Abstract
Sexual reproduction and the specialized cell division it relies upon, meiosis, are biological processes that present an incredible degree of both evolutionary conservation and divergence. One clear example of this paradox is the role of the evolutionarily ancient PCH-2/HORMAD module during meiosis. On one hand, the complex, and sometimes disparate, meiotic defects observed when PCH-2 and/or the meiotic HORMADS are mutated in different model systems have prevented a straightforward characterization of their conserved functions. On the other hand, these functional variations demonstrate the impressive molecular rewiring that accompanies evolution of the meiotic processes these factors are involved in. While the defects observed in pch-2 mutants appear to vary in different systems, in this review, I argue that PCH-2 has a conserved meiotic function: to coordinate meiotic recombination with synapsis to ensure an appropriate number and distribution of crossovers. Further, given the dramatic variation in how the events of recombination and synapsis are themselves regulated in different model systems, the mechanistic differences in PCH-2 and meiotic HORMAD function make biological sense when viewed as species-specific elaborations layered onto this fundamental, conserved role.
1. Introduction
Sexual reproduction relies on the specialized cell division, meiosis, to generate haploid gametes from diploid progenitors for fertilization. Because meiosis evolved from mitosis (Wilkins & Holliday, 2009), they share common mechanisms that drive cell cycle progression. However, since sexual reproduction is subject to a remarkable degree of selection pressure (Swanson & Vacquier, 2002), meiotic mechanisms often exhibit an impressive degree of evolutionary innovation. Studying meiosis in a variety of model systems reinforces this intriguing paradox, suggesting that this specialized cell division presents a unique opportunity for evolution to molecularly tinker with mechanisms that govern cell cycle control and chromosome behavior while maintaining core functions. In this way, our studies of meiosis benefit not only from identifying what is highly conserved but also what has diverged, potentially in response to species-specific requirements.
A good example of a meiotic event that exhibits both extensive conservation and divergence is meiotic recombination. In most organisms, crossover recombination is the ultimate goal of meiosis. During meiotic prophase, homologous chromosomes undergo progressively intimate interactions that ultimately culminate in crossover recombination. After homologs pair, a macromolecular complex, called the synaptonemal complex (SC), is assembled between them in a process called synapsis. Pairing and synapsis are essential for crossover recombination to generate the linkages, or chiasmata, between homologous chromosomes that direct meiotic chromosome segregation (Bhalla & Dernburg, 2008). Because of this interdependence, some organisms, such as budding yeast, mice and plants, mechanistically link pairing, synapsis and recombination in meiotic prophase (Baudat, Manova, Yuen, Jasin, & Keeney, 2000; Giroux, Dresser, & Tiano, 1989; Grelon, Vezon, Gendrot, & Pelletier, 2001; Romanienko & Camerini-Otero, 2000). However, this interdependence does not necessarily require the mechanistic coupling of these events, as C. elegans and Drosophila demonstrate (Dernburg et al., 1998; McKim et al., 1998). Other important differences include the proteins that direct programmed double strand break (DSB) formation, the proteins that promote DNA strand exchange during homologous repair and whether organisms rely on meiosis-specific crossover recombination pathways, mechanisms that are common to both meiosis or mitosis, or both, to produce crossovers (Gray & Cohen, 2016).
In addition to the fundamental role that recombination plays in ensuring accurate chromosome segregation, crossover recombination accomplishes another important function: it generates new haplotypes for natural selection to act upon to drive evolution. Thus, the distribution of crossovers may need to be as tightly regulated as their presence to assure a random assortment of alleles on a population level. Further, recombination’s ability to unlink deleterious combinations of alleles must be balanced with maintaining beneficial ones. The significance of controlling both crossover number and distribution is clearly illustrated by the existence of mechanisms such as crossover assurance, in which every pair of homologous chromosomes gets at least one crossover; crossover homeostasis, in which the number of crossovers remains relatively invariant even if the number of recombination precursors change; and crossover interference, in which the presence of a crossover inhibits the formation of a crossover nearby (Gray & Cohen, 2016).
The AAA-ATPase PCH-2, also known as Pch2 in S. cerevisiae, PCH2 in plants and Drosophila and TRIP13 in mammals, is an evolutionary ancient AAA-ATPase that structurally remodels a family of proteins with HORMA domains (HORMADs) to modify their function (Rosenberg & Corbett, 2015; Vader, 2015). This PCH-2/HORMAD module has been adapted for a wide variety of molecular functions, including meiotic crossover recombination and checkpoint control, the focus of this review. However, attempts to specifically define this module’s “conserved” role in meiotic crossover recombination and checkpoint control have been challenging for the field. In this review, I will argue that the conserved function of the PCH-2/HORMAD module is to coordinate meiotic recombination with synapsis to control the number and distribution of crossovers. Thus, the supposed enigmatic role of PCH-2 in meiosis could be seen as a natural product of the evolutionary variation in how meiotic recombination and/or synapsis is regulated in different model systems.
