Abstract
The human cytomegalovirus (HCMV) glycoprotein B (gB) (also known as gpUL55) homolog is an important mediator of virus entry and cell-to-cell dissemination of infection. To examine the potential ligand-binding properties of gB, a soluble form of gB (gB-S) was radiolabeled, purified, and tested in cell-binding experiments. Binding of gB-S to human fibroblast cells was found to occur in a dose-dependent, saturable, and specific manner. Scatchard analysis demonstrated a biphasic plot with the following estimated dissociation constants (Kd): Kd1, 4.96 × 10−6 M; Kd2, 3.07 × 10−7 M. Cell surface heparan sulfate proteoglycans (HSPGs) were determined to serve as one class of receptors able to facilitate gB-S binding. Both HSPG-deficient Chinese hamster ovary (CHO) cells and fibroblast cells with enzymatically removed HSPGs had 40% reductions in gB-S binding, whereas removal of chondroitin sulfate had no effect. However, a significant proportion of gB-S was able to associate with the cell surface in the absence of HSPGs via an undefined nonheparin component. Binding affinity analysis of gB-S binding to wild-type CHO-K1 cells demonstrated biphasic binding kinetics (Kd1, 9.85 × 10−6 M; Kd2, 4.03 × 10−8 M), whereas gB-S binding to HSPG-deficient CHO-677 cells exhibited single-component binding kinetics (Kd, 7.46 × 10−6 M). Together, these data suggest that gB-S associates with two classes of cellular receptors. The interaction of gB with its receptors is physiologically relevant, as evidenced by an inhibitory effect on HCMV entry when cells were pretreated with purified gB-S. This inhibition was determined to be manifested at the level of virus attachment. We conclude that gB is a ligand for HCMV that mediates an interaction with a cellular receptor(s) during HCMV infection.
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus that is present in approximately 80% of the adult population, as demonstrated by seroreactivity (3, 23). Primary HCMV infection of persons with intact immune systems often results in a self-limiting asymptomatic disease, while HCMV is a significant human pathogen for immunocompromised individuals that is often manifested as severe and debilitating sequelae (2). Despite its importance as a pathogen, limited antiviral therapies exist, due in part to a lack of detailed knowledge of the virus lifecycle.
HCMV infection requires that a viral envelope glycoprotein(s) and the respective cellular receptor(s) engage in a synchronized series of interactions, ultimately resulting in fusion of the viral envelope with the plasma membrane. Initial attachment of HCMV to permissive host cells is dependent upon the presence of cell surface heparan sulfate proteoglycans (HSPGs) (14, 43). Heparin affinity chromatography identified two HCMV glycoprotein complexes that possess the ability to bind immobilized heparin (14, 26). The HCMV glycoprotein complex II (gC-II) was described to be the major HCMV envelope protein complex retained on the heparin matrix, while a lesser proportion of glycoprotein B (gB) (also known as gpUL55) was bound (26). Due to the lack of a manipulable genetic system for HCMV, to date there has been no effective manner by which to evaluate independently the functional relevance of heparin binding for gB or gC-II. This initial heparin-dissociable binding state is rapidly converted to a stable attachment, suggesting that HCMV absorption involves a sequential association with multiple cellular receptors (14). After stable attachment to the cell surface, a direct pH-independent fusion event occurs between the viral envelope and the plasma membrane (13). Two HCMV envelope glycoprotein complexes, gB and gH-gL (also known as gpUL75-gpUL115), are crucial components in mediating fusion events required for subsequent virus entry. The identity of cellular receptors for stable binding or of fusion facilitators is not known, although a number of candidates have been proposed (1, 28, 29, 52, 53).
HCMV gB is a 906-amino-acid protein encoded by the UL55 open reading frame (12, 16). The gB precursor is synthesized as a 105-kDa protein, which matures into a 130- to 160-kDa glycoprotein by acquiring N-linked glycosylation modifications in the endoplasmic reticulum and Golgi network (6, 7). The cellular protease furin cleaves the mature gB into two components, a 93- to 116-kDa amino-terminal fragment and a 55-kDa carboxy-terminal fragment (60). These two fragments have been shown to associate as a disulfide-linked monomer (53, 54) which is presented on the viral envelope as well as on the surface of virus-infected cells as a covalently associated homodimer (9). gB is the most abundant constituent of the viral envelope and is a potent immunogenic HCMV protein (8, 35).
gB has the potential to be a multifunctional regulator of HCMV entry. As described above, HCMV gB is a putative viral ligand in that it possesses heparin-binding capacity (perhaps critical in the initial attachment phase) and is involved in virus penetration and cell-to-cell spread. Neutralizing anti-gB monoclonal antibodies significantly blocked viral fusion events, including penetration and cell-to-cell transmission, while viral attachment remained unaffected (41). Similarly, U373 glioblastoma cells constitutively expressing gB formed multinucleated syncytia, a process which was effectively precluded by the addition of neutralizing anti-gB antibodies (59). In an effort to address the receptor-binding properties of gB, we tested a recombinant soluble form of gB (gB-S) in cellular binding experiments. Previously, we showed that the gB-S protein retained features attributable to the viral protein in that it was dimeric, properly folded, and bound to a heparin affinity matrix (11). Our results presented here demonstrate that gB-S does exhibit conventional ligand properties and may engage more than one class of receptors on the surfaces of both fibroblast and wild-type Chinese hamster ovary (CHO) cells. Cell surface HSPGs were determined to be one receptor for the recombinant gB molecule, since gB-S binding was reduced when these molecules were absent; however, a second HSPG-independent binding site was also implicated. Treatment of cells with gB-S inhibited virus entry and infection, supporting a physiological relevance for the interaction of gB with its cellular receptor(s).
MATERIALS AND METHODS
Cell lines and virus.
