ABSTRACT
The trunk axial skeleton develops from paraxial mesoderm cells. Our recent study demonstrated that conditional knockout of the stem cell factor Sall4 in mice by TCre caused tail truncation and a disorganized axial skeleton posterior to the lumbar level. Based on this phenotype, we hypothesized that, in addition to the previously reported role of Sall4 in neuromesodermal progenitors, Sall4 is involved in the development of the paraxial mesoderm tissue. Analysis of gene expression and SALL4 binding suggests that Sall4 directly or indirectly regulates genes involved in presomitic mesoderm differentiation, somite formation and somite differentiation. Furthermore, ATAC-seq in TCre; Sall4 mutant posterior trunk mesoderm shows that Sall4 knockout reduces chromatin accessibility. We found that Sall4-dependent open chromatin status drives activation and repression of WNT signaling activators and repressors, respectively, to promote WNT signaling. Moreover, footprinting analysis of ATAC-seq data suggests that Sall4-dependent chromatin accessibility facilitates CTCF binding, which contributes to the repression of neural genes within the mesoderm. This study unveils multiple mechanisms by which Sall4 regulates paraxial mesoderm development by directing activation of mesodermal genes and repression of neural genes.
Keywords: Posterior trunk, Presomitic mesoderm, Somites, Sall4, WNT signaling, Chromatin accessibility
Summary: Sall4 regulates posterior trunk mesoderm development by promoting mesodermal gene expression while negatively regulating expression of WNT signaling repressors and neural genes within the mesoderm.
INTRODUCTION
The vertebrate body progressively develops from the anterior to the posterior during body elongation, in which new tissues are generated at the posterior end during the post-gastrulation stages (Henrique et al., 2015). Among mesodermal tissues generated during body elongation of the embryo, the paraxial mesoderm (PM) is located lateral to the neural tube and notochord. Complex genetic systems regulate PM development. In the posterior part of the PM, the tissue is unsegmented and is called the presomitic mesoderm (PSM). Two transcription factor genes, Msgn1 and Tbx6, act as crucial regulators of PM development. Msgn1 acts as a master regulator of PM development (Chalamalasetty et al., 2014; Yoon and Wold, 2000). Tbx6 represses Sox2 to inhibit neural development and specify the PM from bi-potential neuromesodermal progenitors (Chapman and Papaioannou, 1998; Takemoto et al., 2011). The PSM is more mature in the anterior, and periodic segmentation of the anterior end of the PSM leads to somite formation. This process is controlled by an interaction between the segmentation clock and the maturation wavefront (Hubaud and Pourquié, 2014). Specifically, Hes7 plays a central role in generating an oscillatory cycle of Notch signaling, which forms the segmentation clock (Bessho et al., 2003). In the wavefront system, a posterior-high to anterior-low gradient of WNT/β-catenin and fibroblast growth factor (FGF) signaling define specific stages of maturation of the PSM (Dequeant and Pourquié, 2008; Kageyama et al., 2012). The arrest of oscillation at the wavefront leads to boundary formation at the anterior edge of the PSM (Maroto et al., 2012; Pourquié, 2011). During a somite cycle, a complex regulatory system that involves Notch signaling, Tbx6, Mesp2 and Ripply2, regulates precise formation of the somite boundary during somitogenesis (Chan et al., 2007; Yasuhiko et al., 2006; Zhao et al., 2015).
Somites are initially generated as epithelial spheres, consisting of multipotent progenitor cells, which subsequently undergo morphogenetic changes (Brent and Tabin, 2002; Christ et al., 2007). On the ventral side, cells undergo an epithelial-to-mesenchymal transition to form the sclerotome, and subsequently migrate to form the vertebrae primordium. On the dorsal side, the cells in the dermomyotome edges transition to form the myotome (Gros et al., 2004), which contributes to skeletal muscle of the trunk and limbs. During somite differentiation, several genes are well characterized as specific musculoskeletal lineage markers. Among them, Pax9 is a chondrogenic lineage marker and Pax3 is a dermomyotomal myogenic marker (Goulding et al., 1994; Peters et al., 1999; Williams and Ordahl, 1994). Other PM/somite markers include Tcf15 (also known as Paraxis), which is required for somite epithelialization (Burgess et al., 1996). Meox1 is involved in somite morphogenesis, patterning and differentiation, particularly of sclerotome-derived structures (Mankoo et al., 2003; Skuntz et al., 2009). Aldh1a2 (also known as Raldh2) encodes a rate-limiting enzyme in the retinoic acid synthesis pathway. Retinoic acid, which is secreted from the somite, regulates forelimb initiation, somitogenesis and trunk neurogenesis (Cunningham et al., 2013; Diez del Corral et al., 2002; Dubrulle et al., 2001; Zhao et al., 2009).
Sall4 encodes a zinc-finger transcription factor (de Celis and Barrio, 2009; Sweetman and Münsterberg, 2006) that is highly and broadly expressed in the posterior part of the mouse embryo (Akiyama et al., 2015; Kohlhase et al., 2002; Tahara et al., 2018). Conditional knockout (cKO) of Sall4 in the mesoderm lineage and neuromesodermal progenitors using TCre (or Brachyury-Cre) causes tail truncation. Our previous study demonstrated that Sall4 is required for neuromesodermal progenitor maintenance and its differentiation into the mesodermal lineage at the expense of neural differentiation (Tahara et al., 2019). The TCre; Sall4 cKO mutants also exhibited disorganized vertebrae, posterior to the lumbar level (Akiyama et al., 2015; Tahara et al., 2019), which suggests a requirement of Sall4 for multiple aspects of the development of the axial skeleton in the posterior trunk and tail. For example, Sall4 may be involved in cell differentiation in the PSM, in somitogenesis and/or in differentiation of somites. However, our previous study focused on neuromesodermal progenitors, and roles of Sall4 in PM development remain unknown.
In this study, we have examined the expression patterns of genes involved in PM development in the posterior trunk in wild type (WT) and TCre; Sall4 cKO embryos. By revisiting our SALL4 ChIP-seq data (Tahara et al., 2019), we observed SALL4 enrichment in some genes involved in PM development and somite development. Moreover, ATAC-seq analysis of mesoderm tissue from the posterior trunk suggests that SALL4 promotes WNT/β-catenin signaling through regulating accessibility of a set of genes that modulates WNT/β-catenin signaling. Furthermore, our data suggest that Sall4 regulation of chromatin accessibility negatively regulates neural gene expression in the mesoderm. Taken together, our study supports the idea that Sall4 regulates posterior trunk mesoderm development through directly promoting transcription of mesodermal genes while inhibiting genes for WNT/β-catenin signaling repression and neural differentiation in the mesoderm.
RESULTS
Sall4 expression in the posterior trunk and tail
Several studies, including ours, reported the expression pattern of Sall4 during mouse embryonic development by mRNA in situ hybridization. Although Sall4 is broadly expressed in the posterior part of embryos at E9.5-E10.5, including the entire PSM and the somites, Sall4 exhibits a particularly strong expression domain around the PSM-somite boundary (Kohlhase et al., 2002; Tahara et al., 2018). In order to characterize this expression domain in more detail, we performed Sall4 and Uncx4 double in situ hybridization (Fig. 1A). Uncx4 is expressed in the posterior part of the somite and in the area S0, where the somite is forming at the anterior edge of the PSM (Mansouri et al., 1997). Strong Sall4 signals are detected in the entire S0, suggesting that Sall4 might play a role in somite formation (Fig. 1A′-A″″).
Fig. 1.
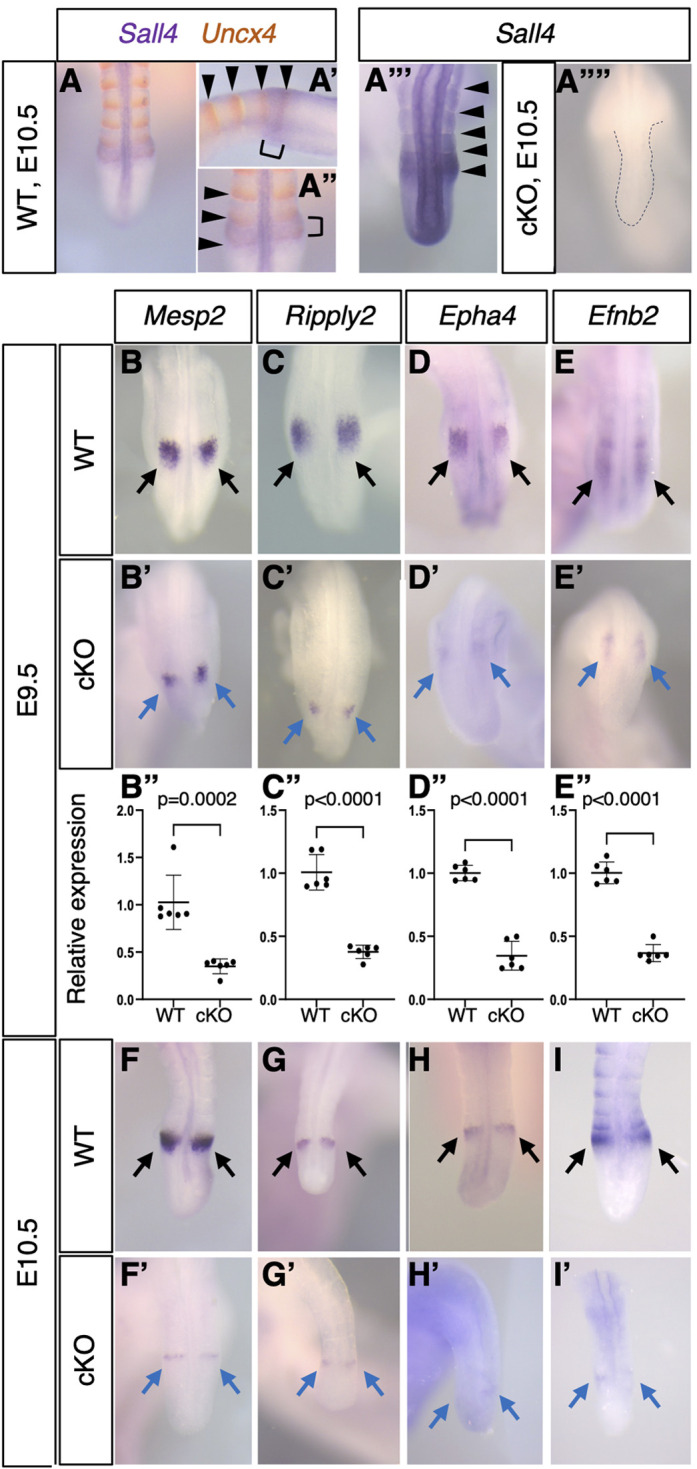
Gene expression patterns in the anterior part of PSM. (A-A″) Double color in situ hybridization of Uncx4 (orange) and Sall4 (purple) at E10.5 (n=3). The Sall4 detection reaction was stopped early to visualize a strong expression domain. A and A″ show dorsal views; A′ shows a lateral view. A′ and A″ show a more-detailed view of the somite-PSM boundary region in A. Arrowheads in A′ and A″ indicate the Uncx4 expression domain. Brackets in A′ and A″ indicate the next somite-forming region at the anterior PSM. (A‴,A⁗) Sall4 in situ hybridization in WT (A‴, n>30) and Sall4 cKO (A⁗, n=3) embryos at E10.5. Arrowheads in A‴ indicate the Sall4 expression domain. The dotted line in A⁗ depicts the tail morphology. (B-E′,F-I′) Expression pattern of Mesp2 [B (n=6), B′ (n=4), F (n=5), F′(n=3)], Ripply2 [C (n=6), C′ (n=5), G (n=6), G′ (n=6)], Epha4 [D (n=6), D′ (n=4), H (n=6), H′ (n=5)] and Efnb2 [E (n=6), E′ (n=4), I (n=6), I′ (n=4)] at E9.5 (B-E′) and E10.5 (F-I′). B-E and F-I show dorsal views of the posterior part of the body or tail in wild-type embryos. B′-E′ and F′-I′ show equivalent regions in Sall4 cKO embryos. Black and blue arrows indicate normal expression in wild type and reduced expression in mutants, respectively. B-E′ and F-I′ are at the same scale. (B″-E″) Relative expression levels of Mesp2 (B″), Ripply2 (C″), Epha4 (D″) and Efnb2 (E″) in the posterior trunk of wild-type and Sall4 cKO embryos at E9.5. Each dot represents an embryo. P values were obtained using an unpaired t-test.
