the event was put in the “Neither splice site has a significantly closer match to consensus sequence” category. We chose to emphasize the -5 position because there is functional evidence of its importance (Itani et al. 2016), and the -3 position because this base interacts with the catalytic core (Dybkov et al. 2023).
CRISPR/Cas9 genome editing
The az192 and az194 alleles of mog-5 were created using the CRISPR/Cas9 genome editing method described in Suzuki et al. (2022). Materials for this experiment and sequences of the new alleles are shown in Supplemental Table 2.
RT-PCR splicing assay
Reverse transcription was performed using AMV RT. PCR was performed with Cy3-labeled reverse primers and Phusion DNA Polymerase (New England Biolabs). Primer sequences are in Supplemental Table 3. PCR products were run on 6% polyacrylamide/urea denaturing gels for 2 h at 42W and imaged using a Typhoon scanner. Average % spliced upstream is calculated as (mean intensity of upper band − background)/[mean intensity of upper band plus the mean intensity of lower band − (background × 2)].
DATA DEPOSITION
Raw mRNA sequencing data for 12 libraries in fastq format, along with.gtf files for all analyzed alternative splicing events, are available at the NCBI Gene Expression Omnibus (GEO—https://www.ncbi.nlm.nih.gov/geo/), accession GSE245899.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We are grateful to Reinhard Luhrmann for sharing results and helpful discussions, and to David Greenstein for generously sharing strains. We thank Catiana Cartwright-Acar, Amy Leslie, and Diana Escalona for technical assistance. We are grateful to our colleagues Manny Ares, Susan Strome, Melissa Jurica, and Joshua Arribere for helpful discussions and comments on the manuscript. This research was funded by the National Institute of General Medical Sciences, R01GM135221, to A.M.Z. O.B. was supported as a fellow of the UCSC MARC Training Program, 5T34GM140956. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079888.123.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Kenneth Osterhoudt.

Meet the First Author(s) is an editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Kenneth Osterhoudt is the first author of this paper, “Spliceosomal helicases DDX41/SACY-1 and PRP22/MOG-5 both contribute to proofreading against proximal 3′ splice site usage.” Kenneth did this work as a PhD student in Alan Zahler's lab at UCSC.
What are the major results described in your paper and how do they impact this branch of the field?
We studied C. elegans mutations in spliceosomal helicases DDX41/SACY-1 and PRP22/MOG-5 that are targeted to disrupt their interaction, or that mimic a human cancer allele; all show increased usage of proximal adjacent 3′ splice sites. Human DDX41 was recently modeled into the C* complex (Dybkov et al. 2023), and our work provides functional results which help establish a role for DDX41/SACY-1 in the spliceosome, and which may be useful in forming hypotheses about how mutation of DDX41 contributes to pathogenesis.
What led you to study RNA or this aspect of RNA science?
My thesis work started with a search for mutations which could alter the splicing of the alternative 3′ splice sites that our laboratory previously discovered. When the Greenstein lab discovered that sacy-1 causes alternative 3′ splice site choice, I began studying that gene.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
While studying the splicing effects of sacy-1 mutation, I hypothesized that DDX41/SACY-1 may be involved in the proofreading activity of PRP22/MOG-5. I was already conducting experiments to test this hypothesis when we learned that the only modeled interaction between DDX41 and the spliceosome was through PRP22. I was then able to design a better test of this hypothesis by creating a targeted disruption based on the modeled interaction.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
I have been enchanted by science since I was a kid, and I loved Star Trek, The Magic School Bus, and Bill Nye the Science Guy in elementary school. When I started college, I did not think that being a scientist was a realistic career option for me. That changed when I was introduced to academic science through my friend's participation in the UCSC Stem Diversity program.
If you were able to give one piece of advice to your younger self, what would that be?
I would tell my younger self to focus your time on what you feel is most meaningful.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
Two researchers that especially inspire me are John Snow and Florence Nightingale. They were able to present their data with such clarity that the unsympathetic establishment could not deny their conclusions and were pushed to make changes that helped prevent huge amounts of suffering. They inspire me to simplify and to always present data in the clearest way possible, and to orient my research toward fighting disease.
What are your subsequent near- or long-term career plans?
I will continue my scientific career by starting a postdoc where I can follow my curiosity, continue to grow as a scientist, and gain new expertise.