Immortalized fibroblasts (IF) (15) were cultured in Dulbecco’s minimal essential medium (DMEM) (BioWhittaker, Walkersville, Md.) supplemented with 5% fetal bovine serum (FBS) (HyClone, Logan, Utah), 1.0% penicillin-streptomycin-fungizone (PSF) (BioWhittaker), 0.3% l-glutamine (BioWhittaker), and 100 μg of geneticin (Gibco BRL, Gaithersburg, Md.) per ml. Wild-type CHO cells (CHO-K1) and glycosaminoglycan-deficient CHO mutants pgsA-745 (CHO-745; xylosyl transferase deficient) and pgsD-677 (CHO-677; N-acetylglucosaminyl [GlcNAc] transferase and glucuronosyl [GlcA] transferase deficient) have been described previously (18, 19). The CHO cell lines were cultured in Ham’s F-12 medium (BioWhittaker) supplemented with 10% FBS, PSF, and l-glutamine. Adherent cultures of Trichoplusia ni (TN-5) insect cells (11) were cultured in ExCell 401 medium (JRH Biosciences, Lenexa, Kans.) supplemented with 5% FBS and PSF. HCMV AD169 was grown and titered on IF cells as previously described (15). A recombinant strain of Autographa californica nuclear polyhedrosis virus encoding amino acids 1 to 692 of the gB homolog from the AD169 strain of HCMV (BV-gB-S) was grown and titered as previously described (11).
Purification of radiolabeled gB-S.
TN-5 cells were infected with BV-gB-S (11) at a multiplicity of infection (MOI) of 8. Two days postinfection, the infected cell monolayer was methionine starved for 1 h in the presence of Insect-Xpress without l-methionine (BioWhittaker). EXPRE35S35S protein labeling mix (DuPont NEN, Boston, Mass.), containing both [35S]methionine and [35S]cysteine, was added at a final concentration of 50 μCi of [35S]methionine per ml. Four days postinfection, the infected cells were harvested and cellular debris was removed via centrifugation at 1,200 × g at 4°C. Secreted gB-S was purified from the tissue culture supernatant by a two-step chromatography protocol. Supernatant containing gB-S was passaged over an immobilized heparin column, and the bound proteins were eluted with 0.65 M sodium chloride. After adjustment of the salt concentration to 0.1 M, the heparin eluate was applied to nickel-nitrilotriacetic acid (NTA) agarose beads (Qiagen, Valencia, Calif.). The nickel column was washed successively with 10 volumes of binding buffer (100 mM NaPO4–10% glycerol, pH 7.8), followed by 3 volumes of pH wash buffer (50 mM NaPO4–10% glycerol, pH 6.0) and 3 volumes of 15 mM imidazole buffer (50 mM NaPO4–300 mM NaCl–10% glycerol–15 mM imidazole, pH 7.0). gB-S was eluted from the column with 0.5 M imidazole (500 mM NaPO4–300 mM NaCl–0.5 M imidazole–10% glycerol, pH 6.0). The samples were dialyzed against 1× phosphate-buffered saline (PBS [10 mM NaPO4–140 mM NaCl, pH 7.4]) overnight at 4°C. The protein concentration was determined by Bradford analysis (Bio-Rad, Hercules, Calif.). The specific activity of the purified [35S]gB-S ranged from 106 to 377 cpm/μg of protein.
SDS-PAGE analysis and immunoblotting.
gB-S samples eluted from the Ni2+-NTA column were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in the absence of reducing agents, as previously described (11, 24). To visualize the radiolabeled protein samples, the gel was fixed in 25% methanol–7.5% acetic acid, washed briefly in 20% methanol, and subsequently incubated in Fluoro-Hance (Research Products International, Mount Prospect, Ill.) prior to drying and exposure to film. Immunoblotting experiments were performed with an anti-gB monoclonal antibody (27-78) (5) followed by incubation with a goat anti-mouse antibody conjugated to horseradish peroxidase (HRP) (Pierce, Rockford, Ill.), and the LumiGLO HRP substrate kit (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.) was used to detect the peroxidase conjugates.
Protein-binding assay.
IF cells were chilled to 4°C and treated with 5 mM ovalbumin diluted in PBS–1% FBS–0.1 mM CaCl2 (PBS-GC) for 30 min to block nonspecific binding. The gB-S protein was diluted in PBS-GC, added to the cell monolayers, and incubated for 90 min at 4°C. Unbound gB-S was removed, and the cells were washed twice with PBS-GC and subsequently lysed in 1% SDS–1% Triton X-100. Both unbound and bound fractions were subjected to scintillation counting. Experiments for all data points were performed in duplicate or triplicate. For some experiments, IF cell monolayers were treated with increasing concentrations of heparinase or chondroitinase ABC (Sigma, St. Louis, Mo.) for 60 min at 37°C prior to the binding assay described above.
Purification of radiolabeled virions and attachment assay.
Virus attachment was quantified by using radiolabeled virions, as previously described (44). Briefly, at 4°C, IF cells were incubated with increasing concentrations of nonradiolabeled gB-S or 10 μg of heparin per ml for 90 min, after which labeled virions were added (approximately 1,000 PFU/cell and 8.7 × 104 cpm/well). Unbound virions were removed, and the cells were subsequently lysed in 1% SDS–1% Triton X-100, and both unbound and bound fractions were subjected to scintillation counting.
Virus entry assay.
IF cells were grown on glass coverslips in 12-well plates. After chilling of the cells at 4°C, increasing concentrations of nonradiolabeled gB-S, bovine serum albumin (BSA) (Sigma), or 10 μg of heparin per ml was added for 60 min. HCMV AD169 was added to the cells (MOI = 0.1) and allowed to adsorb for 90 min. After a 30-min temperature shift to 37°C to allow for virus penetration, a low-pH citrate buffer (40 mM citric acid–10 mM KCl–135 mM NaCl, pH 3.0) was added to inactivate any extracellular virus. The cells were incubated in DMEM–2% FBS for 24 h. Immunofluorescence analysis was performed as previously described (14) with a rabbit anti-IE antibody, followed by detection with a fluorescein-conjugated goat anti-rabbit secondary antibody (Kirkegaard & Perry Laboratories, Inc.) in combination with nuclear staining via 10 μg of Hoechst dye per ml. Experiments for all data points were performed in duplicate with a minimum of 500 cells per coverslip scored.