At the anterior edge of the PSM, several genes are involved in the creation of a new pair of somites. We examined the expression pattern of such genes in wild-type and Sall4 cKO embryos. Although robust recombination by the TCre driver occurs as early as E7.5 in cells that contribute to the post-cranial mesoderm tissue (Akiyama et al., 2015; Perantoni et al., 2005), SALL4 protein depletion in the posterior body becomes evident at E9.5 and is more obvious at E10.5 (Tahara et al., 2019) (Fig. S1). Therefore, we examined gene expression at E9.5-E9.75 (24- to 28-somite stage; we refer to this stage as E9.5, for simplicity) and E10.5 (34- to 39-somite stage) by whole-mount in situ hybridization. Mesp2 and Ripply2 are expressed at the anterior PSM and are involved in somite boundary formation (Biris et al., 2007; Oginuma et al., 2008; Zhao et al., 2015). Expression of both Mesp2 and Ripply2 in Sall4 cKO embryos exhibited comparable spatial distribution to wild type but with reduced signal intensity at E9.5 (Fig. 1B,B′,C,C′). The Eph/ephrin signaling activity participates in the epithelialization of somite boundary at the anterior PSM (Watanabe et al., 2009). In Sall4 cKO embryos, the expression patterns of both Epha4 and Efnb2 were similar to wild type but the signal intensity was reduced at E9.5 (Fig. 1D,D′,E,E′). Quantitative analysis of transcripts in the trunk tissue posterior to the newest somite pair via qRT-PCR confirmed that expression of these genes was reduced in Sall4 cKO embryos (Fig. 1B″-E″). At E10.5, these genes exhibited considerably reduced signals (Fig. 1F,F′,G,G′,H,H′,I,I′).
To determine whether the expression of the above genes is directly regulated by SALL4, we re-visited our previously published data of SALL4 ChIP-seq using mesoderm tissue from the posterior part of the body at E9.5 (Tahara et al., 2019), which showed that ∼17% of SALL4 binding peaks are located at promoters. SALL4 is enriched around the transcription start site (TSS) to exon 1 of Mesp2, Ripply2, Epha4 and Efnb2 (Fig. S2, Tables S1 and S2), suggesting direct regulation of these genes by Sall4.
Sall4 regulates differentiation in PSM
Our previous study showed that Msgn1, a master regulator of PSM differentiation, was bound by SALL4 and was downregulated in Sall4 cKO embryos (Tahara et al., 2019), suggesting the involvement of Sall4 in regulation of PSM differentiation. Because Sall4 is expressed in the entire PSM (Tahara et al., 2018), we followed up this observation and examined other genes expressed in the PSM. Tbx6, another crucial regulator of PM differentiation, was significantly downregulated in Sall4 cKO embryos at E9.5 and was essentially undetectable at E10.5 (Fig. 2A,A′,F,F′). In ChIP-seq data, unlike Msgn1, we did not observe SALL4 enrichment at the Tbx6 gene, suggesting that this regulation is indirect. Expression of Delta1 (Dll1) and Notch1, which are crucial components of Notch signaling in the PSM, were downregulated but detectable in Sall4 cKO embryos at E9.5 (Fig. 2B,B′,C,C′) and E10.5 (Fig. 2G,G′,H,H′). Among the members of the Notch family and the Delta family, SALL4 enrichment is observed around the TSS of the Notch2 gene, introns of Notch3 and Notch4, and 5′ upstream of Dll1 (Table S1, Fig. S2). These results suggest that Sall4 is directly and indirectly involved in regulation of gene expression in the PSM.
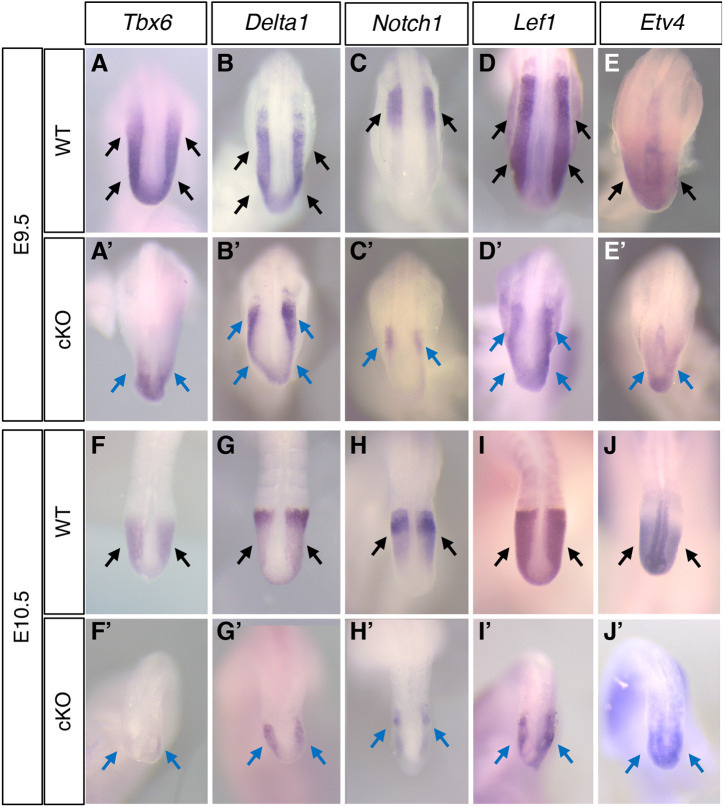
Fig. 2.
Gene expression patterns in the PSM. (A-J′) Expression patterns of Tbx6 [A (n=6), A′ (n=5), F (n=6), F′ (n=6)], Dll1 [B (n=6), B′ (n=3), G (n=6), G′ (n=4)], Notch1 [C (n=6), C′ (n=3), H (n=6), H′ (n=6)], Lef1 [D (n=4), D′(n=3), I (n=4), I′ (n=3)], and Etv4 [E (n=6), E′ (n=3), J (n=6), J′ (n=4)] at E9.5 (A-E′) and E10.5 (F-J′). A-E and F-J show dorsal views of the posterior part of the body or tail in wild-type embryos. B′-E′ and F′-I′ show equivalent regions in Sall4 cKO embryos. Black and blue arrows indicate normal expression in wild type (black) and reduced expression in Sall4 cKO embryos, respectively. A-E′ and F-J′ are at the same scale.
In the determination front model, a posterior-high to anterior-low gradient of WNT/β-catenin and FGF signaling is counteracted by an anterior-high to posterior-low gradient of retinoic acid signaling (Aulehla and Pourquié, 2010; Aulehla et al., 2008; Diez del Corral and Storey, 2004; Dubrulle et al., 2001; Sawada et al., 2001). We examined WNT/β-catenin and FGF signaling through the expression of genes regulated by these signaling pathways. Lef1 is a mediator of and a target of WNT/β-catenin signaling and is strongly expressed in the PSM. Lef1 expression in Sall4 cKO embryos was reduced in comparison with wild-type embryos, and the expression domain along the anterior-posterior axis was shorter than wild type (Fig. 2D,D′,I,I′). Etv4 is downstream of FGF signaling in the PSM. In addition to strong expression in the neural tube, Etv4 is expressed in the posterior PSM (Fig. 2E,J). Compared with wild-type embryos, Etv4 expression in Sall4 cKO embryos was detected in smaller domains in the posterior PSM at E9.5 and E10.5 (Fig. 2E′,J′). These expression patterns suggest that WNT/β-catenin signaling and FGF signaling are present, but their levels are reduced in Sall4 cKO PSM.
We also examined gene expression by qRT-PCR at E8.5, when Sall4 recombination by TCre was detectable in the trunk tissue (Fig. S3A) (Akiyama et al., 2015). We observed downregulation of Mesp2, Ripply2, Epha4 and Efnb2 in Sall4 cKO embryos, when compared with wild type (Fig. S3B-E). The degree of downregulation seems to be milder at E8.5 than E9.5 (Fig. 1B″-E″, Fig. S3B-E). This result suggests that levels of gene expression are reduced in the future thoracic levels at E8.5 in Sall4 cKO, but the skeletal defects become evident at the lumbar level, where downregulation of gene expression is more significant than future thoracic levels. Altogether, these results indicate that expression of genes involved in PSM differentiation is impaired in Sall4 cKO embryos and that the degree of reduction of gene expression correlates with posterior axial skeletal defects.
Sall4 regulates somite development
Next, we examined the expression of genes involved in somite development. The expression domain of Uncx4, a marker of posterior somites, seemed narrower along the medio-lateral axis in Sall4 cKO at E9.5 (Fig. 3A,A′). To clarify whether Uncx4-expressing domains are narrower in Sall4 cKO, we measured the width of the most posterior Uncx4-expressing domain and found that the domain was indeed narrower in Sall4 cKO than in wild type (Fig. S4A). The expression pattern of Uncx4 further narrowed along the medio-lateral and anterior-posterior axes, and the signal became weak in Sall4 cKO at E10.5 (Fig. 3G,G′). These expression patterns indicate severe reduction of the posterior trunk and tail size in Sall4 cKO embryos.
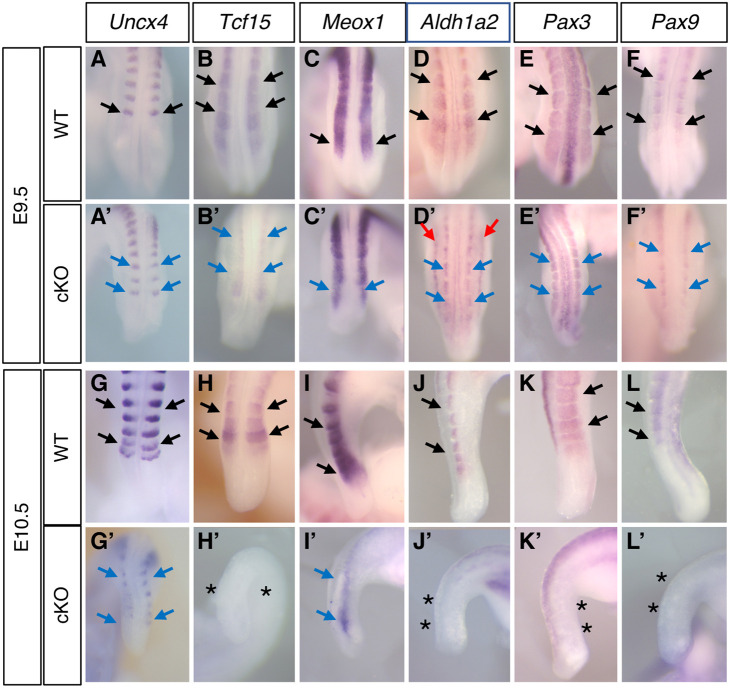
Fig. 3.
