Abstract
The utility of the present generation of recombinant adenovirus vectors for gene therapy applications could potentially be improved by designing targeted vectors capable of gene delivery to selected cell types in vivo. In order to achieve such targeting, we are investigating the possibilities of incorporation of ligands in the adenovirus fiber protein, which mediates primary binding of adenovirus to its cell surface receptor. Based on the proposed structure of the cell-binding domain of the fiber, we hypothesized that the HI loop of the fiber knob can be utilized as a convenient locale for incorporation of heterologous ligands. In this study, we utilized recombinant fiber proteins expressed in baculovirus-infected insect cells to demonstrate that the incorporation of the FLAG octapeptide into the HI loop does not ablate fiber trimerization and does not disturb formation of the cell-binding site localized in the knob. We then generated a recombinant adenovirus containing this modified fiber and showed that the short peptide sequence engineered in the knob is compatible with the biological functions of the fiber. In addition, by using a ligand-specific antibody, we have shown that the peptide incorporated into the knob remains available for binding in the context of mature virions containing modified fibers. These findings suggest that heterologous ligands can be incorporated into the HI loop of the fiber knob and that this locale possesses properties consistent with its employment in adenovirus retargeting strategies.
Recombinant adenovirus vectors have found wide employment for a number of gene therapy applications (22, 36, 40). This fact has derived principally from the high levels of gene transfer achievable with this vector approach both in vitro and in vivo. Indeed, recombinant adenovirus vectors are distinguished from other available systems by their unique ability to accomplish in situ gene delivery to differentiated target cells in a variety of organ contexts (5, 6, 9, 10, 12, 21, 26, 28, 30, 32). Despite this property, specific aspects of the adenovirus biology have prevented the full realization of the potential of such vectors. In this regard, the broad tropism profile of the parent virus for cells of diverse tissues potentially allows unrestricted gene delivery. Thus, for the many gene therapy applications requiring targeted, cell-specific gene delivery, the promiscuous tropism of the adenovirus vector represents a confounding factor. Based on this concept, strategies to modify the native tropism of adenovirus have been developed to allow the derivation of vectors capable of targeted gene delivery.
Strategies to achieve this end are directed at modifying specific steps in the adenovirus infection pathway. Adenoviruses of serotypes 2 and 5 normally achieve initial recognition and binding to target cells by means of interactions between the carboxy-terminal knob domain of the fiber protein and the primary receptor (4, 19, 39). After binding, RGD motifs in the penton base interact with cellular integrins of the αVβ3 and αVβ5 types (1–3, 43, 44). This interaction triggers cellular internalization whereby the virions achieve localization within the endosome. Acidification of the endosome elicits conformational changes in capsid proteins, allowing their interaction with the endosome membrane in a manner that achieves vesicle disruption and particle escape (41). Following endosomolysis, the virion translocates to the nucleus, where the subsequent steps of the viral life cycle occur. This understanding of the key role played by capsid proteins in the viral infectious pathway has suggested strategies to alter this process via modifications of these proteins.
In this regard, genetic retargeting of adenovirus vectors via modification of viral genes encoding coat proteins, if successful, offers a simple way to achieve a significant improvement in the present generation of these gene-delivery vehicles. To this end, several groups have reported genetic modifications to the knob domain of adenovirus fiber protein and incorporation of such chimeric fibers into virions. For instance, Stevenson et al. (37) and Krasnykh et al. (25) reported successful generation of adenovirus type 5 (Ad5) virions containing fibers consisting of the tail and shaft domains of Ad5 fiber and the knob domain of Ad3, respectively. In addition, Michael et al. (31) demonstrated the incorporation of the gastrin-releasing peptide into the carboxy terminus of recombinant Ad5 fiber. This finding was extended by Legrand et al. (30a), who achieved rescue of recombinant adenovirus vectors containing such fibers. Another report published by Wickham et al. (45) described the generation of recombinant virus containing fibers with carboxy-terminal polylysine sequences. These studies have established key feasibility issues with respect to this genetic approach but have also demonstrated a number of potentially limiting factors.
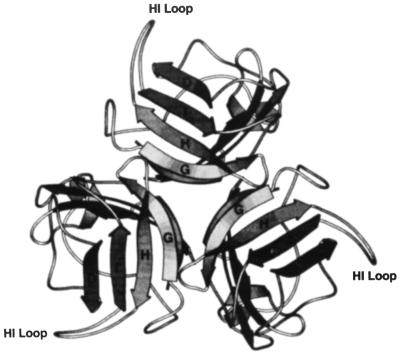
Of note, all the modifications of adenovirus fiber reported so far were directed towards the carboxy terminus of the protein. In addition, these efforts were initiated without prior knowledge of the three-dimensional (3D) structure of the fiber knob. Thus, the employment of the carboxy terminus of the fiber represented a choice of convenience without consideration of the knob tertiary structure. Clearly, 3D structural information has important bearing upon the placement of heterologous protein sequences within the knob for targeting purposes. Such localization of targeting ligands would ideally be achieved in such a manner as to allow their surface presentation and to minimally perturb the fiber quaternary structure. Thus, the recent crystallization of the fiber knob by Xia et al. (47, 48) has provided a level of structural resolution potentially allowing such a rational modification of the fiber protein. According to the proposed 3D model of the knob (Fig. 1), the HI loop possesses a number of features which predict its utility as an alternative site for ligand incorporation. Specifically, the HI loop does not contribute to intramolecular interactions in the knob. Therefore, incorporation of additional protein sequence should not affect the trimerization of the fiber. In addition, the loop consists mostly of hydrophilic amino acid residues and is exposed outside the knob. It thus potentially demonstrates a high degree of flexibility, creating an optimal environment for ligand incorporation. Furthermore, the lengths of HI loops vary significantly in knobs of different adenovirus serotypes. This fact suggests that alterations of the original structure of the loop, such as insertions and deletions, should be compatible with the correct folding of the entire knob domain. Finally, the HI loop is not involved in the formation of the putative cell-binding site localized in the knob.
FIG. 1.
3D model of the Ad5 fiber knob. The trimer forms a propeller-like structure when it is viewed along the threefold-symmetry axis from above. The HI loop, exposed outside the knob, connects the β-strands H and I, which are involved in the formation of the cell-binding site. (Reproduced from reference 47 by permission.)
Based on these considerations, we endeavored to develop a novel approach to modify the adenovirus fiber protein by employing the HI loop of the knob for this purpose. We show in this report that it is possible to incorporate heterologous amino acid sequences into the HI loop without affecting the correct folding of the fiber polypeptide and its biological functions. Further, our results suggest that this locale may offer advantages for strategies designed to achieve tropism modification based on genetic alteration of capsid proteins.
MATERIALS AND METHODS
Cells.
293 cells (15) obtained from Microbix (Toronto, Ontario, Canada) and HeLa human adenocarcinoma cells and A549 human lung carcinoma cells obtained from the American Type Culture Collection (Rockville, Md.) were maintained in Dulbecco’s modified Eagle’s medium (DMEM)-Ham’s F12 from Mediatech (Herndon, Va.) supplemented with 10% fetal calf serum (FCS) (HyClone Laboratories, Logan, Utah) at 37°C and 5% CO2.
