Abstract
Background
Segmenting the whole heart over the cardiac cycle in 4D flow MRI is a challenging and time‐consuming process, as there is considerable motion and limited contrast between blood and tissue.
Purpose
To develop and evaluate a deep learning‐based segmentation method to automatically segment the cardiac chambers and great thoracic vessels from 4D flow MRI.
Study Type
Retrospective.
Subjects
A total of 205 subjects, including 40 healthy volunteers and 165 patients with a variety of cardiac disorders were included. Data were randomly divided into training (n = 144), validation (n = 20), and testing (n = 41) sets.
Field Strength/Sequence
A 3 T/time‐resolved velocity encoded 3D gradient echo sequence (4D flow MRI).
Assessment
A 3D neural network based on the U‐net architecture was trained to segment the four cardiac chambers, aorta, and pulmonary artery. The segmentations generated were compared to manually corrected atlas‐based segmentations. End‐diastolic (ED) and end‐systolic (ES) volumes of the four cardiac chambers were calculated for both segmentations.
Statistical tests
Dice score, Hausdorff distance, average surface distance, sensitivity, precision, and miss rate were used to measure segmentation accuracy. Bland–Altman analysis was used to evaluate agreement between volumetric parameters.
Results
The following evaluation metrics were computed: mean Dice score (0.908 ± 0.023) (mean ± SD), Hausdorff distance (1.253 ± 0.293 mm), average surface distance (0.466 ± 0.136 mm), sensitivity (0.907 ± 0.032), precision (0.913 ± 0.028), and miss rate (0.093 ± 0.032). Bland–Altman analyses showed good agreement between volumetric parameters for all chambers. Limits of agreement as percentage of mean chamber volume (LoA%), left ventricular: 9.3%, 13.5%, left atrial: 12.4%, 16.9%, right ventricular: 9.9%, 15.6%, and right atrial: 18.7%, 14.4%; for ED and ES, respectively.
Data conclusion
The addition of this technique to the 4D flow MRI assessment pipeline could expedite and improve the utility of this type of acquisition in the clinical setting.
Evidence Level
4
Technical Efficacy
Stage 1
Keywords: cardiovascular MRI, 4D flow MRI, segmentation, deep learning, convolutional neural networks
Time‐resolved three‐dimensional flow MRI (4D Flow MRI) is an acquisition technique that allows for full three‐dimensional spatial coverage of the cardiovascular system over time, while also including three‐directional velocity information throughout the cardiac cycle. 1 , 2 , 3 The resulting images can be used for retrospective analysis of blood flow dynamics at any location in the acquired volume. Quantifiable hemodynamic parameters commonly calculated using these images are flow volumes, kinetic energy, wall shear stress, vorticity, relative pressure, stasis, and turbulence, among others. 3 , 4 Additionally, visualization techniques such as streamlines, pathlines, or volume renderings can be utilized to show these parameters over the cardiac cycle, 5 , 6 or to study flow connectivity. 7 , 8
Segmentation is one of the most important, but also most challenging preprocessing step required to accurately separate the regions of interest from the rest of the image and facilitate computation of parameters, in research studies and clinical application of 4D Flow MRI. 4 Therefore, segmentation is often limited to the aorta during systole, utilizing the velocities during this stage to create a phase‐contrast angiographic image. 9 , 10 Segmenting the whole heart over the cardiac cycle is a challenging and time‐consuming process, as there is considerable motion and limited contrast between blood and tissue. Automatic methods aimed at segmenting 4D flow MRI images have achieved good results using atlas‐based techniques to locate the cardiac chambers and great thoracic vessels in all timeframes. 11 , 12 However, these methods make considerable use of computationally expensive registration techniques during execution, which results in relatively slow runtimes. Additionally, atlas‐based segmentation typically relies on a limited number of atlases, which can restrict the amount of morphological variation that it can be handled. 13
More recently, segmentation methods based on deep learning have been applied successfully to a variety of medical imaging tasks. 14 , 15 Training a deep learning convolutional neural network (CNN) to segment the regions of interest in an image requires a large amount of data and computational resources, but runtimes are generally short once trained. In the case of 4D flow MRI, a previous deep learning method has focused mainly on three‐dimensional segmentation of the aorta, 16 while cardiac segmentation approaches have mostly targeted different MR sequences or other imaging techniques such as CT and ultrasound. 17 , 18
Thus, the aim of this study was to develop and evaluate a deep learning‐based segmentation method to automatically segment the cardiac chambers and great thoracic vessels from 4D flow MRI over the cardiac cycle.
Materials and Methods
The study was performed in line with the declaration of Helsinki and was approved by the regional ethics board. The exams included retrospectively in this study were performed specifically for research purposes between 2011 and 2015 and included retrospectively in this study. All subjects gave written informed consent for MRI acquisition and data processing.
MRI Examinations
A total of 205 4D flow MRI datasets were acquired on a clinical 3 T Philips Ingenia scanner (Philips Healthcare, Best, The Netherlands). 4D Flow MRI scans were acquired for each subject directly after a gadolinium contrast agent (0.2 mmol/kg Gadovist, Bayer Schering Pharma AG) was injected into the subjects prior to the acquisition of a late‐enhancement study. Gadolinium administration to the healthy volunteers was approved by the ethical review board.
Free‐breathing, respiratory‐motion‐compensated, time‐resolved, velocity‐encoded 3D gradient echo MRI examinations were acquired using the following scan parameters: Sagittal‐oblique slab covering the whole heart and thoracic aorta, velocity encoding (VENC) 120‐150 cm/sec, flip angle 10°, echo time 2.5–2.6 msec, repetition time 4.2–4.4 msec, SENSE speed up factor 3 (anterior–posterior direction), k‐space segmentation factor 3, acquired temporal resolution of 33.6–52.8 msec reconstructed to 40 timeframes, acquired and reconstructed spatial resolution ~3 mm3, and elliptical k‐space acquisition. Weighted navigator gating with 4 mm in the inner 25% of k‐space and 7 mm in the outer parts of k‐space was used. Typical scan time was 10–15 minutes, with respiratory navigator efficiency of 60%–80%. The 4D flow MRI images were corrected for concomitant gradient fields on the MRI scanner. Phase wraps were corrected offline using a temporal phase unwrapping method, 19 and background phase errors were corrected using a weighted second‐order polynomial fit to the static tissue. 20
Study Population
A set of 212 4D flow MRI acquisitions were used during this study. The group included 40 healthy volunteers with no history of prior or current cardiovascular disease or cardiac medication, and 172 patients with a variety of medical disorders including chronic ischemic heart disease, idiopathic dilated cardiomyopathy, diastolic heart failure, and mild‐to‐moderate mitral valve regurgitation. Detailed subject demographics can be seen in Table 1.
TABLE 1.