Gene expression patterns in the somites. (A-L′) Expression patterns of Uncx4 [A (n=6), A′ (n=3), G (n=6), G′ (n=4)], Tcf15 [B (n=6), B′ (n=5), H (n=6), H′ (n=4)], Meox1 [C (n=4), C′ (n=4), I (n=4), I′ (n=3)], Aldh1a2 [D (n=4), D′ (n=3), J (n=4), J′ (n=3)], Pax3 [E (n=4), E′ (n=3), K (n=4), K′ (n=4)] and Pax9 [F (n=4), F′ (n=3), L (n=4), L′ (n=3)] at E9.5 (A-F′) and E10.5 (G-L′). I-L and I′-L′ show lateral views; all other panels show dorsal views. Black and blue arrows indicate normal expression in wild type (black) and reduced expression in mutants, respectively. Red arrows in D′ indicate ectopic expression of Aldh1a2 in Sall4 cKO embryos. A-F′ and G-L′ are at the same scale. Asterisks indicate lack of expression.
Tcf15 is required for anterior-posterior polarity of somites and its loss causes disorganized vertebrae formation (Burgess et al., 1996; Johnson et al., 2001). In Sall4 cKO, Tcf15 expression is downregulated at E9.5 (Fig. 3B,B′) and was undetectable at E10.5 (Fig. 3H,H′). Meox1 is required for sclerotome polarity and axial skeleton formation (Mankoo et al., 2003; Skuntz et al., 2009). Meox1 expression at E9.5 was not severely affected in Sall4 cKO; however, its expression domain in the somite appears to be narrower than wild type along the medio-lateral axis (Fig. 3C,C′). At E10.5, Meox1 expression in the tail region was severely downregulated in Sall4 cKO embryos (Fig. 3I,I′). Aldh1a2 is expressed in somites (Molotkova et al., 2005), and its expression in Sall4 cKO embryos was restricted to the medial part of somites in Sall4 cKO embryos at E9.5 (Fig. 3D,D′). At E10.5, Aldh1a2 in the tail somites was undetectable (Fig. 3J,J′). Among these genes, SALL4 is enriched near the TSS of Tcf15 and Aldh1a2, and 2.3 kb from the TSS of Meox1 (Tables S1 and S2, Fig. S2), suggesting that SALL4 directly regulates these somite development genes. These results indicate that somites became smaller in Sall4 cKO embryos at E9.5, and that normal somite gene expression is either lost or severely impaired at E10.5.
Consistent with the change in the somite morphology in Sall4 cKO embryos, expression of dermomyotome marker Pax3 and sclerotome marker Pax9 was detected in a narrower region in Sall4 cKO embryos at E9.5 (Fig. 3E-F′, Fig. S4B,C). Expression of both Pax3 and Pax9 was undetectable at E10.5 (Fig. 3K-L′). These analyses support the idea that, as in the PSM, Sall4 regulates somite development, in part, by directly binding to those genes (Fig. S2).
Sall4 regulates trunk-tail transition and tail progenitors
The morphological defects of Sall4 cKO mutants are clearly distinguishable posterior to the lumbar vertebrae, whereas cervical and thoracic levels are unaffected (Tahara et al., 2019). During body elongation, neuromesodermal progenitors are located in the posterior part of the body and contribute to development of the post-cranial body (Aires et al., 2018; Henrique et al., 2015; Steventon and Martinez Arias, 2017; Wilson et al., 2009). Later, the neuromesodermal progenitors relocate to the tail bud and contribute to tail elongation (Aires et al., 2018; Wilson et al., 2009). The defects in Sall4 cKO mutants suggest that Sall4 may have a role in the trunk-tail transition and/or maintenance of neuromesodermal progenitors in the tail bud. These processes rely on a network of Gdf11, Lin28 and Hox13 genes (Aires et al., 2019; Robinton et al., 2019). At E9.5, expression of Gdf11 and Lin28a was downregulated in Sall4 cKO, compared with wild-type embryos (Fig. S5A-B″), and their downregulation was more evident in the tail bud at E10.5 (Fig. S5C-D″). Hoxb13 and Hoxc13 are also downregulated in the tail bud at E10.5 (Fig. S5E-F″). These results support the idea that the trunk-tail transition and neuromesodermal progenitor activity in the tail are disrupted in Sall4 cKO embryos, which may contribute to the defects in the posterior trunk and the tail.
Sall4 is required for accessible chromatin near a subset of genes
A recent study has shown chromatin accessibility analysis to be a useful strategy for investigating molecular and genetic mechanisms of somite development (Mok et al., 2021). To further understand how Sall4 regulates PM development, we examined genome-wide chromatin accessibility via ATAC-seq. We used the mesoderm-enriched fraction of the posterior trunk at E9.5, similar to our previous ChIP-seq experiment (Tahara et al., 2019), from Sall4 cKO and littermate Sall4 conditional heterozygous (cHet) embryos as a control.
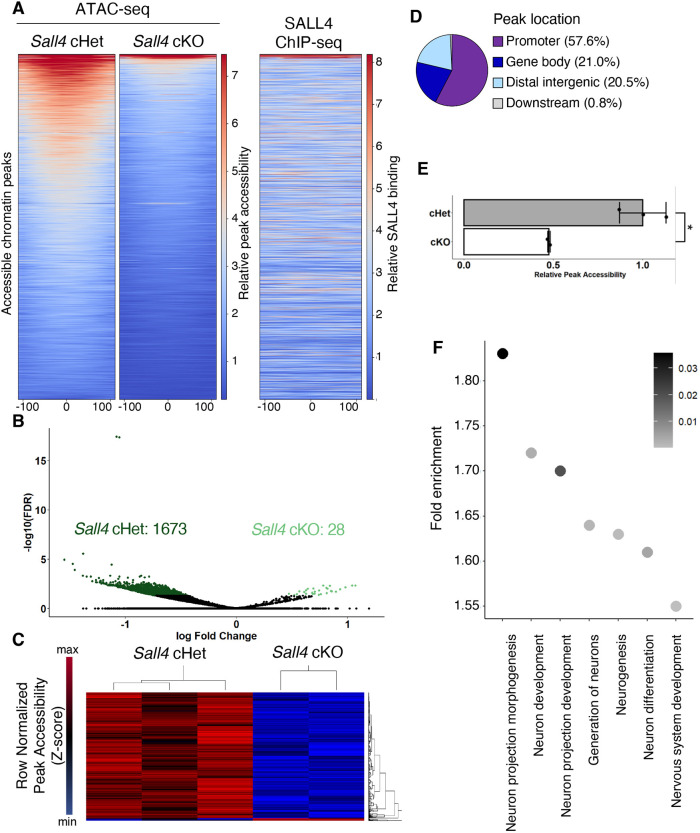
We found that 1673 genomic intervals are more accessible in cHet samples, whereas 28 sites are more accessible in cKO samples (Fig. 4A-C), indicating that Sall4 acts to promote an open chromatin status in the trunk mesoderm. Analysis of the sequences of the differentially accessible regions showed that a large proportion of those regions are located in promoters (57.6% within 2 kb upstream of the TSS to 500 bp downstream of the TSS) or gene bodies (21%) (Fig. 4D, Table S3), suggesting that Sall4 may regulate expression of these genes through changing accessibility for upstream transcription factors. Using deepTools, we scored the mean coverage density at the center of each differentially accessible region±100 bp as a measurement of accessibility (Ramírez et al., 2016). Relative accessibility was reduced ∼50% in Sall4 cKO samples, when compared with cHet samples (Fig. 4E). Comparison of the differentially accessible regions with our SALL4 ChIP-seq data showed that the SALL4 enrichment does not correlate with the differentially accessible regions (Fig. 4A) (Tahara et al., 2019). These data suggest that the differential accessibility in the majority of 1701 sites is a downstream consequence of Sall4-dependent genetic programs. GO analysis of the differential accessible regions included enrichment of multiple GO terms related to neural differentiation (Fig. 4F, Table S4).
Fig. 4.
Chromatin accessibility of mesoderm-enriched tissue from the posterior trunk. (A) Heatmap showing genome-wide accessibility in Sall4 cHet and Sall4 cKO samples, and SALL4 enrichment in the same genomic intervals. The x-axis corresponds to ±100 base pairs from the center of accessible regions (shown as 0). (B) Volcano plot showing differentially accessible genomic intervals between Sall4 cHet and Sall4 cKO samples. (C) Heatmap of the 1701 genomic intervals that showed differential accessibility between Sall4 cHet and Sall4 cKO samples. (D) A pie chart showing distribution of differentially accessible regions. (E) A bar graph showing relative accessibility score between Sall4 cHet and Sall4 cKO samples. *P=0.0186 (Welch′s two-sample t-test). (F) A dot plot showing enriched GOs related to neural development and functions. Gray shading indicates FDR.
Sall4 enhances WNT signaling by regulating chromatin accessibility near positive and negative regulators of WNT signaling
WNT/β-catenin signaling and FGF signaling are major regulators of PSM development (Dequeant and Pourquié, 2008; Kageyama et al., 2012). GO analysis of RNA-seq data obtained from wild-type and Sall4 cKO posterior trunk tissue (Tahara et al., 2019) showed that four WNT signaling-related GO terms are present in the top 100 GO terms (ranked at 3, 4, 5 and 73), whereas no GO terms related to FGF signaling appeared in the analysis (Table S5). Therefore, we examined the correlation between gene expression in the posterior trunk in the RNA-seq data and the current chromatin accessibility data of genes involved in WNT signaling.
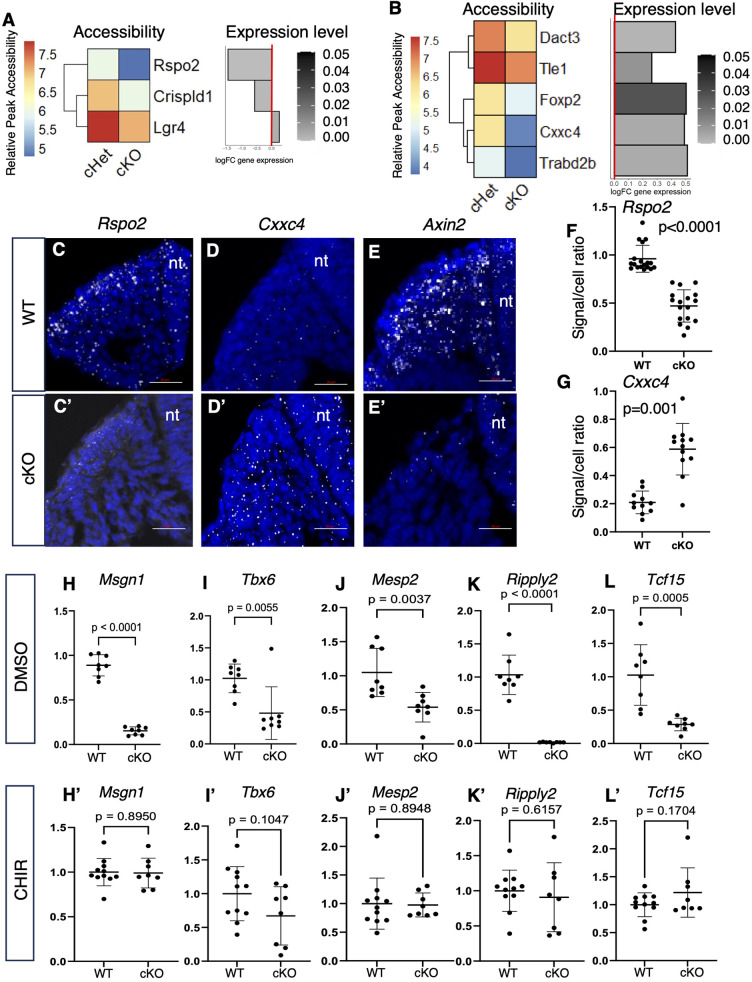
Given that Sall4 primarily promotes an open chromatin status (Fig. 4), we searched for genes whose chromatin status became less accessible in Sall4 cKO embryos. We found Rspo2 (de Lau et al., 2011; Glinka et al., 2011), Crispld1 (Khadjeh et al., 2020) and Lgr4 (de Lau et al., 2011; Glinka et al., 2011), genes known to be positive regulators of WNT signaling, became less accessible in Sall4 cKO (Fig. 5A). Among them, expression of Rspo2 and Crispld1 was downregulated in Sall4 cKO embryos in our RNA-seq data (Fig. 5A). Indeed, downregulation of Rspo2 expression was further supported by RNAscope (Fig. 5C,C′,F). This result suggests that SALL4 promotes expression of these WNT signaling activators by providing accessible TSS chromatin environment for upstream transcriptional activators.
