Tan et al report novel CAG repeat expansion in THAP11 associated with spinocerebellar ataxia (SCA) in two Chinese families. They observed 45–100 repeats, three CAA interruptions, and a long uninterrupted 3’ tail in sequencing of ataxic individuals from a single family. 1
We investigated presence and size of THAP11 expansion in short‐read next‐generation sequencing of individuals with other ancestries in 1000 Genomes and the UK Biobank. 2 Repeat genotypes of up to 50 triplets can be accurately typed with short read sequencing and bioinformatic tools. 3
Methods
We genotyped CAG expansion in THAP11 (GRCh38 position chr16:67842863‐67842950) with ExpansionHunter 5.0.0 and REViewer 4 in 138 individuals with whole‐genome sequencing in the UK Biobank 2 and a history of hereditary ataxia or ataxia of unknown cause (ICD10 G110‐G119; R270). We obtained THAP11 ExpansionHunter genotypes from 2504 unrelated individuals in 1000 Genomes. 5
Results
One European‐ancestry ataxic individual in the UK Biobank has a 46/29 CAG THAP11 genotype, exceeding the proposed pathogenic threshold, 1 and an uninterrupted 22‐repeat CAG repeat expansion in CACNA1A that likely causes spinocerebellar ataxia type 6 (SCA6). 6 REViewer visualization of ExpansionHunter genotyping shows six CAA interruptions to the THAP11 expansion (Fig. 1A) and no interruptions in the SCA6 expansion (Fig. 1B).
FIG. 1.
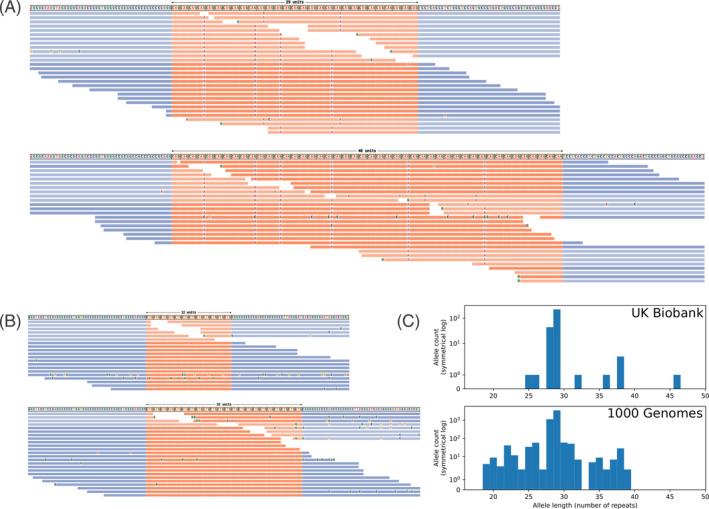
(A) REViewer plot of THAP11 CAG expansion in a European ancestry participant in the UK Biobank. CAA interruptions are present in the expanded allele. (B) REViewer plot of expanded CACNA1A allele in the same individual. (C) Distribution of THAP11 allele sizes in 139 individuals with ataxia in the UK Biobank and 2504 unrelated individuals in the 1000 Genomes cohort.
The individual had primary care diagnoses of hereditary ataxia at 40, Parkinson's disease at 53, and received repeated prescriptions for trihexyphenidyl, a drug used in management of movement disorders. More‐precise age of onset is not available as medical records in the UK Biobank are partial and participants are not re‐contactable.
THAP11 genotypes in the 1000 Genomes cohort were between 19 and 39 repeats. In the UK Biobank ataxia cohort, the range was 25 to 46 repeats. In both cohorts, the median was 29 repeats (IQR, 28–29) (Fig. 1C).
Discussion
We report the first finding of THAP11 CAG expansion in an ataxic individual of European ancestry. Interpretation of this expansion is complicated by detection of an uninterrupted pathogenic full‐penetrance length SCA6 expansion and diagnosis of both ataxia and Parkinson's disease. This individual has six CAA interruptions, with nine uninterrupted 3′ repeats. We also provide length distributions of the THAP11 allele.