RESULTS
Binding of gB-S to fibroblast cells.
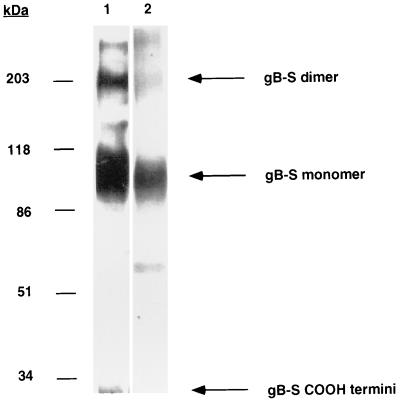
We have previously described the production and structural characterization of a truncated, soluble form of the HCMV gB protein (gB-S) (11). This recombinant form of gB (amino acid residues 1 to 692) lacks the putative transmembrane and cytoplasmic domains, resulting in efficient secretion into the tissue culture media. The purified gB-S retained many features attributable to the viral gB molecule, including proper folding, dimer formation, and the ability to bind immobilized heparin (11). SDS-PAGE analysis and autoradiography demonstrated that the purified gB-S was a heterogeneous preparation consisting of primarily monomeric and dimeric gB structures (Fig. 1). Some reduction of intrachain disulfide bonds occurred during purification, since a small fraction of the gB-S carboxy-terminal fragment (32 kDa) was detected.
FIG. 1.
Purity of [35S]gB-S. Eluates from the Ni2+-NTA column were subjected to SDS-PAGE analysis under nonreducing conditions on a 7.5% acrylamide gel. Lane 1 shows a [35S]gB-S sample that was electrophoresed, transferred to nitrocellulose, probed with an anti-gB monoclonal antibody (27-78), and detected with a secondary goat anti-mouse HRP-conjugated antibody in conjunction with a chemiluminescent substrate. Lane 2 shows a [35S]gB-S sample that was electrophoresed, after which the gel was dried, fixed, and exposed to film.
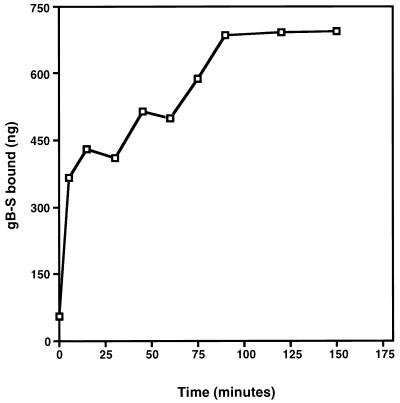
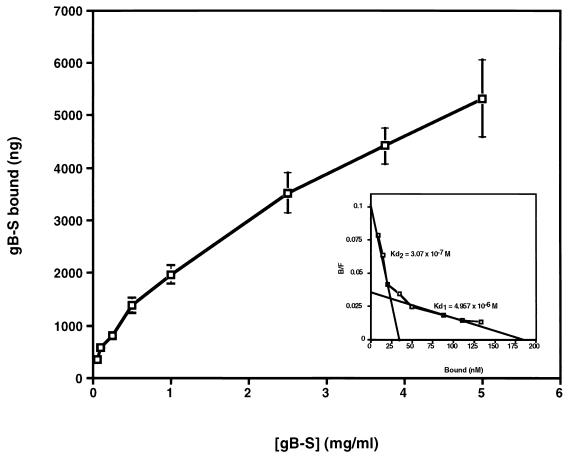
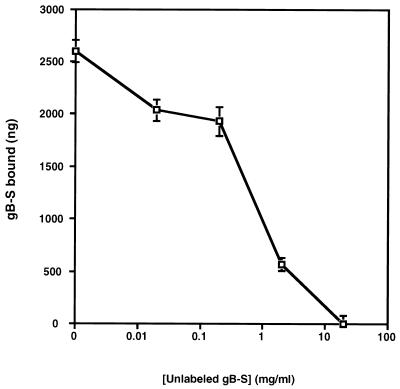
To test potential ligand properties of gB, binding assays were conducted with purified, radiolabeled gB-S protein. Time course analysis demonstrated that binding of gB-S to fibroblast monolayers reached equilibrium by approximately 90 min, a time frame consistent with HCMV adsorption (Fig. 2). Binding of gB-S to fibroblast monolayers was a dose-dependent event (Fig. 3), which approached saturation at high concentrations of protein (5 mg/ml). These results mirror dose response curves generated with HCMV virions (58), suggesting a high abundance of cellular receptors. Transformation of the data from Fig. 3 into a Scatchard plot demonstrated a classic biphasic plot with low and moderate affinities, with the following estimated dissociation constants (Kd): Kd1, 4.96 × 10−6 M; Kd2, 3.07 × 10−7 M (Fig. 3 [inset]). It was estimated that there are 2.8 × 106 low-affinity binding sites per cell and 4.99 × 105 moderate-affinity receptors per cell (Table 1). To address the specificity of gB-S binding to fibroblast monolayers, homologous competition assays were performed with a constant concentration of radiolabeled gB-S and increasing concentrations of nonradiolabeled gB-S. In the presence of a 100-fold molar excess of nonradiolabeled gB-S, binding of radiolabeled gB-S was significantly and reproducibly reduced, indicating that binding of gB-S is specific (Fig. 4).
FIG. 2.
Time course analysis. IF cells were cultured in a 96-well plate and chilled to 4°C, and nonspecific binding sites were blocked for 30 min at 4°C by the addition of 5 mM ovalbumin in PBS-GC. A constant concentration of [35S]gB-S (0.5 mg/ml) was incubated with the cell monolayer for increasing increments of time. At the conclusion of the designated time, unbound [35S]gB-S was removed, the cells were washed twice with PBS-GC, and the cells containing the bound gB-S were lysed by the addition of 1% SDS–1% Triton X-100. Experiments for all data points were performed in triplicate. The specific activity of the [35S]gB-S preparation used for this experiment was 153.55 cpm/μg of protein.
FIG. 3.