Population Demographics
| Group size | Age (mean ± SD) | Gender (f/m) | |
|---|---|---|---|
| Healthy controls | 40 | 64.9 ± 4.5 | 11 |
| Chronic IHD | 80 | 67.3 ± 7.0 | 29 |
| Chronic IHD + MR | 22 | 68.4 ± 6.2 | 6 |
| Chronic IHD + AS | 8 | 68.7 ± 7.3 | 2 |
| DCMP | 30 | 57.7 ± 11.0 | 8 |
| ICMP | 20 | 67.7 ± 7.3 | 3 |
| Diastolic disfunction | 12 | 68.8 ± 10.5 | 6 |
| Total | 212 | 65.7 ± 9.2 | 65 |
IHD = ischemic heart diseases; MR = mitral regurgitation; AS = aortic stenosis; DCMP = nonischemic dilated cardiomyopathy; ICMP = ischemic cardiomyopathy.
Only the magnitude image included in each 4D flow MRI dataset was used as input to the network.
Ground Truth Generation
Segmentations corresponding to the four cardiac chambers: left ventricle (LV), left atrium (LA), right ventricle (RV), right atrium (RA), thoracic aorta (including ascending, proximal, and distal descending aorta), and pulmonary artery (including the pulmonary trunk and main left and right branches) were generated automatically using a previously developed method. 12 In brief, this method uses multiatlas segmentation at two time‐points of the cardiac cycle (end‐diastole and end‐systole) and registration between time‐points to generate time‐resolved segmentations of 4D flow MRI data. All the automatic segmentations were manually checked for defects and corrected when necessary at end‐diastole and end‐systole using the open source tool ITK‐SNAP. 21 Manual corrections were performed by one observer (M.B.) with 10 years of experience in cardiac MR imaging and postprocessing. After correction, the remaining timeframes were regenerated using registration and the results were visually inspected to avoid obvious failures.
Data Preparation
Seven acquisitions were excluded in which the atlas‐based segmentation method failed to generate a viable base segmentation, that is, where the correction would have required an almost completely manual segmentation. As a result, the final data set used was composed of 205 4D flow MR images (40 healthy volunteers and 165 patients).
For the purpose of training the neural network, the group was randomly divided into 70% (144/205, including 35 healthy volunteers) for training, 10% (20/205, including 2 healthy volunteers) for validation, and 20% (41/205, including 3 healthy volunteers) for testing.
Each MRI acquisition was reconstructed to cover the entire cardiac cycle in 40 timeframes, a 3D volume being generated at each timeframe. Accordingly, the CNN was trained using each timeframe as an independent segmentation. This resulted in 5760 3D volumes for training, 800 3D volumes for validation, and 1640 3D volumes for testing.
Network Architecture and Training Details
The model used is based on the 3D U‐net architecture, 22 which is typically used in medical image segmentation and consists of an encoder–decoder CNN with skip‐connections at every level. The implemented architecture includes four resolution steps in each direction, each step consisting of two convolutional blocks composed of a 3 × 3 × 3 kernel convolution with stride of one, followed by batch normalization, and a Parametric Rectified Linear Unit (PReLu) activation function. 23 Max pooling with kernel size 2 × 2 × 2 and stride of two is used to decrease the resolution at every step during the encoding half of the network, while transposed convolution with kernel size 2 × 2 × 2 and stride of two is used to increase the resolution in the decoding phase. The final layer of the network uses convolution with kernel size 1 × 1 × 1 to reduce the number of channels to 7, corresponding to the six regions of interest (LV, RV, LA, RA, Ao, and PA) included in the segmentation plus the background. The architecture of the network is shown in Supplemental Fig. S3.
The model was implemented using PyTorch 24 and was trained for 19 epochs with the following parameters: batch size = 16, AdamW optimizer 25 using Dice Score as loss without including the background label in the calculation, 26 initial learning rate was 0.01 scheduled to reduce by a factor of 0.1 with a patience of 5 epochs. Training took approximately 7 hours on an NVIDIA DGX‐2 server using data parallelism over four 32 GB Tesla V100 GPUs.
The following data augmentation transforms were used during training in order to increase the generalization capabilities of the model: Gaussian smoothing with sigma range [0.5–1.5], addition of Gaussian noise with sigma 0.1, contrast adjustment with gamma range [0.7–1.5], image flipping, addition of MR bias field, addition of MR motion artifacts, addition of ghosting artifacts with strength range [0.5–1.0]. All augmentations were generated on the fly during training with a probability of 0.15 for each transform.
To ease the generation of batches on‐the‐fly during the training process, the input images were resized to a size of 112 × 112 × 48. This volume contained all regions of interest, even in the largest cases.
The MONAI framework and the TorchIO library were used for data preprocessing and augmentation. 27 , 28
Evaluation
VOLUMETRIC ANALYSIS
For each cardiac chamber (LV, RV, LA, RA), the following clinical parameters were calculated for both deep‐learning and ground truth segmentations:
End‐diastolic volume (EDV): Volume of a cardiac chamber at end‐diastole.
End‐systolic volume (ESV): Volume of a cardiac chamber at end‐systole.
KINETIC ENERGY ASSESSMENT
For each region (LV, RV, LA, RA, Ao, PA) the total kinetic energy over the cardiac cycle (tot KE) was calculated for both deep‐learning and ground truth segmentations.
STATISTICAL ANALYSIS
The segmentations generated by the CNN on the test dataset were compared to their corresponding ground truth segmentations at all timeframes using the following metrics 29 :
- Dice score (DS) 26 : Measures the spatial overlap between regions (X and Y), calculated as:
- Hausdorff distance (HD) 30 : Maximum distance from a point in one set to the closest point in the other set. Calculated as:
where:
-
3Average surface distance (ASD) 31 : Computes closest distances from all surface points to the other surface and averages them. Calculated as:
- Sensitivity (true positive rate [TPR]): Proportion of all actual positives that are correctly predicted positive. Calculated using true positives (TP) and false negatives (FN) as:
- Precision (positive predictive value [PPV]): Proportion of all predicted positives that are actually positive. Calculated using true positives (TP) and false positives (FP) as:
- Miss rate (false negative rate [FNR]): Proportion of all actual positives that are wrongly predicted negative. Calculated as:
In addition to the segmentation agreement metrics outlined above, Bland–Altman analysis was used to evaluate agreement between the volumetric parameters determined from the deep learning and ground truth segmentations. The 95% confidence intervals for the mean differences and limits of agreement were calculated.