Enzymes.
Restriction endonucleases, T4 DNA ligase, T4 polynucleotide kinase, and proteinase K were from either New England Biolabs (Beverly, Mass.) or Boehringer Mannheim (Indianapolis, Ind.).
Protein assay.
The concentrations of purified proteins were determined by the Bradford protein assay (Bio-Rad, Hercules, Calif.) with bovine gamma globulin as the standard.
Antibodies.
Antifiber monoclonal antibodies 4D2 (18) and 1D6.14 (14) were generated at the University of Alabama at Birmingham Hybridoma Core Facility. Anti-FLAG monoclonal antibody M2 and M2-affinity gel were purchased from Scientific Imaging Systems (Eastman Kodak, New Haven, Conn.)
Construction of recombinant plasmids.
To generate a gene encoding the Ad5 fiber knob domain with the HI loop deleted, a PCR technique was employed. Two pairs of primers, F1 (5′ TAAGGATCCGGTGCCATTACAGTAGGAAACAAAAATAA 3′) and R1 (5′ CATAGAGTATGCAGATATCGTTAGTGTTACAGGTTTAGTTTTG 3′) and F2 (5′ GTAACACTAACGATATCTGCATACTCTATGTCATTTTCATGG 3′) and R2 (5′ CCCAAGCTTACAATTGAAAAATAAACACGTTGAAACATAAC 3′), were used to amplify portions of the fiber gene corresponding to positions 1159 to 1451 and 1642 to 1747, respectively. In addition, the second fragment also contains 43 bp of Ad5 DNA adjacent to the 3′ end of the fiber gene in the viral genome. The fragments generated were then gel purified, mixed, and joined by the third PCR by using primers F1 and R2. The product obtained contains a unique EcoRV restriction site in place of the deleted portion of the sequence encoding the HI loop, as well as BamHI and HindIII sites inserted into the ends of the molecule to facilitate subsequent cloning. This DNA fragment was cleaved with BamHI and HindIII and cloned into the BamHI- and HindIII-digested bacterial expression vector pQE30 (Qiagen, Santa Clara, Calif.), resulting in plasmid pQE.KNOBΔHI.
To construct an expression plasmid with the FLAG sequence incorporated into the HI loop of the fiber, oligonucleotides TACACTAAACGGTACCCA GGAAACAGGAGACACAACTGACTACAAGGACGACGATGACAAGCC and GGCTTGTCATCGTCGTCCTTGTAGTCAGTTGTGTCTCCTGTTTCCTGGGTACCGTTTAGTGTA were annealed to form a duplex and cloned into EcoRV-digested pQE.KNOBΔHI. The plasmid containing the duplex in the correct orientation was designated pQE.KNOBHIFLAG.
The transfer plasmids for the generation of recombinant baculoviruses expressing chimeric fibers were made as follows: a BglII-MfeI fragment from pQE.KNOBHIFLAG was used to replace the BglII-MfeI fragment in the vector pBS.F5.UTR, which has been described previously (25), thereby generating pBS.F5HIFLAG. A BssHII-XhoI fragment from pBS.F5HIFLAG was then cloned into the BssHII- and XhoI-digested baculovirus transfer vector pFastBac1 (Life Technologies, Gaithersburg, Md.), resulting in pFB.F5HIFLAG. To introduce the six-His purification tag into the amino terminus of the chimeric fiber, the BamHI-BssHII fragment of pFB.F5HIFLAG was replaced with a synthetic duplex made with oligonucleotides GATCCATGCATCACCATCACCATCACAAG and CGCGCTTGTGATGGTGATGGTGATGCATG, which encodes MetHis6Lys. The resultant plasmid, pFB6H.F5HIFLAG, contains the gene coding for a fiber with an amino-terminal six-His tag and FLAG peptide inserted into the HI loop. To derive a similar plasmid containing the fiber gene with the HI loop coding sequence unmodified, the BssHII-MfeI fragment in pFB6H.F5HIFLAG was replaced with the homologous fragment from pNEB.PK3.6 (25), generating pFB6H.F5.
In order to clone the gene encoding the fiber with the FLAG sequence in the HI loop into the fiber shuttle vector pNEB.PK3.6, a BstXI-MfeI fragment of the wild-type fiber gene contained in this plasmid was replaced with a BstXI-MfeI fragment from pQE.KNOBHIFLAG, thereby creating pNEB.F5HIFLAG.
To facilitate the generation of recombinant adenovirus genomes by homologous recombination in Escherichia coli, plasmid pTG3602 (7), obtained from Transgene (Strasbourg, France), was engineered to create a specialized vector suitable for modifications of the fiber gene. To accomplish this end, an NdeI site localized in the fiber gene was employed. Plasmid pTG3602 was partially digested with NdeI and ligated with an NdeI-SwaI linker, TACCCATTTAAATGGG. This plasmid, containing a SwaI site in the fiber gene, was designated pVK50.
A recombinant adenovirus genome containing a gene encoding the fiber-FLAG protein was generated by homologous DNA recombination in E. coli BJ5183 between pVK50 linearized with SwaI and the 3-kb EcoRI fragment from pNEB.F5HIFLAG containing the gene of interest, as described by Chartier et al. (7). The newly generated genome was then excised from the resultant plasmid, pVK300, and employed to rescue the virus.
Viruses.
E1-deleted Ad5 vectors, AdCMVLuc and AdCMVLacZ, which express firefly luciferase and bacterial β-galactosidase (17), respectively, were obtained from R. D. Gerard, the University of Texas Southwestern Medical Center, Dallas, Tex.
Ad5FHIFLAG was generated by transfection of 293 cells with PacI-digested pVK300, as previously described (7).
Adenoviruses were propagated on 293 cells and purified by centrifugation in CsCl gradients according to a standard protocol. Determination of virus particle titer was accomplished spectrophotometrically by the method described by Maizel et al. (29) with a conversion factor of 1.1 × 1012 viral particles per absorbance unit at 260 nm. To determine the titer of infectious viral particles on 293 cells, a plaque assay was employed as described by Mittereder et al. (33).
Recombinant baculoviruses expressing chimeric fibers were generated with a Bac-to-Bac expression kit from Gibco-BRL (Life Technologies) according to the manufacturer’s protocol.
Expression and purification of six-His-tagged recombinant proteins.
Recombinant fibers were expressed in Spodoptera frugiperda Sf9 cells infected with recombinant baculovirus by the method recommended for the Bac-to-Bac system (Life Technologies). Recombinant proteins were then purified by immobilized metal ion affinity chromatography on Ni-nitrilotriacetic acid (NTA)–Sepharose (Qiagen) by following recommendations from the manufacturer.
ELISA.
In order to characterize recombinant fiber proteins, an enzyme-linked immunosorbent assay (ELISA) was employed. The six-His-tagged fibers were immobilized on Ni-NTA HisSorb Strips (Qiagen) essentially as described in the Qiagen manual. Briefly, 200 μl of fiber protein solution at a concentration of 1 μg/ml was added to each well of an Ni-NTA HisSorb Strip and incubated for 1 h at room temperature. After incubation, the wells were washed four times with phosphate-buffered saline (PBS)–Tween buffer, and 200 μl of antifiber antibody (1:2,000 dilution) or anti-FLAG antibody (1:140 dilution) was added. Following incubation at room temperature for 2 h, the wells were washed again and incubated with a 1:10,000 dilution of goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (HRP) for 45 min. The wells were then washed four times with PBS-Tween buffer and developed with 2′,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) (diammonium salt). The ABTS-HRP reaction was read in a microtiter plate reader set at 405 nm.
