Abstract
The membrane-interacting abilities of three sequences representing the putative fusogenic subdomain of the Ebola virus transmembrane protein have been investigated. In the presence of calcium, the sequence EBOGE (GAAIGLAWIPYFGPAAE) efficiently fused unilamellar vesicles composed of phosphatidylcholine, phosphatidylethanolamine, cholesterol, and phosphatidylinositol (molar ratio, 2:1:1:0.5), a mixture that roughly resembles the lipid composition of the hepatocyte plasma membrane. Analysis of the lipid dependence of the process demonstrated that the fusion activity of EBOGE was promoted by phosphatidylinositol but not by other acidic phospholipids. In comparison, EBOEA (EGAAIGLAWIPYFGPAA) and EBOEE (EGAAIGLAWIPYFGPAAE) sequences, which are similar to EBOGE except that they bear the negatively charged glutamate residue at the N terminus and at both the N and C termini, respectively, induced fusion to a lesser extent. As revealed by binding experiments, the glutamate residue at the N terminus severely impaired peptide-vesicle interaction. In addition, the fusion-competent EBOGE sequence did not associate significantly with vesicles lacking phosphatidylinositol. Tryptophan fluorescence quenching by vesicles containing brominated phospholipids indicated that the EBOGE peptide penetrated to the acyl chain level only when the membranes contained phosphatidylinositol. We conclude that binding and further penetration of the Ebola virus putative fusion peptide into membranes might be governed by the nature of the N-terminal residue and by the presence of phosphatidylinositol in the target membrane. Moreover, since insertion of such a peptide leads to membrane destabilization and fusion, the present data would be compatible with the involvement of this sequence in Ebola virus fusion.
Ebola virus belongs to the Filoviridae family (23). This human pathogen occasionally causes epidemics of African hemorrhagic fever with a high rate of mortality (8, 23, 37). Little is known about the viral infectivity mechanism, and there is no specific treatment for Ebola virus hemorrhagic fever as yet. The most prominent pathology of Ebola virus infection includes necrosis of liver parenchyma as a direct consequence of virus replication (23). Ebola virus virions are composed of a helical nucleocapsid containing one linear, negative-sense, single-stranded RNA and surrounded by a lipidic envelope derived from the host cell plasma membrane (8, 23). The envelope contains solely one type of highly glycosylated protein (Ebola GP) arranged into oligomers, most probably trimers, which constitute the spikes that protrude from the virion surface (8, 30, 38, 39).
The mode of entry of Ebola virus into target cells remains unknown. However it seems likely that the single surface protein Ebola GP is responsible for both receptor binding and membrane fusion during entry into the host cells. Homology analysis of its coding gene-derived sequence has identified several structural features that Ebola GP shares with other envelope fusion proteins derived from oncogenic retroviruses (12, 39). Just recently a detailed analysis has detected a high degree of structural homology between Ebola GP and the Rous sarcoma virus transmembrane protein (12). Several structural elements that might be involved in the ectodomain fusogenic function are shared by these viruses. In particular, there exists in both viruses an amino acid region bounded by cysteines that has at its center a sequence of approximately 16 uncharged and hydrophobic residues. Its location with respect to the viral membrane, the presence of a canonical fusion tripeptide (YFG in Ebola virus), and the fact that this sequence exhibits a high degree of identity among the Filoviridae members suggest that this region might constitute in Ebola virus the fusion peptide that is critical for virion-membrane fusion in the Retroviridae and other families (11, 40, 41).
According to the most widely accepted mechanistic model proposed for the initial phase of the viral fusion process, activation of the viral spikes induces the exposure of previously buried hydrophobic fusion peptides in the vicinity of the target cell (5, 43). Further interaction of the viral fusion peptides with the cell membrane would depend mainly on the capacity for binding of these peptides to the membrane lipid components and could eventually trigger the process that brings about the actual merging of the viral and cell membranes via a currently unknown mechanism (41). This fact has justified the development of in vitro studies on the membrane-destabilizing effects of fusion peptides by using representative synthetic peptides of different viruses and model membranes (7, 15, 19, 29).
The membrane environment into which the fusion peptide should partition obviously plays an important role in the process. Previous work from this laboratory has focused on the effect of the target membrane composition on viral fusion. Reports from this and other laboratories indicate the existence of conformational changes induced by lipidic components in the membrane-bound human immunodeficiency virus type 1 (HIV-1) fusion peptide (25, 28, 29), and we have identified a fusogenic conformation of the peptide represented by an extended β-type structure (25, 26, 28). The fusogenic interaction of the HIV-1 fusion peptide is, moreover, sensitive to factors that affect gp41 activity in vivo (27). Modulation of viral fusion by lipids has also been observed for complete virions and reconstituted systems fusing with model membranes (6, 24, 42). These observations indicate that enveloped viruses may optimize host interactions during the entry process, not only at the level of the selective binding to cell receptors but also at the level of the envelope fusion and subsequent capsid penetration.
Our primary objective in this study was to confirm that the proposed fusogenic sequence for Ebola virus might interact with membranes, destabilize them, and eventually induce fusion. Because Ebola virus infects and replicates very efficiently in the liver, we initially employed as target membranes large unilamellar vesicles (LUV) made of a lipidic mixture that represents the hepatocyte plasma membrane composition (18). Our results demonstrate that this Ebola virus peptide interacts with phosphatidylinositol (PI)-containing membranes and induces vesicle fusion. Moreover, we show that the sequence lacking the negatively charged Glu residue at the N terminus interacts more efficiently with membranes. These data suggest that, similarly to the HIV-1 fusion peptide (26–28), the Ebola virus peptide segment under study may be important in viral fusion in vivo.