Results
The CNN was successfully created and trained using the training and validation sets. The resulting model was then applied to the test set to generate four‐dimensional segmentations of the cardiac chambers and great thoracic vessels. The trained network took approximately 6 seconds per subject, resulting in a complete segmentation for a 4D flow MRI dataset, including all timeframes. The best, median, and worst results (DS of 0.96, 0.91, 0.86, respectively) calculated by averaging the DS of each 4D flow MRI over time are shown in Figs. 1, 2, 3. Figure 1 displays the results as isosurface renderings of all the regions of interest at end‐diastole and end‐systole, while Fig. 2 shows similar results superimposed over a four‐chamber image of the heart generated by slicing the 4D flow MRI volume in the four‐chamber direction. Figure 3 depicts flow streamlines at mid‐systole in the segmented regions for the same cases. A limited number of timeframes are included in this section due to space constraints, time‐resolved versions of these images in video format have been included as Supplementary Material (S1).
FIGURE 1.
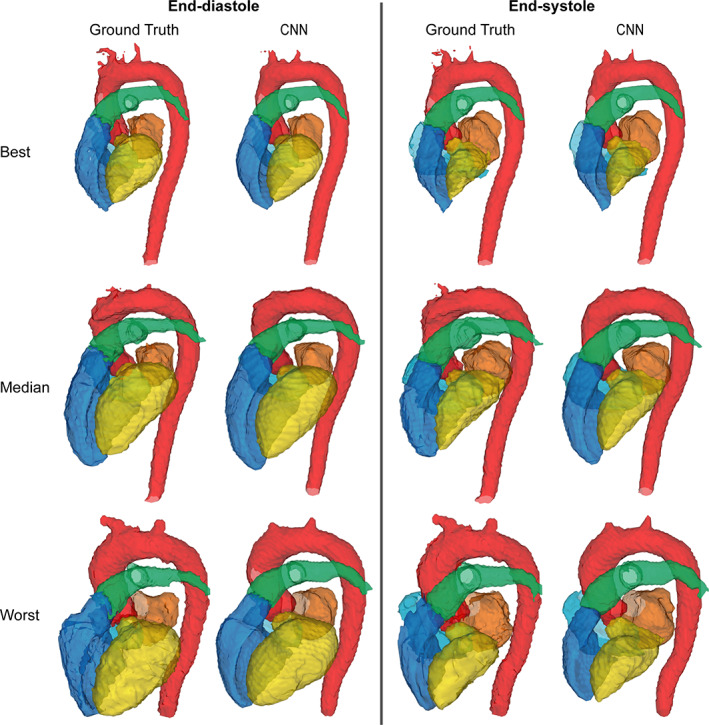
Results obtained in the best, median, and worst cases according to Dice scores visualized as isosurface renderings. Dice scores of 0.96, 0.91, and 0.86, respectively. Each case includes a comparison between the ground truth segmentations and those generated by the CNN at end‐diastole and end‐systole. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta, green: Pulmonary artery.
FIGURE 2.
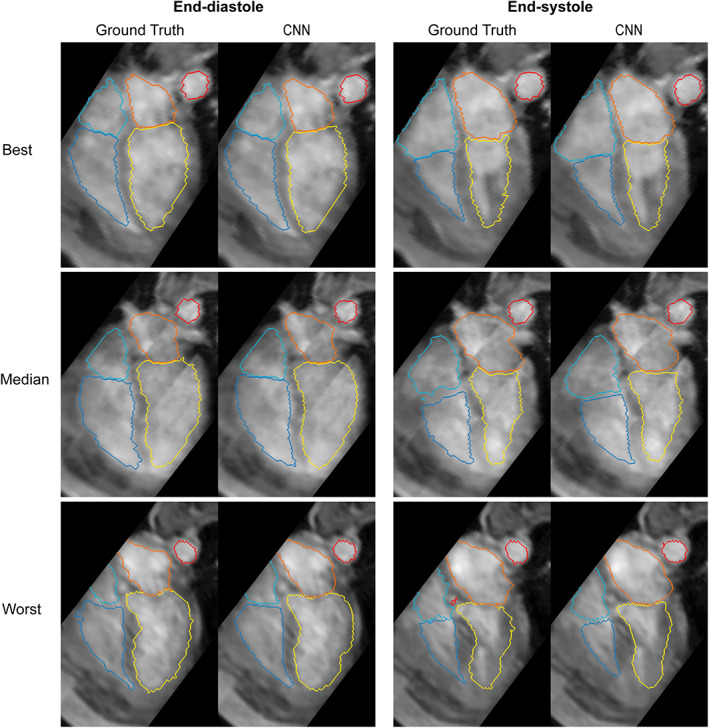
Results obtained in the best, median, and worst cases according to Dice scores. Dice scores of 0.96, 0.91, and 0.86, respectively. The segmentations have been superimposed over a four‐chamber image of the heart. Each case includes a comparison between the ground truth segmentations and those generated by the CNN at end‐diastole and end‐systole. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta.
FIGURE 3.
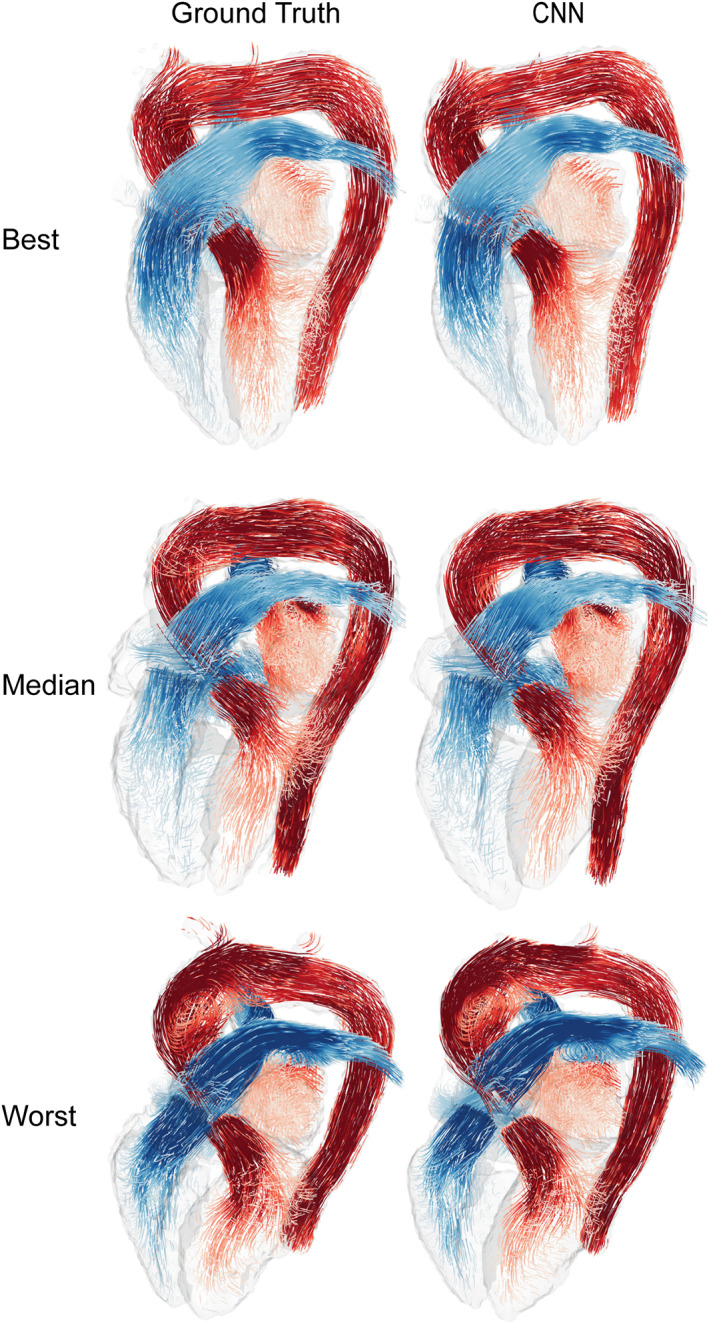
Flow streamlines generated at a mid‐systolic timeframe using the ground truth and CNN results for the best, median, and worst Dice scores. Dice scores of 0.96, 0.91, and 0.86, respectively. Pulmonary flows are depicted using shades of blue, and systemic flows using shades of red. Streamlines were generated independently for each region included in the segmentation.
