Abstract
Porphyromonas gingivalis is an anaerobic Gram‐negative human oral pathogen highly associated with the more severe forms of periodontal disease. Porphyromonas gingivalis utilises the type IX secretion system (T9SS) to transport ∼30 cargo proteins, including multiple virulence factors, to the cell surface. The T9SS is a multiprotein system consisting of at least 20 proteins, and recently, we characterised the protein interactome of these components. Similar to the T9SS, almost all biological processes are mediated through protein‒protein interactions (PPIs). Therefore, mapping PPIs is important to understand the biological functions of many proteins in P. gingivalis. Herein, we provide native migration profiles of over 1000 P. gingivalis proteins. Using the T9SS, we demonstrate that our dataset is a useful resource for identifying novel protein interactions. Using this dataset and further analysis of T9SS P. gingivalis mutants, we discover new mechanistic insights into the formation of the PorQ‐Z complex of the T9SS. This dataset is a valuable resource for studies of P. gingivalis.
Keywords: Porphyromonas gingivalis, PorQ, PorZ, protein interactions, protein network, type IX secretion system
Abbreviations
- A‐LPS
Anionic lipopolysaccharide
- BN‐PAGE
Blue Native PAGE
- DDM
Dodecylmaltoside
- DTT
Dithiothreitol
- iBAQ
intenisty‐based absolute quantitation
- LFQ
Label‐free quantitation
- PPI
Protein‐protein interactions
- SDS
Sodium dodecyl sulfate
- TFA
Trifluoroacetic acid
- T9SS
Type IX secretion system
1. INTRODUCTION
Porphyromonas gingivalis is a Gram‐negative anaerobic bacterium found in subgingival plaque. Various studies have shown that P. gingivalis is a key pathogen associated with severe periodontal disease in humans (Hajishengallis et al., 2011; Holt et al., 1988; Kirst et al., 2015; Socransky et al., 1998; Ximenez‐Fyvie et al., 2000), which is a major cause of tooth loss in industrial nations. Severe forms of periodontal disease have also been linked to an increased risk of cardiovascular diseases, diabetes, pre‐term birth, rheumatoid arthritis and dementia (Dominy et al., 2019; Figuero 2020 et al., 2020; Haraszthy et al., 2000; Linden et al., 2013; Orlandi et al., 2020).
The major virulence factors of P. gingivalis called gingipains are secreted to the cell surface by the type IX secretion system (T9SS) (Lasica et al., 2017; Veith et al., 2017). In addition to the gingipains, this system secretes ∼30 other proteins that have a conserved C‐terminal domain (CTD) (Seers et al., 2006; Veith et al., 2013). The T9SS comprises at least 17 component proteins, namely, PorK, PorL, PorM, PorN, Sov, PorT, PorU, PorW, PorP, PorV, PorQ, PorZ, PorE, PorF, PorG, PorD, Plug, and three transcription regulators PorX, PorY and SigP (Gorasia, Lunar Silva, et al., 2022; Heath et al., 2016; Kadowaki et al., 2016; Lasica et al., 2016; Lauber et al., 2018; Naito et al., 2019; Saiki & Konishi, 2010; Sato et al., 2010). Several subcomplexes of the T9SS have been identified (Gorasia, Lunar Silva, et al., 2022; Gorasia et al., 2020). The PorK/PorN/PorG complex forms a ring‐shaped structure (Gorasia et al., 2016), while PorL and PorM associate to form molecular motors that energise both secretion and motility (Hennell James et al., 2021). The Sov translocon interacts with either PorV or Plug (Lauber et al., 2018), and we recently showed that PorW/PorD forms a bridge between the Sov translocon and the PorK/PorN/PorG complex (Gorasia, Lunar Silva, et al., 2022; Gorasia et al., 2020). We also recently demonstrated that PorP/PorE/PG1035 form a stable subcomplex that may play a role in anchoring the T9SS to the peptidoglycan (Gorasia, Seers, et al., 2022). PorU/PorV/PorQ/PorZ form the attachment complex that anchors the T9SS cargo proteins to the cell surface via A‐LPS using a sortase‐like mechanism (Glew et al., 2012; Gorasia et al., 2015; Veith et al., 2020).
Similar to the T9SS, almost all biological processes are mediated through protein‒protein interactions (PPIs). Therefore, mapping PPIs is essential to understand the biological function of many proteins in P. gingivalis. Herein, we provide native migration profiles of over 1000 P. gingivalis proteins. Using the well‐characterised protein interactome of the T9SS in P. gingivalis, we demonstrate how our dataset can be used to understand the protein interactome of any P. gingivalis subcellular system. Using this dataset and further analysis of the T9SS mutants, we provide new insights into the formation of the PorQ‐PorZ subcomplex.
