Abstract
The intercistronic gene junctions of vesicular stomatitis virus (VSV) contain conserved sequence elements that are important for polyadenylation and transcription termination of upstream transcript as well as reinitiation of transcription of downstream transcript. To examine the role of the putative polyadenylation signal 3′AUACU75′ at the gene junctions in polyadenylation and transcription termination, we constructed plasmids encoding antigenomic minireplicons containing one or two transcription units. In plasmid-transfected cells, analyses of the bicistronic minireplicon containing the wild-type or mutant intercistronic gene junctions for the ability to direct synthesis of polyadenylated upstream, downstream, and readthrough mRNAs showed that the AUACU7 sequence element is required for polyadenylation of VSV mRNA. Deletion of AUAC or U7 resulted in templates that did not support polyadenylation of upstream mRNA. Interestingly, we found that the loss of polyadenylation function led to antitermination of the upstream transcript and resulted in a readthrough transcript that contained the upstream and downstream mRNA sequences. Mutations that blocked polyadenylation also blocked transcription termination and generated mostly readthrough transcript. Reverse transcription-PCR of readthrough transcripts and subsequent nucleotide sequencing of the amplified product revealed no extra adenosine residues at the junction of the readthrough transcript. These results indicate that polyadenylation is required for transcription termination of VSV mRNA. The intergenic dinucleotide GA did not appear to be necessary for transcription termination. Furthermore, we found that insertion of the polyadenylation signal sequence AUACU7 alone was sufficient to direct polyadenylation and efficient transcription termination at the inserted site. Taken together, the data presented here support the conclusion that polyadenylation is the major determinant of transcription termination at the intercistronic gene junctions of VSV.
Vesicular stomatitis virus (VSV) is an enveloped, nonsegmented negative-strand RNA virus belonging to the family Rhabdoviridae and has served as an excellent model virus for many related negative-strand RNA viruses. The viral genomic RNA is 11,161 nucleotides in length (43) and is encapsidated by the viral nucleocapsid protein (N) to form a ribonucleoprotein structure that functions as the template for transcription and replication of the genome. Both transcription and replication are catalyzed by the viral RNA-dependent RNA polymerase complex which is comprised of the large protein (L) and the phosphoprotein (P) (9, 13, 36). Biochemical and genetic studies suggest that the L protein carries all of the enzymatic activities including the polymerization of nucleotides, methyl and guanylyl transferase activities, and poly(A) polymerase activity (17, 20, 45). Although no enzymatic activities have been detected in P protein, the phosphorylation status of this protein has been recently shown to influence the transcriptase and replicase functions of the L protein (38).
The viral genome consisting of five genes encoding the five structural proteins of the virion is organized in a modular fashion. Each gene is flanked by conserved sequence elements that play major roles in initiation and termination of transcription, capping, and polyadenylation. From available evidence, it seems that the VSV RNA polymerase initiates transcription from the very 3′ terminus of the genome (12) and transcribes the genes in a sequential manner (1, 3). In doing so, the viral RNA polymerase first synthesizes a small 47-nucleotide-long leader RNA which is uncapped and nonpolyadenylated and does not encode any viral protein. Following the leader RNA synthesis, five capped and polyadenylated mRNAs encoding each of the individual viral proteins are transcribed sequentially following the order of the genes (3′-N-P-M-G-L-5′) in the genome (1, 3). There exists a gradient in the molar amounts of the mRNAs which also follows the gene order from the 3′ end of the genome so that the 3′-proximal gene is transcribed most frequently and the 3′-distal gene is transcribed least frequently. The gradient in the molar amounts of the mRNAs is believed to be due to attenuation at each of the gene junctions during transcription (23), which may be a result of the inability of the polymerase to reinitiate transcription of the downstream gene following transcription termination and polyadenylation of the upstream mRNA. It is unresolved whether transcription is carried out by the VSV polymerase entering the template only at the 3′ terminus of the genome followed by a stop-start mode of RNA synthesis (12) or by internal initiation by the polymerase (49). Recent studies using PolR1 mutants suggest that VSV mRNA synthesis may be initiated internally (7).
The conserved sequences present at each of the gene junctions of VSV (30, 35, 40) are presumed to be involved in transcription termination and polyadenylation of upstream mRNA and reinitiation of transcription of downstream mRNA. Consistent with this view, it was recently shown that the 23-nucleotide-long conserved intercistronic sequence element 3′-AUACU7(G/C)AUUGUCnnUAG5′ (where n is any nucleotide) can direct expression of a foreign gene in VSV (44). Recent mutational analyses have shown that 3′UUGUC5′ sequence element is required for efficient transcription reinitiation of downstream mRNA following termination and polyadenylation of upstream mRNA (48). The nontranscribed intergenic dinucleotide GA has been shown to play a role in transcription termination (48), although in a separate study (4) the GA dinucleotide has been proposed to function primarily as a spacer element that is required by the VSV polymerase to reinitiate upstream mRNA transcription following termination of downstream mRNA synthesis. Thus, it appears that the dinucleotide may play only an indirect role in transcription termination. The role of 3′AUACU75′ in transcription has not been determined as yet, although this sequence element has been proposed to be the signal that directs polyadenylation of the upstream mRNA by stuttering of the polymerase at U7 residues (46).
In this study, we used transcription- and replication-competent minireplicons of VSV to address the role of the putative polyadenylation signal 3′AUACU75′ in transcription termination and polyadenylation. Plasmids encoding mono- or bicistronic minigenomes of VSV were used in a system developed in our laboratory (27, 37, 39) to examine the effects of mutations in 3′AUACU75′ sequence element on transcription termination and polyadenylation. Our results suggest that deletion of the putative polyadenylation signal abrogates polyadenylation of the upstream transcript, thus establishing that AUACU7 is the polyadenylation signal in the VSV genome. More interestingly, our results show that abrogation of polyadenylation leads to antitermination of transcription, suggesting that polyadenylation of upstream mRNA is the critical requirement for its termination, whereas the dinucleotide GA may not play a direct role in transcription termination. We also show that the AUACU7 sequence alone can induce polyadenylation and termination of transcription at the inserted site in a heterologous context. Our results suggest that VSV RNA polymerase must polyadenylate its mRNAs to generate monocistronic mRNAs.
MATERIALS AND METHODS
Cells, viruses, and VSV protein expression plasmids.
Growth and maintenance of baby hamster kidney (BHK-21) cells and human 143B (thymidine kinase-negative) cells have been described before (27). VSV (Indiana serotype, San Juan strain) was propagated and titrated in BHK-21 cells. Recombinant vaccinia virus (vTF7-3) carrying the bacteriophage T7 RNA polymerase gene (14) was propagated in BHK-21 cells, and titers of stock virus were determined in 143B cells as described previously (27).
Plasmids pN, pP, and pL carrying the coding sequences of VSV proteins N, P, and L respectively, under the control of T7 RNA polymerase promoter have been described elsewhere (39).
Construction of plasmids encoding minireplicons and mutants.