Tan et al suggest toxicity of CAA‐interrupted repeats based on CAG‐pure sequences of ataxic individuals. They report one family where ataxic individuals with THAP11 expansion have three interruptions and 32 to 87 uninterrupted repeats in the 3′ end of the expansion, and unaffected family members have five to six interruptions and shorter tails. They also report THAP11 expansion in an unrelated individual (patient II‐1), with six interruptions and 10 uninterrupted 3′ repeats (Tan et al's supporting information Figure S3), 1 similar to our European‐ancestry individual.
Studies suggest CAA interruptions to CAG expansions stabilize intergenerational variability in repeat length, 7 which may contribute to expansion instability in the family with fewer interruptions. Our findings show further work is necessary to elucidate the role of rare THAP11 expansion and its composition in ataxia and demonstrates that THAP11 expansion is detectable with bioinformatic approaches. It also further highlights the emerging complexity of expansion composition in tandem repeat‐mediated disease.
Financial Disclosures
This work was supported by the National Health and Medical Research Council (NHMRC) grants (GNT2001513, MRFF2007707 and MRFF2007677) to M.B., M.F.B. and H.R. H.R. was supported by a NHMRC Emerging Leadership 1 grant (1194364) and M.B. was supported by a NHMRC Leadership 1 grant (1195236). Additional funding was provided by the Independent Research Institute Infrastructure Support Scheme and the Victorian State Government Operational Infrastructure Program.
Author Roles
(1) Research project: A. Conception, B. Organization, C. Execution; (2) Statistical Analysis: A. Design, B. Execution, C. Review and Critique; (3) Manuscript: A. Writing of the First Draft, B. Review and Critique.
L.G.F.: 1A, 1B, 1C, 2A, 2B, 3A, 3B; H.R. 1A, 2B, 2C, 3B; M.F.B. 1A, 2C, 3B; M.B. 1A, 1B, 2A, 2C, 3B.
Acknowledgments
We thank Professor Paul Lockhart, Dr. Justin Read and Kayli Davies of the Murdoch Children's Research Institute, Melbourne, Australia for comments and feedback. This research was conducted with data from UK Biobank (www.ukbiobank.ac.uk), a major biomedical database, under data use agreement 36610 (PI Bahlo).
Relevant conflicts of interest/financial disclosures: The authors have no conflicts of interest to declare and no financial disclosures to report.
Data Availability Statement
The data that support the findings of this study are available from the UK Biobank. Restrictions apply to the availability of UK Biobank data. Data from the Illumina Repeat Catalog are openly available at https://github.com/Illumina/RepeatCatalogs.
References
- 1. Tan D, Wei C, Chen Z, et al. CAG repeat expansion in THAP11 is associated with a novel spinocerebellar ataxia. Mov Disord 2023;38:1282–1293. 10.1002/mds.29412 [DOI] [PubMed] [Google Scholar]
- 2. Halldorsson BV, Eggertsson HP, Moore KHS, et al. The sequences of 150,119 genomes in the UK biobank. Nature 2022;607:732–740. 10.1038/s41586-022-04965-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ibañez K, Polke J, Hagelstrom RT, et al. Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: a retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol 2022;21(3):234–245. 10.1016/S1474-4422(21)00462-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolzhenko E, Deshpande V, Schlesinger F, et al. ExpansionHunter: a sequence‐graph‐based tool to analyze variation in short tandem repeat regions. Bioinformatics 2019;35(22):4754–4756. 10.1093/bioinformatics/btz431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qiu, Y. , Deshpande, V. , Avdeyev, P. et al. Illumina Repeat Catalogs https://github.com/Illumina/RepeatCatalogs.
- 6. Craig K, Takiyama Y, Soong BW, et al. Pathogenic expansions of the SCA6 locus are associated with a common CACNA1A haplotype across the globe: founder effect or predisposing chromosome? Eur J Hum Genet 2008;16(7):841–847. 10.1038/ejhg.2008.20 [DOI] [PubMed] [Google Scholar]
- 7. Pandey N, Mittal U, Srivastava AK, et al. SMARCA2 and THAP11: potential candidates for polyglutamine disorders as evidenced from polymorphism and protein‐folding simulation studies. J Hum Genet 2004;49:596–602. 10.1007/s10038-004-0194-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the UK Biobank. Restrictions apply to the availability of UK Biobank data. Data from the Illumina Repeat Catalog are openly available at https://github.com/Illumina/RepeatCatalogs.