2. MATERIALS AND METHODS
2.1. Bacterial strains and culture conditions
Porphyromonas gingivalis wild‐type strains W50 and ATCC 33277 were grown in tryptic soy‐enriched brain heart infusion broth (TSBHI) (25 g/L tryptic soy, 30 g/L BHI) supplemented with 0.5 mg/ml cysteine, 5 μg/ml haemin and 5 μg/ml menadione. For blood agar plates, 5% defibrinated horse blood (Equicell, Bayles, Australia) was added to enriched trypticase soy agar. Mutant strains were grown in the same media as above with the appropriate antibiotic selections. All P. gingivalis strains were grown anaerobically (80% N2, 10% H2 and 10% CO2) at 37°C. Mutant P. gingivalis strains, namely, sov, porK, porP, porQ, porT, porW and ABK–, in a ATCC 33277 background were obtained from Professor Koji Nakayama (Sato et al., 2010). Mutant P. gingivalis strains porU, porV and porZ were previously produced in our laboratory in both ATCC 33277 and W50 backgrounds (Chen et al., 2011; Glew et al., 2017, 2012). The mutant porE was also generated in our laboratory in the P. gingivalis W50 background (Heath et al., 2016). Of note, all these mutants have been complemented and shown to return to the wild‐type phenotype; hence, there are no polar effects in these mutants (Chen et al., 2011; Glew et al., 2012; Heath et al., 2016; Sato et al., 2010).
2.2. Blue native gel electrophoresis
Blue native gel electrophoresis was performed essentially as described (Sato et al., 2010). Briefly, P. gingivalis cells were pelleted by centrifugation at 5000 × g for 5 min at 4°C, and the pellet was suspended in native gel buffer containing 1% n‐dodecyl‐β‐d‐maltoside (DDM), complete protease inhibitors and 5 mM MgCl2. After sonication (Sato et al., 2010), the samples were clarified by centrifugation at 16,000 × g for 20 min at 4°C. Coomassie Blue G‐250 was added to the samples at a final concentration of 0.25%, and the samples were electrophoresed on nondenaturing Native PAGE Novex 3–12% Bis‐Tris gels. The proteins in the gels were stained with Coomassie Blue G‐250 and destained until the background was clear. The lane containing the sample was excised into 12 gel bands, and in‐gel digestion was performed. The gel pieces were incubated with 2% sodium dodecyl sulfate and 10 mM dithiothreitol and incubated at 56°C for 1 h. Following incubation, 55 mM iodoacetamide was added to the gel pieces and incubated for 30 min in the dark. In‐gel digestion was performed using sequencing‐grade‐modified trypsin (Promega) and incubated overnight at 37°C, as previously published (Mortz et al., 2001). Tryptic peptides were extracted from the gel pieces using 50% acetonitrile in 0.1% Trifluoroacetic acid and sonicated for 10 min in a sonicator bath. The samples were concentrated in a vacuum centrifuge before analysis using liquid chromatography‐tandem mass spectrometry (LC‐MS/MS).
2.3. Liquid chromatography‐tandem mass spectrometry
The tryptic peptides were analysed by LC‐MS/MS using a Q Exactive Plus Orbitrap mass spectrometer coupled to an Ultimate 3000 UHPLC system (Thermo Fisher Scientific). Buffer A was 2% acetonitrile and 0.1% formic acid, and buffer B consisted of 0.1% formic acid in acetonitrile. Sample volumes of 1 μl were loaded onto a PepMap C18 trap column (75 μM ID X 2 cm X 100 Å) and desalted at a flow rate of 2 μl/min for 15 min using buffer A. The samples were then separated through a PepMap C18 analytical column (75 μM ID X 15 cm X 100 Å) at a flow rate of 300 nl/min, with the percentage of solvent B in the mobile phase changing from 2% to 10% in 1 min, from 10% to 35% in 50 min, from 35% to 60% in 1 min and from 60% to 90% in 1 min. The spray voltage was set at 1.8 kV, and the temperature of the ion transfer tube was 250°C. The S‐lens was set at 50%. The full MS scans were acquired over an m/z range of 300–2000, with a resolving power of 70,000, an automatic gain control (AGC) target value of 3 × 106, and a maximum injection of 30 ms. Dynamic exclusion was set at 90 s. Higher energy collisional dissociation MS/MS scans were acquired at a resolving power of 17500, AGC target value of 5×104, maximum injection time of 120 ms, isolation window of m/z 1.4 and NCE of 25% for the top 15 most abundant ions in the MS spectra. All spectra were recorded in profile mode.
The relative abundances of proteins were quantified by MaxQuant (Ver 1.5.3.30) (Cox et al., 2014). Raw MS/MS files were searched against P. gingivalis W50 or ATCC 33277. The default MaxQuant parameters were used, except the label‐free quantitation (LFQ) min ratio count was set to 1, and the match between runs was selected. MaxQuant normalised the dataset as part of the data processing. LFQ intensities were used for the analysis of the T9SS components, and intensity‐based absolute quantitation (iBAQ) values were used for the overall protein‒protein interaction mapping in P. gingivalis. A heat map of iBAQ intensities of all proteins across gel bands 1–12 was plotted. If the iBAQ intensities of proteins X and Y showed a similar migration profile, then that indicated a potential interaction.