Plasmid p10BN, encoding the antigenomic positive-sense minireplicon containing the coding sequence of the N gene of VSV, was generated by inactivating the unique BglII site that was present immediately downstream of the N gene coding sequence in the previously described plasmid, p9BN (27). When transcribed by T7 RNA polymerase, plasmid p10BN generates a 1,617-nucleotide-long antigenomic-sense RNA containing the first 63 nucleotides from the 5′ terminus of VSV antigenome, the N gene coding sequence (nucleotides 64 to 1339), an extra 11 nucleotides of non-VSV sequences to introduce a unique HpaI site to facilitate further subcloning, and 265 nucleotides from the 3′ terminus of the VSV antigenome.
Plasmid p10BNP, encoding the bicistronic minireplicon, was generated by inserting a PCR-amplified P gene fragment corresponding to nucleotides 1369 to 2217 followed by the sequence CCCGGGCTAAGTG to introduce a unique SmaI site immediately following the P gene coding sequence, at the unique HpaI site of plasmid p10BN. The resulting plasmid, p10BNP, was then digested with SmaI and AflII to delete the L gene sequences and ligated after generation of blunt ends with mung bean nuclease. The resulting plasmid, p11BNP, encoded the bicistronic antigenomic-sense minireplicon in which transcription initiation, termination, and polyadenylation of N and P mRNAs are controlled by their cognate signals. When transcribed by T7 RNA polymerase, plasmid p11BNP generates a 2,238-nucleotide-long transcript containing 63 and 46 nucleotides, respectively, from the 5′ and 3′ termini of the VSV antigenome, flanking the coding sequence of the N, the N-P intercistronic gene junction, and the P gene.
Mutations at the N-P intercistronic gene junctions in p11BNP were introduced by PCR using Pwo polymerase (Boehringer Mannheim, Indianapolis, Ind.). Negative-sense primers containing a unique NdeI site along with the desired mutations that annealed to sequences around the N-P gene junction and a positive-sense primer that annealed to a region in the P gene (nucleotides 1841 to 1851) with the unique PflMI site were used to amplify a DNA fragment by PCR with p11BNP as the template. The PCR product was then digested with NdeI and PflMI and ligated into p11BNP DNA that had been digested with the same enzymes. Following transformation of competent Escherichia coli DH5α cells, bacterial colonies containing mutant p11BNP plasmids were screened and identified by nucleotide sequencing. Insertion mutants of p10BN were also generated similarly by PCR using p10BN as the template. The primers used in this PCR amplification corresponded to the negative-sense primer containing the desired insertion sequences followed by a unique BsmI site (at position 961 within the N gene) and a positive-sense primer that annealed to the beginning of the coding region of the N gene. The resulting PCR product was digested with BglII (unique site at position 160 in the N gene) and BsmI and ligated into p10BN that had been digested with the same enzymes. After transformation, mutant plasmids were identified by nucleotide sequencing. Established methods of DNA manipulation and preparation of plasmids (2, 32) were used.
Virus infections and DNA transfections.
The methods used have been previously described (27, 37, 38). Briefly, BHK-21 cells were grown in 60-mm-diameter plates to about 90% confluency. The cells were infected with the recombinant vaccinia virus (vTF7-3) at a multiplicity of infection of 10. Following virus adsorption at 37°C for 45 min, cells were washed in serum-free Dulbecco’s modified Eagle medium (DMEM) and then transfected with various combinations of plasmid DNAs by using Lipofectin reagent (Gibco/BRL, Bethesda, Md.). Medium from transfected cells was removed at 4 h posttransfection; cells were washed twice in DMEM containing 2% fetal bovine serum (FBS) and incubated with an appropriate volume of the same medium containing 25 μg of cytosine β-d-arabinofuranoside (AraC; Sigma Chemical, St. Louis, Mo.) per ml. For minigenome expression experiments, 5 μg of the plasmid encoding minigenomes was cotransfected along with 3 μg of pN, 0.5 μg of pP, and 1 μg of pL.
Metabolic labeling and analysis of VSV-specific RNAs.
To detect VSV-specific transcription and replication products, transfected cells were pretreated with 15 μg of actinomycin D (Merck and Co., Inc., Rahway, N.J.) and 25 μg of AraC per ml of DMEM containing 2% FBS for 45 min at 16 h posttransfection and then labeled with 15 μCi of [3H]uridine and/or 15 μCi of [3H]adenosine per ml of DMEM-FBS containing actinomycin D and AraC. Labeling of RNA was performed for 6 h. After labeling, cytoplasmic extracts were prepared and total RNA from the extracts were recovered by extraction with phenol and chloroform. When necessary, total labeled or unlabeled RNAs isolated from transfected cells were fractionated into polyadenylated and nonpolyadenylated RNAs by oligo(dT)-cellulose (Gibco/BRL) chromatography following the manufacturer’s protocol. The recovered RNAs were electrophoresed in an acid agarose-urea gel as described before (26, 38) and detected by fluorography (24).
RT-PCR of readthrough transcripts.
Total RNA from cells transfected with wild-type or mutant bicistronic minireplicons were prepared and treated with RNase-free RQ1 DNase (Promega Biotech, Madison, Wis.) to remove any contaminating transfected plasmid DNAs that may have been present in the preparation. The total RNA was then passed through an oligo(dT)-cellulose column to select for polyadenylated RNA. First-strand cDNA synthesis and subsequent PCR amplification using the polyadenylated RNA were performed according to the protocol described in the reverse transcription (RT)-PCR kit from Stratagene (San Diego, Calif.). Briefly, a primer complementary to the positive-sense P gene sequence from nucleotides 1526 to 1515 was used to prime synthesis of first-strand cDNA from polyadenylated RNA fraction, using Moloney murine leukemia virus reverse transcriptase (Gibco/BRL) for 1 h at 37°C. The reaction mixture was then heat inactivated at 90°C for 5 min. One-tenth of the cDNA product was used in PCR amplification using the above P gene primer and another primer that is complementary to nucleotides 1247 to 1280 of the negative-sense N gene sequence. The PCR conditions were denaturation at 94°C for 1 min, annealing at 54°C for 1 min, and extension at 72°C for 90 s for 35 cycles, followed by a final extension reaction at 72°C for 10 min. Identical PCR conditions were used for amplification of DNA using the p11BNP template and the same primers. After amplification, the DNA products were analyzed by electrophoresis in an agarose gel and visualized by staining with ethidium bromide. For sequence analysis, the PCR products were separated in low-melting-point agarose gel, recovered by extraction, and used for DNA sequencing.
RESULTS
Synthesis of mRNAs from mono- and bicistronic minigenomes of VSV.