FLAG accessibility assay.
To demonstrate the binding of the FLAG-tagged fiber protein incorporated into intact virions of Ad5FHIFLAG to anti-FLAG M2 monoclonal antibody, an immunoprecipitation assay was employed. Ad5FHIFLAG and AdCMVLuc purified on CsCl gradients were dialyzed against HEPES buffer (10 mM HEPES, 1 mM MgCl2, 10% glycerol [pH 7.4]) and absorbed onto M2-affinity gel (Eastman Kodak) as follows. Fifty microliters of dialyzed virus containing 1011 viral particles was mixed with 100 μl of M2-affinity gel equilibrated with HEPES buffer containing 50 mM NaCl and 0.5% bovine serum albumin (BSA), and the mixture was then incubated overnight at 4°C on a rotating wheel. Following incubation, the gel was spun down by brief centrifugation in a microcentrifuge. The supernatant was collected for further analysis, and the gel was washed with 0.5 ml of Tris-buffered saline. Virus was eluted at 4°C with 50 μl of Tris-buffered saline containing 400 μg of FLAG peptide per ml. The supernatant containing unbound material, the wash, and the eluate were then employed to detect the presence of the virus. For this, aliquots of these fractions were treated for 1 h at 37°C with sodium dodecyl sulfate (SDS), EDTA, and proteinase K at final concentrations of 1%, 10 mM, and 100 μg/ml, respectively. The samples were analyzed by agarose gel electrophoresis to detect viral DNA.
Purification of the fiber-FLAG protein by immunoprecipitation.
The recombinant fiber-FLAG protein was expressed in baculovirus-infected Sf9 cells as follows. For large-scale expression of the fiber-FLAG protein, monolayers of Sf9 cells in T75 flasks were infected with recombinant baculovirus at a multiplicity of infection of 5 to 10 and then were incubated at 28°C until a complete cytopathic effect was observed. At 2 to 3 days postinfection, the cells were scraped, pelleted by low-speed centrifugation, and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 0.02% sodium azide, 100 μg of phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml). The cells were then incubated on ice for 30 min. The lysate was cleared by centrifugation at 12,000 × g for 5 min in a microcentrifuge. The cleared lysate was mixed with the slurry of M2-affinity gel, and the rest of the procedure was performed as described above for immunoprecipitation of Ad5FHIFLAG.
Trimerization assay of recombinant proteins.
To determine whether the fiber proteins expressed in baculovirus-infected insect cells could form trimers, these proteins were analyzed by SDS-polyacrylamide gel electrophoresis as previously described (31). Proteins were either boiled prior to electrophoresis, to dissociate the trimers, or loaded on the gel without denaturation. The trimeric or monomeric configuration of these molecules was thus determined based on their mobilities in the gel.
Inhibition of virus-mediated gene transfer by recombinant fiber proteins.
The ability of the fiber-FLAG chimera to block adenovirus-mediated gene transfer was evaluated in infection inhibition experiments similar to those described previously (16, 25, 35, 38). Briefly, monolayers of HeLa cells grown in a 24-well plate were preincubated at room temperature with serial 10-fold dilutions of either wild-type fiber or fiber-FLAG protein prior to infection with a replication-defective recombinant adenovirus expressing firefly luciferase, AdCMVLuc. Unbound virus was washed, and the cells were incubated at 37°C to allow internalization of AdCMVLuc and expression of the luciferase gene. A luciferase assay of the lysates of infected cells was performed 30 h postinfection with a luciferase assay system from Promega (Madison, Wis.).
Virus binding assay.
Human lung carcinoma A549 cells were grown in T75 flasks and then harvested with EDTA, washed once with PBS, pelleted, and resuspended to a final concentration of 107 cells/ml in DMEM-Ad medium (DMEM, 20 mM HEPES, 0.5% BSA) as described by Wickham et al. (45). One-hundred-microliter aliquots of the cells were transferred to 5-ml test tubes and incubated for 1 h at 4°C with 100 μl of recombinant fiber protein diluted in DMEM-Ad medium.
The recombinant adenoviruses AdCMVlacZ and Ad5FHIFLAG were purified on a CsCl gradient and dialyzed against buffer containing 10 mM HEPES, 1 mM MgCl2, and 10% glycerol (pH 7.4). Aliquots of both viruses containing 50 μg of viral protein were labeled with 125I with IODO-BEADS iodination reagent (Pierce, Rockford, Ill.) as previously described (20). Labeled viruses were purified from unincorporated 125I by gel filtration on PD-10 columns (Pharmacia, Piscataway, N.J.). Fifty-microliter aliquots of labeled virions with total radioactivities of 105 cpm were then added to A549 cells preincubated with fiber dilutions or PBS and incubated at 4°C for another hour.
The samples were diluted with 4 ml of PBS containing 0.1% BSA, and the cells were pelleted by centrifugation. Supernatant containing unbound virus was aspirated, and the radioactivities of cell pellets were determined in a gamma counter.
RESULTS
Characterization of recombinant fibers expressed in baculovirus-infected insect cells.
To test the concept of the suitability of the HI loop of the fiber knob for incorporation of heterologous protein sequences, we first employed recombinant fiber proteins expressed in a baculovirus expression system. This system has already proved its utility for the expression of functional Ad2, Ad3, and Ad5 fiber proteins, as well as Ad3-Ad5 and Ad5-Ad3 fiber chimeras (13, 27, 34, 38).
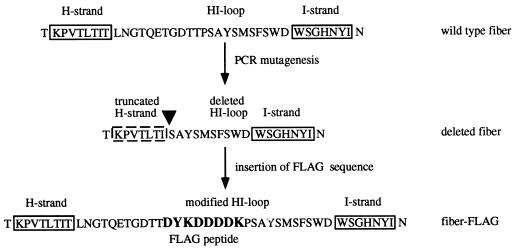
To achieve this aim, we used a PCR approach to derive a gene encoding the Ad5 fiber knob with a partial deletion in the HI loop. This deletion was engineered to remove amino acids TLNGTQETGDTTP from the HI loop of the fiber knob domain and to introduce a unique EcoRV site in place of the deleted sequence, thereby facilitating the cloning of alternative sequences in this region (Fig. 2). The deletion removed the portion of the HI loop which varies most significantly in the fiber knobs of different serotypes of human adenoviruses. The sequence generated by PCR contained an open reading frame corresponding to two segments of the fiber protein including amino acids glycine-387 through isoleucine-534 and serine-548 through glutamine-581 (coordinates given are according to those of the wild-type Ad5 fiber protein sequence). This sequence was cloned into the plasmid vector pQE30. The newly generated plasmid, pQE.KNOBΔHI, was then utilized as a cloning vector to incorporate a fragment of DNA encoding the FLAG octapeptide (DYKDDDDK), which has been widely used as a detection and purification tag in a variety of studies. Thus, we chose to exploit this FLAG peptide in our fiber constructs as a probe to determine whether a heterologous peptide sequence incorporated into the HI loop of the knob was accessible in the context of a trimeric fiber molecule. By incorporating this sequence into the open reading frame of the knob, we also restored the previously deleted codons. Therefore, in the newly generated plasmid, pQE.KNOBHIFLAG, the FLAG coding sequence was introduced as insertions between threonine-546 and proline-547. This plasmid was then employed to construct a full-size recombinant fiber gene in a baculovirus transfer vector. A similar transfer plasmid containing the wild-type fiber gene was designed for control purposes. In order to facilitate subsequent purification of the expression products, we introduced into the designs of both genes a sequence encoding an amino-terminal six-His tag. These plasmids were then utilized to generate two recombinant baculoviruses containing fiber genes encoding wild-type Ad5 fiber and a fiber protein containing FLAG peptide in the HI loop of the knob domain.