Fig. 5.
WNT regulators with differential accessibility and differential expression between Sall4 cHet and Sall4 cKO. (A) Heatmap and bar chart showing differential accessibility and expression of genes involved in activating WNT signaling. Gray shading indicates FDR. (B) Heatmap and bar chart showing differential accessibility and expression of genes involved in negative regulation of WNT signaling. Gray shading indicates FDR. (C-E′) RNA scope images of expression of Rspo2 (C,C′), Cxxc4 (D,D′) and Axin2 (E,E′) in the PM of the posterior trunk in wild-type (C-E) and Sall4 cKO (C′-E′) embryos at E9.5. DAPI signals and transcript signals are shown in blue and white, respectively. nt, neural tube. Scale bars: 20 µm. (F,G) Quantification of Rspo2 (F) and Cxxc4 (G) signals. Each dot represents data from a section. The y-axis shows the ratio between Rspo2 or Cxxc4 signal number and DAPI-positive nuclei number in the PM of the section. Data obtained from five wild-type and five Sall4 cKO embryos. P values are indicated (unpaired t-test). (H-L′) Relative expression levels of Msgn1 (H,H′), Tbx6 (I,I′), Mesp2 (J,J′), Ripply2 (K,K′) and Tcf15 (L,L′) in cultured E10.5 tail of wild-type or Sall4 cKO embryos in the presence of DMSO (H-L) or CHIR (H′-L′). Each dot represents a cultured tail and P values are shown in each panel (unpaired t-test).
We also found that the chromatin became less accessible in Sall4 cKO embryos at sites near Dact3 (Jiang et al., 2008), Tle1 (Arce et al., 2009), Foxp2 (Richter et al., 2021), Cxxc4 (Hino et al., 2001) and Trabd2b (Zhang et al., 2012), which are known negative regulators of WNT signaling (Fig. 5B). Surprisingly, expression of these genes was upregulated in Sall4 cKO embryos in the RNA-seq data (Fig. 5B). Among these genes, we further examined expression of Cxxc4, which is located distal to a region that exhibited the highest fold change in chromatin accessibility we detected. Gene expression analysis by RNAscope showed upregulation of Cxxc4 in the PM (Fig. 5D,D′,G). These results indicate that SALL4 represses expression of these negative regulators of WNT signaling, and produces an accessible chromatin microenvironment at locations that may be of importance to transcriptional repressors.
Consistent with downregulation and upregulation of WNT signaling activators and repressors, respectively, expression of Axin2, which reports WNT/β-catenin signaling activity (Jho et al., 2002), was downregulated in the PM (Fig. 5E,E′). Collectively, these results suggest that Sall4-dependent chromatin accessibility contributes to promotion of WNT signaling.
In order to further examine whether regulation of WNT signaling by Sall4 plays a role in gene expression in the PSM and somites, we sought to test whether activation of WNT signaling could rescue gene expression in Sall4 cKO embryos. For this purpose, we dissected the tail from E10.5 embryos and set up ex vivo tail culture experiments, as previously performed by others (Fig. S6) (Bulusu et al., 2017). We compared responses to CHIR, which inhibits GSK3β and hence activates β-catenin signaling (Silva et al., 2008), at various culture durations. Expression of Axin2 and Lef1 was significantly elevated by CHIR treatment, compared with DMSO treatment, after 4 and 6 h (Fig. S6A,B), indicating that CHIR treatment for 4 h is sufficient to activate β-catenin signaling. With these data in mind, we cultured wild-type and Sall4 cKO E10.5 tail with DMSO or CHIR for 4 h in order to examine whether activation of β-catenin signaling can rescue gene expression defects in the Sall4 cKO tail (Fig. 5H-L′). Expression of Msgn1, Tbx6, Mesp2, Ripply2 and Tcf15 was downregulated in DMSO-treated Sall4 cKO tails compared with DMSO-treated wild-type tails (Fig. 5H-L), consistent with downregulation of these genes in the Sall4 cKO tail without culture (Figs 1, 2, 3) (Tahara et al., 2019). Treatment of cultured tail with CHIR showed that expression of these genes in Sall4 cKO was comparable with that of wild-type tails (Fig. 5H′-L′). This result indicates that activating β-catenin signaling can rescue gene expression in the Sall4 cKO tail and supports our notion that β-catenin signaling acts downstream of Sall4.
Sall4 negatively regulates neural genes in the posterior trunk mesoderm
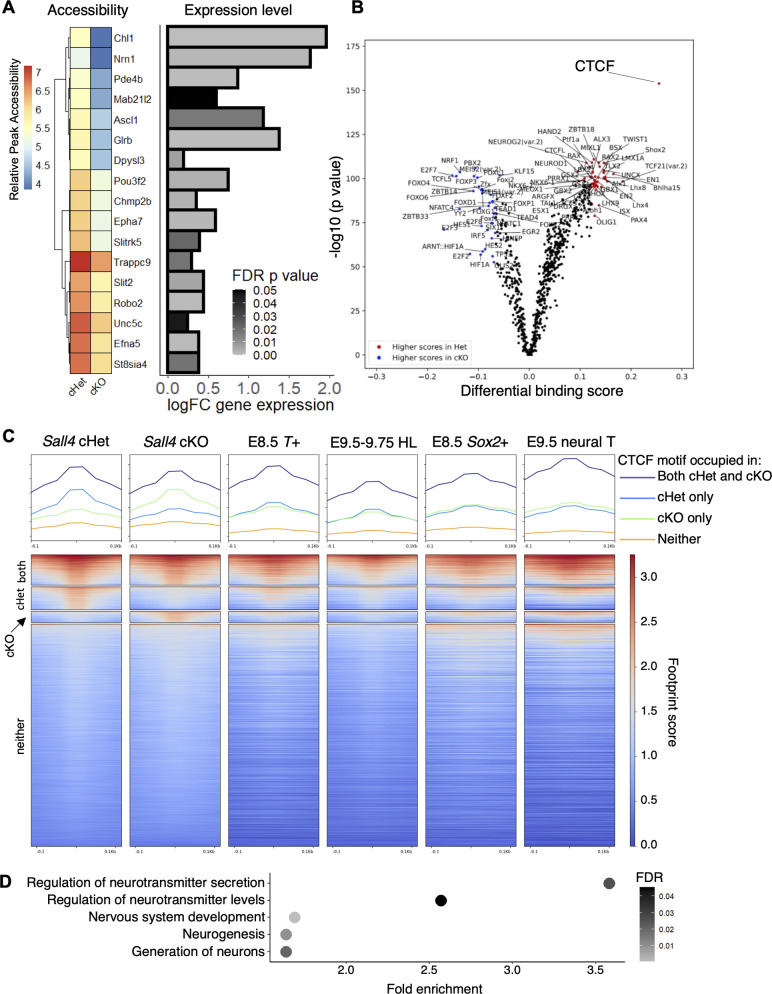
GO analysis of differentially accessible regions showed that GO terms involving neurogenesis and neuronal development were over-represented (Fig. 4F, Table S4). Interestingly, our previous analysis showed that expression of mesodermal and neural genes was downregulated and upregulated, respectively, in Sall4 cKO embryos (Tahara et al., 2019). Therefore, we compared differentially expressed mesodermal and neural genes with the chromatin accessibility data. Upon examination of the nearest genes to the 1701 differentially accessible regions, we did not find genes currently annotated as related to mesoderm development. However, we did find that 17 sequences are located close to genes whose functions are associated with neural lineage development, such as neurogenesis factor Ascl1 (Castro et al., 2011) (Fig. 6A). These genes are upregulated in Sall4 cKO embryos, and all 17 nearby differentially accessible regions are less accessible in Sall4 cKO embryos. These correlations suggest that Sall4 promotes accessibility of chromatin near these neural genes that contributes to repression of their expression.
Fig. 6.
Sall4 represses neural gene expression in mesoderm through regulation of chromatin accessibility. (A) Heatmap and bar chart showing differential accessibility and expression of genes related to neural differentiation. (B) Volcano plot of footprinting of differential accessible regions. The data point for CTCF is indicated. (C) Heatmap of inferred transcription factor occupancy at 11,443 occurrences of CTCF motifs in Sall4 cHet and Sall4 cKO samples. The heatmap shows, from the top to bottom, motifs occupied in both cHet and cKO, followed by motifs significantly more occupied in cHet, motifs significantly more occupied in cKO and motifs not occupied in either cHet or cKO. The x-axis corresponds to ±100 base pairs from the center of accessible regions (shown as 0). Heatmaps of the CTCF motifs of E8.5 T-expressing cells, E9.5-E9.75 nascent hindlimb bud cells (HL), E8.5 Sox2-expressing cells and E9.5 neural tube cells are also shown. (D) Dot plot showing enrichment of neural-related GO terms, obtained from a list of genes closest to the CTCF motifs preferentially bound in Sall4 cHet embryos.
Having characterized differential accessibility in the presence or absence of Sall4, we next wished to determine the activity of which DNA binding factor may be impacted by the altered chromatin landscape. To do this, we performed a genome-wide footprinting analysis with TOBIAS (Bentsen et al., 2020), which can detect evidence of factor binding by characterizing small areas of poor read coverage flanked by relatively accessible regions (Louise Smith et al., 2022; Sung et al., 2016). This analysis showed that some transcription factor-binding motifs exhibited higher occupancy in cHet or cKO samples, indicated by differential binding scores (Fig. 6B). Among all the motifs, the CTCF binding motif exhibited the highest score in cHet samples (Fig. 6B, Table S6), indicating that CTCF binding was strongly enriched in the presence of Sall4.
Although most of the 11,443 CTCF binding motifs in the genome were not occupied in both Sall4 cKO and Sall4 cHet samples, we detected 1570 sites occupied in both Sall4 cKO and Sall4 cHet samples (Fig. 6C, Table S6). We detected 1182 sites occupied in cHet but not cKO, and 569 sites occupied in cKO but not in cHet (Fig. 6C). To determine whether the observed occupancy of CTCF sites is specific to the posterior trunk mesoderm, we performed footprinting analysis of publicly available ATAC-seq data and compared them with our data. We found that CTCF occupancy in mouse embryonic stem cells (Metzis et al., 2018; Tafessu et al., 2023) and mouse embryonic fibroblasts (Wang et al., 2019) exhibited significantly different CTCF occupancy patterns when compared with posterior trunk mesoderm (Fig. S7). Therefore, we next compared our data with cells/tissues obtained from mouse embryos. T-expressing mesoderm progenitors from E8.5 embryos (Koch et al., 2017) and nascent hindlimb buds from E9.5-9.75 embryos, which are derived from the lateral plate mesoderm (Koyano-Nakagawa et al., 2022), shared some sites with detected CTCF occupancy in the posterior trunk mesoderm (Fig. 6C). However, we did not detect a significant correlation between CTCF binding in mesoderm progenitors/nascent hindlimb buds and that of the posterior trunk mesoderm (Fig. 6C). Specifically, areas with the strongest detected CTCF binding signal either only in Sall4 cHet or Sall4 cKO in the posterior trunk mesoderm appeared to show limited CTCF binding activity in mesoderm progenitors and nascent hindlimb buds. We also compared our data with Sox2-expressing neural progenitors from E8.5 embryos and E9.5 neural tube (Koch et al., 2017; Metzis et al., 2018), which also did not exhibit significant correlation to posterior trunk mesoderm (Fig. 6C). Some CTCF occupied sites are shared in E8.5 T+ cells, E8.5 Sox2+ cells, E9.5-9.75 hindlimb bud mesoderm, E9.5 neural tube and in the posterior trunk. These CTCF occupied areas might function commonly in different cell types for fundamental cellular activities. These results suggest that differential occupancy of CTCF binding sites between cHet and cKO is specific to the posterior trunk mesoderm. CTCF is crucial for proper regulation of gene expression and embryonic development, and CTCF is known to repress gene expression (Herold et al., 2012; Kim et al., 2015). Given our observation that Sall4-dependent chromatin accessibility is paired with reduced expression of nearby genes (Fig. 6A), we investigated the fraction of CTCF footprints differentially detected in Sall4 cHet and cKO. Gene ontology analysis of genes closest to these differentially occupied CTCF sites shows enrichment of neural differentiation genes (Fig. 6D, Table S7). These results and the well-characterized repressive function of CTCF collectively suggest that Sall4 contributes to PM development in the posterior trunk by repressing neural gene expression through regulation of CTCF insulator sequence binding.