2. The PCH-2/HORMAD module
The specifics of the PCH-2/HORMAD module have been reviewed elsewhere (Rosenberg & Corbett, 2015; Vader, 2015). In general, this evolutionarily ancient module (Burroughs, Zhang, Schaffer, Iyer, & Aravind, 2015; Tromer, van Hooff, Kops, & Snel, 2019) is defined by the ability of HORMAD proteins to adopt at least two conformations, open and closed, and the requirement of PCH-2 to convert the closed conformation to the open one. HORMADs adopt the closed conformation when they bind a short peptide sequence either present in their own sequence or within other proteins (the closure motif ) and their C-terminal “safety belt” wraps around the closure motif to stabilize this interaction (Fig. 1). This topological interaction requires PCH-2’s ability to consume energy in the form of ATP to remodel and liberate HORMADs (Ye et al., 2015). When free, most HORMADs adopt an open conformation, in which this safety belt is discretely tucked against the HORMA domain (Fig. 1). Given that HORMADs typically participate in a variety of signaling mechanisms, PCH-2 function is essential to recycle HORMADs and make them available for future use.
Fig. 1.
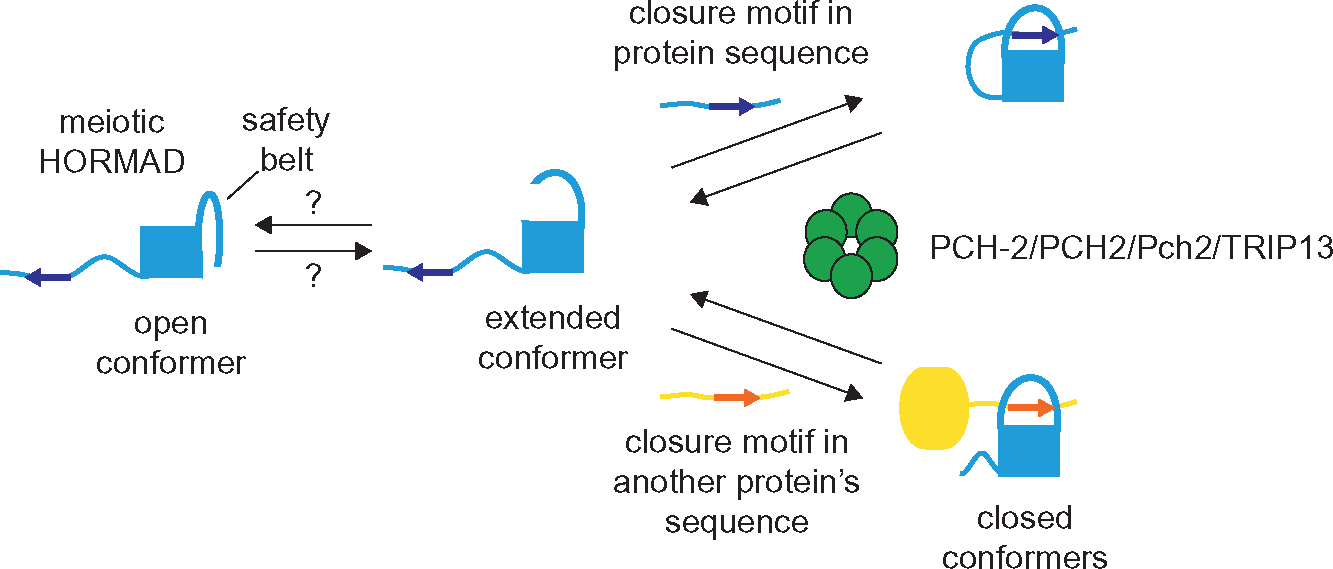
Model for PCH-2/Pch2/PCH2/TRIP13 in remodeling meiotic HORMADs. Schematic of different conformers of meiotic HORMADs that have been demonstrated to exist in vitro (extended and closed) and their remodeling by PCH-2 and its orthologs. Whether meiotic HORMADs also adopt the open conformation is unknown.
Systems from bacteria to humans have exploited the PCH-2/HORMAD module to carry out fundamental biological processes. In most of these cases, the closed version of HORMADs is the active version of the protein. To prevent the spread of bacteriophage, infected bacteria use a HORMAD protein that adopts the closed conformation upon binding foreign phage proteins to initiate a signaling pathway that culminates in death before phage can replicate (Ye et al., 2020). In the eukaryotic spindle checkpoint, the closed version of the HORMAD, Mad2, initiates and amplifies the signal from unattached kinetochores to arrest or delay the cell cycle (Lara-Gonzalez, Westhorpe, & Taylor, 2012). During eukaryotic DNA repair, the closed version of Rev7, another HORMAD, independently regulates both translesion synthesis and DNA repair pathway choice, depending on which protein partner it binds (Clairmont et al., 2020; Malik et al., 2020). And in meiosis, the prophase events of pairing, synapsis and inter-homolog recombination are driven by the closed conformation of meiotic HORMADs assembling on meiotic chromosomes (Kim et al., 2014).