3. RESULTS AND DISCUSSION
3.1. The P. gingivalis protein‒protein interactome
Analysis of PPI networks is a powerful approach to dissect protein function, potential signal transduction, and virulence pathways. In this study, we investigated the global PPI network of the human pathogen P. gingivalis using BN‐PAGE/mass spectrometry. We selected the ABK‐ strain for the PPI study, as the three gingipains are amongst the most abundant proteins in P. gingivalis; hence, their elimination allowed us to detect many less abundant proteins by BN‐PAGE. Furthermore, gingipains are proteases, and their removal minimised protein degradation in the cell lysate. However, one limitation of using this strain is that gingipains process some surface proteins, such as Fim and Mfa pilins, to enable their polymerisation. Our data would not provide accurate polymerisation status of these proteins. BN‐PAGE was performed with P. gingivalis cells (four biological replicates) lysed in 1% DDM. After electrophoresis, each lane was excised into 12 gel bands (Figure 1a) and subjected to tryptic digestion, mass spectrometry and MaxQuant analysis. A total of 1232 proteins were identified, and the protein abundances (iBAQ) were plotted as a heatmap for each gel band (Figure 1b, Table S1). Proteins with a similar native migration profile may be part of the same protein complex. Using this analysis, a large number of potential PPIs were seen. Twelve potential protein complexes consisting of proteins expressed from adjacent genes are shown in Figure 1c and described below. The putative components of a P. gingivalis respiratory chain were predicted from bioinformatic studies and proposed to consist of six subunits (PG2177/PGN_0119‐PG2182/PGN_0114) (Meuric et al., 2010). Previously, two of these, PGN_0119 and PGN_0116, were shown to interact (Glew et al., 2014). Here, we observed all but one of the subunits to peak at bands 8 and 9, suggesting that at least five are part of one complex. Similarly, RagA and RagB (PGN_0293, PGN_0294) co‐migrated by BN‐PAGE, as did Omp40 and Omp41 (PGN_0728, PGN_0729) (Figure 1c). PGN_0557 (HmuR) and PGN_0558 (HmuY) peak at bands 10–11, but HmuY appears to also produce higher molecular weight (MW) complexes, perhaps due to its association with poorly soluble heme. Direct interactions between these protein pairs were reported previously (Nagano et al., 2007; Olczak et al., 2008; Veith et al., 2001). Several other sets of proteins also had migration profiles that suggested they formed complexes: PGN_1124‐PGN_1125, PGN_1172‐PGN_1174, PGN_1529‐PGN_1530, PGN_1890‐PGN_1891, PGN_1930‐PGN_1933 and PGN_1965‐PGN_1966 (Figure 1c). Interactions between the proteins in these groups have not been reported previously, but putative homologues for some of these proteins are thought to interact in other species according to STRING (Szklarczyk et al., 2019) (Table S2). These potential PPIs are a useful resource for further studies of P. gingivalis.
FIGURE 1.
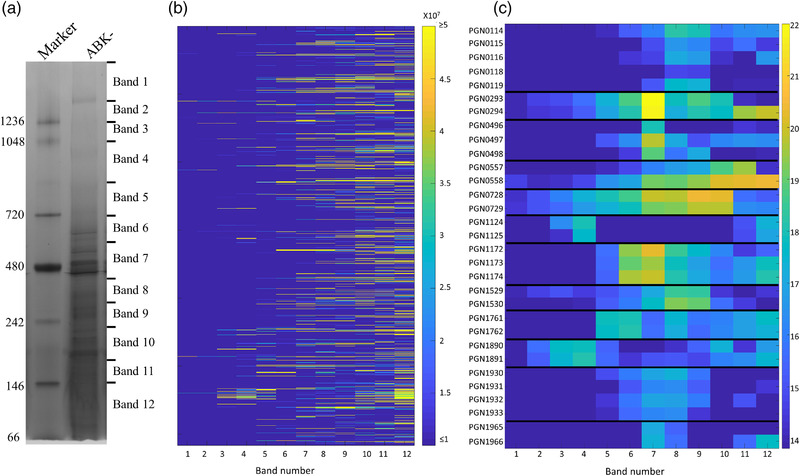
Native migration profiles of P. gingivalis proteins plotted as a heat map. (a) Porphyromonas gingivalis cells lacking the gingipains (ABK‐) were lysed in 1% n‐dodecyl‐β‐d‐maltoside (DDM), electrophoresed on a BN‐PAGE gel and stained with Coomassie brilliant blue G‐250. The gel lane was sliced into 12 bands, and in‐gel tryptic digestion was performed on the gel pieces. Tryptic fragments were analysed by mass spectrometry and identified using MaxQuant software. (b) Migration profile overview of all the proteins identified in each band of the BN‐PAGE gel based on the intensity‐based absolute quantitation (iBAQ) values (average of four replicates). (c) Migration profiles of selected complexes in each band with the iBAQ values expressed on a natural log scale. See also Table S1
Protein interaction networks play key roles in almost all biological processes. A thorough understanding of PPIs should facilitate elucidation of cellular activities and targeted drug design. A study by Glew et al. identified more than 100 proteins forming multisubunit complexes using 2D Blue Native PAGE (Glew et al., 2014). The interactions identified by Glew et al. were also found in this study, and native migration profiles of more than 1000 proteins were mapped, which can be used to identify potential PPIs. Collectively, our study substantially increases the available resources for the protein‒protein interaction networks in P. gingivalis, and therefore, we have generated a valuable dataset for the study of protein‒protein interactions in the periodontal pathogen P. gingivalis and related bacteria.