DISCUSSION
In this study, we have investigated mechanisms by which Sall4 regulates PM development using genetic and genomic approaches. In situ hybridization experiments, coupled with RNA-seq and ChIP-seq data analysis, suggest that Sall4 has a variety of roles in PSM development. In the PSM, Sall4 contributes to regulation of WNT and FGF signaling, and helps to maintain the expression of genes involved in Notch signaling and somite boundary formation (Fig. 7A). Sall4 also promotes somite development (Fig. 7A). Furthermore, ATAC-seq analysis demonstrates that Sall4-dependent chromatin accessibility regulates activation and repression of a subset of genes encoding WNT signaling activators and repressors, respectively (Fig. 7B). Sall4-dependent chromatin accessibility also facilitates repression of neural differentiation gene expression in the PSM (Fig. 7C). These mechanisms act together to regulate posterior trunk development in the developing embryo.
Fig. 7.
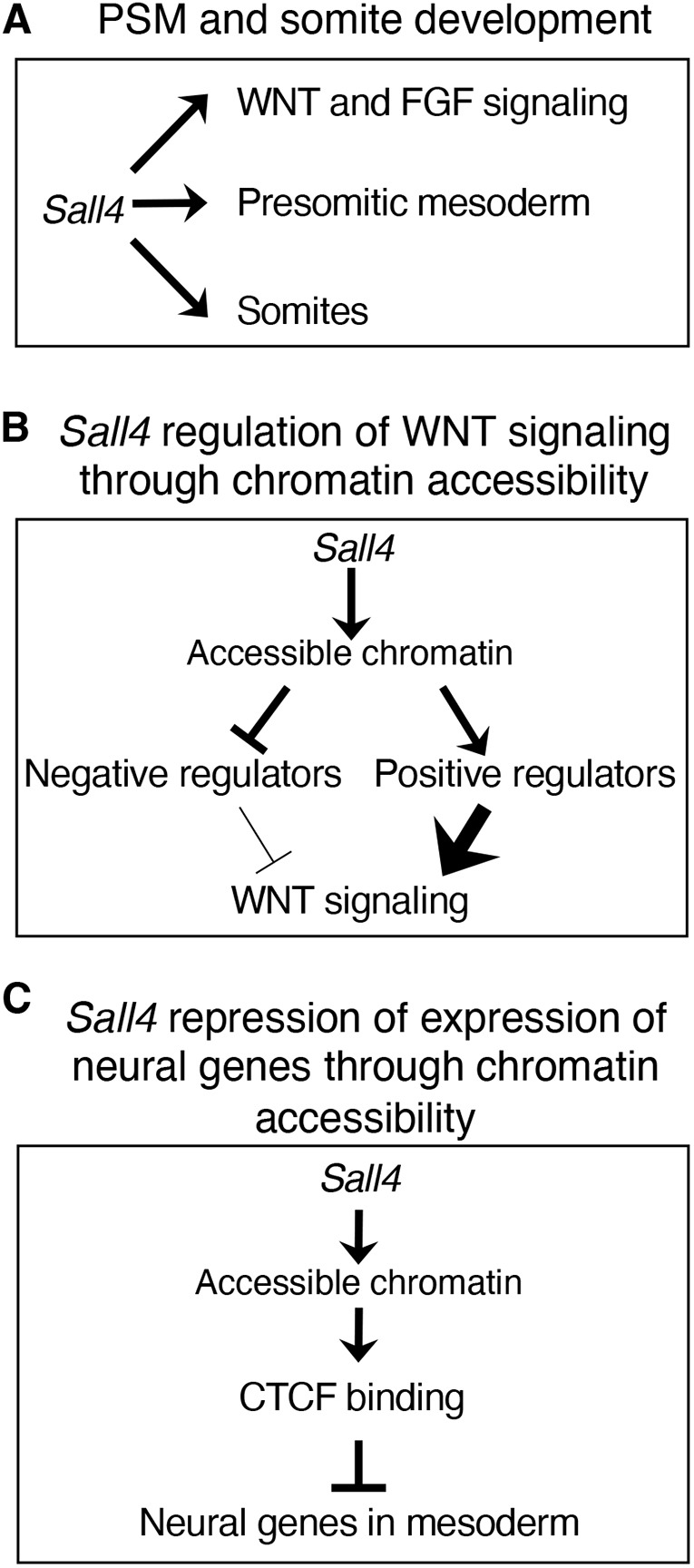
Models for Sall4 functions in PM development. (A) Sall4 contributes to multiple aspects of PSM development and somite development. (B) Sall4-dependent open chromatin status drives repression and activation of negative regulators and positive regulators, respectively, of WNT signaling. (C) Sall4-dependent open chromatin status contributes to repression of neural gene expression in the mesoderm, by facilitating CTCF binding.
Role of Sall4 in PM development
Deleting Sall4 by TCre caused severe axial skeleton defects in the posterior trunk and the tail. Analysis of the gene expression pattern showed defects in multiple genes that are expressed in specific developmental processes. Those include genes expressed in the PSM, during somitogenesis and during somite differentiation. The reduced expression of those genes suggests that Sall4 is broadly involved in PM development. In addition, by re-visiting our recent SALL4 ChIP-seq data, we found that several, but not all, analyzed genes were bound by SALL4 around the TSSs. This suggests that Sall4 directly and indirectly participates in regulation of PM development.
Gene expression analysis in Sall4 cKO embryos demonstrated that the defects were relatively mild at E9.5 and became more severe at E10.5. Several mechanisms could explain this observation. The timing of E9.5 to E10.5 correlates with the transition of trunk to tail development. A simple interpretation is that Sall4 has distinct roles in the development of the trunk and tail, which possess distinct developmental programs (Aires et al., 2019; Jurberg et al., 2013; Robinton et al., 2019). Sall4 function may be more significant in tail development than in posterior trunk development, as observed by downregulation of the Gdf11-Lin28-Hox13 system. A second interpretation is that Sall4 acts on axial skeleton development in a broad region, and the observed differences in the degree of gene expression defects reflect the timing of SALL4 protein depletion. Our previous analysis of SALL4 protein expression in Sall4 cKO embryos showed that SALL4 protein was depleted at E9.5-E10.5 in the posterior part of the body (Tahara et al., 2019), although TCre-mediated recombination occurs as early as E7.5 (Perantoni et al., 2005). The observed timing of the transition to severe defects correlates with this timing of depletion of SALL4, which suggests that milder defects at E9.5 may be due to residual SALL4 protein at this stage. A third mechanism involves possible redundancy with other members of the Sall gene family. For example, our previous study demonstrated that Sall1 and Sall3 redundantly regulate autopod development in mice (Kawakami et al., 2009). A Sall2 knockout; Sall4 gene trap allele exhibits increased neural tube closure defects when compared with Sall2 knockout in mouse embryos (Böhm et al., 2008), which also suggests functional redundancy among Sall genes. Moreover, Sall4+/−; Sall1+/− mice exhibit renal agenesis and anal stenosis, demonstrating genetic interactions between Sall genes (Sakaki-Yumoto et al., 2006). Additionally, Sall4−/−; Sall1−/− mouse embryonic stem cells exhibit more significant spontaneous upregulation of neural gene expression, compared with Sall4−/− or Sall1−/− embryonic stem cells (Miller et al., 2016). It is possible that other Sall members may partially compensate for the loss of Sall4, which might contribute to relatively mild defects at E9.5. These scenarios are not mutually exclusive; it is possible two or all the three scenarios simultaneously act in developing embryos.
Sall4-dependent chromatin accessibility regulates positive and negative regulators of WNT signaling
In addition to the analysis of RNA-seq and ChIP-seq data, our ATAC-seq results suggested additional mechanisms by which Sall4 regulates PM development through regulators of WNT signaling. High levels of WNT signaling in PSM maintains cells in an undifferentiated state (Aulehla and Pourquié, 2010). Our recent study of Sall4 cKO embryos demonstrated that Sall4 is required for promoting WNT signaling in neuromesodermal progenitors (Tahara et al., 2019). A reduced expression domain of Lef1 and downregulation of Axin2, which are targets of WNT signaling (Filali et al., 2002; Jho et al., 2002), suggest that WNT signaling is reduced in the PSM in Sall4 cKO embryos. Rescued expression levels of several genes expressed in the PSM or somites in the Sall4 cKO tail after CHIR treatment also supports the notion that β-catenin signaling acts downstream of Sall4 to regulate expression of PSM and/or somite genes.
Our previous analysis of neuromesodermal progenitors demonstrated downregulation of WNT signaling in Sall4 cKO embryos; however, how Sall4 promotes WNT signaling remained unknown. The analysis of chromatin accessibility in the posterior trunk mesoderm in Sall4 cHet and cKO led us to discover a correlation between upregulation of gene expression and less accessible chromatin status in several negative regulators of WNT signaling. Our observation suggests that Sall4-dependent chromatin accessibility contributes to an open chromatin status that would allow repressor complex access and negatively regulate these negative regulators of WNT signaling. Similarly, our data also suggest that Sall4-dependent chromatin accessibility contributes to activator complex access and upregulates some positive regulators of WNT signaling. WNT signaling is a crucial regulator of development of a variety of tissues and organs, and its level must be tightly regulated by positive and negative regulators (Clevers and Nusse, 2012; Steinhart and Angers, 2018). Sall4-dependent regulation of a set of regulators of WNT signaling may also operate in tissues with high levels of Sall4 expression.
Negative regulation of the neural program in the mesoderm tissue
Similar to WNT signaling, our data support the notion that Sall4 regulates a set of neural genes by creating an open chromatin microenvironment for upstream repressive factors in the posterior trunk mesoderm. Our ATAC-seq analysis uncovered an association between downregulated neural gene expression and CTCF-binding footprints near several neural differentiation genes in the posterior trunk mesoderm. Like the negative regulators of WNT signaling, Sall4 may repress neural genes by making regions of importance accessible to repressor proteins. Interestingly, Sall4 repression of neural gene expression has been also observed in other systems. Specifically, we have reported that Sall4 cKO by TCre caused expanded neural tissue at the expense of mesoderm development from neuromesodermal progenitors (Tahara et al., 2019). The same study also demonstrated an accelerated neural differentiation program evidenced by early expression of motor neuron marker ISL1 in the neural tube in Sall4 cKO embryos, when compared with wild-type embryos. Moreover, loss of Sall4 in mouse embryonic stem cells caused spontaneous upregulation of neural genes, whereas expression of pluripotency-related genes is maintained (Miller et al., 2016). In the study of mouse embryonic stem cells, it has been shown that SALL4 binds to putative enhancer sequences and blocks expression of differentiation-promoting genes (Miller et al., 2016; Ru et al., 2022). These reports and our current study indicate that restricting the neural program seems to be a common function of Sall4.