Structural and biochemical analysis of Mad2’s remodeling by PCH-2 have provided important insight into its molecular mechanics: The hexameric ATPase partially unfolds the N terminus of Mad2, dissociating it from its protein partner containing the closure motif and converting it from the closed to the open state (Alfieri, Chang, & Barford, 2018; Ye et al., 2017). This interaction is likely to be relevant for PCH-2’s remodeling of meiotic HORMADs (Yang, Hu, Portheine, Chuenban, & Schnittger, 2020; Ye et al., 2017). However, instead of an open conformation, meiotic HORMADs adopt an “extended” state in which the safety belt is no longer tucked against the HORMA domain (West, Komives, & Corbett, 2017) (Fig. 1). In contrast to Mad2, attempts to undertake similar structural and biochemical studies with meiotic HORMADs and PCH-2 have been less informative, resulting in major unanswered questions about the role of these proteins during meiosis. For example, what are the biological consequences of PCH-2’s remodeling of meiotic HORMADs? Does this remodeling occur in the nucleoplasm, the cytoplasm, on chromosomes or some combination of the three? What is the biological relevance of the unlocked version of meiotic HORMADs? And are the answers to these questions the same in all systems?
3. The role of meiotic HORMADs
At the outset of meiotic prophase, a combination of proteins, including cohesins, axis core proteins and meiotic HORMADs, assemble on chromosomes and structure them in preparation for the meiosis-specific events of interhomolog pairing, synapsis and recombination (Ur & Corbett, 2021). Cohesins organize meiotic chromosomes into linear loop arrays and axis core proteins scaffold these loop arrays into an axial structure that can support pairing, synapsis and recombination (Ur & Corbett, 2021). In both fission and budding yeasts, plants and mice, axis core proteins also play an essential role in recruiting meiotic HORMADs to the axis. In these systems, axis core proteins, such as Red1 in budding yeast, Rec10/Rec27 in fission yeast, SYCP2/SYCP3 in mice and ASY3/ASY4 in plants, contain closure motifs that allow meiotic HORMADs to adopt the closed conformation on meiotic chromosomes (Ur & Corbett, 2021), driving pairing, synapsis and interhomolog recombination. In C. elegans, axis core proteins appear dispensable for axis formation and meiotic HORMADs are recruited on chromosomes directly by cohesins in this system (Kim et al., 2014).
Despite their presence in almost all systems studied to date, meiotic HORMADs demonstrate some functional divergence of their own. Indeed, since PCH-2 controls the function of multiple HORMADS in some systems, potentially constraining its evolutionary trajectory, evolution may specifically target meiotic HORMADs to test out new functions. Meiotic HORMADs are essential for the formation of DSBs in mice, C. elegans and both fission and budding yeasts (Daniel et al., 2011; Goodyer et al., 2008; Latypov et al., 2010; Mao-Draayer, Galbraith, Pittman, Cool, & Malone, 1996) but not in Arabidopsis (Sanchez-Moran, Santos, Jones, & Franklin, 2007). Meiotic HORMADs are depleted or redistributed from meiotic chromosomes in response to synapsis in mice, budding yeast and plants (Borner, Kleckner, & Hunter, 2004; Cuacos et al., 2021; Lambing et al., 2015; Wojtasz et al., 2009) but not in C. elegans (Couteau & Zetka, 2005; Goodyer et al., 2008; Martinez-Perez & Villeneuve, 2005; Zetka, Kawasaki, Strome, & Muller, 1999). They play a role in signaling meiotic progression in budding yeast, C. elegans and mice (Daniel et al., 2011; Kim, Kostow, & Dernburg, 2015; Kogo, Tsutsumi, Inagaki, et al., 2012; Kogo, Tsutsumi, Ohye, et al., 2012; Martinez-Perez & Villeneuve, 2005; San-Segundo & Roeder, 1999; Shin, McGuire, & Rajkovic, 2013; Silva et al., 2014; Wojtasz et al., 2012) but not in fission yeast (Brown, Jarosinska, & Lorenz, 2018). As another clear example of their divergence, the number of meiotic HORMADS varies between organisms: budding and fission yeast have one meiotic HORMAD (Hop1) (Hollingsworth, Goetsch, & Byers, 1990; Lorenz et al., 2004), plants have two (ASY1 and ASY2) (Caryl, Armstrong, Jones, & Franklin, 2000), mice have two (HORMAD 1 and 2) (Wojtasz et al., 2009), C. elegans have four (HTP-3, HIM-3, HTP-1 and HTP-2) (Couteau & Zetka, 2005; Goodyer et al., 2008; Martinez-Perez & Villeneuve, 2005; Zetka et al., 1999) and Drosophila appear to have none (van Hooff, Tromer, van Wijk, Snel, & Kops, 2017). Highlighting how meiotic HORMADs may diverge in response to species-specific requirements, evolutionary analysis in plants show that meiotic HORMADs experience positive selection in response to increases in ploidy (Hollister et al., 2012; Yant et al., 2013).