MATERIALS AND METHODS
Materials.
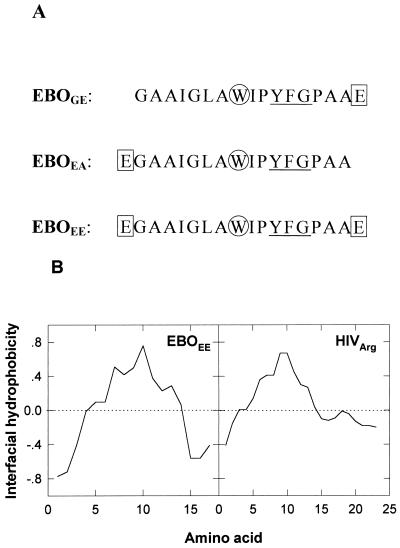
Phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidic acid (PA), and phosphatidylglycerol (PG) were from Lipid Products (South Nutfield, England). PI, the brominated phospholipids 1-palmitoyl-2-stearoyl(6,7)dibromo-sn-glycero-3-phosphocholine (Br6-PSPC) and 1-palmitoyl-2-stearoyl(11,12)dibromo-sn-glycero-3-phosphocholine (Br11-PSPC), and the fluorescent probes N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)phosphatidylethanolamine (N-NBD-PE) and N-(lissamine rhodamine B sulfonyl)phosphatidylethanolamine (N-Rh-PE) were purchased from Avanti Polar Lipids (Birmingham, Ala.) N-(5-Dimethylaminonaphtalene-1-sulfonyl)-1,2-dihexadecanoyl-sn-glyc- ero-3-phosphoethanolamine (d-DHPE) was from Molecular Probes (Junction City, Oreg.). Cholesterol (CHOL), trinitrophenylphosphatidylethanolamine (TNP-PE), and Triton X-100 were obtained from Sigma (St. Louis, Mo.). All other reagents were of analytical grade. The sequences representing the putative fusogenic segment of Ebola GP, i.e., EBOGE, EBOEA, and EBOEE (see Fig. 1A), were synthesized as their C-terminal carboxamides and purified (estimated homogeneity, >90%) by Quality Controlled Biochemicals, Inc. (Hopkinton, Mass.). Peptide stock solutions were prepared in dimethyl sulfoxide (spectroscopy grade).
FIG. 1.
(A) Sequences of the three Ebola GP peptides studied in this work. Positions of Glu (squares) and Trp (circles) residues and the canonical fusion tripeptide (underlined) are indicated. (B) Hydropathy plots for the fusion peptides of Ebola virus (EBOEE) and HIV-1 (HIVarg [26]). A window of 5 amino acids was used with the hydrophobicity scale at membrane interfaces as described by Wimley and White (44).
Vesicle preparation.
LUV were prepared by the extrusion method of Hope et al. (17) in 5 mM HEPES–100 mM NaCl (pH 7.4) buffer. Small unilamellar vesicles (SUV) were obtained by sonication of aqueous lipid dispersions (2). Lipid concentrations in liposome suspensions were determined by phosphate analysis (4).
Fluorimetric assay for vesicle fusion.
All fluorescence measurements were conducted in thermostatically controlled cuvettes (37°C) with a Perkin-Elmer LS50-B spectrofluorimeter. The medium in the cuvettes was continuously stirred to allow the rapid mixing of peptide and vesicles. Membrane lipid mixing was monitored by using the resonance energy transfer assay described by Struck et al. (35). The assay is based on the dilution of N-NBD-PE and N-Rh-PE. Dilution due to membrane mixing results in an increase in N-NBD-PE fluorescence. Vesicles containing each probe at 0.6 mol% were mixed with unlabeled vesicles at a 1:4 ratio (final lipid concentration, 0.1 mM). The NBD emission was monitored at 530 nm, with the excitation wavelength set at 465 nm. A cutoff filter at 515 nm was used between the sample and the emission monochromator to avoid scattering interferences. The fluorescence scale was calibrated such that the zero level corresponded to the initial residual fluorescence of the labeled vesicles and the 100% value corresponded to complete mixing of all the lipids in the system. The latter value was set by the fluorescence intensity of vesicles, labeled with 0.12 mol% of each fluorophore, at the same total lipid concentration as in the fusion assay. A high peptide-to-lipid ratio (1:1.5 unless otherwise stated) was used in these studies in order to maximize vesicle damage, thus facilitating its detection and study.
Peptide binding to vesicles.
Peptide binding to SUV was estimated by using three complementary methods under conditions otherwise similar to those described for the fusion assay. For the centrifugation method, peptides were added to 1 ml of SUV (0.5 mM) prepared in buffer (peptide-to-lipid ratio, 1:500). After incubation of the mixture for 10 min at 37°C, centrifugation of the peptide-lipid complexes in a Beckman Optima TLX ultracentrifuge in a TL120.2 rotor (627,000 × g, 90 min, 25°C) gave rise to a lipidic pellet (>95% of total lipid in the sample). Supernatants depleted of vesicles were carefully removed, and their peptide content was subsequently quantitated by Trp fluorescence (excitation at 280 nm and emission at 350 nm). The percentage of peptide bound to vesicles was estimated by using the following expression:
 |
1 |
where F(s − l) represents Trp fluorescence emission intensity in supernatants of centrifuged peptides in the absence of vesicles and F(s + l) represents Trp fluorescence in supernatants of centrifuged peptide-vesicle mixtures.