Table 2 and Fig. 4 summarize the results for each of the chosen evaluation metrics. The subplots in Fig. 4 include values representing the results for each region of interest independently, together with the mean of all the regions. Note that in this figure, each result corresponds to a single timeframe out of the 40 that compose each 4D flow MRI; consequently, failure to segment one subject could potentially result in 40 low values in each plot. In general, all segments resulted in high scores, where the aorta showed the highest scores and the right atrium showed to be most challenging. Mean DS for all regions and timeframes was 0.908 ± 0.023, with similar values for sensitivity and precision: 0.907 ± 0.032, and 0.913 ± 0.028, respectively. Distance error measures (Hausdorff distance and average surface distance) resulted in lower values for all regions: 1.253 ± 0.293 mm and 0.466 ± 0.136 mm, respectively. Average miss rate for all regions was 0.093 ± 0.032.
TABLE 2.
Metric Results on the Test Dataset
| Metric | Mean | LV | LA | RV | RA | Ao | PA |
|---|---|---|---|---|---|---|---|
| Dice Score | 0.908 ± 0.023 | 0.913 ± 0.035 | 0.923 ± 0.032 | 0.895 ± 0.035 | 0.876 ± 0.048 | 0.934 ± 0.028 | 0.907 ± 0.028 |
| Hausdorff Distance | 1.253 ± 0.293 | 1.260 ± 0.427 | 1.195 ± 0.442 | 1.399 ± 0.696 | 1.571 ± 0.869 | 1.059 ± 0.271 | 1.037 ± 0.131 |
| Avg. Surface Distance | 0.466 ± 0.136 | 0.555 ± 0.203 | 0.450 ± 0.203 | 0.549 ± 0.217 | 0.584 ± 0.252 | 0.305 ± 0.139 | 0.355 ± 0.117 |
| Sensitivity | 0.907 ± 0.032 | 0.913 ± 0.052 | 0.923 ± 0.055 | 0.893 ± 0.061 | 0.880 ± 0.069 | 0.929 ± 0.040 | 0.906 ± 0.050 |
| Precision | 0.913 ± 0.028 | 0.912 ± 0.051 | 0.928 ± 0.047 | 0.902 ± 0.049 | 0.879 ± 0.070 | 0.940 ± 0.027 | 0.912 ± 0.036 |
| Miss Rate | 0.093 ± 0.032 | 0.087 ± 0.052 | 0.078 ± 0.055 | 0.107 ± 0.061 | 0.120 ± 0.069 | 0.071 ± 0.040 | 0.094 ± 0.050 |
Each value corresponds to mean ± standard deviation. LV = left ventricle; LA = left atrium; RV = right ventricle; RA = right atrium; Ao = aorta; PA = pulmonary artery. Distances are expressed in millimeters (mm).
FIGURE 4.
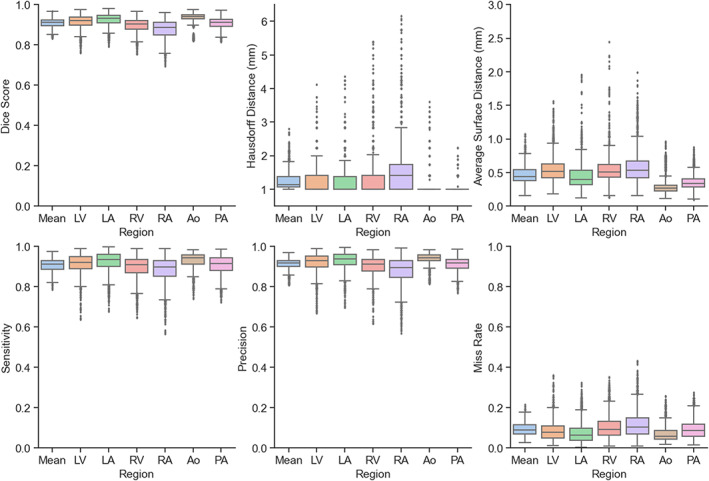
Metric results on the test dataset for each region included in the segmentations. LV = left ventricle; LA = left atrium; RV = right ventricle; RA = right atrium; Ao = aorta; PA = pulmonary artery.
The DS obtained for each timeframe of the 4D flow MRI images can be seen in Fig. 5. The topmost plot combines the DS for all regions, while the remaining plots show each label's result independently. The segmentations resulted in consistently high DS between 0.8 and 1.0 for most regions (LV, LA, Ao, and PA) at all timeframes, with slightly lower scores in a few timeframes corresponding to the right cardiac chambers. The RA seemed to be the most challenging region possibly due to inconsistencies in the acquisition's field of view, which affected its shape and size in 41% of cases in our dataset.
FIGURE 5.
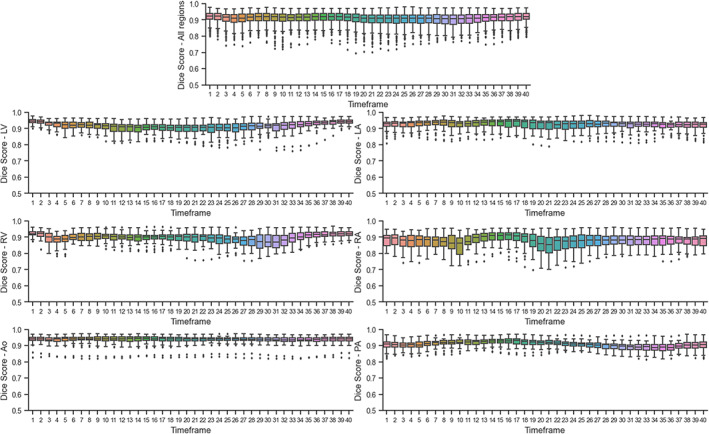
Dice scores calculated per timeframe on the test set. The topmost plot combines all regions. LV = left ventricle; LA = left atrium; RV = right ventricle; RA = right atrium; Ao = aorta; PA = pulmonary artery.