4. PCH-2/Pch2/PCH2/TRIP13 in meiosis
Pch2 was originally identified in S. cerevisiae as a pachytene checkpoint factor that enforced a meiotic arrest when there were defects in synapsis (San-Segundo & Roeder, 1999). This functional link with synapsis was reinforced when work in C. elegans identified PCH-2 as essential for a checkpoint response when there were defects in synapsis but not when there were defects in recombination (Bhalla & Dernburg, 2005) and with the observation that the only model system that appeared to be missing Pch2 was fission yeast, in which homologous chromosomes do not synapse (Wu & Burgess, 2006). However, this simplistic model for Pch2/PCH-2 function was complicated by observations that budding yeast pch2 mutants themselves exhibited recombination defects, including delays in the formation of crossovers (Borner, Barot, & Kleckner, 2008; Wu & Burgess, 2006), an increase in inter-sister recombination (Zanders, Sonntag Brown, Chen, & Alani, 2011), and effects on crossover assurance (Chakraborty et al., 2017), interference and homeostasis (Joshi, Barot, Jamison, & Borner, 2009; Zanders & Alani, 2009). Subsequent experiments in both C. elegans and mice verified PCH-2/TRIP13’s participation in recombination (Deshong, Ye, Lamelza, & Bhalla, 2014; Li & Schimenti, 2007; Roig et al., 2010), namely in crossover assurance and interference, further confounding its identification as simply a synapsis checkpoint protein. Even with the recognition that Pch2/TRIP13 depleted or redistributed meiotic HORMADs as chromosomes synapse (Borner et al., 2008; Wojtasz et al., 2009), the recombination defects observed in pch-2 mutants were difficult to explain.
As the role of PCH-2 in recombination continued to perplex the meiosis community, a more nuanced picture of the regulation of recombination began to emerge. It had always been appreciated that DSBs typically vastly outnumber crossovers. With more careful cytological analysis of crossover formation, there was a growing realization that the process of meiotic crossover recombination is implemented progressively during meiotic prophase (Cole et al., 2012; Joshi, Brown, Bishop, & Borner, 2015; Morgan et al., 2021; Yokoo et al., 2012). Thus, decisions about which DSBs will become crossovers occur gradually over meiotic prophase, with a persistent winnowing of crossover precursors until all aspects of crossover control, including assurance, interference and homeostasis, are satisfied. More importantly, in all systems, no matter if synapsis is dependent on or independent of recombination, this winnowing either overlaps completely (as in C. elegans (Yokoo et al., 2012)) or partially (as in budding yeast, plants, and mice (Capilla-Perez et al., 2021; Cole et al., 2012; Joshi et al., 2015; Morgan et al., 2021)) with a period when chromosomes are synapsed, suggesting that there may be some mechanism to coordinate crossover recombination and synapsis during this progressive implementation of crossover recombination and its control. That the variation of recombination rates correlates better with length of the synaptonemal complex than with the amount of DNA provides additional support for the existence of such a mechanism (Lynn et al., 2002; Ruiz-Herrera et al., 2017).
Altogether, this growing body of knowledge provided a new framework within which to consider PCH-2’s function and its connection to both synapsis and recombination. In this review, I will discuss the wide-ranging literature on PCH-2’s meiotic role and show that PCH-2’s effect on recombination can be explained in some systems through the coordination of recombination with the mechanisms that control pairing and synapsis. Further, I will argue that this role is generalizable to other systems despite the fact that different organisms use different mechanisms to control pairing and synapsis. In this way, what has appeared as disparate and contradictory observations about the role of PCH-2 in different organisms can actually be integrated into a more unified model that describes PCH-2’s conserved function. This role for PCH-2 may have broader implications beyond the regulation of crossover number and control, given the suggestion that variations in pachytene checkpoint stringency may play important roles in genetic incompatibility and speciation (Li, Barringer, & Barbash, 2009).
For the purposes of this review, I will generally use the C. elegans nomenclature for PCH-2. However, when discussing specific model systems, I will use the nomenclature for that organism (Pch2 for yeast, PCH2 for Drosophila and plants and TRIP13 for mammals). Further, as I discuss the details of PCH-2 function in specific contexts (pairing, synapsis, recombination, etc.), I will highlight common themes that provide insight into the core function(s) of PCH-2 across model systems, providing a more complete synthesis at the end of this review.
5. Localization of PCH-2
PCH-2’s localization provides our first illustration of a core, conserved function with species-specific embellishments and very likely reports on where PCH-2 is remodeling meiotic HORMADs. In almost all organisms that it has been characterized in, with the exception of Drosophila ( Joyce & McKim, 2010), PCH-2 localizes to meiotic chromosomes (Deshong et al., 2014; Joshi et al., 2009; Lambing et al., 2015; San-Segundo & Roeder, 1999). When meiotic HORMADs are assembling on chromosomes to structure them for pairing, synapsis and recombination, PCH-2 can be found as puncta on meiotic chromosomes in budding yeast, plants and C. elegans (Deshong et al., 2014; Joshi et al., 2009; Lambing et al., 2015; San-Segundo & Roeder, 1999). In the plant Brassica and budding yeast, these puncta colocalize with other proteins that mark sites where crossover precursors initiate synapsis (Lambing et al., 2015; Joshi et al., 2009), consistent with the link between recombination and synapsis. Later, after synapsis, PCH-2 localizes either as puncta or as linear stretches along the synaptonemal complex in budding yeast, plants and C. elegans (Deshong et al., 2014; Joshi et al., 2009; Lambing et al., 2015; San-Segundo & Roeder, 1999). In C. elegans, this localization to the synaptonemal complex coincides with when chromosomes are competent for crossover recombination (Deshong et al., 2014) and its ejection from the synaptonemal complex depends on the cell cycle polo-like kinase, PLK-2 (Roelens et al., 2019).