An additional method to evaluate binding was based on the variation of the fluorescence emitted by Trp. The change in peptide Trp fluorescence after binding (1 μM peptide) was measured in emission spectra collected in the presence of SUV (0.5 mM). The mixture was incubated for 10 min at 37°C before data acquisition. Excitation was set at 280 nm, and slits of 2.5 nm (excitation) and 10 nm (emission) were used. The signal was corrected for inner filter effects as described previously (3). Binding was also estimated by using an assay of resonance energy transfer from peptide-Trp to the headgroup in TNP-PE as described by Heymann et al. (16). TNP-PE was included in the target vesicle composition to 8.4 mol%. In the presence of TNP-PE-containing vesicles (0.5 mM), the decrease of peptide Trp fluorescence (1 μM peptide) was measured from emission spectra collected under the conditions described above.
Peptide insertion.
The depth of penetration into the membrane was evaluated by two complementary assays. In order to probe the interface, energy transfer from peptide Trp to the surface fluorescent probe d-DHPE was measured as described previously (13). In brief, 6 mol% d-DHPE probe was included in the target vesicle composition (0.25 mM), and its emission spectra at increasing peptide concentrations were collected (maximum emission wavelength, 510 nm). The excitation wavelength was that of the Trp residue (280 nm). For probing the acyl chain region, quenching of peptide Trp fluorescence by the hydrophobic matrix-residing bromolipids was measured as described previously (3). In this case, either Br6-PSPC or Br11-PSPC was used instead of PC in target PC-PI (1:2 molar ratio) vesicles. Trp emission spectra (1 μM peptide) were acquired in the presence of the vesicles (0.5 mM lipid) and corrected as described in reference 3. In both assays, lipid-peptide mixtures were incubated at 37°C for 10 min before data acquisition. In all peptide binding and insertion assays, a low peptide-to-lipid ratio (1:500) was used so that the amount of free peptide would not significantly interfere with the analysis of the experimental results.
RESULTS
Figure 1A displays the three sequences that were used in this work as representative of the proposed Ebola virus fusion peptide. The sequence of the fusion peptide, initially described by Volchkov et al. (39) and Feldmann et al. (8) and whose putative fusogenic character was put forward by Gallaher (12), consists of a stretch of 16 hydrophobic and uncharged amino acids which is flanked by two negatively charged Glu residues. According to the interfacial hydrophobicity scale determined by Wimley and White (44), Glu shows the lowest tendency to partitioning into the bilayer. Therefore, even if the overall interfacial hydrophobicity of the sequence is comparable to that of the HIV-1 fusion peptide (Fig. 1B), flanking Glu residues in the Ebola virus sequence are unlikely to partition into the interface and penetrate into the hydrophobic core of the membrane. This fact prompted us to assess the effect of Glu residues when they are located either at the N terminus, at the C terminus, or at both ends of the sequence. As described below, experimental results in this work indicate that peptide-membrane interaction is severely impaired by Glu located at the N terminus.
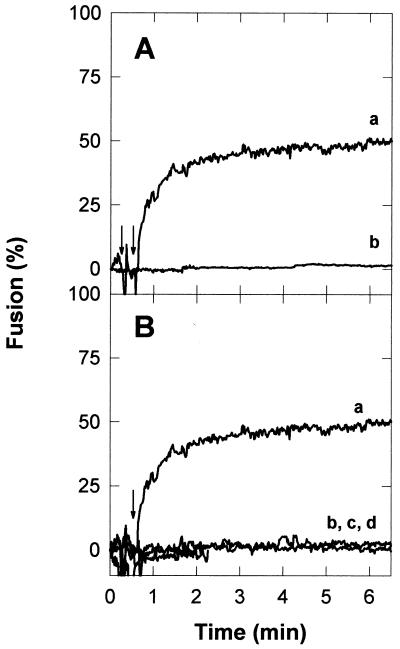
We first assessed the putative fusogenicities of these sequences in a model system. LUV composed of PC, PE, CHOL, and PI (2:1:1:0.5 molar ratio) were selected as membrane targets. As discussed in the introduction, this mixture of lipids roughly mimics the lipid composition of the hepatocyte plasma membrane (18). As shown in Fig. 2, when added to a PC-PE-CHOL-PI vesicle suspension, the EBOGE peptide was able to induce fusion, detected as the mixing of the vesicular membranes. Figure 2A shows that the fusion activity of the peptide could be detected only in the presence of calcium. The cation effect was not dependent on the order of addition; i.e., fusion could be observed when addition of the cation preceded that of the peptide and also when the cation was added after the peptide. The results displayed in Fig. 2B demonstrate that the observed fusion activity was dependent on the presence of PI in the lipidic mixture. Moreover, this phospholipid could not be replaced by other anionic phospholipids such as PG or PA, indicating that the PI effect is not due merely to its electrostatic charge. The acyl chain composition did not appear to be critical, since PI from bovine liver or plant origin sustained the fusion process to the same extent (data not shown). Table 1 summarizes the fusion data obtained with different vesicles as targets. Taken together, the data confirm that PI is necessary for the fusion process. In addition, it can be deduced that both LUV and SUV can be readily fused by the peptide in a PI-dependent fashion.
FIG. 2.
Ebola virus fusion peptide (EBOGE)-induced LUV fusion. The peptide was added at the times indicated by the arrows to PC-PE-CHOL-PI (2:1:1:0.5) LUV suspensions (0.1 mM) at a peptide-to-lipid ratio of 1:1.5. (A) Influence of calcium. Trace a, sample containing both calcium (10 mM) and peptide; trace b, control without calcium. (B) Influence of bilayer composition. Molar ratios: trace a, PC-PE-CHOL-PI, 2:1:1:0.5; trace b, PC-PE-CHOL, 2:1:1; trace c, PC-PE-CHOL-PG, 2:1:1:0.5; trace d, PC-PE-CHOL-PA, 2:1:1:0.5.