Sall4 is broadly and highly expressed in the neural tube (Kohlhase et al., 2002; Tahara et al., 2018); however, neural genes are expressed in the neural tube. This observation raises a question: does Sall4 have a role in neural cells? Our previous study demonstrated that Sall4 cKO in neuromesodermal progenitors by TCre caused accelerated neural differentiation in the neural tube in the posterior trunk (Tahara et al., 2019). This observation indicates that SALL4 also represses neural program in the neural tube, but it does not completely suppress high levels of neural gene network when compared with the mesoderm tissue. Such repression in neural cells may involve SALL4-dependent chromatin accessibility, as we observed in the mesoderm tissue in this study. It is also possible that SALL4 function may require cell type-specific molecular partners. For example, while SALL4 directly binds to AT-rich sequences in mouse embryonic stem cells (Pantier et al., 2021; Ru et al., 2022), SALL4 indirectly binds to DNA via its interaction partner PLZF in spermatogonial stem cells (Lovelace et al., 2016). The presence or absence of SALL4 molecular partners may explain the different degrees of repression of neural gene expression in mesodermal versus neural cells.
We highlighted differential CTCF binding of potential insulators near neural genes as one mechanism by which this regulation occurs. Footprinting analysis of ATAC-seq data indicated the highest score for CTCF motifs in the Sall4-dependent differentially accessible regions. Given that a major role of CTCF is regulating gene expression, the reduced CTCF score in Sall4 cKO embryos would be expected to change gene expression in Sall4 cKO embryos. This mechanism may contribute to changes of global gene expression in Sall4 cKO embryos, including neural genes in the posterior trunk mesoderm. These reports and our current study support the idea that Sall4 functions to negatively regulate neural development.
MATERIALS AND METHODS
Animal breeding and in situ hybridization
Embryos were collected by timed mating of Sall4fl/fl females and TCreTg/Tg; Sall4+/fl males (Akiyama et al., 2015). Animal breeding was performed according to the approval by the Institutional Animal Care and Use Committee of the University of Minnesota. After fixation in 4% paraformaldehyde in PBS-Tween 20 overnight, embryos were washed and dehydrated in graded series of PBT/methanol and stored in 100% methanol. Three to six embryos per probe per stage were examined by whole-mount in situ hybridization.
RNAscope
In situ hybridization was carried out with a RNAscope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostics). Embryos were fixed in 4% paraformaldehyde overnight at 4°C and cryosectioned at 14 µm. After the sections were dried, OCT compound around the sections was removed with 1×PBS for 3 min, followed by dehydration in an ascending ethanol series (50%, 70%, 100% and 100%). After applying hydrophobic pen around sections, slides were placed in 3% hydrogen peroxide in methanol for 15 min, followed by washes in water for 3 min. After this step, we used the manufacturer's protocol for fresh frozen sections, including the step with Protease IV at room temperature for 30 min. Hybridization was carried out with the mouse Axin2 probe (400331), mouse Cxxc4 probe (1184501-C2) and mouse Rspo2 probe (402001-C2 (all from Advanced Cell Diagnostics). Detection of the probes was performed with Opal dyes 520 (Axin2), 650 (Cxxc4) and 570 (Rspo2). Fluorescent images were acquired with a Zeiss LSM710 with Zen software.
Quantitative PCR analysis
E9.5 embryos were cut at the somite-PSM boundary using a carbon steel scalpel (10316-14, Fine Science Tools) and the posterior tissue was collected. E10.5 embryos were cut at the 4th and 5th somite boundary for collecting the tail tissue. E8.5 embryos were cut at the anterior edge of the first somite for collecting the trunk tissue. Total RNA was isolated using the Direct-zol RNA MicroPrep kit (Zymo Research) and cDNA was synthesized using iScript cDNA synthesis kit (BioRad) according to the manufacturers' instructions. qPCR was performed using PowerUp SYBR Green Master Mix (A25742, ThermoFisher) and primers in Table S8. Gene of interest expression was normalized to expression of the housekeeping gene Actb (β-actin).
Tail ex vivo culture
Mouse embryos were dissected in PBS with 1% FBS. The tail was cut at the 4th and 5th somite boundary from E10.5 embryos, and culture experiments were carried out according to a published procedure (Bulusu et al., 2017). The tails were placed on Transwell (3470, Corning), and cultured at the liquid-air interface. The culture medium was DMEM/F12, supplemented with 20% (v/v) rat serum and DMSO or CHIR99021 (10 µM, R&D Systems). The culture plate was incubated in a humidified chamber at 37°C in 5% CO2, 60% O2 and 35% N2.
ATAC-seq
Embryos at E9.5-9.75 were collected by timed mating of Sall4fl/fl females and TCreTg/Tg; Sall4+/fl males, and Sall4 cKO embryos were identified by short and thin posterior trunks. Sall4 cHet and cKO embryos were separately pooled and processed. Each embryo was genotyped by genomic PCR to confirm the genotype after the ATAC reaction. Tissues posterior to the boundary of PSM and the somite were collected and kept in PBS on ice during dissection. The dissected tissues were treated by dispase (1.5 mg/ml, Roche, 4942078001) at 37°C for 5 min, followed by removal of the neural tube using a tungsten needle in ice-cold PBS, as previously described (Tahara et al., 2019). The remaining tissue (the posterior mesoderm tissue) was dissociated by TrypLE (Invitrogen) at 37°C for 5 min, neutralized by DMEM+10% FBS, and collected by low-speed centrifugation (500 g for 3 min at room temperature). The ATAC reaction was performed as described previously (Buenrostro et al., 2015), and libraries were created using dual indexed Nextera reagent kits. The sequencing was performed by NextSeq Midoutput 2×75 bp Run and generated 18.4 M mean read-pairs per sample.
ATAC-seq data processing and analysis
Reads were trimmed using TrimGalore (0.6.0) (https://doi.org/10.5281/zenodo.5127899) and mapped to the GRCm38 genome using bwa mem (v 0.7.17) (https://doi.org/10.48550/arXiv.1303.3997). Mapped reads were sorted with samtools (v1.9) (Danecek et al., 2021) and duplicates were marked with Picard MarkDuplicates (2.18.16; https://broadinstitute.github.io/picard/). ATAC peaks were identified with MACS (v2.1.1.20160309; https://doi.org/10.5281/zenodo.3748809) using the following command line options: callpeak –broad -q 0.05 –nomodel –shift -100 –extsize 20.
Differential accessibility testing was performed with DiffBind using default parameters (Ross-Innes et al., 2012; Stark and Brown, 2011). Differentially accessible peaks were annotated using default parameters in ChIPpeakAnno (Zhu et al., 2010) with Ensembl annotation data from release 79 (Cunningham et al., 2022). Local chromatin accessibility was scored using the computeMatrix function in deepTools2 (Ramírez et al., 2016) via Galaxy (Afgan et al., 2018) on genomic regions of interest. Footprinting analysis was caried out with the TOBIAS snakemake pipeline (Bentsen et al., 2020) using transcription factor binding motifs from the JASPAR database (Fornes et al., 2020). GO analysis was performed using PANTHER (Mi et al., 2019; Thomas et al., 2022). The BioProject accession number of ATAC-seq in the GEO database is GSE217671.
ChIP-seq data processing and analysis
Sequencing data was obtained from the Sequence Read Archive database (BioProject accession number PRJNA525663). Reads were mapped to the mm10 genome with Bowtie2 (Langmead and Salzberg, 2012). Potential PCR duplicates were removed with Samtools rmdup (Danecek et al., 2021). Peaks were called on replicates with Macs2 (Jeon et al., 2020) and differential binding testing was carried out with DiffBind (Ross-Innes et al., 2012).
RNA-seq data processing and analysis
Sequencing data were obtained from the Sequence Read Archive database (BioProject accession number PRJNA525663). Analysis used the previously published CHURP pipeline (https://doi.org/10.1145/3332186.3333156). Briefly, quality control was carried out with fastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and reads were trimmed with Trimmomatic (Bolger et al., 2014). Hisat2 (Kim et al., 2019) was used to align reads to the GRCm38 genome. Aligned reads were sorted with SAMtools (Danecek et al., 2021) and gene counts were determined with featureCounts (Liao et al., 2014). Differential expression testing with edgeR revealed differentially expressed genes (Robinson et al., 2010).
Supplementary Material
Acknowledgements
We are grateful to Jennifer Kim, Nhi Hyunh, Naoyuki Tahara and Justin Wang for their excellent technical assistance. We are also grateful to Drs Juan Carlos Izpisúa Belmonte, Alexandra Joyner, Michael O'Connor, Virginia Papaioannou and Terry Yamaguchi for sharing materials or equipment. We acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this article.
Footnotes
Author contributions
Conceptualization: Y.K.; Formal analysis: M.P.P., D.C., E.P.S., Y.N., Y.K.; Investigation: M.P.P., H.K., D.C., K.Q.C., E.P.S., J.W., M.D.G., Y.N., Y.K.; Resources: R.N., Y.K.; Writing - original draft: M.P.P., Y.K.; Writing - review & editing: M.P.P., H.K., D.C., K.Q.C., E.P.S., J.W., M.D.G., R.N., Y.N., Y.K.; Supervision: Y.N., Y.K.; Project administration: Y.K.; Funding acquisition: Y.K.
Funding
This study was supported by grants from the National Institutes of Health to Y.K. (R01AR064195) and by a University of Minnesota, Grant-in-Aid of Artistry, Research and Scholarship (378836) to Y.K. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Open access funding provided by the University of Minnesota. Deposited in PMC for immediate release.