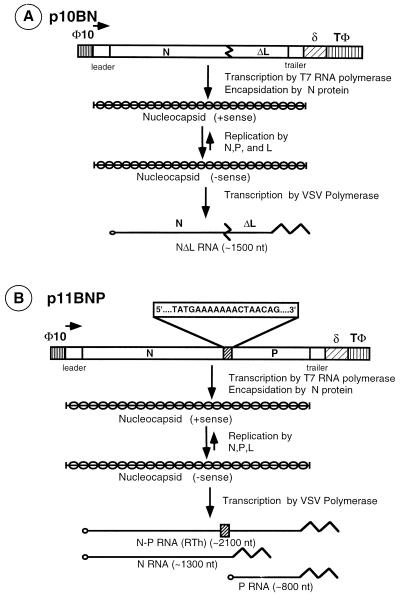
To investigate the role of conserved sequences at the N-P gene junction of VSV in transcription termination and polyadenylation of mRNA, we designed two plasmids that encode antigenomic positive-sense minireplicons of VSV containing either one (N alone) or two (N and P) transcription units (Fig. 1). The 10BN antigenomic minireplicon contains the coding sequences for the N gene (nucleotides 64 to 1339) flanked by the first 63 nucleotides of the 5′ terminus and the last 265 nucleotides of the 3′ terminus of VSV antigenome. The 11BNP antigenomic minireplicon contains coding sequences for the N gene (nucleotides 64 to 1339), the conserved intergenic sequences of the N-P gene junction (nucleotides 1370 to 1395), and the coding sequences for the P gene (nucleotides 1396 to 2218), which are flanked by the first 63 nucleotides of the 5′ terminus and the last 46 nucleotides of the 3′ terminus of the VSV antigenome. In cells transfected with plasmids encoding the minireplicons, the minireplicon RNAs are synthesized by T7 RNA polymerase with two additional guanosine (G) residues at the 5′ terminus (37), and the 3′ terminus of these transcripts, which is generated as a result of autolytic cleavage by the hepatitis delta virus ribozyme (37), is identical in nucleotide sequence to that of the VSV antigenome.
FIG. 1.
Diagrammatic representation of plasmids encoding the monocistronic (A) and bicistronic (B) minireplicons of VSV. Synthesis of antigenomic positive-sense minireplicons from the plasmids by T7 RNA polymerase is initiated at the T7 RNA polymerase promoter (φ10) and terminated at the T7 RNA polymerase terminator (Tφ), resulting in transcripts with two extra guanosine residues at the 5′ terminus. The 3′ terminus of the transcripts, which is generated due to autolytic cleavage by the hepatitis delta virus ribozyme (δ) (37), is identical in nucleotide sequence to that of the VSV antigenome. Various replication and transcription products with approximate sizes (in nucleotides [nt]) predicted to be generated from the minireplicon templates by VSV RNA polymerase are shown. RTh, readthrough product of transcription.
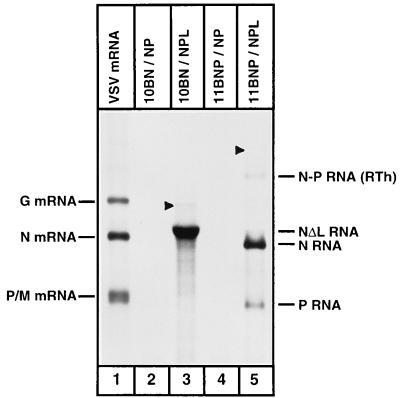
To examine various RNA species synthesized from these minireplicons by VSV RNA polymerase, we cotransfected the plasmids encoding the VSV proteins N, P, and L and either plasmid p10BN or plasmid p11BNP into cells infected with vTF7-3. We then metabolically labeled the RNAs in the presence of actinomycin D, a drug that inhibits RNA synthesis from DNA templates without affecting RNA synthesis by VSV RNA polymerase. Total RNAs from cytoplasmic extracts of these cells were then analyzed by agarose-urea gel electrophoresis. A fluorogram of such an analysis is shown in Fig. 2. The major RNA species (NΔL RNA) synthesized from the 10BN minireplicon (lane 3) migrated with slightly slower electrophoretic mobility than the authentic N mRNA synthesized in VSV-infected cells (lane 1). This was expected since the mRNA generated from this minireplicon is predicted to be about 170 nucleotides longer [excluding the poly(A) tail] than the authentic N mRNA (Fig. 1). The faint slower-migrating band (indicated by an arrowhead in lane 3) represents the genomic-sense minireplicon, which is the product of replication of the antigenomic-sense minireplicon. This RNA is immunoprecipitable by anti-N protein antibody and also does not bind to oligo(dT)-cellulose column (data not shown). The antigenomic-sense minireplicon, which migrates slightly faster than the genomic-sense minireplicon, was not detectable under these conditions, since the level of the antigenomic RNA is less than 10% of that of the genomic RNA (27). None of these RNA species were synthesized in cells that did not express the polymerase protein L (lane 2). The bicistronic minireplicon 11BNP directed synthesis of two major species of RNA (lane 5) with sizes corresponding to the predicted sizes of N and P RNAs. An additional transcript (N-P RNA) was also clearly detected (lane 5). The size of this transcript [approximately 2,100 nucleotides, excluding the poly(A) tail] is consistent with it being the readthrough product of transcription containing the entire sequences of both transcription units and is generated when the transcription termination signals at the N-P gene junction are ignored by the VSV RNA polymerase. In addition to these transcripts, the genomic-sense minireplicon of 11BNP (faint band indicated by an arrowhead in lane 5) was also consistently detected. The corresponding antigenomic-sense minireplicon was not detectable.
FIG. 2.
Transcription and replication of the minireplicons. BHK-21 cells were infected with vTF7-3 and cotransfected with plasmids encoding the minireplicons as well as the N, P, and/or L proteins (as shown above each lane). Cells were labeled with [3H]uridine in the presence of actinomycin D as described in Materials and Methods. Total labeled RNAs from cytoplasmic extracts of cells were analyzed in an acid-agarose urea gel and detected by fluorography. Lane 1, VSV mRNAs from infected cells. Transcription products from minireplicons are shown on the right. Arrowheads in lanes 3 and 5 show the genomic-sense minireplicons. RTh is the readthrough product of transcription.
Deletion of polyadenylation signal(s) abrogates transcription termination.
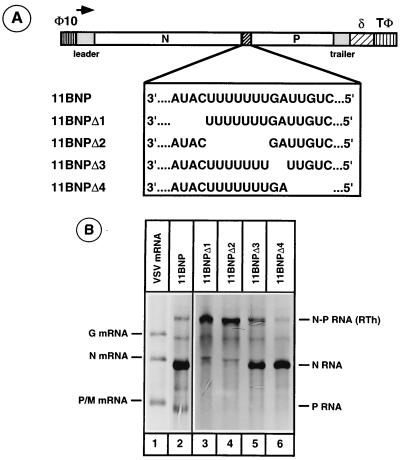
The intercistronic gene junctions of VSV contain highly conserved sequence elements 3′AUACU7(G/C)AUUGUC5′ that play key roles in polyadenylation and transcription termination of upstream mRNA and reinitiation of downstream mRNA transcription. To examine the role of these conserved sequences, we generated a series of deletion mutants of the 11BNP minireplicon (Fig. 3A): 11BNPΔ1, in which AUAC was deleted; 11BNPΔ2, in which U7 was deleted; 11BNPΔ3, in which the GA dinucleotide was deleted; and 11BNPΔ4, in which the UUGUC sequence was deleted. The ability of these mutant minireplicon templates to direct transcription of upstream, downstream, and readthrough mRNAs was examined in cells transfected with plasmids encoding the minireplicons and the plasmids encoding VSV proteins N, P, and L (Fig. 3B). Compared to the template containing the wild-type N-P gene junction sequences (lane 2), the deletion mutant minireplicon templates exhibited very different abilities to synthesize various RNA species. Deletion of downstream transcription initiation signal sequence UUGUC resulted in ablation of downstream P RNA synthesis but had little or no effect on upstream N RNA or readthrough N-P RNA synthesis (lane 6). Deletion of intergenic dinucleotide GA sequence not only abrogated downstream P RNA synthesis but also increased the amount of readthrough RNA (lane 5). This is consistent with a recent finding (4) which suggests that deletion of intergenic dinucleotide GA results in increased readthrough at the gene junction. Interestingly, the termination of upstream RNA was not affected. In contrast, the template with deletion of AUAC sequence supported synthesis of large amounts of readthrough RNA and significantly low levels of upstream N RNA (lane 3). Upon longer exposure of the gel, the downstream P RNA could also be detected. Deletion of the U7 sequence element, on the other hand, resulted in the synthesis of only the readthrough RNA (lane 4). Synthesis of upstream or downstream mRNAs could not be detected, even upon very long exposure of the fluorogram.