TABLE 1.
Influence of bilayer composition on EBOGE-induced vesicle fusion
| Composition (PC-PE-CHOL-PI, molar ratio) | Type of vesicle | Initial rate (% min−1)a,b | Extent (%)a,c |
|---|---|---|---|
| 2:1:1:0.5 | LUV | 52 ± 3.6 (4) | 56 ± 3.6 (4) |
| 2:1:1:0 | LUV | ≈0 (2) | ≈0 (2) |
| 1.5:1:1:1 | LUV | 20 ± 1.5 (2) | 32 ± 2.6 (2) |
| 2:1:1:0.5d | LUV | ≈0 (2) | ≈0 (2) |
| 2:1:1:0.5e | LUV | ≈0 (2) | ≈0 (2) |
| 2:1:1:0.5 | SUV | 13 ± 2.0 (4) | 19 ± 2.1 (4) |
| 2:1:1:0 | SUV | ≈0 (2) | ≈0 (2) |
| 1:0:0:2 | SUV | 10 ± 2.9 (3) | 10 ± 3.2 (3) |
| 1:0:0:0 | SUV | ≈0 (2) | ≈0 (2) |
The data are means ± standard errors. The number of experiments is in parentheses. The peptide-to-lipid ratio was 1:1.5 in all cases.
Expressed as the slope of the curve of percent fluorescence intensity versus time.
Fusion percentage at 10 min.
PG was included instead of PI.
PA was included instead of PI.
The fusion capacities of the three peptides under study are compared in Fig. 3. The results reveal that all three sequences induced fusion in a dose-dependent manner and that EBOGE was more efficient than EBOEE and EBOEA sequences. This observation suggests that a negative net charge or a polar residue at the peptide N terminus somehow hinders the membrane fusion process. However, net charge might not be the only cause of the impairing effect exerted by the Glu residue. Experiments conducted at pH 4.5 indicate that the fusion process mediated by the three sequences was not influenced by the low pH (data not shown).
FIG. 3.
Influence of the terminal sequence on Ebola virus fusion peptide-induced liposome fusion. (A) Kinetics of fusion as a function of time. Peptides were added at the time indicated by the arrow to PC-PE-CHOL-PI (2:1:1:0.5) LUV suspensions (0.1 mM) at a peptide-to-lipid ratio of 1:1.5. (B) Extents of fusion (NBD fluorescence after 10 min) as a function of increasing concentrations of Ebola virus fusion peptides. The vesicle concentration was 0.1 mM in all cases. •, EBOGE; ▪, EBOEA; ▴, EBOEE.
A decreased binding of the peptide to the vesicles could be at the origin of the lack of fusion detected in the absence of PI or could be the cause of the reduced fusion activity observed for the sequences bearing Glu at the N terminus. Therefore, we decided to estimate the amount of peptide bound to vesicles and the depth of its penetration into the membrane. To that end, we made use of the single Trp fluorescent residue existing in the sequences (Fig. 1A). For these experiments we employed SUV as targets, since they interfere less than LUV with the fluorescence measurements because of their low light-scattering properties. We also selected the simplest vesicle composition supporting fusion that was suitable for carrying out the penetration assays (PC-PI, 1:2 molar ratio).
Figure 4 illustrates the binding capacities of the three sequences as detected through changes in the intrinsic peptide fluorescence emission. Trp emission for EBOGE changed significantly in the presence of PC-PI SUV (Fig. 4A). The emission intensity was enhanced and the maximum was shifted to a lower wavelength, from 351 to 348 nm. Both effects suggest that the Trp residue in the presence of PC-PI vesicles senses a less polar environment, which is indicative of binding to and subsequent penetration of the peptide into the hydrophobic core of the bilayer. In contrast, in the presence of PC SUV, EBOGE Trp emission did not change significantly. EBOEA and EBOEE Trp emissions show only a very small increase in the presence of PC-PI vesicles (Fig. 4B and C). The fact that the Trp emission fluorescence changed little in the latter samples could be a consequence of the lack of peptide association to vesicles or of a surface binding phenomenon that would not imply changes in the polarity of the Trp environment. We therefore complemented these observations with binding results obtained by two other methods. First, unbound peptide was physically separated from peptide bound to the vesicles by centrifugation and subsequently was quantitated in supernatants. Second, we also measured partitioning in the intact system. To that end we employed an assay based on the quenching of Trp fluorescence by TNP-PE incorporated into vesicles. The TNP headgroup quenches by such a long range that, in principle, quenching is observed for Trp residues inserted at any level in the bilayer (16). The data obtained by the three methods used to estimate binding correlate very well (Table 2). Maximum binding was found by all three methods to occur for EBOGE peptide and PC-PI vesicles. The results in Table 2 further indicate that under the same conditions, EBOEA and EBOEE peptides bound to vesicles to a lesser extent. This limited binding could be the cause for the reduced fusion induced by these sequences (Fig. 2). From the results in Table 2 it can also be concluded that EBOGE hardly bound to pure PC vesicles. Again, this phenomenon could be correlated to the absence of fusion when PC vesicles are used as targets for this peptide (Table 1). The presence of 10 mM Ca2+ in the samples did not affect the binding capacity of the peptides in any of the samples; e.g., under conditions similar to those for Table 2, EBOGE displayed a Δλmax of 4.4 nm and a fluorescence intensity of 1.30, while the effect of EBOEE was clearly smaller, with a Δλmax of 2.2 nm and an intensity of 1.14.