Figure 6 shows Bland–Altman plots corresponding to the cardiac volumetric parameters (EDV and ESV) for each cardiac chamber. A detailed summary of the results regarding the mean differences and limits of agreement can be seen in Table 3. Bland–Altman analysis using ED and ES volumes resulted in good agreements between the segmentations with average differences between −2.30 and 1.27 mL, while the limits of agreement expressed as percentage of the mean chamber volume ranged from 9.34% in the best case to 18.76% in the worst.
FIGURE 6.
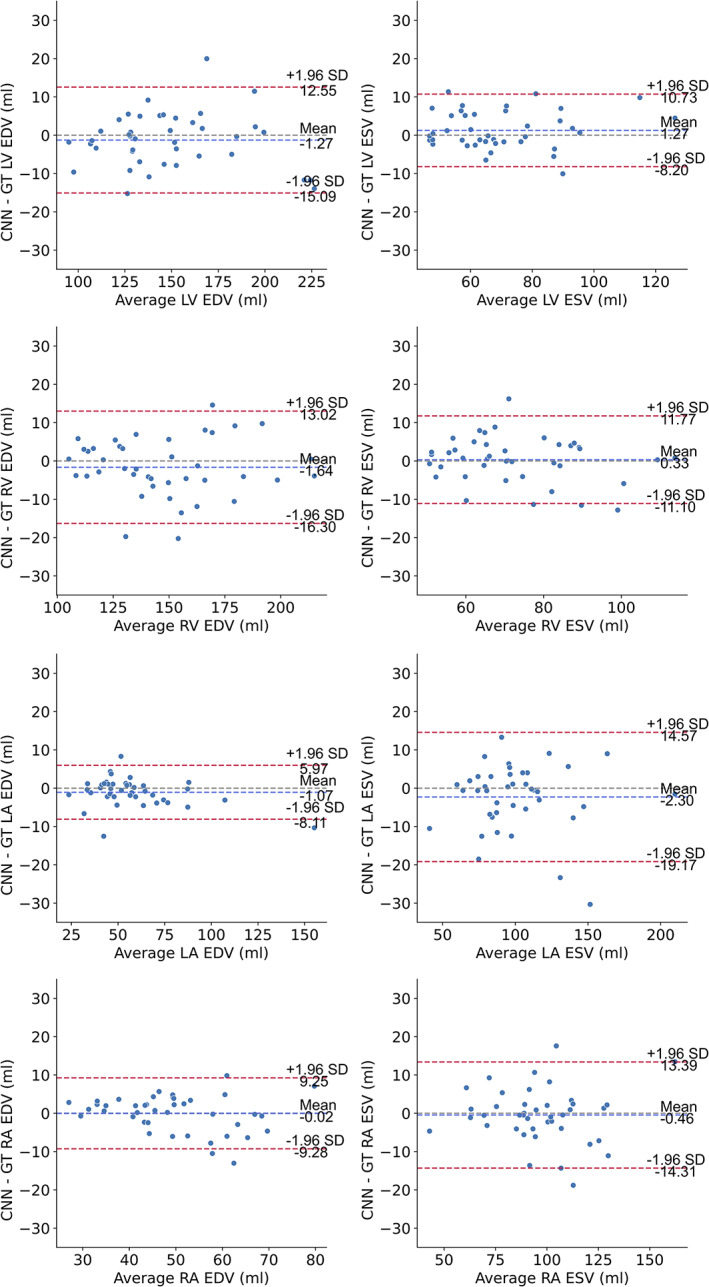
Bland–Altman plots of end‐diastolic volume (EDV) and end‐systolic volume (ESV) for the cardiac chambers. The dashed blue line shows the mean difference, while the dashed red lines denote the 95% limits of agreement (± 1.96 * standard deviation). GT = ground truth; LV = left ventricle, LA = left atrium; RV = right ventricle; RA = right atrium;EDV = end‐diastolic volume; ESV = end‐systolic volume.
TABLE 3.
Bland–Altman Analysis Results on the Test Dataset
| Region | LoA | LoA (% of mean volume/tot KE) | LoA 95% confidence interval | Mean diff. 95% confidence interval |
|---|---|---|---|---|
| LV EDV | 13.82 mL | 9.34 | (−18.65, 17.12) mL | (−3.02, 1.48) mL |
| LV ESV | 9.46 mL | 13.52 | (−10.23, 14.25) mL | (0.47, 3.55) mL |
| LA EDV | 7.04 mL | 12.40 | (−10.88, 7.33) mL | (−2.92, −0.62) mL |
| LA ESV | 16.87 mL | 16.91 | (−22.67, 20.97) mL | (−3.60, 1.90) mL |
| RV EDV | 14.66 mL | 9.98 | (−20.35, 17.57) mL | (−3.78, 1.00) mL |
| RV ESV | 11.44 mL | 15.69 | (−13.01, 16.56) mL | (−0.09, 3.64) mL |
| RA EDV | 9.27 mL | 18.76 | (−13.73, 10.23) mL | (−3.26, −0.24) mL |
| RA ESV | 13.85 mL | 14.41 | (−19.22, 16.60) mL | (−3.57, 0.95) mL |
| LV tot KE | 5.68 mJ | 7.08 | (−8.85, 5.83) mJ | (−2.43, −0.58) mJ |
| LA tot KE | 3.88 mJ | 11.07 | (−5.58, 4.54) mJ | (−1.09, 0.16) mJ |
| RV tot KE | 8.56 mJ | 13.78 | (−13.82, 8.30) mJ | (−4.15, −1.36) mJ |
| RA tot KE | 5.34 mJ | 14.54 | (−8.04, 5.76) mJ | (−2.01, −0.27) mJ |
| Ao tot KE | 17.28 mJ | 7.02 | (−28.93, 15.74) mJ | (−9.41, −3.78) mJ |
| PA tot KE | 7.98 mJ | 10.25 | (−10.47, 10.16) mJ | (−1.45, 1.15) mJ |
Limits of agreement (LoA) centered on the mean difference (in milliliters and millijoule) and expressed as a percentage of the mean volume or total kinetic energy. 95% confidence intervals are given for the limits of agreement and the mean differences. LV = left ventricle; LA = left atrium; RV = right ventricle; RA = right atrium; Ao = aorta; PA = pulmonary artery; EDV, end‐diastolic volume; ESV, end‐systolic volume; tot KE, total kinetic energy.