A series of elegant experiments in budding yeast have highlighted a more complicated picture of Pch2 localization, some of which may be unique to this system. Before its localization to the synaptonemal complex, Pch2 is found at the nucleolus, the region of the nucleus where repetitive ribosomal (r)DNA is localized (San-Segundo & Roeder, 1999). Pch2’s enrichment at the nucleolus appears to be specifically required to limit DSB formation at this repetitive sequence in budding yeast (Vader et al., 2011) and is not functionally connected to its role regulating meiotic progression (Herruzo, Santos, Freire, Carballo, & San-Segundo, 2019). Further, targeting Pch2 to the cytoplasm supports its ability to arrest the cell cycle when there are synapsis defects (Herruzo et al., 2021), indicating that Pch2’s remodeling of meiotic HORMADs and effect on meiotic progression can take place away from meiotic chromosomes. Plants exhibit a similar phenomenon, in which PCH2’s ability to remodel one of its meiotic HORMADs, ASY1, is essential to shuttle it into meiotic nuclei to structure meiotic chromosomes in preparation for pairing, synapsis and recombination (Yang et al., 2020).
Altogether, these data suggest that PCH-2’s primary meiotic role in most systems is to remodel meiotic HORMADs on meiotic chromosomes, both prior to and during synapsis, to regulate pairing, synapsis, recombination and meiotic progression, potentially coordinating these events. In some systems, such as in budding yeast and plants, this remodeling of meiotic HORMADs can also happen away from chromosomes with functional implications for both meiotic progression and interhomolog recombination. Meanwhile, in Drosophila, where PCH2 localizes to the nuclear periphery, PCH2’s role in regulating recombination and meiotic progression appears to only occur away from meiotic chromosomes, potentially presenting one extreme of evolutionary innovation in PCH2’s meiotic role.
6. Role in pairing and synapsis
As mentioned above, PCH-2’s localization to meiotic chromosomes in most systems suggests a role for PCH-2 in controlling pairing and synapsis. The clearest case for this role for PCH-2 is in C. elegans. In this organism, both pairing and synapsis are accelerated in pch-2 mutants (Deshong et al., 2014), indicating that PCH-2 actively inhibits these events. This inhibition involves the destabilization of incorrect homolog interactions during pairing and synapsis, since a mutant version of PCH-2 that can bind its substrates but is not able to remodel them results in non-homologous synapsis (Giacopazzi, Vong, Devigne, & Bhalla, 2020). Thus, PCH-2 proofreads homolog interactions that underlie pairing and synapsis in C. elegans, contributing both to their accuracy and completion.
It is currently unclear whether PCH-2’s role in proofreading homolog interactions that underlie accurate homolog pairing and synapsis is conserved in other systems, particularly those that use recombination to initiate these events, but preliminary data is promising. In plants and mice, synapsis is incomplete in pch2/Trip13 mutants (Lambing et al., 2015; Miao et al., 2013; Roig et al., 2010). However, in plants, this phenotype is likely to be an indirect result of the requirement for PCH2 to shuttle meiotic HORMADs into the nucleus during meiotic prophase (Yang et al., 2020). In mice, the defects in synapsis are limited to regions that might present a challenge for synapsis, such as pericentric regions (Roig et al., 2010), suggesting a more direct role in regulating synapsis. Further, the localization of Pch2/PCH2 in budding yeast and plants as puncta that colocalize with proteins involved in recombination and synapsis (Lambing et al., 2015; Joshi et al., 2009) supports this proposed role in regulating pairing and synapsis.
7. Role in recombination
Soon after Pch2’s identification as a checkpoint factor, a role for Pch2 in recombination began to emerge in multiple systems but the observations did not easily lend themselves to the development of a coherent model. Two clear themes materialize from the analysis of pch-2 mutants in different systems: (1) there is an effect on the number and distribution of crossovers; and (2) this effect is variable and non-uniform. In budding yeast pch2 mutants, there is a general increase in the number of crossovers on mid-sized and large chromosomes (Zanders & Alani, 2009), loss of assurance on small chromosomes (Chakraborty et al., 2017), and a loss of interference in some genetic intervals but not others ( Joshi et al., 2009; Zanders & Alani, 2009). In plant and Drosophila pch2 mutants, recombination is elevated in some genetic intervals and reduced and even unaffected in others ( Joyce & McKim, 2009; Lambing et al., 2015), leading to similar variable effects on interference. And in murine Trip13 mutants, this variable effect on crossover number and distribution manifests as a reduction in crossover assurance, a reduction in the spacing between crossovers on large chromosomes and changes in the position of crossovers on shorter chromosomes (Roig et al., 2010). The variability of these mutant phenotypes, and the lack of a regular pattern, has definitely contributed to the difficulty in developing a unified, integrated model of PCH-2 function in the field.