FIG. 2.
Modifications of the HI loop of the fiber knob. PCR-based mutagenesis was employed to delete a portion of the fiber gene encoding the hypervariable region of the HI loop. A unique EcoRV restriction site was incorporated in place of the deletion to allow the cloning of segments of DNA coding for heterologous protein sequences. In the fiber-FLAG protein, deleted amino acids of the HI loop were restored, and FLAG octapeptide was incorporated between threonine-546 and proline-547. The site of deletion is indicated by a filled triangle.
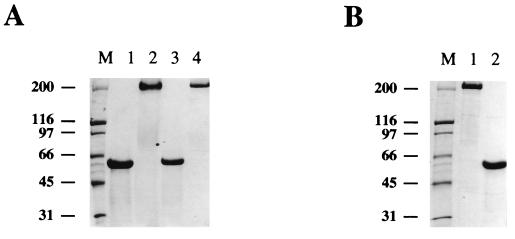
Recombinant fibers were recovered from the lysates of baculovirus-infected insect cells with Ni-NTA–Sepharose designed for purification of the six-His-tagged proteins. The yield of purified fibers was in the range of 10 μg of protein per 106 infected cells. Analysis by SDS-polyacrylamide gel electrophoresis of both recombinant proteins showed that they formed stable trimers which, when boiled in the gel loading buffer, dissociated into monomers of the expected molecular mass of 63 kDa (Fig. 3A). This result demonstrated that the incorporation of a short peptide sequence in the HI loop of the knob does not ablate trimerization of the fiber. Therefore, by using the baculovirus expression system we were able to obtain preparative amounts of the recombinant fibers of interest which were suitable for subsequent assays.
FIG. 3.
Analysis of recombinant fiber proteins by polyacrylamide gel electrophoresis. Fiber proteins expressed in insect cells were analyzed by gel electrophoresis to confirm their trimeric configurations. To dissociate trimers to monomers, the proteins were denatured by boiling them in the sample buffer prior to loading them on a 7.5% polyacrylamide gel. The bands were visualized by Coomassie blue staining. (A) Six-histidine-tagged fiber proteins purified on an Ni-NTA–Sepharose column. Lane 1, wild-type fiber, boiled; lane 2, wild-type fiber, unboiled; lane 3, fiber-FLAG, boiled; lane 4, fiber-FLAG, unboiled; lane M, broad-range protein standards. (B) Fiber-FLAG protein purified by immunoprecipitation with anti-FLAG M2-affinity gel. Lane 1, unboiled protein; lane 2, boiled protein; lane M, broad-range protein standards. The numbers on the left indicate molecular masses of marker proteins in kilodaltons.
Accessibility of the FLAG peptide in the context of trimeric fiber.
To find out whether the FLAG peptide introduced into the HI loop of the fiber was available for binding, we used an assay based on the specific interaction of the FLAG-tagged proteins with an affinity matrix containing anti-FLAG monoclonal antibody. For these experiments, the recombinant fiber protein with the FLAG sequence in the HI loop (fiber-FLAG) was purified on an Ni-NTA–Sepharose column and then immunoprecipitated with M2-affinity gel. Protein bound to the matrix was then specifically eluted with FLAG peptide and analyzed on an SDS-containing polyacrylamide gel (Fig. 3B). According to this analysis, the fiber-FLAG protein efficiently bound to M2-affinity gel, demonstrating the availability of the FLAG epitope for interaction with an anti-FLAG monoclonal antibody in the context of the trimeric fiber molecule. Importantly, this interaction did not affect the stability of the trimer, suggesting that a recombinant virion containing a novel ligand incorporated in the HI loop of the fiber knob will maintain its structural integrity throughout the binding step of the infection.
Inhibition of adenovirus infection by recombinant fiber-FLAG protein.
Since we did not expect the FLAG peptide to possess the ability to target adenovirus to a novel cellular receptor, it was necessary to determine whether the incorporation of this peptide in the HI loop affected proper folding of the cell-binding site localized in the fiber knob. If the hypothesis that the HI loop is not involved in the formation of this site were incorrect and if fiber-FLAG could not bind to the fiber receptor on the cell surface, further attempts to rescue the virus containing this recombinant fiber would inevitably fail. To address these issues, we employed a fiber-FLAG recombinant protein to block adenovirus infection in the in vitro setting. This established assay is based on the fact that recombinant adenovirus fiber proteins are capable of blocking infection by the adenovirus from which they were derived. In addition, this inhibition of viral infection takes place in a dose-dependent manner.
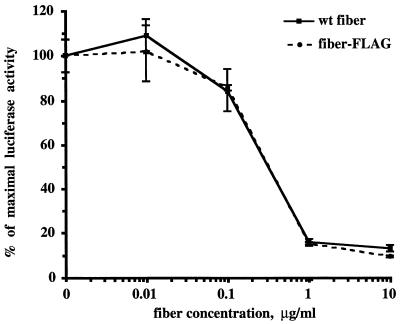
In our experiments, HeLa cells seeded in 12-well tissue culture plates were preincubated with various concentrations of the wild-type Ad5 fiber or fiber-FLAG protein prior to infection with the recombinant Ad5 vector AdCMVLuc, which expresses firefly luciferase as a reporter. Previously, we showed that this assay, based on gene transfer by the viral vector, generates data correlating well with a classic binding assay accomplished with radiolabeled virus (25). Thirty hours postinfection, the cells were lysed and the lysates were utilized for the luciferase activity assay (Fig. 4). According to this assay, both fiber proteins blocked infection by AdCMVLuc in a dose-dependent manner and demonstrated identical profiles of infection inhibition. These data confirmed that incorporation of heterologous peptide sequences into the HI loop of the fiber knob does not affect the correct folding of the cell-binding site formed by the carboxy-terminal portion of the fiber protein.
FIG. 4.
Inhibition of adenovirus infectivity by recombinant fiber proteins. HeLa cells were preincubated with either the wild-type (wt) fiber or fiber-FLAG at the indicated concentrations for 10 min at room temperature. AdCMVLuc was then added at a multiplicity of infection of 10, and incubation was continued for another 30 min at room temperature. The unbound virus was aspirated, complete medium was added, and the cells were transferred to 37°C. After 30 h, the cells were lysed and luciferase activity was determined. Luciferase activities are given as percentages of the activity in the absence of blocking fiber protein. Each point represents the mean of four determinations obtained in one experiment.