Dose response binding curve. IF cells cultured in a 24-well plate were chilled and blocked at 4°C. Increasing concentrations of [35S]gB-S, diluted in PBS-GC, were added to the cell monolayers and incubated for 90 min at 4°C. Unbound and bound fractions were collected and subjected to scintillation counting. Experiments for all data points were performed in duplicate. Bars represent standard deviations. The specific activity of the [35S]gB-S preparation used for this experiment was 267 cpm/μg of protein. On the basis of the specific activity of the [35S]gB-S preparation, the amounts of total input and free and bound proteins were determined and subjected to Scatchard analysis (49). Shown in the inset are lines representing the best fits as determined by a linear regression analysis. The experiment was repeated several times with consistent results.
TABLE 1.
Binding affinity constants and numbers of binding sites/cell
| Cell type | Kd (10−7 M) | No. of binding sites/cell (106) |
|---|---|---|
| IF | 49.6a | 2.8 |
| 3.07b | 0.499 | |
| CHO-K1 | 98.5a | 41.5 |
| 0.403b | 3.46 | |
| CHO-677 | 74.6 | 91.9 |
Kd1.
Kd2.
FIG. 4.
Homologous competition assay. A constant concentration of [35S]gB-S (0.2 mg/ml) was added to IF cells in the presence of increasing concentrations of nonradiolabeled gB-S for 90 min at 4°C. Each data point represents the average of duplicate wells. Bars represent standard deviations. The specific activity of the [35S]gB-S preparation used for this experiment was 230 cpm/μg of protein. Nonspecific binding was determined by the addition of a 100-fold molar excess of nonradiolabeled gB-S and was determined to be 21%. Nonspecific binding was deducted from each data point prior to graphing.
Cell surface HSPG molecules are receptors for gB-S.
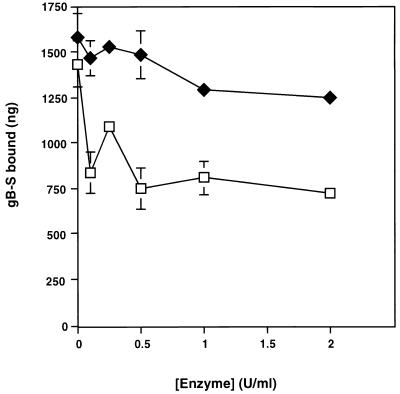
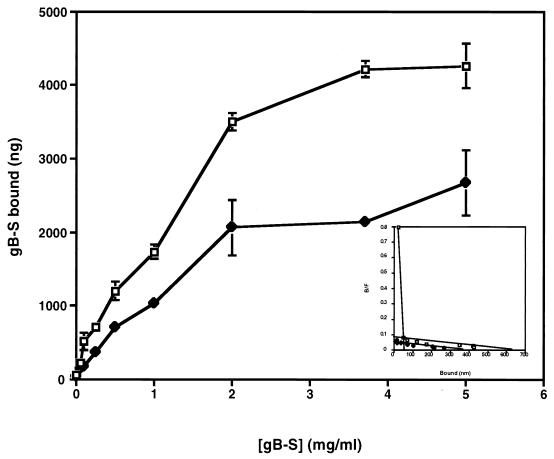
HSPGs are known HCMV attachment receptors (14, 43), and it is likely that gB and a component of the gC-II complex are responsible for this interaction. Heparin affinity chromatography analysis indicated that these two HCMV envelope glycoprotein complexes are capable of binding to immobilized heparin, with the gC-II complex being the primary HSPG ligand, while it is postulated that gB is only a minor heparin-binding protein. (14, 26). However, it has never been experimentally demonstrated that either of these two glycoprotein complexes bind to cell surface HSPGs. To determine if cellular HSPGs are engaged by gB-S, fibroblast monolayers were treated with increasing concentrations of heparinase (removes heparin) or chondroitinase (removes chondroitin sulfate [CS]). As shown in Fig. 5, enzymatic removal of HSPGs diminished binding of gB-S by approximately 40 to 50%, while treatment with chondroitinase had no appreciable affect on gB-S binding. Parallel virus entry assays were performed under the identical conditions. In heparinase-treated cells, virus infection was reduced by 95% compared to untreated cells, whereas virus entry was unaffected in chondroitinase-treated cells (data not shown). These results demonstrate that the enzymatic treatment was effective and suggests that cellular HSPGs, but not CS proteoglycans (CSPGs), are indeed one class of cellular receptors capable of engaging the gB-S ligand. However a nonheparin component is additionally recognized, as demonstrated by the retention of gB-S binding in the absence of heparin molecules. We also examined gB-S binding to CHO cell line mutants which are defective in the biosynthesis of various glycosaminoglycan chains. The CHO-745 cell line, due to a defect in xylosyl transferase activity, expresses only about 1% of the normal levels of glycosaminoglycans compared to wild-type CHO-K1 cells, whereas the CHO-677 cell line (GlcNAc and GlcA deficient) produces no detectable levels of heparan sulfate but elevated levels of CS compared to CHO-K1 cells (18, 19). Binding of gB-S to both the CHO-K1 and CHO-677 cells was found to occur in a dose-dependent and saturable manner (Fig. 6). Overall, gB-S binding to the CHO-745 and CHO-677 cell lines was maximally reduced approximately 40 to 50% compared to wild-type CHO-K1 cells (data not shown), consistent with the results obtained by enzymatic removal of cell surface heparan sulfate (Fig. 5). Also similar to the fibroblast cells, gB-S binding to CHO-K1 cells demonstrated dual binding kinetics, with the following estimated dissociation constants: Kd1, 9.85 × 10−6 M; Kd2, 4.03 × 10−8 M. Conversely, gB-S binding to CHO-677 cells exhibited single binding kinetics, with an estimated Kd of 7.46 × 10−6 M (Fig. 6 [inset]). It was estimated that on CHO-K1 cell surfaces, there are 4.15 × 107 low-affinity receptors per cell and 3.46 × 106 high-affinity receptors per cell (Table 1). On CHO-677 cell surfaces, it was estimated that there are 9.15 × 107 gB-S receptors per cell.