FIG. 4.
Fluorescence emission spectra of EBOGE (A), EBOEA (B), and EBOEE (C) in buffer and incubated with SUV (0.5 mM). Spectra: 1, buffer alone; 2, PC-PI (1:2) SUV; 3, PC SUV. The peptide-to-lipid ratio was 1:500.
TABLE 2.
Binding of peptides to PC-PI (1:2) SUVa
| Peptide | Δλmax (nm)b | Intensityb (arbitrary units) | Bound peptide (%)c | Peptide fluores- cence in vesi- cles containing TNP-PE (%)d |
|---|---|---|---|---|
| EBOGE | 2.7 ± 0.94 (4) | 1.49 ± 0.13 (4) | 32.5 ± 0.2 (4) | 54 ± 6 (4) |
| EBOEA | 1.5 ± 1.27 (4) | 1.14 ± 0.06 (4) | 11.9 ± 1.5 (3) | 78 ± 11 (4) |
| EBOEE | 1.2 ± 0.83 (4) | 1.10 ± 0.08 (4) | 9.9 ± 1.2 (3) | 84 ± 4 (4) |
| EBOGE (PC)e | 1.0 ± 0.40 (4) | 0.99 ± 0.02 (4) | 3.2 ± 0.4 (3) | 99 ± 6 (4) |
Values are means ± standard errors. The number of experiments is in parentheses. The peptide-to-lipid ratio was 1:500 in all cases.
Determined from emission spectra by using the software supplied with the fluorimeter.
Centrifugation assay; the percentage of bound peptide was deduced by using equation 1 (see Materials and Methods).
Expressed as the percentage of fluorescence observed for peptides in pure PC-PI (1:2) SUV.
PC vesicles were used as targets in this case.
Finally, the results displayed in Fig. 5 and Table 3 prove that the Ebola virus fusion peptide penetrates into the membrane to the level of the acyl chains in the presence of PI. The experiments illustrated in Fig. 5 were carried out to determine the location of fusogenic EBOGE-Trp in the vesicle bilayer. In Fig. 5A emission spectra of SUV containing d-DHPE are displayed. The spectra were acquired in the presence of increasing amounts of EBOGE with the excitation wavelength set at 280 nm. The results demonstrate that peptide addition does not affect significantly the d-DHPE fluorescence under those conditions. This would mean that the Trp residue is unable to effectively transfer energy by resonance to this surface-residing group. However, both Br6-PSPC and Br11-PSPC efficiently quenched EBOGE-Trp fluorescence (Fig. 5B). Bromine atoms quench by a short-range process that requires a close approach to the fluorophore (3). Consequently, the Trp residue must be located close to the quenchers in the hydrophobic matrix of the bilayer.
FIG. 5.
Depth of penetration into the bilayer of the EBOGE Trp residue. (A) Fluorescence emission spectra of PC–PI–d-DHPE (1:2:0.19) SUV (0.25 mM) incubated with increasing amounts of EBOGE. Spectra: 1, SUV alone; 2, peptide-to-lipid ratio of 1:250; 3, peptide-to-lipid ratio of 1:125. Dashed line, positive control containing the colicin A thermolytic fragment surface bound to PG SUV (for comparison, the emission spectrum of SUV alone in this sample was normalized to spectrum 1). (B) Fluorescence emission spectra of EBOGE incubated with SUV (0.5 mM) containing brominated phospholipids. Spectra: 1, buffer alone; 2, PC-PI (1:2) SUV; 3, Br6-PSPC–PI (1:2) SUV; 4, Br11-PSPC–PI (1:2) SUV. The peptide-to-lipid ratio was 1:500.
TABLE 3.
Level of peptide insertion into PC-PI (1:2) SUV bilayersa
| Peptide | Fluorescence of PC–PI–d-DHPE vesiclesb (peptide/lipid ratio, 1:250) | Peptide fluorescence in vesicles containing bromolipidsc (peptide/lipid ratio, 1:500)
|
|
|---|---|---|---|
| Br6-PSPC | Br11-PSPC | ||
| EBOGE | 99 ± 1.8 (4) | 54 ± 3 (4) | 49 ± 3 (3) |
| EBOEA | 99 ± 0.1 (2) | 85 ± 1 (3) | 90 ± 2 (3) |
| EBOEE | 100 ± 0.1 (2) | 88 ± 7 (3) | 88 ± 1 (3) |
| EBOGE (PC)d | 99 ± 2.2 (2) | 93 ± 1 (3) | 97 ± 5 (3) |
Values are means ± standard errors. The number of experiments is in parentheses.
Expressed as the percentage of fluorescence observed in the absence of peptide.
Expressed as the percentage of fluorescence observed for peptides in pure PC-PI (1:2) SUV.
PC vesicles were used as targets in this case.
Table 3 summarizes the results obtained by the two methods, i.e., use of d-DHPE and brominated lipids, for the three peptides and for EBOGE in the presence of PC vesicles. d-DHPE fluorescence was unaffected in the presence of any of the peptides. Nevertheless, peptide fluorescence was quenched by the bromolipids. The results indicate that for the three peptides interacting with PC-PI vesicles and for EBOGE in the presence of PC vesicles, the quenching effect parallels the extent of binding (Table 2). In the absence of PI in the target vesicle, even the most binding-competent EBOGE sequence is unable to associate with and penetrate into the vesicle bilayer.