Characterization of the fiber-FLAG protein by ELISA.
To obtain additional evidence supporting the functional utility of the fiber-FLAG protein, we analyzed this recombinant protein by ELISA, employing several monoclonal antibodies specific for the FLAG epitope and different conformations of the Ad5 fiber. To achieve this end, wild-type fiber and fiber-FLAG proteins expressed in insect cells were absorbed on HisSorb ELISA strips covered with Ni-NTA (Qiagen) and probed with antifiber antibody 4D2 or 1D6.14 or anti-FLAG antibody M2. Antibody 4D2 reacts with Ad5 fiber monomers and trimers and was used in this assay as a positive control, whereas antibody 1D6.14 binds to an as yet unidentified conformational epitope in the fiber knob and is trimer specific. The ELISA strips were then developed with goat anti-mouse antibody–HRP conjugate.
As seen in Table 1, both fiber proteins efficiently reacted with antifiber antibodies 4D2 and 1D6.14, thereby suggesting that the 3D structure of the knob in the fiber-FLAG molecule is identical to that of the wild-type fiber. In addition, the fiber-FLAG chimera specifically reacted with anti-FLAG antibody M2, confirming the availability of this epitope for binding in the context of a trimeric fiber molecule. Thus, these results validated the data generated earlier by gel electrophoresis analysis of Ni-NTA- or M2-affinity gel-purified fiber-FLAG protein, providing the rationale for the incorporation of the fiber-FLAG chimera into the adenovirus virion for further characterization.
TABLE 1.
Characterization of recombinant fiber proteins by ELISA
| Fiber protein |
A405a for monoclonal antibody:
|
|||
|---|---|---|---|---|
| None | 4D2 | 1D6.14 | M2 | |
| Wild-type fiber | −0.001 ± 0.000 | 0.444 ± 0.050 | 1.225 ± 0.131 | 0.003 ± 0.001 |
| Fiber-FLAG | 0.010 ± 0.002 | 0.239 ± 0.021 | 0.904 ± 0.013 | 1.860 ± 0.204 |
Values are means ± standard deviations of results obtained in two independent experiments.
Generation of Ad5FHIFLAG.
Despite the fact that the data obtained with the recombinant fiber-FLAG protein supported the concept of its functional utility in the context of the adenovirus virion, only successful generation of the recombinant virus would prove our hypothesis regarding the compatibility of the modifications of the HI loop of the fiber knob with viral functions. Therefore, we undertook the task of incorporating the fiber-FLAG chimera into the adenovirus virion.
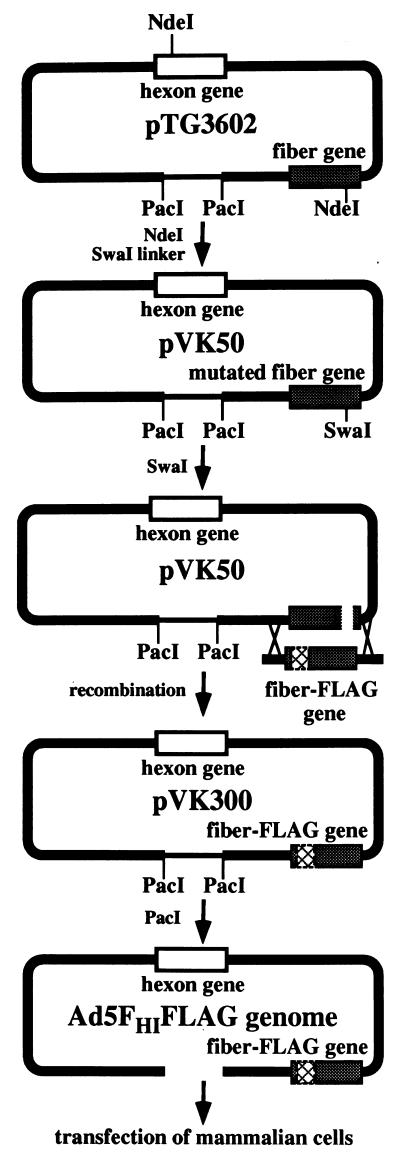
In order to derive this virus, a novel genetic method based on homologous DNA recombination in E. coli cells was utilized (7). In brief, this method involves recombination between two linear DNA molecules cotransformed into bacterial cells to generate a recombinant adenovirus genome. One of these molecules is plasmid pTG3602, or its derivative, containing the full-size adenovirus genome cloned in the bacterial vector and flanked with two PacI sites. The second partner in this recombination schema is the genetic construct of interest flanked with two segments of adenovirus genomic DNA which dictate the localization of this construct in the adenovirus genome generated as a result of the recombination. This DNA sequence can be either a transgene or the original Ad5 gene, modified by traditional methods of genetic engineering in the context of small recombinant plasmids. To reduce the nonrecombinant background generated by pTG3602, prior to transformation this plasmid was cleaved with a restriction enzyme within or near the region of the genome where the final construct was going to be inserted. Although this method has numerous advantages compared to traditional generation of recombinant adenovirus genomes by homologous recombination in mammalian cells, it requires the existence of unique restriction sites within the regions of the adenovirus genome to be modified. However, Ad5 genomic DNA in pTG3602 does not contain any unique restriction sites in the fiber gene, which limits its utility for modifications of fiber. Thus, to overcome this limitation, we modified this plasmid by inserting a unique cleavage site for the restriction endonuclease SwaI into the fiber gene. To this end, one of the two NdeI sites present in Ad5 DNA and localized 47 bp downstream from the fiber gene’s 5′ end was converted into an SwaI site by insertion of an SwaI linker (Fig. 5). The plasmid generated, pVK50, was then utilized for homologous recombination with the fragment of DNA containing the gene encoding fiber-FLAG flanked with viral DNA adjacent to the fiber gene in the Ad5 genome. As a result of this recombination, a plasmid, pVK300, containing a modified fiber gene in the context of the complete adenovirus genome was derived. Adenovirus DNA was released from pVK300 by PacI digestion and used for transfection of 293 cells to rescue the virus as described previously (7).
FIG. 5.
Generation of Ad5FHIFLAG. The master plasmid, pTG3602, was modified to incorporate a unique SwaI restriction site in the fiber gene, thereby creating plasmid pVK50, suitable for fiber modifications. The genome of Ad5FHIFLAG was generated by homologous DNA recombination in E. coli between the DNA fragment containing the fiber-FLAG gene and plasmid pVK50 linearized by SwaI digestion. To rescue the virus, the resulting plasmid, pVK300, which contains the complete adenovirus genome with a modified fiber gene, was cleaved with PacI and was then used to transfect 293 cells.
DNA isolated from CsCl gradient-purified virions of the newly generated virus, Ad5FHIFLAG, was subjected to PCR analysis and cycle sequencing to confirm the presence of the FLAG coding sequence in the fiber gene incorporated in the genome. According to both analyses, Ad5FHIFLAG indeed contained the fiber gene of interest.
Characterization of Ad5FHIFLAG by a cell-binding assay.