FIG. 5.
Binding of [35S]gB-S in the absence of HSPGs. IF cell monolayers were treated with increasing concentrations of heparinase (□) or chondroitinase ABC (⧫) for 60 min at 37°C. Binding assays were conducted for 90 min with 0.5 mg of [35S]gB-S per ml diluted in PBS-GC. Each data point represents the average of duplicate wells. Bars represent standard deviations. The specific activity of the [35S]gB-S preparation used for this experiment was 368.05 cpm/μg of protein.
FIG. 6.
Saturable binding of gB-S to CHO cell monolayers. CHO-K1 (□) and CHO-677 (⧫) cells were cultured in a 96-well plate, chilled, and blocked at 4°C. Increasing concentrations of [35S]gB-S were added to the cell monolayers, which were incubated for 90 min at 4°C. Unbound and bound fractions were collected and subjected to scintillation counting. Experiments for all data points were performed in triplicate. Bars represent standard deviations. The specific activity of the [35S]gB-S preparation used for this experiment was 320.5 cpm/μg of protein. On the basis of the specific activity of the [35S]gB-S preparation, the amounts of total input and free and bound proteins were determined and subjected to Scatchard analysis (49). Shown in the inset are lines representing the best fits as determined by a linear regression analysis.
Inhibition of virus entry by gB-S.
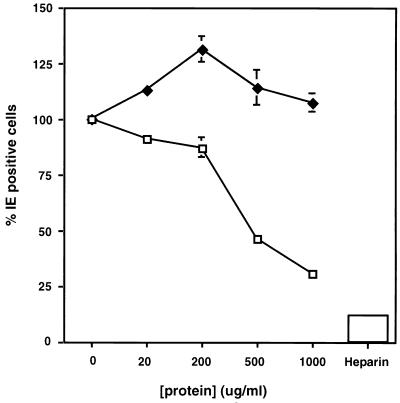
In efforts to determine if the interaction of gB-S with cell surfaces is biologically relevant, fibroblast cells were cultured on glass coverslips and treated with increasing concentrations of nonradiolabeled gB-S at 4°C for 1 h. HCMV was allowed to incubate with the gB-S-pretreated cells for 90 min at 4°C; the cells were then warmed to 37°C, after which a low-pH buffer was added to the cells. Twenty-four hours postinfection, the cells were processed for immunofluorescence to assess the expression of the major immediate early protein p72, the first protein to be synthesized in infected cells. As increasing concentrations of gB-S were added, the ability of HCMV to initiate viral infection was diminished by approximately 70% compared to the untreated control (Fig. 7). Treatment of cells with identical concentrations of BSA had no effect on virus entry, implying that the block in HCMV entry is specifically attributed to the engagement of a necessary HCMV cellular receptor by gB-S and is not simply protein-mediated interference.
FIG. 7.
HCMV infection is decreased in the presence of gB-S. IF monolayers cultured on glass coverslips in a 12-well plate were chilled to 4°C. Increasing concentrations of nonradiolabeled gB-S (□), BSA (⧫), or 10 μg of heparin per ml were allowed to incubate with the cells for 60 min at 4°C. Virus was added to the cells (MOI = 0.1) and allowed to absorb for 90 min at 4°C. After a 30-min temperature shift to 37°C, a low-pH citrate buffer was added to inactivate any extracellular virus. At 24 h postinfection, immunofluorescence staining was performed with a rabbit anti-IE antibody followed by the secondary goat anti-rabbit–fluorescein conjugate in combination with Hoechst dye. Experiments for all data points were performed in duplicate, and a minimum of 500 cells per coverslip were scored. Bars represent standard deviations.
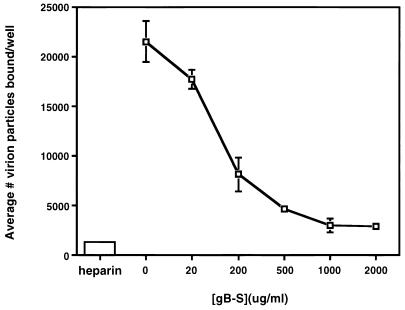
To assess if the block in entry occurred at the initial attachment phase or the subsequent penetration event, cells were incubated with increasing concentrations of nonradiolabeled gB-S prior to and during the addition of radiolabeled HCMV virions. As depicted in Fig. 8, HCMV adsorption is hindered in a dose-dependent fashion as the concentration of gB-S increases. This implies that gB-S recognizes and occupies a cell surface molecule(s) that functions as an HCMV attachment receptor. Although gB has been identified as functioning in the penetration and fusion of the virion, its function in early binding events is a novel role.
FIG. 8.
Soluble form of gB blocks HCMV attachment. IF cells were treated with increasing concentrations of gB-S or 10 μg of heparin per ml for 60 min, after which [35S]methionine gradient-purified HCMV virions were added for 90 min. All samples were tested in triplicate.
DISCUSSION
A gB homolog has been identified in all herpesviruses examined to date. This conservation of gB is manifested at both a structural and a functional level. However, concerning the sequences of the gB proteins of the different subfamilies, it is predicted that unique features also exist. For example, between the three subclasses of herpesviruses (alpha, beta, and gamma), the level of gB homology is only 30 to 40% at the amino acid level (45). Neighbor-joining analysis revealed that the gBs of the alphaherpesviruses clustered very closely together in a dendrogram while the two HCMV gB molecules (from strains AD169 and Towne) were clustered close to each other but quite distinct from both the alphaherpesviruses and the gammaherpesviruses (17). Most of the significant homology is clustered at the carboxy termini of the molecules, while the amino termini are very divergent (17, 45, 48). A second reason to predict unique functional activities for the HCMV gB protein pertains to distinct biological patterns of infection in humans. In contrast to the alphaherpesviruses, HCMV infects deep tissues, such as liver, spleen, kidney, and lung (2). Thus, each gB protein must be evaluated independently before prescribed functional properties can be defined.