REFERENCES
- Burgess SM, Guthrie C. 1993. A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391. 10.1016/0092-8674(93)90363-U [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium. 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2: 1415–1425. 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K, Reed R. 1999. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature 402: 207–210. 10.1038/46086 [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B, Colla S, et al. 2015. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep 13: 1033–1045. 10.1016/j.celrep.2015.09.053 [DOI] [PubMed] [Google Scholar]
- De Bortoli F, Espinosa S, Zhao R. 2021. DEAH-box RNA helicases in pre-mRNA splicing. Trends Biochem Sci 46: 225–238. 10.1016/j.tibs.2020.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybkov O, Preußner M, El Ayoubi L, Feng VY, Harnisch C, Merz K, Leupold P, Yudichev P, Agafonov DE, Will CL, et al. 2023. Regulation of 3′ splice site selection after step 1 of splicing by spliceosomal C* proteins. Sci Adv 9: eadf1785. 10.1126/sciadv.adf1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. 10.1038/nature12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Kimble J. 1993. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133: 919–931. 10.1093/genetics/133.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Schedl T, Kimble J. 1993. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev Genet 14: 471–484. 10.1002/dvg.1020140608 [DOI] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet 36: 1255–1257. 10.1038/ng1469 [DOI] [PubMed] [Google Scholar]
- Howard JM, Sanford JR. 2015. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA 6: 93–110. 10.1002/wrna.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itani OA, Flibotte S, Dumas KJ, Guo C, Blumenthal T, Hu PJ. 2016. N-Ethyl-N-Nitrosourea (ENU) mutagenesis reveals an intronic residue critical for Caenorhabditis elegans 3′ splice site function in vivo. G3 (Bethesda) 6: 1751–1756. 10.1534/g3.116.028662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadono M, Kanai A, Nagamachi A, Shinriki S, Kawata J, Iwato K, Kyo T, Oshima K, Yokoyama A, Kawamura T, et al. 2016. Biological implications of somatic DDX41 p.R525H mutation in acute myeloid leukemia. Exp Hematol 44: 745–754.e4. 10.1016/j.exphem.2016.04.017 [DOI] [PubMed] [Google Scholar]
- Kim S, Govindan JA, Tu ZJ, Greenstein D. 2012. SACY-1 DEAD-Box helicase links the somatic control of oocyte meiotic maturation to the sperm-to-oocyte switch and gamete maintenance in Caenorhabditis elegans. Genetics 192: 905–928. 10.1534/genetics.112.143271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, Bork P. 2021. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49: D458–D460. 10.1093/nar/gkaa937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R, Lacroute F. 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci 76: 5134–5137. 10.1073/pnas.76.10.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima H, Bowman TV, Godley LA. 2023. DDX41-associated susceptibility to myeloid neoplasms. Blood 141: 1544–1552. 10.1182/blood.2022017715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayas RM, Maita H, Staley JP. 2006. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13: 482–490. 10.1038/nsmb1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2022. ColabFold: making protein folding accessible to all. Nat Methods 19: 679–682. 10.1038/s41592-022-01488-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar LR, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, Fischer C, Gonzalez JN, Hinrichs AS, Lee BT, et al. 2023. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res 51: D1188–D1195. 10.1093/nar/gkac1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C. 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239. 10.1016/0092-8674(87)90564-2 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82. 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragle JM, Katzman S, Akers TF, Barberan-Soler S, Zahler AM. 2015. Coordinated tissue-specific regulation of adjacent alternative 3′ splice sites in C. elegans. Genome Res 25: 982–994. 10.1101/gr.186783.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitzová J, Cretu C, Dienemann C, Urlaub H, Pena V. 2023. Structural basis of catalytic activation in human splicing. Nature 617: 842–850. 10.1038/s41586-023-06049-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Blanco MR, Walter NG, Staley JP. 2016. Spliceosomal DEAH-Box ATPases remodel pre-mRNA to activate alternative splice sites. Cell 164: 985–998. 10.1016/j.cell.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinriki S, Hirayama M, Nagamachi A, Yokoyama A, Kawamura T, Kanai A, Kawai H, Iwakiri J, Liu R, Maeshiro M, et al. 2022. DDX41 coordinates RNA splicing and transcriptional elongation to prevent DNA replication stress in hematopoietic cells. Leukemia 36: 2605–2620. 10.1038/s41375-022-01708-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Ahmed D, Dolatshad H, Tatwavedi D, Schulze U, Sanchi A, Ryley S, Dhir A, Carpenter L, Watt SM et al. 2020. SF3B1 mutations induce R-loop accumulation and DNA damage in MDS and leukemia cells with therapeutic implications. Leukemia 34: 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RS, Vidhyasagar V, Yang S, Arna AB, Yadav M, Aggarwal A, Aguilera AN, Shinriki S, Bhanumathy KK, Pandey K, et al. 2022. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep 39: 110856. 10.1016/j.celrep.2022.110856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Porro EB, Patton JG, Nadal-Ginard B. 1989. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342: 243–247. 10.1038/342243a0 [DOI] [PubMed] [Google Scholar]
- Stiernagle T. 2006. WormBook, ed. The C. elegans Research Community (February 11, 2006), WormBook, doi/10.1895/wormbook.1.7.1, http://www.wormbook.org.
- Suzuki J, Osterhoudt K, Cartwright-Acar CH, Gomez DR, Katzman S, Zahler AM. 2022. A genetic screen in C. elegans reveals roles for KIN17 and PRCC in maintaining 5′ splice site identity. PLoS Genet 18: e1010028. 10.1371/journal.pgen.1010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Gearhart MD, Kim S, Mekonnen G, Spike CA, Greenstein D. 2020. Insights into the involvement of spliceosomal mutations in myelodysplastic disorders from analysis of SACY-1/DDX41 in Caenorhabditis elegans. Genetics 214: 869–893. 10.1534/genetics.119.302973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Charenton C, Nagai K. 2020. RNA splicing by the spliceosome. Annu Rev Biochem 89: 359–388. 10.1146/annurev-biochem-091719-064225 [DOI] [PubMed] [Google Scholar]
- Zhan X, Lu Y, Zhang X, Yan C, Shi Y. 2022. Mechanism of exon ligation by human spliceosome. Mol Cell 82: 2769–2778.e4. 10.1016/j.molcel.2022.05.021 [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. 10.1038/45597 [DOI] [PubMed] [Google Scholar]