FIG. 3.
(A) 11BNP and mutant minireplicons encoding deletion of various regions of the intercistronic gene junction. (B) Analysis of RNAs generated from various deletion mutant minireplicons. Cells were infected with vTF7-3 and cotransfected with plasmids encoding the minireplicons as well as the N, P, and L proteins. Cells were labeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D and AraC as described in Materials and Methods. Total labeled RNAs from cytoplasmic extracts of cells were electrophoresed in an acid-agarose urea gel and detected by fluorography. Lane 1 shows VSV mRNAs from infected cells. VSV RNA polymerase catalyzed transcription products are shown on the right. RTh, readthrough product of transcription.
Since the AUACU7 sequence forms the polyadenylation signal of VSV mRNAs (22), the results presented above suggest that polyadenylation may be the primary determinant of termination of VSV mRNA transcription. Removal of any part of the polyadenylation signal severely impairs termination of upstream mRNA as well as increases the amount of readthrough mRNA. The intergenic dinucleotide sequence GA and/or the initiating pentanucleotide sequence UUGUC may not play a direct role in mediating the upstream mRNA transcription termination since deletion of GA or UUGUC sequence elements did not affect termination of upstream N RNA. The mutant template in which the AUAC sequence has been deleted still generated low levels of upstream RNA. We reason that this template supports low levels of polyadenylation of upstream RNA and therefore synthesizes low levels of terminated upstream RNA, since the four nucleotides upstream of the U7 sequence in this deletion mutant may still support low level of polyadenylation (22).
Readthrough transcripts are polyadenylated at the 3′ terminus and do not contain additional adenosine residues at the N-P junction.
Results shown in Fig. 3B suggested that polyadenylation of VSV mRNA appeared to be a prerequisite for termination of transcription at the intercistronic gene junctions. However, we could not rule out the possibility that the readthrough RNAs which are generated due to the inability of upstream RNA to terminate at the gene junction may still contain extra adenosine residues at the junction of the readthrough transcripts. Such a possibility would then argue against the involvement of polyadenylation in transcription termination.
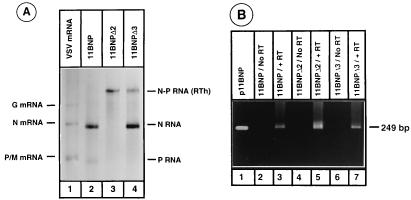
To investigate this possibility, we first analyzed the polyadenylated RNAs from cells transfected with plasmids encoding various 11BNP mutant minireplicons, using oligo(dT)-cellulose chromatography (Fig. 4A). Both N and P transcripts generated from the wild-type 11BNP minireplicon were polyadenylated, as evidenced by their ability to bind to oligo(dT)-cellulose column (lane 2). This was expected since both transcription units contained intact polyadenylation signals. A small amount of readthrough RNAs also bound to oligo(dT)-cellulose column. This readthrough product most likely used the downstream P RNA polyadenylation signal to generate its poly(A) tail (see below). The mutant in which the U7 sequence was deleted generated only the polyadenylated readthrough RNA (lane 3). This polyadenylated readthrough RNA was also the major product from AUAC deletion mutant, although a very low level of the polyadenylated upstream N RNA was detected (data not shown). Deletion of intergenic dinucleotide GA led to the synthesis of the polyadenylated readthrough RNA as well as the polyadenylated upstream mRNA (lane 4). To determine whether nonpolyadenylated N RNA transcripts were being synthesized from the mutant minireplicon templates, the RNAs present in the unbound fraction from oligo(dT)-cellulose column were also analyzed. However, we were unable to detect nonpolyadenylated N RNA transcripts from the wild-type or mutant minireplicon templates (data not shown).
FIG. 4.
(A) Oligo(dT) selection of RNAs produced in cells transfected with 11BNP minireplicons containing wild-type or mutated intercistronic gene junctions (as shown above each lane). Total labeled RNAs from transfected cells were obtained as described in the legend to Fig. 3. Polyadenylated RNA species were selected by oligo(dT)-cellulose chromatography and analyzed as for Fig. 3. Oligo(dT)-selected VSV mRNAs are shown in lane 1. (B) RT-PCR amplification of polyadenylated readthrough (RTh) RNAs. The template RNAs for RT-PCR were obtained as described in Materials and Methods and subjected to RT-PCR amplification. The amplified products were electrophoresed in a 1.8% agarose gel and visualized by staining with ethidium bromide. The 249-bp amplified product is the predicted-size DNA from p11BNP template DNA (lane 1).
Although the readthrough RNA product is polyadenylated, it is possible that the intervening sequences between the upstream and downstream RNA sequences of the readthrough transcript contain additional adenosine residues at the N-P junction as a result of polyadenylation that could help binding of readthrough RNAs to oligo(dT)-cellulose. To examine this, we used RT-PCR of polyadenylated readthrough transcripts to determine the size of the readthrough junctions. Total RNA from cytoplasmic extracts of transfected cells were extracted and treated with RNase-free DNase to remove the contaminating transfected plasmid DNAs which could be used as templates during subsequent PCR amplification. This was followed by selection of only the polyadenylated transcripts by oligo(dT)-cellulose. The polyadenylated transcripts were reverse transcribed by using a primer that annealed to a region (nucleotides 1526 to 1515) of the positive-sense P gene sequence. The cDNA was then subjected to PCR using the same primer along with another primer that annealed to a region (nucleotides 1247 to 1280) of negative-sense N gene sequence. The amplified product from the p11BNP template is predicted to be 249 bp in length. In this reaction, only the readthrough transcripts containing the junction sequences between the upstream and downstream transcripts will be amplified, and the presence of extra sequences at the junction may be inferred by comparing the size of the amplified DNA fragment to that of a product amplified from the parental plasmid p11BNP.