3.2. Native migration profiles of the T9SS proteins
Very recently, with the help of this dataset, we reported the protein interactome of the T9SS components (Gorasia, Lunar Silva, et al., 2022; Gorasia, Seers, et al., 2022) and the native MW profiles of outer membrane proteins in general (Veith et al., 2021). For example, to show how this dataset can be used, the native migration profiles of the T9SS proteins are analysed. The LFQ intensities of the T9SS proteins were normalised and plotted (Figure 2a–d). The proteins are grouped based on their migration profiles as well as previous knowledge of the subcomplexes of the T9SS. PorK and PorN were the predominant T9SS proteins observed in gel bands 1, 2 and 3. As mentioned above, we have shown that PorK and PorN form large ring‐shaped structures (Gorasia et al., 2016; Song et al., 2022). In gel bands 4 and 5, PorK and PorN intensities were increased and joined by PorL and PorM (Figure 2a). PorL and PorM have also been shown to form a complex (Gorasia et al., 2016; Hennell James et al., 2021; Vincent et al., 2017). PorT was found in gel bands 1–5 following a similar profile to PorK and PorN, suggesting that it may interact with the PorK‐PorN complex (Figure 2a). Further studies are needed to validate this finding. Figure 2b shows the native migration profiles of the Sov and PorW proteins. The Sov and PorW proteins had similar native profiles, showing increased amounts between gel bands 5 and 7, suggesting that these proteins may interact to form a complex (Figure 2b). Using affinity purification and mass spectrometry (AP‐MS), we recently showed that Sov, PorW and PorD form a complex (Gorasia, Lunar Silva, et al., 2022). Figure 2c shows the native migration profiles of PorP, PorE and PorF. PorE, PorP and PorF were mainly detected in gel bands 9–10 (Figure 2c). Although PorF had a similar migration profile, there were no changes to its migration profile in the absence of PorE or PorP, suggesting that its similar native profile may be coincidental (Gorasia, Seers, et al., 2022). Again, using AP‐MS, we showed that PorP and PorE interact (Gorasia, Seers, et al., 2022). Finally, Figure 2d shows the native migration profile of the well‐characterised attachment complex comprising PorU, PorV, PorQ and PorZ. A peak of PorU, PorV, PorQ and PorZ consistent with the complete complex was observed in gel band 7 in the BN‐PAGE analysis of P. gingivalis ABK‐ (Figure 2d). We also observed PorQ and PorZ peaks at gel band 10 (Figure 2d), suggesting the presence of a PorQ‐PorZ subcomplex. Together, the native migration profiles provide an indication of whether two or more proteins of interest interact. Further insights into the protein interactions can be obtained by methods such as AP‐MS.
FIGURE 2.
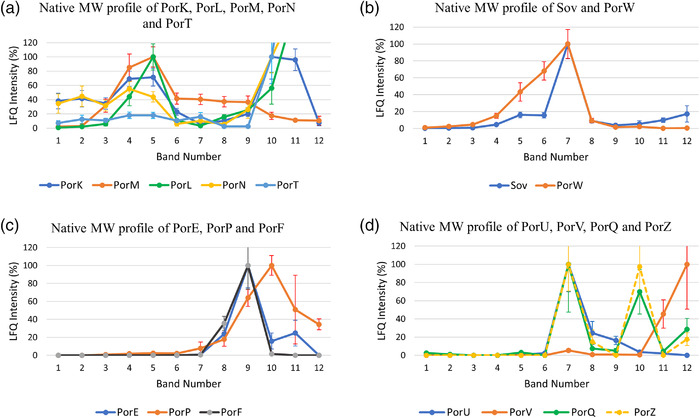
Subcomplexes of the T9SS identified by BN‐PAGE analysis of P. gingivalis ABK‐. The label‐free quantitation (LFQ) intensities for the T9SS proteins identified from each BN‐PAGE band were converted to a percentage with the highest LFQ intensity being assigned 100% for each protein in the group except for PorT, PorL and PorN, where the intensity at band 10 was assigned 100% to aid visualisation of the high molecular weight bands. Proteins that comigrated on a BN‐PAGE gel were grouped and plotted together (a) PorK, PorL, PorM, PorN and PorT, (b) Sov and PorW, (c) PorE, PorP and PorF and (d) PorU, PorV, PorQ and PorZ
3.3. The PorQ‐PorZ subcomplex is formed independently of PorU and PorV
PorQ is an outer membrane β‐barrel protein, while PorZ is a cell surface protein that is also a substrate of the T9SS (Lasica et al., 2017). Previously, PorZ was shown to localise to the cell surface in the absence of PorU. To investigate which other T9SS components are essential for the formation of the PorQ‐PorZ subcomplex, we performed BN‐PAGE analysis on P. gingivalis T9SS mutants (Figure 3). The attachment complex was not observed in any of the mutants, but the putative PorQ‐PorZ subcomplex was observed in the porU and porV mutants, suggesting that the PorQ‐PorZ subcomplex can form in the absence of PorU and PorV.