For bilayer and hydrocarbon chain region thickness of ≈50 and 30 Å, respectively, McIntosh and Holloway (21) determined the bromine distances from the bilayer center to be 11 Å for 6,7 labels and 6.5 Å for 11,12 labels. The Förster distances (R0) for energy transfer estimated for these labels, ≈6 to 9 Å (1, 3), would allow quenching of Trp residues immersed in the methylene chain region of the bilayer. On the other hand, the dansyl moiety in the d-DHPE probe is probably located at the edge of the lipid-water interface in the vesicles. Given that the R0 distance calculated for the couple dansyl-Trp is approximately 17 Å (36), quenching of Trp fluorescence should have been observed if this residue remained associated with the interface and/or with the acyl chain region close to it. We can conclude that in the peptide population bound to the vesicles (Table 2), Trp fluorescence is completely quenched when bromolipid-containing vesicles are used as targets and, therefore, that these residues must be buried deep into the hydrophobic core of the bilayer, far from the vesicle surface, under conditions in which the peptide is fusion competent.
DISCUSSION
Our results in this work show that the Ebola virus putative fusion peptide is able to interact with lipid vesicles and destabilize them in the direction of membrane fusion. These observations imply that if, as a consequence of the activation of Ebola virus fusion process, the fusion peptide is exposed to the aqueous medium, this protein sequence would actually be capable of partitioning into a target membrane. This behavior is identical to that found for various sequences arising from different enveloped viruses that have been proposed to act as fusion peptides (7, 15, 19, 25–29). Moreover, our results also demonstrate that this sequence would not act as an inert anchor of the spike to the target membrane; rather, the peptide ability to induce fusion indicates that when effectively bound to membranes, this sequence might be involved in the induction of some kind of bilayer perturbation leading to fusion. Taken together, our findings therefore support the implication of this sequence in Ebola virus fusion.
The fusion activity of the Ebola virus peptide could be observed only in the presence of calcium. The fact that the cation did not influence the binding process or the depth of penetration of the bound peptides implies that its action must be confined to the fusion process itself. The molecular mechanism behind this phenomenon is not clear at the moment. The combined action of the cation and the peptide could be due to a specific interaction between both of them or else could be the result of an electrostatic effect, namely, the neutralization of the peptide net charge, that would help bring about the peptide-induced fusion process. The joint requirement of calcium ions and PI could even suggest the implication of a PI-Ca-Glu salt bridge. Several studies are under way in our laboratory to discern among these possibilities.
A specific feature of Ebola virus peptide-induced fusion that distinguishes this process from other so-far-described liposome fusion events induced by fusion peptides is its dependence on the presence of a particular phospholipid, PI, in the target vesicles. According to our experimental results, PI is necessary for the initial association of the peptide to the vesicles. The interfacial hydrophobicity values for the Ebola virus fusion peptide (Fig. 1B) indicate a general tendency for partitioning into electrically neutral zwitterionic phospholipid membranes (44). However, our results demonstrate that the peptide does not partition into neutral interfaces unless PI is included in the membrane composition. Several facts argue against the existence of a purely electrostatic interaction at the origin of this effect. The only residues containing net charged side chains in the sequence are the negatively charged Glu residues. It seems unlikely that the anionic PI in vesicles would promote peptide interaction at neutral pH. Moreover, other anionic phospholipids at neutral pH, such as PA or PG, could not sustain the fusion process. Finally, the high ionic strength used in the experiments, which is close to the physiological values in serum, would also weaken putative electrostatic interactions between the sequences and charged vesicle surfaces.
Among the possible explanations for the PI effect detected in our system is that PI might be involved in inducing some kind of change in the physical properties of the bulk membrane, other than altering the vesicle surface charge, that would be necessary for peptide association. However, there are no indications of any such physical properties specific to PI. Alternatively, there might exist a specific interaction of the peptide with this particular phospholipid, and, as mentioned above, calcium ions could be involved in this phenomenon. The existence of stereospecific interactions has been invoked to explain the ceramide dependence of the Semliki Forest virus fusion with membranes (22, 24). Vesicular stomatitis virus also binds preferentially to phosphatidylserine (31), and the fusogenic activity of the reconstituted fusion protein depends on the presence of this lipid in the target vesicles (6). In addition to their structural function in membranes, inositol lipids are well known because they play specific roles in cell signalling (33) and membrane protein anchoring (9). Several findings involve this class of lipids in viral entry and infectivity as well. Stimulation of certain viral receptors results in the activation of signal transduction pathways involving the enzyme PI 3-kinase (32). It has also been suggested that glycan PI hydrolysis might be involved in the initiation of HIV-1 replication (20). Just recently, it has been found that envelope PI is essential for a correct epitope presentation to neutralizing antibodies in African swine fever virions (14).
Our results also indicate that the N-terminal composition might be a limiting factor for the destabilizing interaction of this sequence with membranes. Glu residues at the N terminus appear to interfere with the initial interaction of the peptide with vesicles. A correct residue at the fusion peptide N terminus appears to be crucial for the fusogenic function of certain spike proteins (10, 34). We have reported that a polar substitution, Val2xGlu, in the HIV-1 fusion peptide renders a sequence unable to induce liposome fusion (26–28). However, the Glu effect in the case of the HIV-1 peptide seems to be different, since this residue does not interfere with binding to or penetration into the vesicle bilayer (26). Moreover, the three peptides used in this study are representative of the correct putative Ebola virus fusion sequence (12). Therefore, our results should be taken as indicative of the mechanism of action of the correct sequence within the wild-type protein. The fact that even the bound sequences containing Glu at the N terminus were also able to insert and induce some vesicle fusion points to the internal amino acid sequence as the promoter of the fusogenic activity. Given the sequence homology with the Rous sarcoma virus transmembrane protein, it is also likely that Ebola GP contains the fusion peptide in internal regions of the glycoprotein (12, 40, 41). If this is the case, we conclude that masking the Glu residue located at the N terminus would optimize the partition of the putative fusogenic sequence into the target membrane. It should also be noted that our experiments are designed for detecting a maximum of membrane fusion. Under physiological conditions, however, what is seen here as an impairment of fusion could be beneficial for viral replication, e.g., by avoidance of cell-cell fusion.