As can be seen from Fig. 4B, amplification of 11BNP template generated the expected-length (249-bp) fragment (lane 1). RT-PCR amplification of polyadenylated RNAs from cells transfected with the wild-type or mutant minireplicons also generated DNA fragments (lanes 3, 5, and 7) that comigrated with the 249-bp DNA fragment obtained from p11BNP template DNA. Such DNA fragments were not generated in the absence of RT (lanes 2, 4, and 6), indicating that these DNA products were generated only from the readthrough transcripts. Since the RT-PCR products of readthrough transcripts of wild-type, U7 deletion, and GA deletion mutants comigrated with the 249-bp fragments, the data suggested that no extra adenosine residues were incorporated at the junction of the polycistronic transcript as the polymerase read through the junction. To unequivocally ascertain this, we also determined the nucleotide sequence of the junction region of the RT-PCR amplification products. For the wild-type template and each of the mutant templates producing readthrough transcripts, an exact copy of the sequence corresponding to its cognate plasmid template was found at the junction of the readthrough transcripts. The lack of extra adenosine residues at the junction of the readthrough RNAs suggests that deficiency in polyadenylation leads to antitermination of upstream RNA and production of readthrough RNA. These results together with the earlier results suggest that the ability of an mRNA to be terminated depends on its ability to be polyadenylated.
Further evidence that polyadenylation is required for transcription termination.
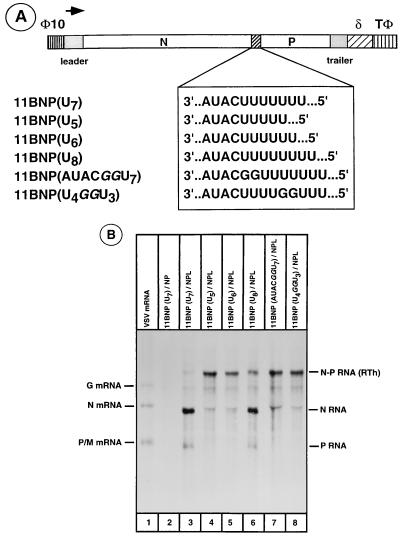
Recent data from our laboratory (22) have suggested that changing the length of uridine residues in the U7 stretch of the polyadenylation signal AUACU7 or by inserting guanosine residues into this signal results in changes in polyadenylation of mRNA by VSV polymerase. Changing U7 to U5 or AUACU7 to AUACGGU7 or to AUACU4GGU3, in which the inserted nucleotides are shown in boldface, completely abrogated polyadenylation, whereas the template with U8 supported polyadenylation like the wild-type template. The U6 template supported polyadenylation less efficiently. We were therefore interested to determine whether such changes in the polyadenylation signal at the intercistronic gene junction of the bicistronic minireplicon 11BNP would influence termination of transcription at the gene junction.
Accordingly, a series of mutant 11BNP minireplicons with altered length of uridine residues (U5, U6, and U8) in the U7 stretch of the polyadenylation signal was generated (Fig. 5A). In addition, two other insertion mutants in which the wild-type polyadenylation signal AUACU7 was changed to AUACGGU7 or AUACU4GGU3 were also generated. The ability of the mutant templates to generate polyadenylated and terminated upstream RNA or readthrough RNA was analyzed (Fig. 5B). The U8 template generated the upstream, the downstream, and the readthrough transcripts (lane 6), although the relative amounts of the readthrough transcripts from this mutant were consistently higher than those from the wild-type template (lane 3). The U6 template produced significantly higher amounts of the readthrough transcript (lane 5). A very low level of upstream transcript was also detected, possibly due to inefficient polyadenylation of the upstream transcript from the U6 template. An RNA band migrating slightly slower than the N RNA band is present in all lanes and is most likely a background band of cellular origin. This band becomes more clearly visible when the N RNA band is absent (lanes 4, 7, and 8) or significantly reduced (lane 5). The U5, AUACGGU7, and AUACU4GGU3 templates produced much higher levels of readthrough transcript (lanes 4, 7, and 8), and the upstream transcript was not detectable even upon longer exposure of the fluorogram. It should be noted that by varying the length of exposure time of fluorogram, we would have been able to detect levels of N and P RNAs from the mutant minireplicon templates that are greater than 5 to 7% of those obtained from the wild-type template. Thus, these results provide further evidence that transcription termination at the intercistronic gene junction of VSV is directly linked to the ability of the upstream RNA to be polyadenylated.
FIG. 5.
(A) 11BNP and mutant minireplicons encoding different-length uridine residues at the intercistronic gene junction. (B) RNA transcripts generated from various mutant minireplicon templates. The experiment was performed as described for Fig. 3, using plasmids encoding the mutant minireplicons as shown above each lane. Lane 1 shows VSV mRNAs from infected cells. RTh, readthrough product of transcription.
The AUACU7 sequence element is the minimum requirement for transcription termination.
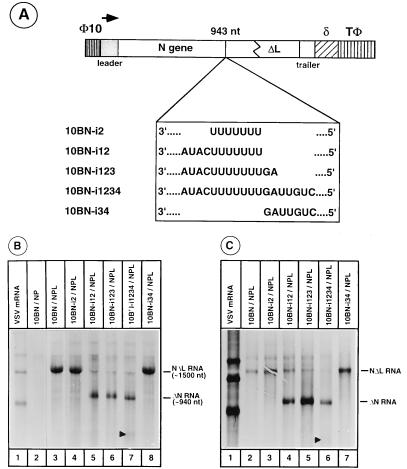
To unequivocally demonstrate that polyadenylation is the critical requirement for transcription termination at the intercistronic gene junctions, we wanted to determine whether insertion of the polyadenylation signal AUACU7 alone upstream of the normal polyadenylation site in the 10BN minireplicon template would result in the synthesis of a prematurely terminated and polyadenylated transcript of predicted length. To perform this experiment, we generated the following series of mutant 10BN minireplicons (Fig. 6A) in which various regions of the intercistronic junction sequences were inserted at nucleotide 943 from the N mRNA start site: 10BN-i2, in which only U7 sequence was inserted; 10BN-i12, in which AUACU7 was inserted; 10BN-i123, in which AUACU7GA was inserted; 10BN-i1234, in which AUACU7GAUUGUC was inserted; and 10BN-i34, in which GAUUGUC was inserted.
FIG. 6.
(A) 10BN and the mutant minireplicons with insertion of various intercistronic junction sequences. nt, nucleotides (B) Effect of insertion of polyadenylation signal on transcription termination. Plasmids encoding 10BN or various 10BN insertion mutants as shown above each lane were transfected along with plasmids encoding N, P, and L proteins into cells infected with vTF7-3. Cells were labeled with [3H]uridine and [3H]adenosine in the presence of actinomycin D and AraC. Total labeled RNAs from the transfected cells were analyzed in an acid-agarose urea gel and detected by fluorography. The transcription products are shown on the right. The arrowhead in lane 7 shows the reinitiated transcription product of approximately 550 nucleotides. (C) Oligo(dT) selection of RNAs from various 10BN insertion mutants. Total labeled RNAs from cells transfected with plasmids encoding 10BN or mutant minireplicons (as shown above each lane) were subjected to oligo(dT)-cellulose chromatography, and the oligo(dT)-selected RNAs were analyzed as described above. The arrowhead in lane 6 shows the reinitiated transcription product of approximately 550 nucleotides.