This challenge may be addressed by experiments in C. elegans, which benefits from a clearer understanding of synapsis initiation and the distribution of DSB formation across chromosomes, allowing distinct patterns to appear from the array of observed recombination defects in pch-2 mutants. Once again, in this system, mutation of pch-2 produces a loss of crossover assurance and non-uniform effects on recombination on two chromosomes tested, an autosome and the sex chromosome (Deshong et al., 2014). However, when analyzed in the context of pairing, synapsis and DSB formation, these recombination defects appear less mysterious. First, recombination is less severely affected in pch-2 mutants at pairing center ends of chromosomes (Deshong et al., 2014). Pairing Centers are cis-acting sites near one end of each chromosome that promote homolog pairing and synapsis (MacQueen et al., 2005). Therefore, these regions are often the first to pair, the first to synapse and possibly, the first to initiate early events in crossover recombination. Altogether, these data suggest that PCH-2 limits this temporal bias in crossover formation, potentially to more widely distribute recombination events to regions distal to pairing centers.
Second, recombination is less severely affected in pch-2 mutants in chromosomal regions where DSBs are enriched (Deshong et al., 2014). C. elegans autosomes do not experience DSB formation, and therefore crossover formation, uniformly: chromosome ends are enriched for DSB formation and crossovers, relative to the central regions of the chromosome (Nadarajan, Altendorfer, Saito, Martinez-Garcia, & Colaiacovo, 2021; Yu, Kim, & Dernburg, 2016). Therefore, in addition to its possible role limiting the temporal bias in crossover formation, PCH-2 may also be limiting the effect of DSB enrichment to more widely distribute crossovers across the genome. Since the central regions of autosomes are also gene rich, in comparison to chromosome ends, PCH-2’s role in redistributing crossovers to this region may specifically be an attempt to create new haplotypes for evolution to act on, despite the paucity of DSBs. It remains an open question, however, whether these patterns are seen on the remaining four autosomes in C. elegans.
In addition to this hypothesized role during unperturbed meiosis, additional work in Drosophila and C. elegans recombination mutants provides further evidence for PCH2/PCH-2 controlling crossover distribution. In both of these systems, defects in recombination alters the distribution of crossovers across chromosomes and this redistribution partially relies on PCH2/PCH-2 (Deshong et al., 2014; Joyce & McKim, 2010). This effect on crossover distribution is accompanied by protracted PCH-2 activity: In C. elegans, PCH-2 persists on meiotic chromosomes while in Drosophila, PCH-2’s remains at the nuclear periphery longer (Deshong et al., 2014; Joyce & McKim, 2010). In Drosophila, overexpression of PCH2 is sufficient to recapitulate these effects on crossover distribution ( Joyce & McKim, 2010). Thus, both the absence and the persistence of PCH-2 activity during meiotic prophase has consequences on the distribution of crossovers, consistent with a direct role in this process.
This context now provides a useful way to frame and potentially reanalyze the puzzling phenotypes observed in budding yeast, plants and mice. In these systems, DSBs both initiate crossover recombination and act as potential sites for synapsis initiation (Baudat et al., 2000; Giroux et al., 1989; Grelon et al., 2001; Romanienko & Camerini-Otero, 2000). Similar to C. elegans, they are also not uniformly distributed and specific regions, known as “hot spots,” tend to be more favorable for DSB formation (Gray & Cohen, 2016) and presumably, the resulting downstream events, such as the decision to become a crossover and initiation of synapsis. Therefore, given the observation that PCH-2 limits crossovers where pairing and synapsis initiate and DSBs are enriched in C. elegans, an analogous role for Pch2/TRIP13/PCH2 in systems such as budding yeast, mice and plants might be to similarly limit the frequency with which these “hot spots” would become crossovers to ensure a wider distribution of crossovers throughout the genome and on a population level. Since a reduction in the number of DSBs may exacerbate the bias for “hot spots” to become crossovers, this role may also explain why Pch2 is essential for crossover homeostasis ( Joshi et al., 2009; Zanders & Alani, 2009).