Data availability
Raw and processed data files have been deposited in GEO under accession number GSE217671.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.202649.reviewer-comments.pdf
References
- Afgan, E., Baker, D., Batut, B., van den Beek, M., Bouvier, D., Čech, M., Chilton, J., Clements, D., Coraor, N., Grüning, B. A.et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 46, W537-w544. 10.1093/nar/gky379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires, R., Dias, A. and Mallo, M. (2018). Deconstructing the molecular mechanisms shaping the vertebrate body plan. Curr. Opin. Cell Biol. 55, 81-86. 10.1016/j.ceb.2018.05.009 [DOI] [PubMed] [Google Scholar]
- Aires, R., de Lemos, L., Nóvoa, A., Jurberg, A. D., Mascrez, B., Duboule, D. and Mallo, M. (2019). Tail Bud progenitor activity relies on a network comprising Gdf11, Lin28, and Hox13 genes. Dev. Cell 48, 383-395.e388. 10.1016/j.devcel.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Akiyama, R., Kawakami, H., Wong, J., Oishi, I., Nishinakamura, R. and Kawakami, Y. (2015). Sall4-Gli3 system in early limb progenitors is essential for the development of limb skeletal elements. Proc. Natl. Acad. Sci. USA 112, 5075-5080. 10.1073/pnas.1421949112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce, L., Pate, K. T. and Waterman, M. L. (2009). Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer 9, 159. 10.1186/1471-2407-9-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulehla, A. and Pourquié, O. (2010). Signaling gradients during paraxial mesoderm development. Cold Spring Harb. Perspect. Biol. 2, a000869. 10.1101/cshperspect.a000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulehla, A., Wiegraebe, W., Baubet, V., Wahl, M. B., Deng, C., Taketo, M., Lewandoski, M. and Pourquié, O. (2008). A β-catenin gradient links the clock and wavefront systems in mouse embryo segmentation. Nat. Cell Biol. 10, 186-193. 10.1038/ncb1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen, M., Goymann, P., Schultheis, H., Klee, K., Petrova, A., Wiegandt, R., Fust, A., Preussner, J., Kuenne, C., Braun, T.et al. (2020). ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11, 4267. 10.1038/s41467-020-18035-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho, Y., Hirata, H., Masamizu, Y. and Kageyama, R. (2003). Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 17, 1451-1456. 10.1101/gad.1092303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biris, K. K., Dunty, W. C., Jr and Yamaguchi, T. P. (2007). Mouse Ripply2 is downstream of Wnt3a and is dynamically expressed during somitogenesis. Dev. Dyn. 236, 3167-3172. 10.1002/dvdy.21342 [DOI] [PubMed] [Google Scholar]
- Böhm, J., Buck, A., Borozdin, W., Mannan, A. U., Matysiak-Scholze, U., Adham, I., Schulz-Schaeffer, W., Floss, T., Wurst, W., Kohlhase, J.et al. (2008). Sall1, sall2, and sall4 are required for neural tube closure in mice. Am. J. Pathol. 173, 1455-1463. 10.2353/ajpath.2008.071039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A. M., Lohse, M. and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114-2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent, A. E. and Tabin, C. J. (2002). Developmental regulation of somite derivatives: muscle, cartilage and tendon. Curr. Opin. Genet. Dev. 12, 548-557. 10.1016/S0959-437X(02)00339-8 [DOI] [PubMed] [Google Scholar]
- Buenrostro, J. D., Wu, B., Chang, H. Y. and Greenleaf, W. J. (2015). ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.21-21.29.29. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulusu, V., Prior, N., Snaebjornsson, M. T., Kuehne, A., Sonnen, K. F., Kress, J., Stein, F., Schultz, C., Sauer, U. and Aulehla, A. (2017). Spatiotemporal analysis of a glycolytic activity gradient linked to mouse embryo mesoderm development. Dev. Cell 40, 331-341.e334. 10.1016/j.devcel.2017.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess, R., Rawls, A., Brown, D., Bradley, A. and Olson, E. N. (1996). Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature 384, 570-573. 10.1038/384570a0 [DOI] [PubMed] [Google Scholar]
- Castro, D. S., Martynoga, B., Parras, C., Ramesh, V., Pacary, E., Johnston, C., Drechsel, D., Lebel-Potter, M., Garcia, L. G., Hunt, C.et al. (2011). A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 25, 930-945. 10.1101/gad.627811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalamalasetty, R. B., Garriock, R. J., Dunty, W. C., Jr., Kennedy, M. W., Jailwala, P., Si, H. and Yamaguchi, T. P. (2014). Mesogenin 1 is a master regulator of paraxial presomitic mesoderm differentiation. Development 141, 4285-4297. 10.1242/dev.110908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, T., Kondow, A., Hosoya, A., Hitachi, K., Yukita, A., Okabayashi, K., Nakamura, H., Ozawa, H., Kiyonari, H., Michiue, T.et al. (2007). Ripply2 is essential for precise somite formation during mouse early development. FEBS Lett. 581, 2691-2696. 10.1016/j.febslet.2007.05.017 [DOI] [PubMed] [Google Scholar]
- Chapman, D. L. and Papaioannou, V. E. (1998). Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature 391, 695-697. 10.1038/35624 [DOI] [PubMed] [Google Scholar]
- Christ, B., Huang, R. and Scaal, M. (2007). Amniote somite derivatives. Dev. Dyn. 236, 2382-2396. 10.1002/dvdy.21189 [DOI] [PubMed] [Google Scholar]
- Clevers, H. and Nusse, R. (2012). Wnt/β-catenin signaling and disease. Cell 149, 1192-1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Cunningham, T. J., Zhao, X., Sandell, L. L., Evans, S. M., Trainor, P. A. and Duester, G. (2013). Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 3, 1503-1511. 10.1016/j.celrep.2013.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, F., Allen, J. E., Allen, J., Alvarez-Jarreta, J., Amode, M. R., Armean, I. M., Austine-Orimoloye, O., Azov, A. G., Barnes, I., Bennett, R.et al. (2022). Ensembl 2022. Nucleic Acids Res. 50, D988-d995. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., Whitwham, A., Keane, T., McCarthy, S. A., Davies, R. M.et al. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. 10.1093/gigascience/giab008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis, J. F. and Barrio, R. (2009). Regulation and function of Spalt proteins during animal development. Int. J. Dev. Biol. 53, 1385-1398. 10.1387/ijdb.072408jd [DOI] [PubMed] [Google Scholar]
- de Lau, W., Barker, N., Low, T. Y., Koo, B.-K., Li, V. S. W., Teunissen, H., Kujala, P., Haegebarth, A., Peters, P. J., van de Wetering, M.et al. (2011). Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293-297. 10.1038/nature10337 [DOI] [PubMed] [Google Scholar]
- Dequeant, M.-L. and Pourquié, O. (2008). Segmental patterning of the vertebrate embryonic axis. Nat. Rev. Genet. 9, 370-382. 10.1038/nrg2320 [DOI] [PubMed] [Google Scholar]
- Diez del Corral, R. and Storey, K. G. (2004). Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. BioEssays 26, 857-869. 10.1002/bies.20080 [DOI] [PubMed] [Google Scholar]
- Diez del Corral, R., Breitkreuz, D. N. and Storey, K. G. (2002). Onset of neuronal differentiation is regulated by paraxial mesoderm and requires attenuation of FGF signalling. Development 129, 1681-1691. 10.1242/dev.129.7.1681 [DOI] [PubMed] [Google Scholar]
- Dubrulle, J., McGrew, M. J. and Pourquié, O. (2001). FGF signaling controls somite boundary position and regulates segmentation clock control of spatiotemporal Hox gene activation. Cell 106, 219-232. 10.1016/S0092-8674(01)00437-8 [DOI] [PubMed] [Google Scholar]
- Filali, M., Cheng, N., Abbott, D., Leontiev, V. and Engelhardt, J. F. (2002). Wnt-3A/beta-catenin signaling induces transcription from the LEF-1 promoter. J. Biol. Chem. 277, 33398-33410. 10.1074/jbc.M107977200 [DOI] [PubMed] [Google Scholar]
- Fornes, O., Castro-Mondragon, J. A., Khan, A., van der Lee, R., Zhang, X., Richmond, P. A., Modi, B. P., Correard, S., Gheorghe, M., Baranašić, D.et al. (2020). JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87-D92. 10.1093/nar/gkz1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka, A., Dolde, C., Kirsch, N., Huang, Y. L., Kazanskaya, O., Ingelfinger, D., Boutros, M., Cruciat, C. M. and Niehrs, C. (2011). LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 12, 1055-1061. 10.1038/embor.2011.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding, M., Lumsden, A. and Paquette, A. J. (1994). Regulation of Pax-3 expression in the dermomyotome and its role in muscle development. Development 120, 957-971. 10.1242/dev.120.4.957 [DOI] [PubMed] [Google Scholar]
- Gros, J., Scaal, M. and Marcelle, C. (2004). A two-step mechanism for myotome formation in chick. Dev. Cell 6, 875-882. 10.1016/j.devcel.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Henrique, D., Abranches, E., Verrier, L. and Storey, K. G. (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864-2875. 10.1242/dev.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, M., Bartkuhn, M. and Renkawitz, R. (2012). CTCF: insights into insulator function during development. Development 139, 1045-1057. 10.1242/dev.065268 [DOI] [PubMed] [Google Scholar]
- Hino, S., Kishida, S., Michiue, T., Fukui, A., Sakamoto, I., Takada, S., Asashima, M. and Kikuchi, A. (2001). Inhibition of the Wnt signaling pathway by Idax, a novel Dvl-binding protein. Mol. Cell. Biol. 21, 330-342. 10.1128/MCB.21.1.330-342.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubaud, A. and Pourquié, O. (2014). Signalling dynamics in vertebrate segmentation. Nat. Rev. 15, 709-721. 10.1038/nrm3891 [DOI] [PubMed] [Google Scholar]
- Jeon, H., Lee, H., Kang, B., Jang, I. and Roh, T.-Y. (2020). Comparative analysis of commonly used peak calling programs for ChIP-Seq analysis. Genomics Informat. 18, e42. 10.5808/GI.2020.18.4.e42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho, E.-H., Zhang, T., Domon, C., Joo, C.-K., Freund, J.-N. and Costantini, F. (2002). Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22, 1172-1183. 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Tan, J., Li, J., Kivimäe, S., Yang, X., Zhuang, L., Lee, P. L., Chan, M. T. W., Stanton, L. W., Liu, E. T.et al. (2008). DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13, 529-541. 10.1016/j.ccr.2008.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J., Rhee, J., Parsons, S. M., Brown, D., Olson, E. N. and Rawls, A. (2001). The anterior/posterior polarity of somites is disrupted in paraxis-deficient mice. Dev. Biol. 229, 176-187. 10.1006/dbio.2000.9969 [DOI] [PubMed] [Google Scholar]
- Jurberg, A. D., Aires, R., Varela-Lasheras, I., Nóvoa, A. and Mallo, M. (2013). Switching axial progenitors from producing trunk to tail tissues in vertebrate embryos. Dev. Cell 25, 451-462. 10.1016/j.devcel.2013.05.009 [DOI] [PubMed] [Google Scholar]
- Kageyama, R., Niwa, Y., Isomura, A., González, A. and Harima, Y. (2012). Oscillatory gene expression and somitogenesis. Wiley Interdiscip. Rev. Dev. Biol. 1, 629-641. 10.1002/wdev.46 [DOI] [PubMed] [Google Scholar]
- Kawakami, Y., Uchiyama, Y., Rodriguez Esteban, C., Inenaga, T., Koyano-Nakagawa, N., Kawakami, H., Marti, M., Kmita, M., Monaghan-Nichols, P., Nishinakamura, R.et al. (2009). Sall genes regulate region-specific morphogenesis in the mouse limb by modulating Hox activities. Development 136, 585-594. 10.1242/dev.027748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadjeh, S., Hindmarsh, V., Weber, F., Cyganek, L., Vidal, R. O., Torkieh, S., Streckfuss-Bömeke, K., Lbik, D., Tiburcy, M., Mohamed, B. A.et al. (2020). CRISPLD1: a novel conserved target in the transition to human heart failure. Basic Res. Cardiol. 115, 27. 10.1007/s00395-020-0784-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Yu, N.-K. and Kaang, B.-K. (2015). CTCF as a multifunctional protein in genome regulation and gene expression. Exp. Mol. Med. 47, e166. 10.1038/emm.2015.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Paggi, J. M., Park, C., Bennett, C. and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907-915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, F., Scholze, M., Wittler, L., Schifferl, D., Sudheer, S., Grote, P., Timmermann, B., Macura, K. and Herrmann, B. G. (2017). Antagonistic activities of Sox2 and brachyury control the fate choice of neuro-mesodermal progenitors. Dev. Cell 42, 514-526.e517. 10.1016/j.devcel.2017.07.021 [DOI] [PubMed] [Google Scholar]