The ability of the above series of insertion mutants to direct synthesis of various transcripts was analyzed in transfected cells. The results (Fig. 6B) show that insertion of U7 alone (lane 4) or GAUUGUC (lane 8) did not produce the predicted-length transcript of approximately 940 nucleotides. Instead, the normal-length NΔL RNA transcript of approximately 1,500 nucleotides was synthesized and comigrated with the transcript from wild-type 10BN template (lane 3). However, insertion of AUACU7 resulted in the synthesis of the predicted shorter transcript, ΔN RNA (lane 5). Low levels of full-length transcript NΔL RNA were also synthesized. Similar patterns of transcripts were also synthesized from the AUACU7GA insertion mutant (lane 6). These results along with the result from the GAUUGUC insertion mutant (lane 8) indicate that the intergenic dinucleotide GA does not play a direct role in transcription termination of upstream RNA. Interestingly, insertion of the entire intercistronic junction sequence AUACU7GAUUGUC resulted in the synthesis not only of the prematurely terminated transcript ΔN RNA but also of another transcript approximately 550 nucleotides in length (indicated by the arrowhead in lane 7). We reason that this transcript is the result of reinitiation by VSV RNA polymerase after termination, since this particular insertion mutant contained the initiation signal UUGUC following the polyadenylation signal AUACU7 and the spacer dinucleotide GA. These results suggest that insertion of AUACU7 sequence element alone is sufficient to terminate transcription of upstream mRNA.
To determine whether the prematurely terminated transcript ΔN RNA generated from these minireplicons is polyadenylated, total RNA from transfected cells were prepared as for Fig. 6B and the polyadenylated RNAs were selected by oligo(dT)-cellulose. Analysis of oligo(dT)-selected RNAs (Fig. 6C) show that the prematurely terminated ΔN RNA transcripts were polyadenylated (lanes 4 to 6). The smaller transcript generated from AUACU7GAUUGUC insertion mutant was also polyadenylated, as seen by its ability to bind to oligo(dT)-cellulose (the faint band indicated by an arrowhead in lane 6). Taken together, the results shown in Fig. 6 strongly support the conclusion that the AUACU7 motif can induce both polyadenylation and transcription termination and that polyadenylation is the major determining factor in termination of upstream RNA transcription.
DISCUSSION
The conserved intercistronic sequences have been proposed to play key roles in polyadenylation and termination of upstream mRNA as well as reinitiation and capping of downstream mRNA. In this article, we addressed the role of sequences found at the intercistronic gene junctions of VSV in directing polyadenylation of VSV mRNA and termination of transcription. Using a variety of mutants that encode bicistronic minireplicons with altered gene junction sequences, our study provides evidence that polyadenylation of upstream mRNA is necessary and sufficient for its termination. Deletion of sequences that signal polyadenylation results in the viral polymerase reading through the mutated junction rather than terminating transcription. The intergenic dinucleotide GA, which was recently proposed to play a role in transcription termination (48), may play only an indirect role, if any, in directing the polymerase to terminate transcription. Although the data presented here cannot rule out the possibility that termination of transcription is required for VSV mRNA polyadenylation, considering the current proposal for VSV mRNA polyadenylation in which the viral transcriptase may carry out polyadenylation by repeated chattering at the stretch of uridine residues on the template (46), we suggest that polyadenylation of VSV mRNA occurs prior to transcription termination and is the determining factor for termination. The conclusion that polyadenylation is required for transcription termination is further strengthened by the demonstration that insertion of the polyadenylation signal sequence AUACU7 alone was sufficient to direct the polymerase to polyadenylate the transcript and induce efficient transcription termination at the inserted site. Furthermore, our data support the conclusion that termination of transcription is independent of reinitiation whereas transcription reinitiation is dependent on prior termination.
Previous studies by Masters and Samuel (33) have shown that polyadenylated polycistronic (or readthrough) transcripts from VSV-infected cells do not contain intervening poly(A) sequences. In addition, polycistronic transcripts lacking the intervening poly(A) sequences have also been demonstrated in cells infected with Sendai virus (16) and Newcastle disease virus (50). Our observation that the readthrough transcripts generated from the minireplicons of VSV do not contain intervening poly(A) sequences supports the contention that when poly(A) sequences are not added to the 3′ end of the upstream transcript, the behavior of VSV RNA polymerase at the intercistronic junction under the circumstance changes dramatically. Instead of terminating transcription of the upstream mRNA and reinitiating the downstream mRNA, the polymerase just reads through the intercistronic junction sequences and generates a bicistronic transcript. Our results suggesting the requirement for polyadenylation in VSV mRNA transcription termination at the intercistronic junction were surprising in the light of other previous observations (18, 19) that readthrough transcripts of VSV containing two adjacent mRNAs possess intervening poly(A) sequences. These observations lead to the suggestion that VSV transcriptase polyadenylates the upstream transcript, perhaps by chattering on the U7 sequence on the template, and instead of terminating, it resumes transcription by adding nucleotides onto the 3′ end of the poly(A) sequence (46). Studies using the temperature-sensitive mutant tsG16(I) with an aberrant polyadenylation phenotype have also suggested that increased synthesis of polycistronic mRNA is associated with increased length of the poly(A) tail (21). However, it must be pointed out that such polycistronic transcripts containing intervening poly(A) sequences are synthesized under in vitro transcription conditions at very low frequency (19) and most probably do not result from normal transcription events that occur at the gene junctions of VSV. Readthrough transcripts containing intervening poly(A) sequences synthesized in vitro may be artifactual since abnormal transcription products containing heterogeneous-length poly(A) tail or leader RNA linked N mRNA have been shown to be generated in vitro (6, 21, 42).
The role of conserved sequence elements at the intercistronic junctions of VSV has been recently addressed by using similar minireplicon systems. In one study (48), it was found that nucleotide changes at the intergenic dinucleotide GA led to an increase in readthrough transcription, thus prompting the authors to conclude that the intergenic dinucleotide plays a role in transcription termination, although the intergenic dinucleotide changes did not significantly influence the termination of the upstream mRNA. However, using saturation mutagenesis, it was shown in a separate study (4) that changes in the intergenic dinucleotide GA, although affecting the levels of downstream mRNA as well as the readthrough bicistronic mRNA significantly, did not influence termination of upstream mRNA appreciably. Our results (Fig. 3 and 6) are consistent with the interpretation that the dinucleotide GA does not play a direct role in transcription termination of the upstream mRNA. It may be required simply as a spacer for the polymerase to initiate transcription of the downstream mRNA only after polyadenylation and termination of the upstream mRNA. The observation that insertion of the polyadenylation signal AUACU7 alone was sufficient to allow the polymerase to polyadenylate and terminate transcription at the inserted site whereas insertion of GAUUGUC was not sufficient to mediate termination provides the strongest support for the interpretation that polyadenylation alone is sufficient to dictate transcription termination of the upstream mRNA effectively.