Two distinct, complementary experimental approaches are used to study the structure and function of herpesvirus genes. Genetic deletions, or knockouts, have proven very useful in determining the essentialness of individual genes. To this end, all gB molecules examined to date, including that of HCMV, have been demonstrated to be essential components in the replication cycle (4, 10, 21, 41). More recently, viral recombinants containing different gBs have been used to identify cross-complementation. For example, within the alphaherpesviruses, some complementation has been demonstrated (30, 37–39, 47). Analysis of herpes simplex virus type 1 (HSV-1) recombinants containing the gBs of both HSV-1 and bovine herpesvirus 1 (BHV-1) have indicated that BHV-1 gB may function in HSV-1 infectivity (39). HSV-1 containing null mutations in gB can also be complemented by pseudorabies virus (PrV) gB (37), whereas the reverse is not true. PrV containing a gB null mutation can be complemented by BHV-1 gB either by growth on a BHV-1 gB-expressing cell line or by insertion of BHV-1 gB into the PrV viral genome (30, 46). In contrast, a BHV-1 gB null mutant is unable to be complemented by PrV gB (38). These findings suggest that even within this group of highly conserved gBs, unique functional features exist in conjunction with shared properties. When complementation between the gB molecules from different subfamilies was examined, no complementation at all was found (33). Specifically, neither the HCMV gB nor the HSV gB gene sequences are able to restore the infectivity of an Epstein-Barr virus (EBV) gB-null strain, nor is gB from EBV or HCMV able to complement a strain of HSV lacking gB (33). These results suggest that although gB proteins are conserved among the family, each herpesvirus contains a unique gB protein that functionally fills a particular niche.
The use of recombinant soluble versions of putative viral ligands is a powerful tool for assessing the role of cellular receptor-viral ligand interactions. Perhaps the most well characterized is the specificity of human immunodeficiency virus gp120 for T-lymphocyte CD4 molecules (31, 32, 36). A soluble recombinant version of gp120 binds with high affinity to CD4-expressing cells displayed in a single-component binding curve with an approximate Kd of 4 × 10−9 M (32). Similar values for the gp120-CD4 dissociation constant were obtained when soluble secreted CD4 was reacted with recombinant gp120 (51). For the human herpesviruses, it is well established that the EBV gp350/220 is a ligand for the B-cell-specific CR2 receptor and that the interaction between these two components is essential and sufficient for B-cell infection (42, 56, 57). Use of a recombinant truncated EBV gp350/220 molecule generated a biphasic Scatchard plot with an estimated Kd1 of 1.2 × 10−8 M and a Kd2 of 3.3 × 10−7 M, with the number of high-affinity gp350/220 binding sites approximating the number of anti-CR2 monoclonal antibody binding sites (57). Similarly, there are a several reports on the construction, characterization, and biochemical analysis of both truncated and full-length forms of three HSV envelope glycoproteins, gB, gC, and gD (20, 50, 55). Characterization of a soluble HSV-1 gC demonstrated that gC functions solely during the attachment phase, that it interacts with cell surface HSPG molecules, and that it binds in a dose-dependent, saturable manner (55). In contrast, incubation of cells with soluble HSV-1 or HSV-2 gD (gDt-1 and gDt-2, respectively) provided no protection from viral attachment; however, viral penetration was strictly inhibited (25). Binding of these recombinant proteins occurred in a dose-dependent, saturable, and specific manner, with estimated dissociation constants of 2.6 × 10−7 M for gDt-1 and 2.3 × 10−7 for gDt-2 (25). These early data support the recent finding that recombinant gD can associate with a soluble version of a cellular protein that can mediate HSV entry (HVEM) (40, 61). Enzyme-linked immunosorbent assay analysis demonstrated that gD binds to HVEM with an affinity in the micromolar range, a rate of kinetics similar to that demonstrated with our recombinant HCMV gB.
Specific analysis of recombinant forms of various gB molecules reflects the diversity observed with cross-complementation experiments. For example, soluble HSV-2 is unable to block HSV infection and has yet to demonstrate any ligand characteristics (25), in spite of the fact that gB can engage HSPGs when gC is absent (22). A recombinant BHV-1 gB homolog which possesses the transmembrane and cytoplasmic regions had a major inhibitory effect in blocking virus entry, whereas the soluble form was inert in this capacity, implying at least for BHV-1, that the gB homolog has the potential to function in a multitude of interactions between the virus and the cell surface (34). Our results on HCMV gB also reflected dual binding properties, with some notable differences from those observed with the BHV-1 homolog. For instance, dual affinities were detected with the soluble form of HCMV gB, suggesting that a membrane orientation is not needed to engage its receptor(s) or to block infection.
With its proposed roles in both virus attachment and penetration, it is not unexpected that HCMV gB possesses properties of a true viral ligand. Binding of gB-S to fibroblast monolayers exhibited criteria of known receptor-ligand interactions: saturability, specificity, affinity, and induction of a measurable physiological response. At least two classes of cellular receptors are able to mediate binding of gB-S. Although it has been shown previously that gB binds immobilized heparin (14, 26), this is the first description of the affinity of gB for HSPG molecules localized on the cell surface. Our data suggest that binding of gB is specific for heparan sulfate moieties, not all glycosaminoglycans in general, since removal of CS (Fig. 5) had no effect on binding, and that elevated levels of CS on CHO-677 cell surfaces do not affect gB-S binding ability (data not shown). In addition, binding curves to the GAG-deficient CHOs demonstrated that gB-S retained its propensity to act as a viral ligand and that the binding kinetics of the recombinant protein are shifted to a single-component binding curve, indicating that gB-S binds to at least one class of heparin-independent molecules. Interestingly, the ability of gB-S to bind in the absence of HSPGs does not mirror HCMV virions’ absolute requirement for HSPG for attachment (14, 43), supporting the conclusion that the HCMV envelope complex, gC-II, is the principle mediator of virus attachment to HSPGs (26, 27). Since the gC-II complex is incompletely defined genetically, confirmation of the independent contributions of gB and gC-II await further genetic analysis. Further efforts are also aimed at the identification of cellular gB-binding partners. Work from our laboratory has recently demonstrated a direct interaction between HCMV gB and cellular annexin II (46), and experiments are currently under way to determine if this interaction is relevant in binding of gB to cells in the absence of HSPGs.