FIGURE 3.
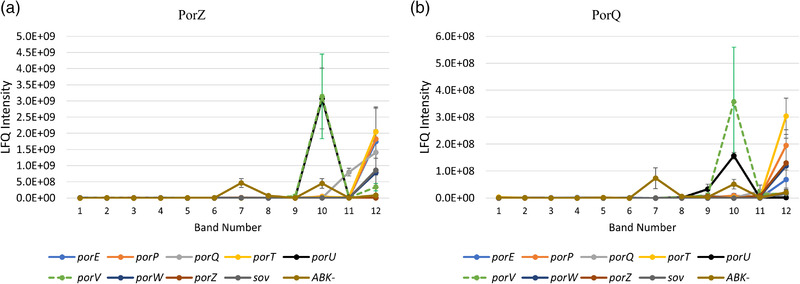
The PorZ and PorQ complex is formed independently of PorU and PorV. Porphyromonas gingivalis T9SS mutants (porT, porQ, porP, porE, porW, sov, porZ, porU and porV) were lysed in 1% n‐dodecyl‐β‐d‐maltoside (DDM) and electrophoresed on a 3%–12% BN‐PAGE gel. The gel lanes were sliced at the same positions as in Figure 1a. The tryptic fragments were analysed by mass spectrometry and MaxQuant software. The label‐free quantitation (LFQ) intensities of (a) PorZ and (b) PorQ in each T9SS mutant and ABK– were plotted.
Glew et al. identified a complex of PorZ with the OM β‐barrel protein PorQ in the PorUC690A catalytic mutant, providing a mechanism for PorZ to be anchored to the cell surface (Glew et al., 2017). Previously, Lasica et al. showed that PorZ is on the cell surface in the absence of PorU (Lasica et al., 2017). Therefore, it is likely that PorZ is on the surface bound to PorQ in the porV mutant as well. Together, it appears that PorZ can translocate through the T9SS secretion channel in the absence of PorU or PorV. PorU and PorZ both have conserved CTD domains and are known to be T9SS substrates as well as components. However, unlike other CTD proteins, the CTD signals of PorU and PorZ are not cleaved (Glew et al., 2012; Lasica et al., 2017). Kulkarni et al. have shown that there are at least two types of CTD signals, types A and B (Kulkarni et al., 2017). Flavobacterium johnsoniae type A CTD proteins are dependent on PorV for secretion, whereas many type B CTD proteins are not (Kharade & McBride, 2015; Kulkarni et al., 2019). A similar observation was also noted in P. gingivalis, as the type B CTD protein PG1035 was found to be PorV independent of complex formation with PorP and PorE (Gorasia, Seers, et al., 2022). PorZ appears to have a CTD that is neither type A nor type B (Lasica et al., 2017). Since PorV forms a complex with Sov (Lauber et al., 2018) and binds to newly secreted CTD proteins before shuttling them to the attachment complex (Glew et al., 2017), we suggest that PorQ, particularly in the absence of PorV, can target the same Sov binding site as PorV and recruit its respective substrate accordingly (Figure 4).
FIGURE 4.
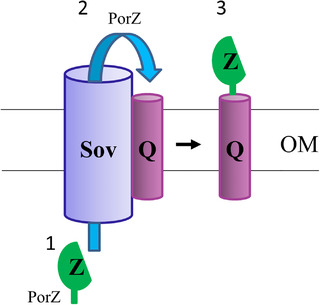
Proposed mechanism of PorQ‐Z complex formation. Schematic diagram showing the proposed association of PorQ with the Sov translocon. (1) PorZ in the periplasm passes through the Sov translocon. (2) PorQ collects PorZ from the Sov translocon. (3) The PorQ‐Z subcomplex dissociates from the Sov translocon. Q: PorQ, Z: PorZ. Abbreviation: OM, outer membrane
In conclusion, we generated a useful dataset for the study of protein interactions in P. gingivalis. Using this dataset and further analysis of the T9SS mutants, we provide mechanistic insight into the formation of the PorQ‐PorZ subcomplex of the T9SS.
CONFLICT OF INTEREST
The authors declare no competing interests.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1111/omi.12383.