- Kohlhase, J., Heinrich, M., Liebers, M., Fröhlich Archangelo, L., Reardon, W. and Kispert, A. (2002). Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet Genome Res. 98, 274-277. 10.1159/000071048 [DOI] [PubMed] [Google Scholar]
- Koyano-Nakagawa, N., Gong, W., Das, S., Theisen, J. W. M., Swanholm, T. B., Van Ly, D., Dsouza, N., Singh, B. N., Kawakami, H., Young, S.et al. (2022). Etv2 regulates enhancer chromatin status to initiate Shh expression in the limb bud. Nat. Commun. 13, 4221. 10.1038/s41467-022-31848-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Y., Smyth, G. K. and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Louise Smith, E., Mok, G. F. and Münsterberg, A. (2022). Investigating chromatin accessibility during development and differentiation by ATAC-sequencing to guide the identification of cis-regulatory elements. Biochem. Soc. Trans. 50, 1167-1177. 10.1042/BST20210834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelace, D. L., Gao, Z., Mutoji, K., Song, Y. C., Ruan, J. and Hermann, B. P. (2016). The regulatory repertoire of PLZF and SALL4 in undifferentiated spermatogonia. Development 143, 1893-1906. 10.1242/dev.132761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankoo, B. S., Skuntz, S., Harrigan, I., Grigorieva, E., Candia, A., Wright, C. V. E., Arnheiter, H. and Pachnis, V. (2003). The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development 130, 4655-4664. 10.1242/dev.00687 [DOI] [PubMed] [Google Scholar]
- Mansouri, A., Yokota, Y., Wehr, R., Copeland, N. G., Jenkins, N. A. and Gruss, P. (1997). Paired-related murine homeobox gene expressed in the developing sclerotome, kidney, and nervous system. Dev. Dyn. 210, 53-65. [DOI] [PubMed] [Google Scholar]
- Maroto, M., Bone, R. A. and Dale, J. K. (2012). Somitogenesis. Development 139, 2453-2456. 10.1242/dev.069310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzis, V., Steinhauser, S., Pakanavicius, E., Gouti, M., Stamataki, D., Ivanovitch, K., Watson, T., Rayon, T., Mousavy Gharavy, S. N., Lovell-Badge, R.et al. (2018). Nervous system regionalization entails axial allocation before neural differentiation. Cell 175, 1105-1118.e1117. 10.1016/j.cell.2018.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan, A., Huang, X., Ebert, D., Mills, C., Guo, X. and Thomas, P. D. (2019). Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703-721. 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A., Ralser, M., Kloet, S. L., Loos, R., Nishinakamura, R., Bertone, P., Vermeulen, M. and Hendrich, B. (2016). Sall4 controls differentiation of pluripotent cells independently of the Nucleosome Remodelling and Deacetylation (NuRD) complex. Development 143, 3074-3084. 10.1242/dev.139113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok, G. F., Folkes, L., Weldon, S. A., Maniou, E., Martinez-Heredia, V., Godden, A. M., Williams, R. M., Sauka-Spengler, T., Wheeler, G. N., Moxon, S.et al. (2021). Characterising open chromatin in chick embryos identifies cis-regulatory elements important for paraxial mesoderm formation and axis extension. Nat. Commun. 12, 1157. 10.1038/s41467-021-21426-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkova, N., Molotkov, A., Sirbu, I. O. and Duester, G. (2005). Requirement of mesodermal retinoic acid generated by Raldh2 for posterior neural transformation. Mech. Dev. 122, 145-155. 10.1016/j.mod.2004.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oginuma, M., Niwa, Y., Chapman, D. L. and Saga, Y. (2008). Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis. Development 135, 2555-2562. 10.1242/dev.019877 [DOI] [PubMed] [Google Scholar]
- Pantier, R., Chhatbar, K., Quante, T., Skourti-Stathaki, K., Cholewa-Waclaw, J., Alston, G., Alexander-Howden, B., Lee, H. Y., Cook, A. G., Spruijt, C. G.et al. (2021). SALL4 controls cell fate in response to DNA base composition. Mol. Cell 81, 845-858.e848. 10.1016/j.molcel.2020.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perantoni, A. O., Timofeeva, O., Naillat, F., Richman, C., Pajni-Underwood, S., Wilson, C., Vainio, S., Dove, L. F. and Lewandoski, M. (2005). Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132, 3859-3871. 10.1242/dev.01945 [DOI] [PubMed] [Google Scholar]
- Peters, H., Wilm, B., Sakai, N., Imai, K., Maas, R. and Balling, R. (1999). Pax1 and Pax9 synergistically regulate vertebral column development. Development 126, 5399-5408. 10.1242/dev.126.23.5399 [DOI] [PubMed] [Google Scholar]
- Pourquié, O. (2011). Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell 145, 650-663. 10.1016/j.cell.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan, D. P., Grüning, B., Bhardwaj, V., Kilpert, F., Richter, A. S., Heyne, S., Dündar, F. and Manke, T. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160-W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, G., Gui, T., Bourgeois, B., Koyani, C. N., Ulz, P., Heitzer, E., von Lewinski, D., Burgering, B. M. T., Malle, E. and Madl, T. (2021). β-catenin regulates FOXP2 transcriptional activity via multiple binding sites. FEBS J. 288, 3261-3284. 10.1111/febs.15656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M. D., McCarthy, D. J. and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton, D. A., Chal, J., Lummertz da Rocha, E., Han, A., Yermalovich, A. V., Oginuma, M., Schlaeger, T. M., Sousa, P., Rodriguez, A., Urbach, A.et al. (2019). The Lin28/let-7 pathway regulates the mammalian caudal body axis elongation program. Dev. Cell 48, 396-405.e393. 10.1016/j.devcel.2018.12.016 [DOI] [PubMed] [Google Scholar]
- Ross-Innes, C. S., Stark, R., Teschendorff, A. E., Holmes, K. A., Ali, H. R., Dunning, M. J., Brown, G. D., Gojis, O., Ellis, I. O., Green, A. R.et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389-393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru, W., Koga, T., Wang, X., Guo, Q., Gearhart, M. D., Zhao, S., Murphy, M., Kawakami, H., Corcoran, D., Zhang, J.et al. (2022). Structural studies of SALL family protein zinc finger cluster domains in complex with DNA reveal preferential binding to an AATA tetranucleotide motif. J. Biol. Chem. 298, 102607. 10.1016/j.jbc.2022.102607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki-Yumoto, M., Kobayashi, C., Sato, A., Fujimura, S., Matsumoto, Y., Takasato, M., Kodama, T., Aburatani, H., Asashima, M., Yoshida, N.et al. (2006). The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133, 3005-3013. 10.1242/dev.02457 [DOI] [PubMed] [Google Scholar]
- Sawada, A., Shinya, M., Jiang, Y.-J., Kawakami, A., Kuroiwa, A. and Takeda, H. (2001). Fgf/MAPK signalling is a crucial positional cue in somite boundary formation. Development 128, 4873-4880. 10.1242/dev.128.23.4873 [DOI] [PubMed] [Google Scholar]
- Silva, J., Barrandon, O., Nichols, J., Kawaguchi, J., Theunissen, T. W. and Smith, A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, e253. 10.1371/journal.pbio.0060253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuntz, S., Mankoo, B., Nguyen, M.-T. T., Hustert, E., Nakayama, A., Tournier-Lasserve, E., Wright, C. V. E., Pachnis, V., Bharti, K. and Arnheiter, H. (2009). Lack of the mesodermal homeodomain protein MEOX1 disrupts sclerotome polarity and leads to a remodeling of the cranio-cervical joints of the axial skeleton. Dev. Biol. 332, 383-395. 10.1016/j.ydbio.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R. and Brown, G. (2011). DiffBind: differential binding analysis of ChIP-Seq peak data. 10.18129/B9.bioc.DiffBind [DOI]
- Steinhart, Z. and Angers, S. (2018). Wnt signaling in development and tissue homeostasis. Development 145, dev146589. 10.1242/dev.146589 [DOI] [PubMed] [Google Scholar]
- Steventon, B. and Martinez Arias, A. (2017). Evo-engineering and the cellular and molecular origins of the vertebrate spinal cord. Dev. Biol. 432, 3-13. 10.1016/j.ydbio.2017.01.021 [DOI] [PubMed] [Google Scholar]
- Sung, M.-H., Baek, S. and Hager, G. L. (2016). Genome-wide footprinting: ready for prime time? Nat. Methods 13, 222-228. 10.1038/nmeth.3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman, D. and Münsterberg, A. (2006). The vertebrate spalt genes in development and disease. Dev. Biol. 293, 285-293. 10.1016/j.ydbio.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Tafessu, A., O'Hara, R., Martire, S., Dube, A. L., Saha, P., Gant, V. U. and Banaszynski, L. A. (2023). H3.3 contributes to chromatin accessibility and transcription factor binding at promoter-proximal regulatory elements in embryonic stem cells. Genome Biol. 24, 25. 10.1186/s13059-023-02867-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara, N., Kawakami, H., Zhang, T., Zarkower, D. and Kawakami, Y. (2018). Temporal changes of Sall4 lineage contribution in developing embryos and the contribution of Sall4-lineages to postnatal germ cells in mice. Sci. Rep. 8, 16410. 10.1038/s41598-018-34745-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara, N., Kawakami, H., Chen, K. Q., Anderson, A., Yamashita Peterson, M., Gong, W., Shah, P., Hayashi, S., Nishinakamura, R., Nakagawa, Y.et al. (2019). Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos. Development 146, dev177659. 10.1242/dev.177659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, T., Uchikawa, M., Yoshida, M., Bell, D. M., Lovell-Badge, R., Papaioannou, V. E. and Kondoh, H. (2011). Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394-398. 10.1038/nature09729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P. D., Ebert, D., Muruganujan, A., Mushayahama, T., Albou, L. P. and Mi, H. (2022). PANTHER: making genome-scale phylogenetics accessible to all. Protein Sci. 31, 8-22. 10.1002/pro.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B., Wu, L., Li, D., Liu, Y., Guo, J., Li, C., Yao, Y., Wang, Y., Zhao, G., Wang, X.et al. (2019). Induction of pluripotent stem cells from mouse embryonic fibroblasts by Jdp2-Jhdm1b-Mkk6-Glis1-Nanog-Essrb-Sall4. Cell Rep. 27, 3473-3485.e3475. 10.1016/j.celrep.2019.05.068 [DOI] [PubMed] [Google Scholar]
- Watanabe, T., Sato, Y., Saito, D., Tadokoro, R. and Takahashi, Y. (2009). EphrinB2 coordinates the formation of a morphological boundary and cell epithelialization during somite segmentation. Proc. Natl. Acad. Sci. USA 106, 7467-7472. 10.1073/pnas.0902859106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. A. and Ordahl, C. P. (1994). Pax-3 expression in segmental mesoderm marks early stages in myogenic cell specification. Development 120, 785-796. 10.1242/dev.120.4.785 [DOI] [PubMed] [Google Scholar]
- Wilson, V., Olivera-Martinez, I. and Storey, K. G. (2009). Stem cells, signals and vertebrate body axis extension. Development 136, 1591-1604. 10.1242/dev.021246 [DOI] [PubMed] [Google Scholar]
- Yasuhiko, Y., Haraguchi, S., Kitajima, S., Takahashi, Y., Kanno, J. and Saga, Y. (2006). Tbx6-mediated Notch signaling controls somite-specific Mesp2 expression. Proc. Natl. Acad. Sci. USA 103, 3651-3656. 10.1073/pnas.0508238103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J. K. and Wold, B. (2000). The bHLH regulator pMesogenin1 is required for maturation and segmentation of paraxial mesoderm. Genes Dev. 14, 3204-3214. 10.1101/gad.850000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Abreu, J. G., Yokota, C., MacDonald, B. T., Singh, S., Coburn, K. L. A., Cheong, S.-M., Zhang, M. M., Ye, Q.-Z., Hang, H. C.et al. (2012). Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell 149, 1565-1577. 10.1016/j.cell.2012.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Sirbu, I. O., Mic, F. A., Molotkova, N., Molotkov, A., Kumar, S. and Duester, G. (2009). Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr. Biol. 19, 1050-1057. 10.1016/j.cub.2009.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W., Ajima, R., Ninomiya, Y. and Saga, Y. (2015). Segmental border is defined by Ripply2-mediated Tbx6 repression independent of Mesp2. Dev. Biol. 400, 105-117. 10.1016/j.ydbio.2015.01.020 [DOI] [PubMed] [Google Scholar]
- Zhu, L. J., Gazin, C., Lawson, N. D., Pagès, H., Lin, S. M., Lapointe, D. S. and Green, M. R. (2010). ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237. 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.