ACKNOWLEDGMENTS
These studies were supported by Public Health Service grant RO1 AI-34998 and a Basic Research grant from the March of Dimes Birth Defects Foundation.
REFERENCES
- 1.Adlish J D, Lahijani R S, St. Jeor S C. Identification of a putative cell receptor for human cytomegalovirus. Virology. 1990;176:337–345. doi: 10.1016/0042-6822(90)90003-a. [DOI] [PubMed] [Google Scholar]
- 2.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1981–2010. [Google Scholar]
- 3.Berry N J, Burns D M, Wannamethee G, Grundy J E, Lui S F, Prentice H G, Griffiths P D. Seroepidemiologic studies on the acquisition of antibodies to cytomegalovirus, herpes simplex virus, and human immunodeficiency virus among general hospital patients and those attending a clinic for sexually transmitted diseases. J Med Virol. 1988;24:385–393. doi: 10.1002/jmv.1890240405. [DOI] [PubMed] [Google Scholar]
- 4.Boyle, K. A., and T. Compton. Unpublished results.
- 5.Britt W J. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology. 1984;135:369–378. doi: 10.1016/0042-6822(84)90193-4. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J, Auger D. Synthesis and processing of the envelope gp55-116 complex of human cytomegalovirus. J Virol. 1986;58:185–191. doi: 10.1128/jvi.58.1.185-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britt W J, Vugler L G. Processing of the gp55-116 envelope glycoprotein complex (gB) of human cytomegalovirus. J Virol. 1989;63:403–410. doi: 10.1128/jvi.63.1.403-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britt W J, Vugler L, Butfiloski E J, Stephens E B. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britt W J, Vugler L G. Oligomerization of the human cytomegalovirus major envelope glycoprotein complex gB (gp55-116) J Virol. 1992;66:6747–6754. doi: 10.1128/jvi.66.11.6747-6754.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson C, Britt W J, Compton T. Expression, purification and characterization of a soluble form of human cytomegalovirus glycoprotein B. Virology. 1997;239:198–205. doi: 10.1006/viro.1997.8892. [DOI] [PubMed] [Google Scholar]
- 12.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 13.Compton T, Nepomuceno R R, Nowlin D M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 14.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 15.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cranage M P, Kouzarides T, Bankier A T, Satchwell S C, Weston K M, Tomlinson P, Barrell B G, Hart H, Bell S E, Minson A C, Smith G L. Identification of the human cytomegalovirus glycoprotein B gene and induction of neutralizing antibodies via its expression in recombinant vaccina virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberle R, Black D. Sequence analysis of herpes simplex virus gB gene homologs of two platyrrhine monkey alpha-herpesviruses. Arch Virol. 1993;129:167–182. doi: 10.1007/BF01316893. [DOI] [PubMed] [Google Scholar]
- 18.Esko J D, Stewart T E, Taylor W H. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esko J D, Rostand K S, Weinke J L. Tumor formation dependent on proteoglycan biosynthesis. Science. 1988;241:1092–1094. doi: 10.1126/science.3137658. [DOI] [PubMed] [Google Scholar]
- 20.Ghiasi H, Kaiwar R, Nesburn A B, Wechsler S L. Expression of herpes simplex virus type 1 glycoprotein B in insect cells. Virus Res. 1991;22:25–39. doi: 10.1016/0168-1702(92)90087-p. [DOI] [PubMed] [Google Scholar]
- 21.Harrold R E, Marchini A, Fruehing S, Longnecker R. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J Virol. 1996;70:2049–2054. doi: 10.1128/jvi.70.3.2049-2054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold B C, Visalli R J, Susmarski N, Brandt C R, Spear P G. Glycoprotein C-independent binding of herpes simplex to cells requires cell surface heparan sulfate and glycoprotein B. J Gen Virol. 1994;75:1211–1222. doi: 10.1099/0022-1317-75-6-1211. [DOI] [PubMed] [Google Scholar]
- 23.Ho M. Epidemiology of cytomegalovirus infection. Rev Infect Dis. 1990;12:701S–710S. doi: 10.1093/clinids/12.supplement_7.s701. [DOI] [PubMed] [Google Scholar]
- 24.Huber M, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–5398. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson D C, Burke R L, Gregory T. Soluble forms of herpes simples virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kari B, Gehrz R. Structure, composition and heparin binding properties of human cytomegalovirus glycoprotein complex designated gC-II. J Gen Virol. 1993;74:255–264. doi: 10.1099/0022-1317-74-2-255. [DOI] [PubMed] [Google Scholar]
- 28.Keay S, Baldwin B. The human fibroblast receptor for gp86 of human cytomegalovirus is a phosphorylated glycoprotein. J Virol. 1992;66:4834–4838. doi: 10.1128/jvi.66.8.4834-4838.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keay S, Baldwin B. Update on the 92.5 kDa putative HCMV fusion receptor. Scand J Infect Dis. 1995;99:32–33. [PubMed] [Google Scholar]
- 30.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein I of bovine herpesvirus 1. J Virol. 1992;66:2754–2764. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau N R, Warton M, Littman D R. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature. 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 32.Lasky L A, Nakamura G, Smith D H, Fennie C, Shimasaki C, Patzer D, Berman P, Gregory T, Capon D J. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 33.Lee S K, Compton T, Longnecker R. Failure to complement infectivity of EBV and HSV-1 glycoprotein B (gB) deletion mutants with gBs from different human herpesvirus subfamilies. Virology. 1997;237:170–181. doi: 10.1006/viro.1997.8765. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, van Drunen Littel-van den Hurk S, Babiuk L A, Liang X. Characterization of cell-binding properties of bovine herpesvirus 1 glycoproteins B, C, and D: identification of a dual cell-binding function of gB. J Virol. 1995;69:4758–4768. doi: 10.1128/jvi.69.8.4758-4768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marshall G S, Rabalais G P, Stout G G, Waldeyer S L. Antibodies to recombinant-derived glycoprotein B after natural human cytomegalovirus infection correlate with neutralizing activity. J Infect Dis. 1992;165:381–384. doi: 10.1093/infdis/165.2.381. [DOI] [PubMed] [Google Scholar]