Supporting information
Supplementary information
Supplementary Table 2: Predicted protein interactions identified by STRING
ACKNOWLEDGEMENTS
We acknowledge the use of the Mass Spectrometry and Proteomics Facility, The University of Melbourne, Australia. This work was supported by the Australian Government Department of Industry, Innovation and Science (Grant ID 20080108), the Australian National Health and Medical Research Council (Grant IDs 1193647 and 1123866), the Australian Research Council (Grant ID DP200100914) and the Australian Dental Research Foundation (Grant ID: 349–2018).
Open access publishing facilitated by The University of Melbourne, as part of the Wiley ‐ The University of Melbourne agreement via the Council of Australian University Librarians.
Gorasia, D. G. , Veith, P. D. , & Reynolds, E. C. (2023). Protein interactome mapping of Porphyromonas gingivalis provides insights into the formation of the PorQ‐Z complex of the type IX secretion system. Molecular Oral Microbiology, 38, 34–40. 10.1111/omi.12383
Contributor Information
Paul D. Veith, Email: pdv@unimelb.edu.au.
Eric C. Reynolds, Email: e.reynolds@unimelb.edu.au.
DATA AVAILABILITY STATEMENT
The raw MS data are available upon request.
REFERENCES
- Chen, Y. Y. , Peng, B. , Yang, Q. , Glew, M. D. , Veith, P. D. , Cross, K. J. , Goldie, K. N. , Chen, D. , O'Brien‐Simpson, N. , Dashper, S. G. , & Reynolds, E. C. (2011). The outer membrane protein LptO is essential for the O‐deacylation of LPS and the co‐ordinated secretion and attachment of A‐LPS and CTD proteins in Porphyromonas gingivalis . Molecular Microbiology, 79, 1380–1401. [DOI] [PubMed] [Google Scholar]
- Cox, J. , Hein, M. Y. , Luber, C. A. , Paron, I. , Nagaraj, N. , & Mann, M. (2014). Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular and Cellular Proteomics, 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominy, S. S. , Lynch, C. , Ermini, F. , Benedyk, M. , Marczyk, A. , Konradi, A. , Nguyen, M. , Haditsch, U. , Raha, D. , Griffin, C. , Holsinger, L. J. , Arastu‐Kapur, S. , Kaba, S. , Lee, A. , Ryder, M. I. , Potempa, B. , Mydel, P. , Hellvard, A. , Adamowicz, K. , … Potempa, J. (2019). Porphyromonas gingivalis in Alzheimer's disease brains: Evidence for disease causation and treatment with small‐molecule inhibitors. Science Advances, 5, eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figuero, E. , Han, Y. W. , & Furuichi, Y. (2020). Periodontal diseases and adverse pregnancy outcomes: Mechanisms. Periodontology 2000, 83, 175–188. [DOI] [PubMed] [Google Scholar]
- Glew, M. D. , Veith, P. D. , Chen, D. , Gorasia, D. G. , Peng, B. , & Reynolds, E. C. (2017). PorV is an outer membrane shuttle protein for the type IX secretion system. Scientific Reports, 7, 8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glew, M. D. , Veith, P. D. , Chen, D. , Seers, C. A. , Chen, Y. Y. , & Reynolds, E. C. (2014). Blue native‐PAGE analysis of membrane protein complexes in Porphyromonas gingivalis . Journal of Proteomics, 110, 72–92. [DOI] [PubMed] [Google Scholar]
- Glew, M. D. , Veith, P. D. , Peng, B. , Chen, Y. Y. , Gorasia, D. G. , Yang, Q. , Slakeski, N. , Chen, D. , Moore, C. , Crawford, S. , & Reynolds, E. C. (2012). PG0026 is the C‐terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis . Journal of Biological Chemistry, 287, 24605–24617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorasia, D. G. , Lunar Silva, I. , Butler, C. A. , Chabalier, M. , Doan, T. , Cascales, E. , Veith, P. D. , & Reynolds, E. C. (2022). Protein interactome analysis of the type IX secretion system identifies PorW as the missing link between the PorK/N ring complex and the Sov translocon. Microbiology Spectrum, 10, e0160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorasia, D. G. , Seers, C. A. , Heath, J. E. , Glew, M. D. , Soleimaninejad, H. , Butler, C. A. , McBride, M. J. , Veith, P. D. , & Reynolds, E. C. (2022). Type B CTD proteins secreted by the type IX secretion system associate with PorP‐like proteins for cell surface anchorage. International Journal of Molecular Sciences, 23, 5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorasia, D. G. , Veith, P. D. , Chen, D. , Seers, C. A. , Mitchell, H. A. , Chen, Y. Y. , Glew, M. D. , Dashper, S. G. , & Reynolds, E. C. (2015). Porphyromonas gingivalis type IX secretion substrates are cleaved and modified by a sortase‐like mechanism. PLoS Pathogens, 11, e1005152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorasia, D. G. , Veith, P. D. , Hanssen, E. G. , Glew, M. D. , Sato, K. , Yukitake, H. , Nakayama, K. , & Reynolds, E. C. (2016). Structural insights into the PorK and PorN components of the Porphyromonas gingivalis type IX secretion system. PLoS Pathogens, 