Sequence analysis of the intercistronic gene junctions of VSV (30, 35, 40, 46) have suggested that AUACU7 may signal polyadenylation of VSV mRNAs. Our results from mutant minireplicons with deletion of AUAC or U7 elements provide direct evidence that AUACU7 forms the intact polyadenylation signal of VSV. The demonstration that AUACU7 alone can signal polyadenylation efficiently in a heterologous context further strengthens our conclusion. Mutagenic analysis to understand the role of this element for efficient polyadenylation of VSV mRNA is currently under investigation. Similar sequence elements containing a stretch of uridine residues have been shown to exist in other negative-strand viruses, and these sequences have been proposed to signal polyadenylation in these viruses (15, 31). In case of influenza virus, a stretch of five to seven uridine residues followed by an RNA duplex structure has been shown to be required for efficient polyadenylation of mRNA by stuttering at the uridine stretch when the polymerase encounters the double-stranded RNA barrier next to the stretch of uridines (28, 31). The mechanism by which AUACU7 sequence mediates polyadenylation in VSV is not known. It has been proposed that the viral transcriptase may carry out polyadenylation by repeated chattering at the stretch of uridine residues on the template (46). Such a process may require the polymerase to pause or slow down at the AUACU7 or nearby sequences. Evidence for VSV RNA polymerase to pause or slow down has been experimentally provided by Iverson and Rose (23), although it is not known what factors cause the polymerase to pause or slow down.
The average length of poly(A) sequences in VSV mRNAs has been shown to be approximately 100 to 200 nucleotides (11, 47), although smaller poly(A) sequences have also been reported (41). In addition, different VSV mRNAs have been shown to contain different average-length poly(A) sequences (41). Whether the variability in average length of poly(A) on different mRNAs is due to the upstream and/or downstream sequences around the poly(A) signal at various intercistronic gene junctions is purely speculative and remains to be determined.
The relationship between polyadenylation and transcription termination in higher eukaryotes and yeast has been well documented. It has been shown that a bipartite signal consisting of a functional polyadenylation element as the upstream signal (8, 29) and various types of downstream elements (DSE) as the downstream signal are required for mediating efficient termination of RNA polymerase II (pol II) transcription. Furthermore, the strength of the polyadenylation signal has been shown to correlate with termination efficiency (10). A very similar relationship is also found in yeast, where removal of 3′-end formation signals or DSE caused transcription to proceed beyond the normal termination signal (5). Although it is tempting to draw some similarities between what happens in yeast and eukaryotes to the situation in VSV, a eukaryotic virus, the process of polyadenylation in yeast and eukaryotes is significantly different from what has been proposed for VSV. mRNA ends in higher eukaryotes are formed by endonucleolytic cleavage and subsequent polyadenylation by a complex of 3′-end processing factors, whereas in VSV and other negative-strand viruses, it has been proposed that the L protein catalyzes the production of the poly(A) tail by stuttering at the uridine stretch of the polyadenylation signal on the template. In yeast, polymerase molecules have been shown to accumulate over the DSE, and the DSE has been proposed to act as the pausing site for the elongating polymerase to allow proper termination (5). A scenario similar to this could be envisioned for VSV, where the VSV RNA polymerase uses the AUACU7 signal to polyadenylate the transcript but the action of stuttering and/or pausing at the U7 stretch possibly enables the polymerase to terminate transcription. In a recent study, the carboxy-terminal domain of pol II was shown to be required for 3′-end processing and termination (34). The authors proposed a mechanism linking polyadenylation event and transcription termination in which polyadenylation machinery associated with the carboxy-terminal domain of pol II dissociates from the transcription complex at the polyadenylation signal, therefore causing the polymerase to become termination competent. Whether a similar model for VSV mRNA polyadenylation and transcription termination is operative is difficult to envision at the present time, and further experimentation is required to understand how mRNA polyadenylation leads to transcription termination in VSV.
The significance of polyadenylation prior to VSV mRNA transcription termination is unknown. We reason that for VSV RNA polymerase to generate stable and functional mRNAs, it must polyadenylate the mRNAs. Nonpolyadenylated mRNAs may be rapidly degraded in the cellular milieu such that they may not act as efficient templates for translation to generate the viral proteins. Therefore, it is in the best interest of the virus to ensure that the mRNAs it generates are polyadenylated and stable before they are terminated. This is consistent with our observation that polyadenylation is closely linked to termination of VSV mRNA transcription. In this regard, it is also interesting that the leader RNA, which is not polyadenylated, is not a stable RNA species in virus-infected cells (25).
In conclusion, our results provide evidence that polyadenylation of upstream mRNA is required for its termination at the intercistronic gene junction. We suggest that VSV uses this mechanism to generate stable and functional monocistronic mRNAs. Further experimentation is necessary to understand exactly how polyadenylation leads to transcription termination of VSV mRNAs.
ACKNOWLEDGMENTS
We thank Michelle Perez for excellent preparation of the manuscript. We also thank Merck and Co., Inc., Rahway, N.J., for a gift of actinomycin D.
This investigation was supported by Public Health Service grant AI34956 from the National Institutes of Health. L.N.H. was supported by a predoctoral fellowship from training grant T32EY07129 from the National Eye Institute, NIH.
ADDENDUM
While this report was under review, a paper by Barr et al. (4a) with similar conclusions was published.