- 36.McDougal J, Kennedy M, Sligh J, Cort S, Mawle A, Nicholson J. Binding of the HTLVIII/LAV to T4+ T cells by a complex of the 110K viral protein and the T4 molecule. Science. 1986;231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 37.Mettenleiter T C, Spear P G. Glycoprotein B (gII) of pseudorabies virus can functionally substitute for glycoprotein gB in herpes simplex virus type 1. J Virol. 1994;68:500–504. doi: 10.1128/jvi.68.1.500-504.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miethke A, Keil G M, Weiland F, Mettenleiter T C. Unidirectional complementation between glycoprotein B homologues of pseudorabies virus and bovine herpesvirus 1 is determined by the carboxy-terminal part of the molecule. J Gen Virol. 1995;76:1623–1635. doi: 10.1099/0022-1317-76-7-1623. [DOI] [PubMed] [Google Scholar]
- 39.Misra V, Blewett E L. Construction of herpes simplex viruses that are pseudodiploid for the glycoprotein B gene: a strategy for studying the function of an essential herpesvirus gene. J Gen Virol. 1991;72:385–392. doi: 10.1099/0022-1317-72-2-385. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 41.Navarro D, Paz P, Tugizov S, Topp K, La Vail J, Pereira L. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology. 1993;197:143–158. doi: 10.1006/viro.1993.1575. [DOI] [PubMed] [Google Scholar]
- 42.Nemerow G R, Mold C, Keivens Schwend V, Tollefson V, Cooper N R. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol. 1987;61:1416–1420. doi: 10.1128/jvi.61.5.1416-1420.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neyts J, Snoeck R, Schols D, Balzarini J, Esko J D, Van S A, De C E. Sulfated polymers inhibit the interaction of human cytomegalovirus with cell surface heparan sulfate. Virology. 1992;189:48–58. doi: 10.1016/0042-6822(92)90680-n. [DOI] [PubMed] [Google Scholar]
- 44.Nowlin D M, Cooper N R, Compton T. Expression of a human cytomegalovirus receptor correlates with infectibility of cells. J Virol. 1991;65:3114–3121. doi: 10.1128/jvi.65.6.3114-3121.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pereira L. Function of glycoprotein B homologues of the family Herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 46.Pietropaolo R L, Compton T. Direct interaction between human cytomegalovirus glycoprotein B and cellular annexin II. J Virol. 1997;71:9803–9807. doi: 10.1128/jvi.71.12.9803-9807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauh I, Weiland F, Fehler F, Keil G, Mettenleiter T C. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J Virol. 1991;65:621–631. doi: 10.1128/jvi.65.2.621-631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross L J, Sanderson M, Scott S D, Binns M M, Doel T, Milne B. Nucleotide sequence and characterization of the Marek’s disease virus homologue of glycoprotein B of herpes simplex virus. J Gen Virol. 1989;70:1789–1804. doi: 10.1099/0022-1317-70-7-1789. [DOI] [PubMed] [Google Scholar]
- 49.Scatchard G. The attraction of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660–672. [Google Scholar]
- 50.Sisk W P, Bradley J D, Leipold R J, Stoltzfus A M, Ponce de Leon M, Hilf M, Peng C, Cohen G H, Eisenberg R J. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J Virol. 1994;68:766–775. doi: 10.1128/jvi.68.2.766-775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith D, Byrn R, Marsters S, Gregory T, Groopman J, Capon D. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science. 1987;238:1704–1707. doi: 10.1126/science.3500514. [DOI] [PubMed] [Google Scholar]
- 52.Soderberg C, Giugni T D, Zaia J A, Larsson S, Walhberg J M, Moller E. CD13 (human aminopeptidase) mediates human cytomegalovirus infection. J Virol. 1993;67:6576–6585. doi: 10.1128/jvi.67.11.6576-6585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spaete R R, Sazena A, Scott P I, Song G J, Probert W S, Britt W J, Gibson W, Rasmussen L, Pachl C. Sequence requirements of proteolytic processing of glycoprotein B of human cytomegalovirus strain Towne. J Virol. 1990;64:2922–2931. doi: 10.1128/jvi.64.6.2922-2931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spaete R R, Thayer R M, Probert W S, Masiarz F R, Chamberlain S H, Rasmussen L, Merigan T C, Pachl C. Human cytomegalovirus strain Towne glycoprotein B is processed by proteolytic cleavage. Virology. 1988;167:207–225. doi: 10.1016/0042-6822(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 55.Tal-Singer R, Peng C, Ponce de Leon M, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanner J, Weis J, Fearon D T, Whang Y, Kieff E. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping and endocytosis. Cell. 1987;50:203–213. doi: 10.1016/0092-8674(87)90216-9. [DOI] [PubMed] [Google Scholar]
- 57.Tanner J, Whang Y, Sample J, Sears A, Kieff E. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus absorption to lymphocytes. J Virol. 1988;62:4452–4464. doi: 10.1128/jvi.62.12.4452-4464.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor H P, Cooper N R. The human cytomegalovirus receptor on fibroblasts is a 30-kilodalton membrane protein. J Virol. 1990;64:2484–2490. doi: 10.1128/jvi.64.6.2484-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tugizov S, Navarro D, Paz P, Wang Y, Qadri I, Pereira L. Function of human cytomegalovirus glycoprotein B: syncytium formation in cells constitutively expressing gB is blocked by virus-neutralizing antibodies. Virology. 1994;201:263–276. doi: 10.1006/viro.1994.1291. [DOI] [PubMed] [Google Scholar]
- 60.Vey M, Schafer W, Reis B, Ohuchi R, Britt W J, Garten W, Klenk H D, Radsak K. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology. 1995;206:746–749. doi: 10.1016/s0042-6822(95)80002-6. [DOI] [PubMed] [Google Scholar]
- 61.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]