Investigations of synthetic fusion peptides and model membranes have delineated the structural requirements of these sequences and their membrane activities (7, 15, 19, 25–29). In addition, it has recently been demonstrated for the HIV-1 fusion peptide that its lipid-mixing activity measured in vitro is sensitive to factors of physiological relevance during the virus entry into host cells (27). Those findings could open the possibility of using in vitro assays for assessing in an easy manner the potencies of antiviral agents targeted to these sequences; this would stress the importance of studies using model systems. Taking into account the strict lipid dependence for the fusion peptide-induced fusion event detected here, our observations might in the future be important in the design of specific therapeutic approaches for the treatment of Ebola virus-infected individuals.
ACKNOWLEDGMENTS
We thank J. M. González-Mañas for helpful discussions and for the provision of colicin A (thermolytic fragment).
This work was supported by the EC Concerted Action Programme “Interaction of HIV Proteins with Cell Membranes,” the Basque Government (PI 94-53; PI 96-46), and the University of the Basque Country (UPV 042.310-EA085/97).
REFERENCES
- 1.Abrams F S, London E. Calibration of the parallax fluorescence quenching method for determination of membrane penetration depth: refinement and comparison of quenching by spin-labeled and brominated lipids. Biochemistry. 1992;31:5312–5322. doi: 10.1021/bi00138a010. [DOI] [PubMed] [Google Scholar]
- 2.Alonso A, Saez R, Villena A, Goñi F M. Increase in size of sonicated phospholipid vesicles in the presence of detergents. J Membr Biol. 1982;67:55–62. doi: 10.1007/BF01868647. [DOI] [PubMed] [Google Scholar]
- 3.Bolen E J, Holloway P W. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 1990;29:9638–9643. doi: 10.1021/bi00493a019. [DOI] [PubMed] [Google Scholar]
- 4.Böttcher C S F, van Gent C M, Fries C. A rapid and sensitive sub-micro phosphorus determination. Anal Chim Acta. 1961;24:203–204. [Google Scholar]
- 5.Carr C, Kim P S. A spring-loaded mechanism for the conformational change of Influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 6.Eidelman O, Schlegel R, Tralka T S, Blumenthal R. pH-dependent fusion induced by vesicular stomatitis virus glycoprotein reconstituted into phospholipid vesicles. J Biol Chem. 1984;259:4622–4628. [PubMed] [Google Scholar]
- 7.Epand R M, Cheetham J, Epand P F, Yeagle P L, Richardson C D, DeGrado W F. Peptide models for the membrane destabilizing actions of viral fusion proteins. Biopolymers. 1992;32:309–314. doi: 10.1002/bip.360320403. [DOI] [PubMed] [Google Scholar]
- 8.Feldmann H, Klenk H D, Sanchez A. Molecular biology and evolution of filoviruses. Arch Virol Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson M A J, Williams A F. Cell surface anchoring of proteins via glycosylphosphatidylinositol structures. Annu Rev Biochem. 1988;57:285–320. doi: 10.1146/annurev.bi.57.070188.001441. [DOI] [PubMed] [Google Scholar]
- 10.Freed E O, Delwart E L, Buchschacher G L, Panganiban A T. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallaher W R. Detection of a fusion peptide sequence in the transmembrane protein of the human immunodeficiency virus. Cell. 1987;50:327–328. doi: 10.1016/0092-8674(87)90485-5. [DOI] [PubMed] [Google Scholar]
- 12.Gallaher W R. Similar structural models of the transmembrane proteins of Ebola and avian sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert G E, Furie B C, Furie B. Binding of human factor VIII to phospholipid vesicles. J Biol Chem. 1990;265:815–822. [PubMed] [Google Scholar]
- 14.Gómez-Puertas P, Oviedo J M, Rodríguez F, Coll J, Escribano J M. Neutralization susceptibility of African swine fever virus is dependent on the phospholipid composition of viral particles. Virology. 1997;228:180–189. doi: 10.1006/viro.1996.8391. [DOI] [PubMed] [Google Scholar]
- 15.Gray C, Tatulian S A, Wharton S A, Tamm L K. Effect of the N-terminal glycine on the secondary structure, orientation and interaction of the influenza hemagglutinin fusion peptide with lipid bilayers. Biophys J. 1996;70:2275–2286. doi: 10.1016/S0006-3495(96)79793-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heymann J B, Zakharov S D, Zhang Y L, Cramer W A. Characterization of electrostatic and nonelectrostatic components of protein-membrane binding interactions. Biochemistry. 1996;35:2717–2725. doi: 10.1021/bi951535l. [DOI] [PubMed] [Google Scholar]
- 17.Hope M J, Bally M B, Webb G, Cullis P R. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim Biophys Acta. 1985;812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]
- 18.Jain M K. Introduction to biological membranes. 2nd ed. New York, N.Y: John Wiley & Sons; 1988. [Google Scholar]
- 19.Lear J D, DeGrado W F. Membrane binding and conformational properties of peptides representing the NH2 terminus of influenza HA-2. J Biol Chem. 1987;262:2500–2505. [PubMed] [Google Scholar]