REFERENCES
- 1.Abraham G, Banerjee A K. Sequential transcription of the genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:1504–1508. doi: 10.1073/pnas.73.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Greene Publishing Associates and John Wiley & sons, Inc.; 1988. [Google Scholar]
- 3.Ball L A, White C N. Order of transcription of genes of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1976;73:442–446. doi: 10.1073/pnas.73.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Barr J N, Whelan S P J, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birse C E, Lee B A, Hansen K, Proudfoot N J. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinchar V G, Amesse L, Portner A. Linked transcripts of gene for leader and N message are synthesized in vitro by vesicular stomatitis virus. Biochem Biophys Res Commun. 1982;105:1296–1302. doi: 10.1016/0006-291x(82)90927-5. [DOI] [PubMed] [Google Scholar]
- 7.Chuang J L, Perrault J. Initiation of vesicular stomatitis virus mutant polRI transcription internally at the N gene in vitro. J Virol. 1997;12:1395–1400. doi: 10.1128/jvi.71.2.1466-1475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly S, Manley J L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 9.De B P, Banerjee A K. Requirements and functions of vesicular stomatitis virus L and NS proteins in the transcription process in vitro. Biochem Biophys Res Commun. 1985;126:40–49. doi: 10.1016/0006-291x(85)90568-6. [DOI] [PubMed] [Google Scholar]
- 10.Edwards-Gilber G, Prescott J, Falck-Pedersen E. 3′ RNA processing efficiency plays a primary role in generating termination competent RNA polymerase II elongation complexes. Mol Cell Biol. 1993;13:3472–3480. doi: 10.1128/mcb.13.6.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrenfeld E, Summers D. Adenylate-rich sequences in vesicular stomatitis virus messenger ribonucleic acid. J Virol. 1972;10:683–688. doi: 10.1128/jvi.10.4.683-688.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson S U. Reconstitution studies detect a single RNA polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982;31:635–642. doi: 10.1016/0092-8674(82)90319-1. [DOI] [PubMed] [Google Scholar]
- 13.Emerson S U, Yu Y H. Both NS and L proteins are required for in vitro RNA synthesis by vesicular stomatitis virus. J Virol. 1975;15:1348–1356. doi: 10.1128/jvi.15.6.1348-1356.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta K C, Kingsbury D W. Conserved polyadenylation signals in two negative-strand RNA virus families. Virology. 1982;120:518–523. doi: 10.1016/0042-6822(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 16.Gupta K C, Kingsbury D W. Polytranscripts of Sendai virus do not contain intervening polyadenylate sequences. Virology. 1985;141:102–109. doi: 10.1016/0042-6822(85)90186-2. [DOI] [PubMed] [Google Scholar]
- 17.Hercyk N, Horikami S M, Moyer S A. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology. 1988;163:222–225. doi: 10.1016/0042-6822(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 18.Herman R, Adler S, Lazzarini R, Colonno R, Banerjee A, Westphal W. Intervening polyadenylate sequences in RNA transcripts of vesicular stomatitis virus. Cell. 1978;15:587–596. doi: 10.1016/0092-8674(78)90027-2. [DOI] [PubMed] [Google Scholar]
- 19.Herman R, Schubert M, Keene J, Lazzarini R. Polycistronic vesicular stomatitis virus RNA transcripts. Proc Natl Acad Sci USA. 1980;77:4662–4665. doi: 10.1073/pnas.77.8.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt D M, Smith E G, Buckley D W. Aberrant polyadenylation by a vesicular stomatitis mutant is due to an altered L protein. J Virol. 1984;52:515–521. doi: 10.1128/jvi.52.2.515-521.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson K L, Herman R C, Hunt D M. Increased synthesis of polycistronic mRNA associated with increased polyadenylation by vesicular stomatitis virus. Virology. 1992;189:67–78. doi: 10.1016/0042-6822(92)90682-f. [DOI] [PubMed] [Google Scholar]
- 22.Hwang, L., and A. K. Pattnaik. 1997. Unpublished data.
- 23.Iverson L E, Rose J K. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- 24.Laskey R. The use of intensifying screens or organic scintillators for visualizing radioactive molecules resolved by gel electrophoresis. Methods Enzymol. 1980;65:363–371. doi: 10.1016/s0076-6879(80)65047-2. [DOI] [PubMed] [Google Scholar]
- 25.Leppert M, Rittenhouse L, Perrault J, Summers D, Kolakofsky D. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell. 1979;18:735–747. doi: 10.1016/0092-8674(79)90127-2. [DOI] [PubMed] [Google Scholar]
- 26.Lerach H, Diamond D, Wozney J, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical examination. Biochemistry. 1977;16:4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 27.Li T, Pattnaik A K. Replication signals in the genome of vesicular stomatitis virus and its defective interfering particles: identification of a sequence element that enhances DI RNA replication. Virology. 1997;232:248–259. doi: 10.1006/viro.1997.8571. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Palese P. Characterization of the polyadenylation signal of influenza virus RNA. J Virol. 1994;68:1245–1249. doi: 10.1128/jvi.68.2.1245-1249.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan J, Falck-Pedersen E, Darnell J, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β-globin gene. Proc Natl Acad Sci USA. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luk D, Masters P, Gill D, Banerjee A. Intergenic sequences of the vesicular stomatitis virus genome (New Jersey Serotype): evidence for two transcription initiation sites within the L gene. Virology. 1987;160:88–94. doi: 10.1016/0042-6822(87)90048-1. [DOI] [PubMed] [Google Scholar]
- 31.Luo G, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 33.Masters P S, Samuel C E. Detection of in vivo synthesis of polycistronic mRNAs of vesicular stomatitis virus. Virology. 1984;134:277–286. doi: 10.1016/0042-6822(84)90297-6. [DOI] [PubMed] [Google Scholar]
- 34.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 35.McGeoch D J. Structure of the gene N gene NS intercistronic junction in the genome of vesicular stomatitis virus. Cell. 1979;17:673–681. doi: 10.1016/0092-8674(79)90274-5. [DOI] [PubMed] [Google Scholar]
- 36.Naito S, Ishihama A. Function and structure of RNA polymerase from vesicular stomatitis virus. J Biol Chem. 1976;251:4307–4314. [PubMed] [Google Scholar]
- 37.Pattnaik A K, Ball L A, Legrone A W, Wertz G W. Infectious defective interfering particles of vesicular stomatitis virus from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 38.Pattnaik A K, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee A K. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J Virol. 1997;71:8167–8175. doi: 10.1128/jvi.71.11.8167-8175.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pattnaik A K, Wertz G W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990;64:2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose J K. Complete intergenic and flanking gene sequence from the genome of vesicular stomatitis virus. Cell. 1980;19:415–421. doi: 10.1016/0092-8674(80)90515-2. [DOI] [PubMed] [Google Scholar]
- 41.Rose J K, Knipe D. Nucleotide sequence complexities, molecular weights, and poly(A) content of the vesicular stomatitis virus mRNA species. J Virol. 1975;15:994–1003. doi: 10.1128/jvi.15.4.994-1003.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose J K, Lodish H, Brock M. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J Virol. 1977;21:683–693. doi: 10.1128/jvi.21.2.683-693.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose J, Schubert M. Rhabdovirus genomes and their products. In: Wagner R R, editor. The rhabdoviruses. New York, N.Y: Plenum Publishing Corp.; 1987. pp. 129–166. [Google Scholar]
- 44.Schnell M J, Buonocore L, Whitt M A, Rose J K. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol. 1996;70:2318–2323. doi: 10.1128/jvi.70.4.2318-2323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubert M, Harmison G G, Richardson C D, Meier E. Expression of a cDNA encoding a functional 241-kilodalton vesicular stomatitis virus RNA polymerase. Proc Natl Acad Sci USA. 1985;82:7984–7988. doi: 10.1073/pnas.82.23.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert M, Keene J D, Herman R C, Lazzarini R A. Site on the vesicular stomatitis virus genome specifying polyadenylation at the end of the L gene. J Virol. 1980;34:550–559. doi: 10.1128/jvi.34.2.550-559.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soria M, Huang A. Association of polyadenylic acid with messenger RNA of vesicular stomatitis virus. J Mol Biol. 1973;77:449–455. doi: 10.1016/0022-2836(73)90450-6. [DOI] [PubMed] [Google Scholar]
- 48.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and transcriptional start sequences of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Testa D, Chanda P I, Banerjee A K. Unique mode of transcription in vitro by vesicular stomatitis virus. Cell. 1980;21:267–275. doi: 10.1016/0092-8674(80)90134-8. [DOI] [PubMed] [Google Scholar]
- 50.Wilde A, Morrison T. Structural and functional characterization of Newcastle disease virus polycistronic RNA species. J Virol. 1984;51:71–76. doi: 10.1128/jvi.51.1.71-76.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]