How PCH-2 mechanistically ensures the correct number and distribution of crossovers is currently unclear but in budding yeast and mice, it involves both the regulation of DSB formation and interhomolog bias via meiotic HORMADs. A direct interaction between meiotic HORMADs and an essential member of the DSB forming machinery (Mer2 in budding yeast and IHO1 in mice) explains how PCH-2 controls DSB formation through its removal and redistribution of meiotic HORMADs (Panizza et al., 2011; Stanzione et al., 2016). PCH-2’s participation in the installation of homolog bias is less clear. During crossover recombination, DNA repair is deliberately directed away from the sister chromatid, the preferred template during homologous recombination in mitosis, toward the homolog. Meiotic HORMADs are essential for homolog bias (Schwacha & Kleckner, 1994). More specifically, in budding yeast, phosphorylation of the HORMAD, Hop1, by conserved DNA damage kinases promotes homolog bias (Carballo, Johnson, Sedgwick, & Cha, 2008). Pch2 contributes to this phosphorylation event, and homolog bias, likely in part by regulating the availability of Hop1 (Herruzo et al., 2016). However, since Pch2 interacts with the N-terminus of the DNA repair protein Xrs2 in a two-hybrid assay and removal of this region in Xrs2 recapitulates phenotypes observed in pch2 mutants (Ho & Burgess, 2011), Pch2’s regulation of interhomolog bias might be more direct and occur on chromosomes. Given the knowledge that recombination is progressively implemented, Pch2/TRIP13/PCH2 may participate in destabilizing some crossover precursors involving meiotic HORMADs on chromosomes to favor the stabilization of others and promote this redistribution, both before and during synapsis. Whether Pch2 contributes to interhomolog bias indirectly, directly, or some combination of the two, this role likely explains the increase in inter-sister recombination observed in budding yeast pch2 mutants (Zanders et al., 2011).
C. elegans differs from budding yeast and mice in that the regulation of DSB formation appears to be distinct from that of interhomolog recombination (Deshong et al., 2014). This partition has been clearly demonstrated in Drosophila ( Joyce & McKim, 2009) and suggests that in both of these ecdysozoans, PCH-2/PCH2’s role in controlling crossover number and distribution is primarily through control of interhomolog recombination (Deshong et al., 2014). In C. elegans, this control is likely through the remodeling of meiotic HORMADs on chromosomes, given their conserved role in promoting interhomolog crossover recombination (Couteau, Nabeshima, Villeneuve, & Zetka, 2004; Couteau & Zetka, 2011; Hayashi, Chin, & Villeneuve, 2007; Martinez-Perez & Villeneuve, 2005) and PCH-2’s localization to chromosomes when they are competent for meiotic recombination (Deshong et al., 2014). This functional partition is not because these two systems have uncoupled DSB formation from synapsis (Dernburg et al., 1998; McKim et al., 1998) since Arabidopsis, which needs DSBs for synapsis (Grelon et al., 2001), also does not rely on meiotic HORMADs for DSB formation (Sanchez-Moran et al., 2007). Instead, this separation of function may reflect yet another set of species-specific requirements for meiotic HORMADs and their regulation by PCH-2 in meiotic progression and crossover recombination in these systems. The major question in Drosophila is how PCH2 accomplishes this effect on crossover distribution from the nuclear periphery ( Joyce & McKim, 2010) and in the apparent absence of a meiotic HORMAD (van Hooff et al., 2017).
8. Role in regulating meiotic progression
Pch2 was first identified in budding yeast as a checkpoint factor that restored meiotic progression when there were defects in synapsis, providing a clear link between Pch2 function and a role in regulating meiotic progression (San-Segundo & Roeder, 1999). Unfortunately, deciphering this role was complicated by observations that pch2 single mutants produced defects in meiotic progression (Hochwagen, Tham, Brar, & Amon, 2005; Wu & Burgess, 2006). These contradictory phenotypes can be reconciled when we recognize that Pch2 remodels Hop1 both on and away from chromosomes to promote meiotic progression (Herruzo et al., 2021; Raina & Vader, 2020). As had been suggested by dosage experiments with Hop1 (Bailis, Smith, & Roeder, 2000; Herruzo et al., 2016; Raina & Vader, 2020), Pch2 ensures there is enough Hop1 available for functional checkpoint signaling and this can occur away from chromosomes (Herruzo et al., 2021). However, once synapsis is complete, Pch2 is recruited to synapsed chromosomes to remove or redistribute meiotic HORMADs from chromosomes as an important signal for meiotic progression to continue (Raina & Vader, 2020).