The yield of Ad5FHIFLAG grown on 293 cells, approximately 1011 PFU per preparation obtained from 20 75-cm2 tissue culture flasks, was comparable to what we normally obtain when growing the wild-type Ad5. Also, we did not see any delay in infection dynamics when rescuing the virus or when expanding it. These observations suggested that the introduction of the FLAG peptide in the HI loop of the knob did not significantly affect the correct folding of the fiber molecule and its biological functions.
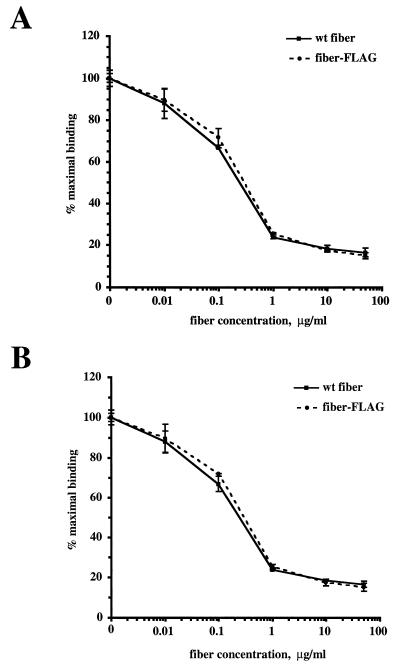
In order to prove this, we employed radiolabeled Ad5FHIFLAG to investigate its ability to bind the fiber receptors on the cell surface. In this assay, 125I-labeled Ad5FHIFLAG was allowed to bind A549 human lung carcinoma cells, which are known to express high levels of Ad5 fiber receptors. Baculovirus-expressed wild-type Ad5 fiber and fiber-FLAG were used as competitors to selectively block cellular receptors and inhibit virus binding. The recombinant adenovirus vector Ad5CMVLacZ containing wild-type fibers was used as a control. The results of this experiment (Fig. 6A and B) clearly showed that, as expected, both viruses demonstrate identical dose responses when competing with fibers of either type. Thus, incorporation of the heterologous peptide in the HI loop of the fiber-FLAG protein did not have any negative effect on the formation of the cell-binding site localized in the knob and, therefore, did not affect virus infectivity.
FIG. 6.
Adenovirus binding assay. Aliquots of A549 cells containing 105 cells per sample were incubated for 1 h at 4°C with serial dilutions of either wild-type (wt) Ad5 fiber or fiber-FLAG (see Materials and Methods). Virions of Ad5CMVLacZ (A) and Ad5FHIFLAG (B) labeled with 125I were added to samples, and incubation was continued for an additional hour. The cells were washed with 4 ml of PBS containing 0.1% BSA and pelleted by low-speed centrifugation. Radioactivities of samples were determined with a gamma counter. Each point represents the mean of two determinations obtained in one experiment.
FLAG accessibility in the context of the Ad5FHIFLAG virion.
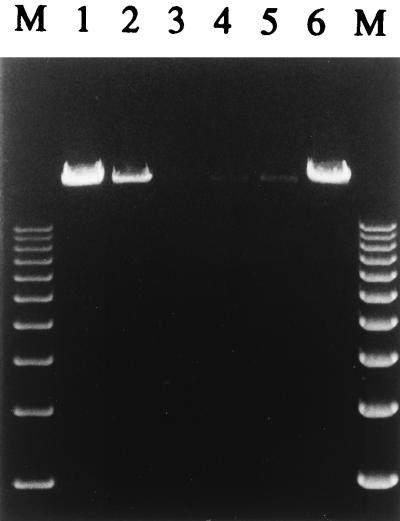
Since the ultimate goal of our efforts is the insertion of a targeting ligand into the knob, it was necessary to determine whether such a ligand would be available for interaction with its target cell surface receptor after its incorporation into the adenovirus virion. To this end, we employed the FLAG sequence incorporated into the fiber of Ad5FHIFLAG to test the accessibility of the HI loop of the knob in the context of an intact adenovirus particle. This was accomplished in an assay similar to the one used to evaluate FLAG accessibility in the recombinant fiber-FLAG protein expressed in insect cells. Virions purified on a CsCl gradient were dialyzed against HEPES buffer and incubated with M2-affinity gel to allow interaction between the FLAG peptide and an anti-FLAG monoclonal antibody conjugated to the gel matrix. Similarly prepared virions of AdCMVLuc containing wild-type fibers were utilized in this experiment as a negative control. After incubation, the buffer containing unbound material was collected and the gel was washed with the buffer to remove the traces of free virus. Finally, the viruses were eluted from the gel with soluble FLAG peptide. Aliquots of the samples collected were treated with proteinase K to release viral DNA from virions, which was then visualized by agarose gel electrophoresis (Fig. 7). As expected, virions of AdCMVLuc did not react with M2 antibody and were detected only in the fraction containing unbound virus and in the wash. In marked contrast, Ad5FHIFLAG particles efficiently bound to the M2-affinity gel, since viral DNA was present primarily in the FLAG peptide eluate. Thus, our findings have established that the heterologous ligand sequence engineered into the HI loop of the knob domain of the fiber incorporated in the intact Ad5 virion remains accessible for interaction with the relevant receptor structure, thereby providing the rationale for the generation of genetically targeted adenovirus vectors on this basis.
FIG. 7.
Accessibility of the FLAG peptide in the context of intact Ad5FHIFLAG virions. Virions of Ad5FHIFLAG purified on a CsCl gradient were dialyzed, immunoprecipitated with anti-FLAG M2-affinity gel as described in Materials and Methods, and eluted from the gel with free FLAG peptide. Recombinant adenovirus vector Ad5CMVLuc containing unmodified fiber was used as a negative control for binding. Aliquots of all the fractions collected throughout the purification procedure were treated with DNase I to digest traces of the cellular DNA and then treated with SDS, EDTA, and proteinase K to release adenovirus DNA from the virions. The samples obtained were analyzed on a 0.8% agarose gel, and DNA was detected by ethidium bromide staining. Lanes 1 through 3, AdCMVLuc in the supernatant containing unbound material, buffer wash, and FLAG-eluate, respectively; lanes 4 through 6, Ad5FHIFLAG in the supernatant, buffer wash, and FLAG-eluate, respectively; lanes M, DNA molecular weight standards (the bands corresponding to marker fragments ranging from 3 to 12 kb are seen on the gel).
DISCUSSION
Despite the numerous attempts which have been made to improve adenovirus as a vector for gene therapy applications, it still suffers from a number of important disadvantages, one of them being the promiscuous tropism of this virus. Genetic modification of adenovirus coat proteins to target novel cell surface receptors is the most radical and, if successful, potentially the most efficient way to overcome this limitation. In this regard, fiber, penton base, and hexon proteins are candidates for such genetic modifications. While modifications of the penton base (42, 46) and the hexon (8, 11) have been reported, these alterations were limited to the introduction of short peptide sequences into the exterior domains of these components of the adenovirus virion. In contrast, a larger number of studies have attempted functional modifications of the fiber protein. These attempts to modify the fiber protein have an obvious explanation: in contrast to the hexon and penton base proteins, the fiber protein mediates the primary interaction of the virus with its cognate cellular receptor and therefore dictates the tropism of the virus. In addition, due to its rod-like structure, the fiber can optimally expose a novel binding ligand engineered into its structure, thus providing efficient binding to an alternative cellular receptor. Thus, alterating to the carboxy-terminal knob domain of the fiber normally containing the cell-binding site is a logical approach to modifying viral tropism.