Figure 7 shows Bland–Altman plots corresponding to the kinetic energy (tot KE) for each region. The Bland–Altman analysis showed good agreements between the segmentation for all regions, with the mean of the differences ranging from −3.40 to 1.64 mJ, with the limits of agreements expressed as percentage of the mean kinetic energy for each region ranging from 7.02% to 13.78%. A detailed summary of the Bland–Altman analysis results is presented in Table 3.
FIGURE 7.
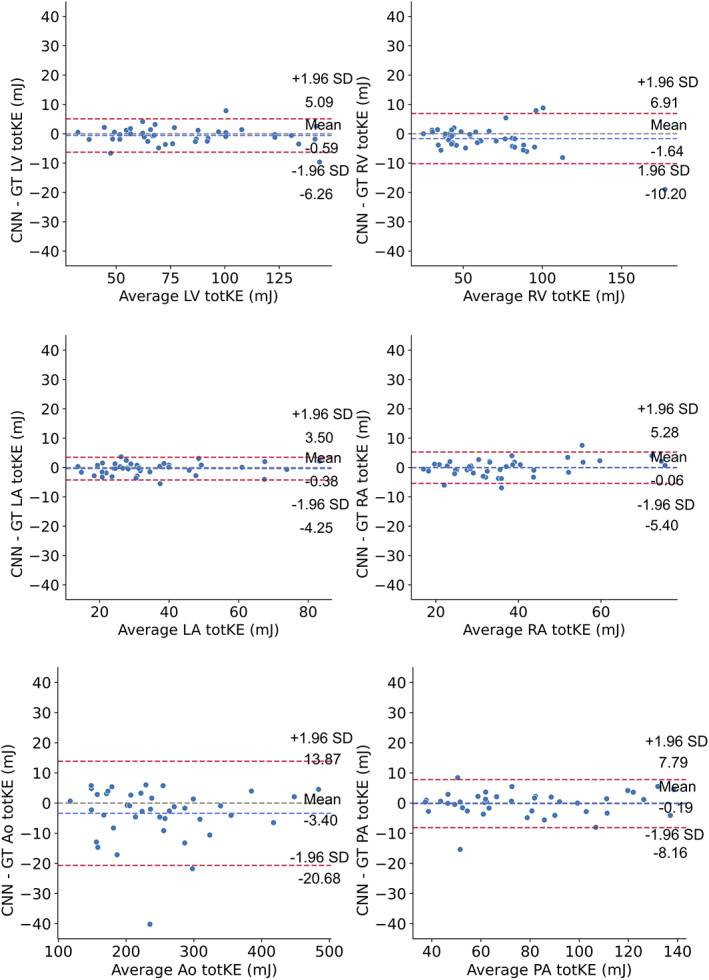
Bland–Altman plots of the total kinetic energy over the cardiac cycle, for all regions. The dashed blue line shows the mean difference, while the dashed red lines denote the 95% limits of agreement (± 1.96 * standard deviation). GT = ground truth; LV = left ventricle; LA = left atrium; RV = right ventricle; RA = right atrium; Ao = aorta; PA = pulmonary artery; tot KE: total kinetic energy.
Discussion
In this study, we have developed and evaluated a deep learning‐based method to segment the cardiac chambers, aorta, and pulmonary artery from 4D flow MRI data. Once generated, the segmentations can be used to expedite and automate flow assessment using the velocity data included in the acquisitions.
Evaluation of the trained network using segmentation metrics resulted in good scores overall. The best values were obtained in the aorta, probably due to its generally high contrast, and simple and consistent shape over the cardiac cycle. The left chambers of the heart and pulmonary artery also resulted in DS of over 0.9 when averaged over all timeframes, and favorable values on the remaining metrics. The model was able to adapt to the different shapes and sizes of the LV and LA, even though these change throughout the cardiac cycle. The right chambers of the heart had high but relatively lower DS, which could be explained by the more irregular shapes of these regions and their variability over time when compared to the more rounded left chambers. The RA in particular can present an extra challenge in these images since part of the chamber was left outside the image in a number of cases (83/205) which resulted in a wider variety of possible shapes and sizes for this label.
Very little variation was observed between the Dice scores obtained for the different timeframes corresponding to regions with the highest scores overall (LV, LA, aorta, and pulmonary artery), while some differences were visible in the RV and RA results at certain points during the cardiac cycle. In addition to the already discussed challenging shapes of these regions, we speculate that these small differences could be a consequence of the way the ground truth data was generated, by only manually correcting the atlas‐based segmentation at two timepoints and using registration to generate the remaining timeframes. Nevertheless, these results demonstrate the capability of the CNN to adapt to the different shapes and sizes of the cardiac chambers over time. In this setting, training the network with variety related to time in the data could be seen as a type of augmentation, which is a technique commonly used to improve neural network performance.
A further evaluation of the results of the network was performed by comparing the total kinetic energy for each region. The results of the Bland–Altman analysis showed generally a good agreement between ground truth and CNN segmentations for all regions. For the aorta, there were a few cases with a larger difference, which seem to be related to some slight undersegmentation in regions with high velocities due to an aortic valve stenosis. Only few cases with aortic stenosis were present in the training data, and we expect the results to improve by adding more.
Segmentation of whole heart 4D flow MRI over the cardiac cycle is challenging and few attempts have been published so far. A previously published CNN‐based segmentation method aimed at labeling the systolic anatomy of the aorta in 4D flow MRI resulted in similar metrics as the ones obtained by the proposed technique for the same region, with average DS of 0.951, and average Hausdorff Distance of 2.8 mm. 16 Additionally, a recent study used a fine‐tuned CNN originally trained on cine balanced steady‐state free precession (bSSFP) cardiac MR images to segment the ventricles in 4D Flow MRI resulting in average DS of 0.92 for the LV, and 0.86 for the RV. 18 These results are also comparable to the ones generated by the proposed method, with the added limitation of being restricted by the anatomy included in the bSSFP MR images, which typically only cover the cardiac ventricles with sufficient resolution to generate a complete segmentation.
A total of 8200 three‐dimensional segmentations were used during this study (205 acquisitions, 40 timeframes in each), with all segmentations including the four cardiac chambers, aorta, and pulmonary artery. It would be unfeasible to generate all the ground truth necessary to successfully train the model in a completely manual way; thus, an already developed atlas‐based automatic method was used to generate the segmentations and facilitate their correction. This could perhaps be seen as a form of transfer learning from one method to another. 32 One of the main disadvantages of atlas‐based segmentation is that the results depend on the ability of the registration technique to generate a suitable transformation between the atlases and the image to be segmented. The difficulty of this challenge increases as the image to segment differs from the atlases; consequently, the technique might fail to generate satisfactory results on images that are markedly different from the atlases in their morphology, that is, in cases of extremely enlarged ventricles, altered cardiac shapes, highly tortuous vessels, and in congenital heart disease. In the case of the deep learning‐based segmentation, the results should not be as strongly affected by morphological differences since the network has been trained on a larger, more diverse dataset. In comparison, the atlas‐based method included only eight 4D flow MR images as atlases (six healthy volunteers and two patients with normal left ventricular function at rest). Additionally, augmentation was used during training of the current model, which helps reduce overfitting and further increases the variation in the training dataset.