- 20.Leung D W, Peterson P K, Weeks R, Gekker G, Chao C C, Kaplan A H, Balantac N, Tompkins C, Underiner G E, Bursten S, Harris W, Bianco J A, Singer J W. CT-2576, an inhibitor of phospholipid signaling, suppresses constitutive and induced expression of human immunodeficiency virus. Proc Natl Acad Sci USA. 1995;92:4813–4817. doi: 10.1073/pnas.92.11.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIntosh T J, Holloway P W. Determination of the depth of bromine atoms in bilayers formed from bromolipid probes. Biochemistry. 1987;26:1783–1788. doi: 10.1021/bi00380a042. [DOI] [PubMed] [Google Scholar]
- 22.Moesby L, Corver J, Erukulla R K, Bittman R, Wilschut J. Sphingolipids activate membrane fusion of Semliki Forest virus in a stereospecific manner. Biochemistry. 1995;34:10319–10324. doi: 10.1021/bi00033a001. [DOI] [PubMed] [Google Scholar]
- 23.Murphy F A, Kiley M P, Fisher-Hoch S P. Filoviridae. Marburg and Ebola viruses. In: Fields B N, Knipe D M, et al., editors. Virology. New York, N.Y: Raven Press; 1990. pp. 933–942. [Google Scholar]
- 24.Nieva J L, Bron R, Corver J, Wilschut J. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 1994;13:2797–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieva J L, Nir S, Muga A, Goñi F M, Wilschut J. Interaction of the HIV-1 fusion peptide with phospholipid vesicles: Different structural requirements for leakage and fusion. Biochemistry. 1994;33:3201–3209. doi: 10.1021/bi00177a009. [DOI] [PubMed] [Google Scholar]
- 26.Pereira F B, Goñi F M, Muga A, Nieva J L. Permeabilization and fusion of uncharged lipid vesicles induced by the HIV-1 fusion peptide adopting an extended conformation: dose and sequence effects. Biophys J. 1997;73:1977–1986. doi: 10.1016/S0006-3495(97)78228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira F B, Goñi F M, Nieva J L. Membrane fusion induced by the HIV type 1 fusion peptide: modulation by factors affecting glycoprotein 41 activity and potential anti-HIV compounds. AIDS Res Hum Retroviruses. 1997;13:1203–1211. doi: 10.1089/aid.1997.13.1203. [DOI] [PubMed] [Google Scholar]
- 28.Pereira F B, Goñi F M, Nieva J L. Liposome destabilization induced by the HIV-1 fusion peptide. Effect of a single amino acid substitution. FEBS Lett. 1995;362:243–246. doi: 10.1016/0014-5793(95)00257-a. [DOI] [PubMed] [Google Scholar]
- 29.Rafalski M, Lear J, DeGrado W F. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez A, Trappier S G, Mahy B W J, Peters C J, Nichol S T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlegel R, Tralka T S, Willingham M C, Pastan I. Inhibition of VSV binding and infectivity by phosphatidylserine: is phosphatidylserine a VSV-binding site? Cell. 1983;32:639–646. doi: 10.1016/0092-8674(83)90483-x. [DOI] [PubMed] [Google Scholar]
- 32.Sinclair A J, Farrell P J. Host cell requirements for efficient infection of quiescent primary B lymphocytes by Epstein-Barr virus. J Virol. 1995;69:5461–5468. doi: 10.1128/jvi.69.9.5461-5468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiegel S, Foster D, Kolesnick R. Signal transduction through lipid second messengers. Curr Opin Cell Biol. 1996;8:159–167. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- 34.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struck D K, Hoekstra D, Pagano R E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 36.Vaz W L C, Schoellmann G. A fluorescence study on some specifically modified derivatives of chymotrypsin, trypsin and subtilisin. Biochim Biophys Acta. 1976;439:206–218. doi: 10.1016/0005-2795(76)90176-8. [DOI] [PubMed] [Google Scholar]
- 37.Volchkov V, Volchkova V, Eckel C, Klenk H-D, Bouloy M, Leguenno B, Feldmann H. Emergence of subtype Zaire Ebola virus in Gabon. Virology. 1997;232:139–144. doi: 10.1006/viro.1997.8529. [DOI] [PubMed] [Google Scholar]
- 38.Volchkov V E, Becker S, Volchkova V A, Ternovoj V A, Kotov A N, Netesov S V, Klenk H D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and Vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 39.Volchkov V E, Blinov V M, Netesov S V. The envelope glycoprotein of Ebola virus contains an immunosuppressive-like domain similar to oncogenic retroviruses. FEBS Lett. 1992;305:181–184. doi: 10.1016/0014-5793(92)80662-z. [DOI] [PubMed] [Google Scholar]
- 40.White J. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 41.White J. Membrane fusion. Science. 1992;258:917–923. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]
- 42.White J, Helenius A. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc Natl Acad Sci USA. 1980;77:3273–3277. doi: 10.1073/pnas.77.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wild C, Dubay J W, Greenwell T, Baird T, Oas T G, McDanal C, Hunter E, Matthews T. Propensity for a leucine zipper-like domain of human immunodeficiency virus type 1 gp41 to form oligomers correlates with a role in virus-induced fusion rather than assembly of the glycoprotein complex. Proc Natl Acad Sci USA. 1994;91:12676–12680. doi: 10.1073/pnas.91.26.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wimley W, White S H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]