Since the time this idea was originally employed (31), several groups of investigators have proved its utility. To this end, recombinant adenoviruses containing chimeric Ad5-Ad3 (25, 37) fiber were derived, demonstrating the possibility of creating functional fiber chimeras. In addition, it was shown that by replacing the knob domain of the fiber, one can alter the receptor specificity of the virus. Furthermore, in an elegant study by Wickham et al. (45), addition of a carboxy-terminal polylysine sequence to the fiber polypeptide resulted in expanded tropism of the adenovirus vector. Recently, recombinant adenoviruses with fibers containing carboxy-terminal gastrin-releasing peptide (30a), somatostatin, E-selectin-binding peptide, and six-His sequence (24a) have been generated. However, it should be noted that none of these efforts has addressed the goal of ablating the native tropism of the adenovirus vector; in these approaches, novel tropism distinct from the preexisting natural tropism of the vector was engineered.
Until recently, the ability to accomplish the practical design of retargeted adenovirus vectors was limited by two major problems: lack of knowledge of the structure of the fiber knob domain and difficulty in manipulating the fiber gene in the context of the adenovirus genome. In this regard, publication of the 3D model of the Ad5 fiber knob by Xia et al. (47, 48) and the development of a novel genetic method by Chartier et al. (7), which allow modification of virtually any region of the adenovirus genome, will dramatically facilitate efforts to retarget the adenovirus via alterations to the knob domain of the fiber. Thus, the present study is the first attempt to exploit our expanded knowledge regarding the fiber structure, as well as new technological breakthroughs in generating recombinant adenovirus genomes, to derive adenovirus vectors with modified fibers containing novel peptide ligands. In this report we describe the utilization of the HI loop of the fiber knob as an alternative site for incorporation of heterologous peptide sequences. According to the 3D model of the Ad5 fiber knob, the HI loop does not contribute to interactions within the knob which stabilize its trimeric configuration and is not involved in the formation of the receptor-binding site. Importantly, due to the prevalence of hydrophilic amino acid residues in its primary sequence, the HI loop is exposed outside the knob, thereby facilitating the interaction of potential ligand with the cellular receptor.
For initial proof of this concept, we incorporated a FLAG coding sequence into the region of the fiber gene corresponding to the HI loop and expressed this modified gene in baculovirus-infected insect cells. An amino-terminal six-His tag incorporated into the design was used for simple chromatographic purification of recombinant fiber protein. Baculovirus-directed expression of this recombinant full-size fiber was efficient, and according to our gel analysis and ELISA with the trimer-specific anti-fiber monoclonal antibody, the product of expression was trimeric.
To further characterize the fiber-FLAG protein produced in insect cells, we demonstrated the accessibility of FLAG in the context of the fiber trimer. For this we employed an assay based on the specific interaction of FLAG-tagged proteins with M2-affinity gel containing anti-FLAG monoclonal antibody. This analysis confirmed that the FLAG peptide is localized on the surface of the trimeric knob and is available for binding, thereby supporting the hypothesis about surface localization of the HI loop. By employing the fiber-FLAG chimera to block adenovirus infection, we have also shown that insertion of the FLAG peptide into the HI loop of the knob does not affect the correct folding of the cell-binding domain localized in the knob. This is a significant finding, considering that the HI loop connects β-strands H and I, which are hypothesized to be involved in binding to the cellular receptor (47, 48).
To incorporate fiber-FLAG chimeras into the adenovirus virion, we generated a recombinant adenovirus genome by using a novel method described recently (7). To reach this end, we modified a master plasmid, pTG3602, obtained from Transgene to engineer a vector which greatly facilitates modifications of the fiber gene in the adenovirus genome. By using this plasmid, we generated a recombinant genome and rescued the virus of interest, Ad5FHIFLAG. Importantly, this new virus is produced in high yields and demonstrates dynamics of infection identical to those of the wild-type Ad5. Successful rescue of Ad5FHIFLAG, as well as subsequent characterization of the virion, confirmed our conclusions based on the results obtained with fiber-FLAG protein expressed in baculovirus-infected insect cells, thereby making baculovirus an expression system of choice for further fiber-modeling experiments.
Thus, we have proved that the HI loop of the fiber knob is a convenient site for incorporation of heterologous peptide ligands which may be successfully utilized in order to target adenovirus vectors for gene therapy applications. This location in the knob can be used either as an alternative site or in addition to carboxy-terminal modifications of the fiber protein, offering a unique loop-like environment, which may be required for proper biological functioning of some ligand sequences. For example, this structure may be beneficial for peptide ligands obtained from phage display libraries containing random peptide sequences flanked with two cysteine residues forming a disulfide bridge (23, 24). In addition, ligands with the loop-like configuration may be less susceptible to degradation by cellular carboxypeptidases than ligands positioned at the carboxy terminus of the fiber. To realize the full potential of the HI loop for ligand incorporation, we have experiments in progress to make recombinant adenoviruses containing different targeting moieties in this locale. Generation of recombinant adenoviruses containing fibers with targeting ligands incorporated into the HI loop of the knob will facilitate further efforts towards an improved adenovirus vector for gene therapy applications. Although the development of novel methods for the purification of adenoviruses was not the focus of our study, successful use of the FLAG epitope in our binding experiments suggests that this or a similar purification tag can be incorporated into an adenovirus virion to facilitate its purification. This simple purification technique does not require expensive laboratory equipment such as ultracentrifuges or high-pressure liquid chromatography systems and can be easily scaled up if needed. The utility of this method for purification of recombinant adenoviruses remains to be examined.
ACKNOWLEDGMENTS
This study was supported in part by NIH grants RO1-HL50255 and RO1-CA74242, a grant from the Muscular Dystrophy Association, and a grant from the American Lung Association.
We thank Majid Mehtali for making plasmid pTG3602 available and Robert Gerard for his permission to reproduce Fig. 1, which shows a 3D model of the fiber knob. Jesus Gomes Navarro, Claudine Rancourt, and Joanne Douglas are thanked for critically reading and evaluating the manuscript. We are indebted to Becky Brazeel for editorial assistance and typing the manuscript.