As an additional method of evaluation, we tried to segment some of the images that could not be included in this study since the atlas‐based method failed to generate a result that could be manually corrected in a relatively short amount of time. The proposed method managed to generate acceptable segmentations in these cases, a comparison of three of the results can be seen in Supplemental Figs. S1 and S2.
Specifically in the case of the aorta and pulmonary artery, the focus of the segmentation was to include the main structures of the vessels. The supra aortic branches, and the distal branches of the pulmonary artery were not expected to be fully segmented since they were not included in the ground truth segmentations in all cases.
The proposed method reduces the time required to generate a full four‐dimensional segmentation when compared to the previously published atlas‐based technique from 25 minutes to approximately 6 seconds per acquisition while running on the same hardware. 12 This improvement could make real‐time flow assessment feasible in the clinic.
Limitations
The model was trained using datasets where an extracellular Gadolinium contrast agent was used. The proposed technique is expected to handle a certain level of variation in blood‐tissue contrast due to the original differences already present in the training set, in addition to the augmentation transforms related to contrast adjustment and artifact simulation. However, if the goal is to perform similar segmentations on noncontrasted data, the model should be retrained using these data. Alternatively, previous studies have shown promising results using generative adversarial networks in order to emulate the use of contrast agents in medical images. 33 , 34
In a similar manner, all the data included in this study were acquired using one MR scanner, with a similar field of view, and no subjects with exceedingly atypical cardiovascular anatomies were included. The model might require some retraining, probably in the form of transfer learning, to perform at the same level using data from a wide range of MRI scanners and patient diseases.
We attempted to include as much variation as possible in our training distribution by manually correcting most of the available images, even those with the most diverging anatomies within our set. However, there was a few where the correction would have required almost a fully manual segmentation, and these cases were not included. The cause for atlas‐segmentation failure could have been related to anatomical reasons and consequently could have affected the variability in our training set. However, it is important to note that there are quite a few other reasons related to image quality that can influence the atlas‐based method such as blood‐tissue contrast, noise, and the presence of artifacts. As mentioned previously, manual correction of each 3D image used would have been an extremely time‐consuming task, even when the segmentations did not have to be created from scratch. In this study, manual corrections were performed only on end diastolic and end systolic frames of the atlas‐based segmentations, which might have affected the ground truths and consequently the CNN results. Nevertheless, visual assessment of the final time‐resolved ground truth segmentations showed no major issues due to this.
The proposed CNN takes 3D images including the complete field‐of‐view contained in the flow MRI acquisitions as inputs; therefore, the model requires a relatively large amount of GPU memory during training (approximately 70 GB with batches of size 16). Adding the velocity data during training could potentially improve the performance, given that the current model used only the magnitude image included in the 4D flow MR acquisitions. However, the increase in memory requirements for the network would be significant. To reduce these requirements, in addition to reducing the batch size, the model could be trained on 3D patches of smaller size; however, this will make the problem more challenging for the network and could affect the performance negatively. The model was designed to be completely automatic, as manual correction of the target images is quite time‐consuming. However, if desired, the method could be adapted to receive interactive annotations that could also help refine the network's training to improve future results.
Conclusion
The proposed method can generate a full time‐resolved segmentation of a 4D flow MRI image within seconds. The segmentations include all the regions of interest within the acquisition and might be used to perform a comprehensive assessment of the blood‐flow hemodynamics. Addition of this technique to the 4D flow MRI assessment pipeline may expedite and significantly improve the utility of this type of acquisition in the clinical setting.
Supporting information
Supplemental Figure 1 Three cases in which the atlas‐based segmentation method failed to generate a viable segmentation to use as ground truth, compared with the result of the proposed method at end‐diastole and end‐systole. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta.
Supplemental Figure 2 Three cases in which the atlas‐based segmentation method failed to generate a viable segmentation to use as ground truth, compared with the result of the proposed method at end‐diastole and end‐systole. The segmentations have been superimposed over a four‐chamber image of the heart. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta.
Supplemental Figure 3 Architecture of the U‐net network.
supplementary material
Description: Ground truth vs. CNN result for the best case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the median case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the worst case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the best case in the test set superimposed over a four‐chamber cardiac image.
Description: Ground truth vs. CNN result for the median case in the test set superimposed over a four‐chamber cardiac image.
Description: Ground truth vs. CNN result for the worst case in the test set superimposed over a four‐chamber cardiac image.
Description: Flow streamlines using the ground truth vs. CNN result for the best case.
Description: Flow streamlines using the ground truth vs. CNN result for the median case.
Description: Flow streamlines using the ground truth vs. CNN result for the worst case.
Acknowledgments
The authors would like to express their gratitude to Omkar Bhutra for his valuable support to this study. This work was funded by Sweden's Innovation Agency Vinnova, grant number 2017‐02447; the Swedish Research Council, grant number 2018‐04454; the Swedish Medical Research Council, grant number 2018‐02779; the Swedish Heart and Lung Foundation, grants number 20180657 and 20170440; and ALF Grants Region Östergötland, number LIO‐797721.