12, e1005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorasia, D. G. , Veith, P. D. , & Reynolds, E. C. (2020). The type IX secretion system: Advances in structure, function and organisation. Microorganisms, 8, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G. , Liang, S. , Payne, M. A. , Hashim, A. , Jotwani, R. , Eskan, M. A. , McIntosh, M. L. , Alsam, A. , Kirkwood, K. L. , Lambris, J. D. , Darveau, R. P. , & Curtis, M. A. (2011). Low‐abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe, 10, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraszthy, V. I. , Zambon, J. J. , Trevisan, M. , Zeid, M. , & Genco, R. J. (2000). Identification of periodontal pathogens in atheromatous plaques. Journal of Periodontology, 71, 1554–1560. [DOI] [PubMed] [Google Scholar]
- Heath, J. E. , Seers, C. A. , Veith, P. D. , Butler, C. A. , Nor Muhammad, N. A. , Chen, Y. Y. , Slakeski, N. , Peng, B. , Zhang, L. , Dashper, S. G. , Cross, K. J. , Cleal, S. M. , Moore, C. , & Reynolds, E. C. (2016). PG1058 is a novel multidomain protein component of the bacterial type IX secretion system. PLoS One, 11, e0164313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennell James, R. , Deme, J. C. , Kjr, A. , Alcock, F. , Silale, A. , Lauber, F. , Johnson, S. , Berks, B. C. , & Lea, S. M. (2021). Structure and mechanism of the proton‐driven motor that powers type 9 secretion and gliding motility. Nature Microbiology, 6, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, S. C. , Ebersole, J. , Felton, J. , Brunsvold, M. , & Kornman, K. S. (1988). Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science, 239, 55–57. [DOI] [PubMed] [Google Scholar]
- Kadowaki, T. , Yukitake, H. , Naito, M. , Sato, K. , Kikuchi, Y. , Kondo, Y. , Shoji, M. , & Nakayama, K. (2016). A two‐component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Scientific Reports, 6, 23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharade, S. S. , & McBride, M. J. (2015). Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. Journal of Bacteriology, 197, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst, M. E. , Li, E. C. , Alfant, B. , Chi, Y. Y. , Walker, C. , Magnusson, I. , & Wang, G. P. (2015). Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Applied and Environmental Microbiology, 81, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. S. , Johnston, J. J. , Zhu, Y. , Hying, Z. T. , & McBride, M. J. (2019). The carboxy‐terminal region of Flavobacterium johnsoniae SprB facilitates its secretion by the type ix secretion system and propulsion by the gliding motility machinery. Journal of Bacteriology, 201, e00218–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, S. S. , Zhu, Y. , Brendel, C. J. , & McBride, M. J. (2017). Diverse C‐terminal sequences involved in Flavobacterium johnsoniae protein secretion. Journal of Bacteriology, 199, e00884–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica, A. M. , Goulas, T. , Mizgalska, D. , Zhou, X. , de Diego, I. , Ksiazek, M. , Madej, M. , Guo, Y. , Guevara, T. , Nowak, M. , Potempa, B. , Goel, A. , Sztukowska, M. , Prabhakar, A. T. , Bzowska, M. , Widziolek, M. , Thogersen, I. B. , Enghild, J. J. , Simonian, M. , … Gomis‐Ruth, F. X. (2016). Structural and functional probing of PorZ, an essential bacterial surface component of the type‐IX secretion system of human oral‐microbiomic Porphyromonas gingivalis . Scientific Reports, 6, 37708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasica, A. M. , Ksiazek, M. , Madej, M. , & Potempa, J. (2017). The type ix secretion system (T9SS): Highlights and recent insights into its structure and function. Frontiers in Cellular and Infection Microbiology, 7, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, F. , Deme, J. C. , Lea, S. M. , & Berks, B. C. (2018). Type 9 secretion system structures reveal a new protein transport mechanism. Nature, 564, 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden, G. J. , Lyons, A. , & Scannapieco, F. A. (2013). Periodontal systemic associations: Review of the evidence. Journal of Periodontology, 84, S8–S19. [DOI] [PubMed] [Google Scholar]
- Meuric, V. , Rouillon, A. , Chandad, F. , & Bonnaure‐Mallet, M. (2010). Putative respiratory chain of Porphyromonas gingivalis . Future Microbiology, 5, 717–734. [DOI] [PubMed] [Google Scholar]
- Mortz, E. , Krogh, T. N. , Vorum, H. , & Gorg, A. (2001). Improved silver staining protocols for high sensitivity protein identification using matrix‐assisted laser desorption/ionization‐time of flight analysis. Proteomics, 1, 1359–1363. [DOI] [PubMed] [Google Scholar]
- Nagano, K. , Murakami, Y. , Nishikawa, K. , Sakakibara, J. , Shimozato, K. , & Yoshimura, F. (2007). Characterization of RagA and RagB in Porphyromonas gingivalis: Study using gene‐deletion mutants. Journal of Medical Microbiology, 56, 1536–1548. [DOI] [PubMed] [Google Scholar]