REFERENCES
- 1.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bai M, Harfe B, Freimuth P. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J Virol. 1993;67:5198–5205. doi: 10.1128/jvi.67.9.5198-5205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belin M T, Boulanger P. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1993;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 5.Bout A, Imler J L, Schultz H, Perricaudet M, Zurcher C, Herbrink P, Valerio D, Pavirani A. In vivo adenovirus-mediated transfer of human CFTR cDNA to rhesus monkey airway epithelium: efficacy, toxicity and safety. Gene Ther. 1994;1:385–394. [PubMed] [Google Scholar]
- 6.Bout A, Perricaudet M, Baskin G, Imler J L, Scholte B J, Pavirani A, Valerio D. Lung gene therapy: in vivo adenovirus-mediated gene transfer to rhesus monkey airway epithelium. Hum Gene Ther. 1994;5:3–10. doi: 10.1089/hum.1994.5.1-3. [DOI] [PubMed] [Google Scholar]
- 7.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crompton J, Toogood C I, Wallis N, Hay R T. Expression of a foreign epitope on the surface of the adenovirus hexon. J Gen Virol. 1994;75:133–139. doi: 10.1099/0022-1317-75-1-133. [DOI] [PubMed] [Google Scholar]
- 9.Crystal R G, McElvaney N G, Rosenfeld M A, Chu C S, Mastrangeli A, Hay J G, Brody S L, Jaffe H A, Eissa N T, Danel C. Administration of an adenovirus containing the human CFTR cDNA to the respiratory tract of individuals with cystic fibrosis. Nat Genet. 1994;8:42–51. doi: 10.1038/ng0994-42. [DOI] [PubMed] [Google Scholar]
- 10.Csete M E, Benhamou P Y, Drazan K E, Wu L, McIntee D F, Afra R, Mullen Y, Busuttil R W, Shaked A. Efficient gene transfer to pancreatic islets mediated by adenoviral vectors. Transplantation. 1995;59:263–268. [PubMed] [Google Scholar]
- 11.Curiel D T, Wagner E, Cotten M, Birnstiel M L, Agarwal S, Li C-M, Loechel S, Hu P-C. High-efficiency gene transfer mediated by adenovirus coupled to DNA-polylysine complexes. Hum Gene Ther. 1992;3:147–154. doi: 10.1089/hum.1992.3.2-147. [DOI] [PubMed] [Google Scholar]
- 12.DeMatteo R P, Raper S E, Ahn M, Fisher K J, Burke C, Radu A, Widera G, Claytor B R, Barker C F, Markmann J F. Gene transfer to the thymus. A means of abrogating the immune response to recombinant adenovirus. Ann Surg. 1995;222:229–239. doi: 10.1097/00000658-199509000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Guilmi A M, Barge A, Kitts P, Gout E, Chroboczek J. Human adenovirus serotype 3 (Ad3) and the Ad3 fiber protein bind to a 130-kDa membrane protein on HeLa cells. Virus Res. 1995;38:71–81. doi: 10.1016/0168-1702(95)00043-p. [DOI] [PubMed] [Google Scholar]
- 14.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 16.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herz J, Gerard R D. Adenovirus-mediated transfer of low density lipoprotein receptor gene acutely accelerates cholesterol clearance in normal mice. Proc Natl Acad Sci USA. 1993;90:2812–2816. doi: 10.1073/pnas.90.7.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong J S, Engler J A. The amino terminus of the adenovirus fiber protein encodes the nuclear localization signal. Virology. 1991;185:758–767. doi: 10.1016/0042-6822(91)90547-o. [DOI] [PubMed] [Google Scholar]
- 19.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Kamata T, Takada Y, Ruggeri Z M, Nemerow G R. Adenovirus interaction with distinct integrins mediates separate events in cell entry and gene delivery to hematopoietic cells. J Virol. 1996;70:4502–4508. doi: 10.1128/jvi.70.7.4502-4508.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffe H A, Danel C, Longenecker G, Metzger M, Setoguchi Y, Rosenfeld M A, Gant T W, Thorgeirsson S S, Stratford-Perricaudet L D, Perricaudet M, et al. Adenovirus-mediated in vivo gene transfer and expression in normal rat liver. Nat Genet. 1992;1:372–378. doi: 10.1038/ng0892-372. [DOI] [PubMed] [Google Scholar]
- 22.Jolly D. Viral vector systems for gene therapy. Cancer Gene Ther. 1994;1:51–64. [PubMed] [Google Scholar]
- 23.Koivunen E, Wang B, Dickinson C, Rouslahti E. Peptides in cell adhesion research. Methods Enzymol. 1994;245:346–369. doi: 10.1016/0076-6879(94)45019-6. [DOI] [PubMed] [Google Scholar]
- 24.Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the alpha 5 beta 1 integrin from a phage display library. J Cell Biol. 1994;124:373–380. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Krasnykh, V., and I. Dmitriev. Unpublished results.
- 25.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Gal La Salle G, Robert J J, Berrard S, Ridoux V, Stratford-Perricaudet L D, Perricaudet M, Mallet J. An adenovirus vector for gene transfer into neurons and glia in the brain. Science. 1993;259:988–990. doi: 10.1126/science.8382374. [DOI] [PubMed] [Google Scholar]
- 27.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda H, Danel C, Crystal R G. Adenovirus-mediated transfer of human lipase complementary DNA to the gallbladder. Gastroenterology. 1994;106:1638–1644. doi: 10.1016/0016-5085(94)90421-9. [DOI] [PubMed] [Google Scholar]
- 29.Maizel J V, Jr, White D O, Scharff M D. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 30.Mastrangeli A, O’Connell B, Aladib W, Fox P C, Baum B J, Crystal R G. Direct in vivo adenovirus-mediated gene transfer to salivary glands. Am J Physiol. 1994;266:G1146–G1155. doi: 10.1152/ajpgi.1994.266.6.G1146. [DOI] [PubMed] [Google Scholar]
- 30a.Mehtali, M. Personal communication.
- 31.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 32.Mitani K, Graham F L, Caskey C T. Transduction of human bone marrow by adenoviral vector. Hum Gene Ther. 1994;5:941–948. doi: 10.1089/hum.1994.5.8-941. [DOI] [PubMed] [Google Scholar]
- 33.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novelli A, Boulanger P A. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 35.Roelvink P W, Kovesdi I, Wickham T J. Comparative analysis of adenovirus fiber-cell interaction: adenovirus type 2 (Ad2) and Ad9 utilize the same cellular fiber receptor but use different binding strategies for attachment. J Virol. 1996;70:7614–7621. doi: 10.1128/jvi.70.11.7614-7621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegfried W. Perspectives in gene therapy with recombinant adenoviruses. Exp Clin Endocrinol. 1993;101:7–11. doi: 10.1055/s-0029-1211201. [DOI] [PubMed] [Google Scholar]
- 37.Stevenson S C, Rollence M, Marshall-Neff J, McClelland A. Selective targeting of human cells by a chimeric adenovirus vector containing a modified fiber protein. J Virol. 1997;71:4782–4790. doi: 10.1128/jvi.71.6.4782-4790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell B, Gorziglia M. Gene therapy using adenoviral vectors. Curr Opin Biotechnol. 1994;5:617–625. doi: 10.1016/0958-1669(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 41.Varga M J, Weibull C, Everitt E. Infectious entry pathway of adenovirus type 2. J Virol. 1991;65:6061–6070. doi: 10.1128/jvi.65.11.6061-6070.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 43.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 45.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 46.Wickham T J, Segal D M, Roelvink P W, Carrion M E, Lizonova A, Lee G M, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia D, Henry L, Gerard R D, Deisenhofer J. Structure of the receptor binding domain of adenovirus type 5 fiber protein. Curr Top Microbiol Immunol. 1995;199:39–46. doi: 10.1007/978-3-642-79496-4_3. [DOI] [PubMed] [Google Scholar]
- 48.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]