References
- 1. Markl M, Frydrychowicz A, Kozerke S, Hope M, Wieben O. 4D flow MRI. J Magn Reson Imaging 2012;36:1015‐1036. [DOI] [PubMed] [Google Scholar]
- 2. Stankovic Z, Allen BD, Garcia J, Jarvis KB, Markl M. 4D flow imaging with MRI. Cardiovasc Diagn Ther 2014;4:173‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Soulat G, McCarthy P, Markl M. 4D flow with MRI. Annu Rev Biomed Eng 2020;22:103‐126. [DOI] [PubMed] [Google Scholar]
- 4. Dyverfeldt P, Bissell M, Barker AJ, et al. 4D flow cardiovascular magnetic resonance consensus statement. J Cardiovasc Magn Reson 2015;17:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Köhler B, Born S, van Pelt RFP, Hennemuth A, Preim U, Preim B. A survey of cardiac 4D PC‐MRI data processing. Comput Graph Forum 2017;36:5‐35. [Google Scholar]
- 6. Julio G, J BA, Michael M. The role of imaging of flow patterns by 4D flow MRI in aortic stenosis. JACC Cardiovasc Imaging 2019;12:252‐266. [DOI] [PubMed] [Google Scholar]
- 7. Eriksson J, Carlhäll C, Dyverfeldt P, Engvall J, Bolger AF, Ebbers T. Semi‐automatic quantification of 4D left ventricular blood flow. J Cardiovasc Magn Reson 2010;12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fredriksson AG, Zajac J, Eriksson J, et al. 4‐D blood flow in the human right ventricle. Am J Physiol Heart Circ Physiol 2011;301:H2344‐H2350. [DOI] [PubMed] [Google Scholar]
- 9. Bock J, Frydrychowicz A, Stalder AF, et al. 4D phase contrast MRI at 3 T: Effect of standard and blood‐pool contrast agents on SNR, PC‐MRA, and blood flow visualization. Magn Res Med 2010;63:330‐338. [DOI] [PubMed] [Google Scholar]
- 10. Juffermans JF, Westenberg JJM, van den Boogaard PJ, et al. Reproducibility of aorta segmentation on 4D flow MRI in healthy volunteers. J Magn Reson Imaging 2021;53:1268‐1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bustamante M, Petersson S, Eriksson J, et al. Atlas‐based analysis of 4D flow CMR: Automated vessel segmentation and flow quantification. J Cardiovasc Magn Reson 2015;17:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bustamante M, Gupta V, Forsberg D, Carlhäll CJ, Engvall J, Ebbers T. Automated multi‐atlas segmentation of cardiac 4D flow MRI. Med Image Anal 2018;49:128‐140. [DOI] [PubMed] [Google Scholar]
- 13. Iglesias JE, Sabuncu MR. Multi‐atlas segmentation of biomedical images: A survey. Med Image Anal 2015;24:205‐219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60‐88. [DOI] [PubMed] [Google Scholar]
- 15. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019;29:102‐127. [DOI] [PubMed] [Google Scholar]
- 16. Berhane H, Scott M, Elbaz M, et al. Fully automated 3D aortic segmentation of 4D flow MRI for hemodynamic analysis using deep learning. Magn Reson Med 2020;84:2204‐2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C, Qin C, Qiu H, et al. Deep learning for cardiac image segmentation: A review. Front Cardiovasc Med 2020;7:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Corrado PA, Wentland AL, Starekova J, Dhyani A, Goss KN, Wieben O. Fully automated intracardiac 4D flow MRI post‐processing using deep learning for biventricular segmentation. Eur Radiol 2022. Advance online pubblication. 10.1007/s00330-022-08616-7. [DOI] [PubMed] [Google Scholar]
- 19. Xiang QS. Temporal phase unwrapping for CINE velocity imaging. J Magn Reson Imaging 1995;5:529‐534. [DOI] [PubMed] [Google Scholar]
- 20. Ebbers T, Haraldsson H, Dyverfeldt P. Higher order weighted least‐squares phase offset correction for improved accuracy in phase‐contrast MRI. In: Proceedings of the 16th annual meeting of ISMRM. Toronto, 2008. p 1367. [Google Scholar]
- 21. Yushkevich PA, Piven J, Hazlett HC, et al. User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006;31:1116‐1128. [DOI] [PubMed] [Google Scholar]
- 22. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U‐net: Learning dense volumetric segmentation from sparse annotation. Proceeding of the 19th medical image computing and computer‐assisted intervention conference (MICCAI 2016), Vol 9901. Cham: Springer; 2016. p 424‐432. [Google Scholar]
- 23. He K, Xiangyu Z, Shaoqing R, Jian S. Delving deep into rectifiers: Surpassing human‐level performance on ImageNet classification. IEEE Int Conf Comput Vis (ICCV) 2015;2015:1026‐1034. [Google Scholar]
- 24. Paszke A, Gross S, Chintala S, et al. Automatic differentiation in PyTorch. NIPS‐W; 2017. https://openreview.net/forum?id=BJJsrmfCZ [Google Scholar]
- 25. Loshchilov I, Hutter F. Decoupled weight decay regularization. International conference on learning representations (ICLR). New Orleans; 2019. 10.48550/arXiv.1711.05101 [DOI] [Google Scholar]
- 26. Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297‐302. [Google Scholar]
- 27. Ma N, Li W, Brown R, et al.: Project‐MONAI/MONAI: 0.5.0; 2021. 10.5281/zenodo.4679866 [DOI]
- 28. Pérez‐García F, Sparks R, Ourselin S. TorchIO: A python library for efficient loading, preprocessing, augmentation and patch‐based sampling of medical images in deep learning. Comput Methods Prog Biomed 2021;208:106236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med Imaging 2015;15:1‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gerig G, Jomier M, Chakos M. Valmet: A new validation tool for assessing and improving 3D object segmentation. In: Niessen WJ, Viergever MA, editors. Medical image computing and computer‐assisted intervention ‐‐ MICCAI, Vol 2001. Berlin, Heidelberg: Springer; 2001. p 516‐523. [Google Scholar]
- 31. Khotanlou H, Colliot O, Atif J, Bloch I. 3D brain tumor segmentation in MRI using fuzzy classification, symmetry analysis and spatially constrained deformable models. Fuzzy Set Syst 2009;160:1457‐1473. [Google Scholar]
- 32. Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. Proceeding of the 21st medical image computing and computer‐assisted intervention conference (MICCAI 2018), Vol 11141. Cham: Springer; 2018. p 270‐279. [Google Scholar]
- 33. Goodfellow IJ, Pouget‐Abadie J, Mirza M, et al. Generative adversarial nets. Adv Neural Inf Process Syst (NIPS) 2014;27:2672‐2680. [Google Scholar]
- 34. Bustamante M, Viola F, Carlhäll CJ, Ebbers T. Using deep learning to emulate the use of an external contrast agent in cardiovascular 4D flow MRI. J Magn Reson Imaging 2021;54:777‐786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Three cases in which the atlas‐based segmentation method failed to generate a viable segmentation to use as ground truth, compared with the result of the proposed method at end‐diastole and end‐systole. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta.
Supplemental Figure 2 Three cases in which the atlas‐based segmentation method failed to generate a viable segmentation to use as ground truth, compared with the result of the proposed method at end‐diastole and end‐systole. The segmentations have been superimposed over a four‐chamber image of the heart. Yellow: Left ventricle, orange: Left atrium, dark blue: Right ventricle, light blue: Right atrium, red: Aorta.
Supplemental Figure 3 Architecture of the U‐net network.
supplementary material
Description: Ground truth vs. CNN result for the best case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the median case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the worst case in the test set visualized as an isosurface rendering.
Description: Ground truth vs. CNN result for the best case in the test set superimposed over a four‐chamber cardiac image.
Description: Ground truth vs. CNN result for the median case in the test set superimposed over a four‐chamber cardiac image.
Description: Ground truth vs. CNN result for the worst case in the test set superimposed over a four‐chamber cardiac image.
Description: Flow streamlines using the ground truth vs. CNN result for the best case.
Description: Flow streamlines using the ground truth vs. CNN result for the median case.
Description: Flow streamlines using the ground truth vs. CNN result for the worst case.