- Naito, M. , Tominaga, T. , Shoji, M. , & Nakayama, K. (2019). PGN_0297 is an essential component of the type IX secretion system (T9SS) in Porphyromonas gingivalis: Tn‐seq analysis for exhaustive identification of T9SS‐related genes. Microbiology and Immunology, 63, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olczak, T. , Sroka, A. , Potempa, J. , & Olczak, M. (2008). Porphyromonas gingivalis HmuY and HmuR: Further characterization of a novel mechanism of heme utilization. Archives of Microbiology, 189, 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi, M. , Graziani, F. , & D'Aiuto, F. (2020). Periodontal therapy and cardiovascular risk. Periodontology 2000, 83, 107–124. [DOI] [PubMed] [Google Scholar]
- Saiki, K. , & Konishi, K. (2010). Identification of a novel Porphyromonas gingivalis outer membrane protein, PG534, required for the production of active gingipains. FEMS Microbiology Letters, 310, 168–174. [DOI] [PubMed] [Google Scholar]
- Sato, K. , Naito, M. , Yukitake, H. , Hirakawa, H. , Shoji, M. , McBride, M. J. , Rhodes, R. G. , & Nakayama, K. (2010). A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 107, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seers, C. A. , Slakeski, N. , Veith, P. D. , Nikolof, T. , Chen, Y. Y. , Dashper, S. G. , & Reynolds, E. C. (2006). The RgpB C‐terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C‐terminal‐domain family found in Porphyromonas gingivalis . Journal of Bacteriology, 188, 6376–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky, S. S. , Haffajee, A. D. , Cugini, M. A. , Smith, C. , & Kent, R. L. Jr. (1998). Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25, 134–144. [DOI] [PubMed] [Google Scholar]
- Song, L. , Perpich, J. D. , Wu, C. , Doan, T. , Nowakowska, Z. , Potempa, J. , Christie, P. J. , Cascales, E. , Lamont, R. J. , & Hu, B. (2022). A unique bacterial secretion machinery with multiple secretion centers. Proceedings of the National Academy of Sciences of the United States of America, 119, e2119907119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D. , Gable, A. L. , Lyon, D. , Junge, A. , Wyder, S. , Huerta‐Cepas, J. , Simonovic, M. , Doncheva, N. T. , Morris, J. H. , Bork, P. , Jensen, L. J. , & Mering, C. V. (2019). STRING v11: Protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Research, 47, D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith, P. D. , Glew, M. D. , Gorasia, D. G. , & Reynolds, E. C. (2017). Type ix secretion: The generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Molecular Microbiology, 106, 35–53. [DOI] [PubMed] [Google Scholar]
- Veith, P. D. , Gorasia, D. G. , & Reynolds, E. C. (2021). Towards defining the outer membrane proteome of Porphyromonas gingivalis . Molecular Oral Microbiology, 36, 25–36. [DOI] [PubMed] [Google Scholar]
- Veith, P. D. , Nor Muhammad, N. A. , Dashper, S. G. , Likic, V. A. , Gorasia, D. G. , Chen, D. , Byrne, S. J. , Catmull, D. V. , & Reynolds, E. C. (2013). Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C‐terminal secretion signal, extensive post‐translational modification, and cell‐surface attachment. Journal of Proteome Research, 12, 4449–4461. [DOI] [PubMed] [Google Scholar]
- Veith, P. D. , Shoji, M. , O'Hair, R. A. J. , Leeming, M. G. , Nie, S. , Glew, M. D. , Reid, G. E. , Nakayama, K. , & Reynolds, E. C. (2020). Type ix secretion system cargo proteins are glycosylated at the C terminus with a novel linking sugar of the Wbp/Vim pathway. mBio, 11, e01497–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith, P. D. , Talbo, G. H. , Slakeski, N. , & Reynolds, E. C. (2001). Identification of a novel heterodimeric outer membrane protein of Porphyromonas gingivalis by two‐dimensional gel electrophoresis and peptide mass fingerprinting. European Journal of Biochemistry, 268, 4748–4757. [DOI] [PubMed] [Google Scholar]
- Vincent, M. S. , Canestrari, M. J. , Leone, P. , Stathopulos, J. , Ize, B. , Zoued, A. , Cambillau, C. , Kellenberger, C. , Roussel, A. , & Cascales, E. (2017). Characterization of the Porphyromonas gingivalis type ix secretion trans‐envelope PorKLMNP core complex. Journal of Biological Chemistry, 292, 3252–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenez‐Fyvie, L. A. , Haffajee, A. D. , & Socransky, S. S. (2000). Comparison of the microbiota of supra‐ and subgingival plaque in health and periodontitis. Journal of Clinical Periodontology, 27, 648–657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary Table 2: Predicted protein interactions identified by STRING
Data Availability Statement
The raw MS data are available upon request.