Mechanistically, Pch2’s depletion of Hop1 from budding yeast chromosomes in response to synapsis promotes meiotic progression by reducing the activity of Mek1 kinase, a key regulator of inter-homolog recombination (Niu et al., 2005; Subramanian et al., 2016; West et al., 2017). When Mek1 activity is down regulated, its inhibition of the transcription factor, Ndt80 is relieved and Ndt80 is capable of driving the transcription of factors essential for exit from pachytene (Chen et al., 2018; Prugar, Burnett, Chen, & Hollingsworth, 2017). By contrast, the depletion of meiotic HORMADs from murine meiotic chromosomes by TRIP13 during synapsis promotes meiotic progression by effectively silencing the DNA damage response. However, what this checkpoint response looks like is different based on sex. During spermatogenesis, when the X and Y chromosomes fail to completely synapse, components of the DNA damage response participate in a whole scale remodeling of these chromosomes to silence them in a process called meiotic sex chromosomes inactivation (MSCI) (Turner, 2007). MSCI is essential for the progression of spermatogenesis and events that disrupt it, even indirectly, lead to an early block in meiotic progression (Turner, 2007). For example, defects in recombination and/or synapsis produce persistent DNA damage on autosomes, preventing the effective accumulation of the DNA repair factors on sex chromosomes and early steps in MSCI (Mahadevaiah et al., 2008). TRIP13’s removal of meiotic HORMADs during autosomal synapsis downregulates DSB formation and alleviates the block to sister repair (Daniel et al., 2011; Li & Schimenti, 2007; Wojtasz et al., 2009), reducing the number of persistent DSBs on autosomes that may compete for DNA repair factors with unsynapsed sex chromosomes. In this way, MSCI is an indirect readout for meiotic progression and quality control. Since oocytes have two X chromosomes and do not experience MSCI, meiotic progression is assessed directly through the completion of DNA repair, although it appears that there are dose-dependent responses depending on the load of DNA damage: Defects in recombination that produce many persistent repair intermediates block meiosis earlier than defects that only disrupt synapsis or produce fewer repair intermediates (Rinaldi, Bolcun-Filas, Kogo, Kurahashi, & Schimenti, 2017). Thus, despite the mechanistic differences in how meiotic progression is promoted, synapsis, and the concomitant removal/redistribution of meiotic HORMADs by Pch2/TRIP13, is an essential reporter of effective quality control during meiosis in both budding yeast and mice.
What about in systems in which synapsis precedes and does not rely on DSB formation and the initiation of recombination? The role of meiotic HORMADs in C. elegans in regulating meiotic progression shares some similarities and some differences with budding yeast and mice. In this system, meiotic HORMADs also assemble on meiotic chromosomes to drive pairing, synapsis and recombination but do not get removed in response to synapsis (Couteau & Zetka, 2005; Goodyer et al., 2008; Martinez-Perez & Villeneuve, 2005; Zetka et al., 1999). In contrast to what is observed in budding yeast and mice and inferred in plants, it is the binding of three of the four meiotic HORMADs, HIM-3, HTP-1 and HTP-2, on chromosomes that promotes meiotic progression (Kim et al., 2015). Some evidence suggests that a soluble version of HTP-1 that fails to assemble on meiotic chromosomes, and presumably is present as an extended conformer, delays meiotic prophase (Silva et al., 2014). This model for how meiotic HORMADs might regulate meiotic progression in C. elegans, in which a soluble version that fails to bind meiotic chromosomes delays meiosis, is in direct contrast to the model presented in budding yeast and mice, in which the removal of meiotic HORMADs from meiotic chromosomes by Pch2/TRIP13 drives meiotic progression and may signal that the downstream mechanisms that impact meiotic progression will be specific to C. elegans.
From this assortment of checkpoint mechanisms in budding yeast, mice and C. elegans, one coherent picture that emerges is that PCH-2 and/or meiotic HORMADs consistently coordinates the cell cycle with the events of recombination and synapsis in multiple systems but in ways that are layered on species-specific regulation of meiotic progression. Indeed, the situation in Drosophila, in which PCH2 delays meiosis in response to defects ( Joyce & McKim, 2009, 2010) despite the apparent absence of a meiotic HORMAD in its genome (van Hooff et al., 2017), may be one extreme example that highlights this point. Given that defects in meiosis produce varying effects on meiotic progression, both within species and between species, it is entirely possible that this variation in PCH-2 dependent checkpoint responses is a defining feature in the evolution of this process.
9. Requirement for a cofactor
Another major difference that highlights the differences between PCH-2’s role in different meiotic systems is the requirement of a cofactor. Structural and biochemical studies with Mad2 have reinforced that PCH-2/TRIP13 uses a cofactor, CMT-1/p31comet, to mediate interaction with its HORMAD substrates (Alfieri et al., 2018; Ye et al., 2015). The expansion of the HORMAD/PCH2 module to include an adaptor is conserved in plants and C. elegans (Balboni, Yang, Komaki, Brun, & Schnittger, 2020; Giacopazzi et al., 2020; Ji et al., 2016). In Arabidopsis, COMET facilitates PCH2’s ability to shuttle the meiotic HORMAD, ASY1, into the nucleus in early prophase and PCH2’s depletion of ASY1 from meiotic chromosomes in response to synapsis (Balboni et al., 2020). In rice, mutation of P31comet function produces defects in DSB formation, homolog pairing and crossover recombination ( Ji et al., 2016). In C. elegans, unlike in Arabidopsis, mutation of cmt-1 does not affect PCH-2’s localization to meiotic chromosomes but recapitulates the phenotype of a mutant version of PCH-2 that binds substrates but cannot remodel them (Giacopazzi et al., 2020), implicating CMT-1 in PCH-2’s ability to hydrolyze ATP and remodel HORMADs but not in its PCH-2’s ability to bind its substrates. By contrast, other organisms, such as budding yeast, do not have an obvious CMT-1/p31comet ortholog (van Hooff et al., 2017), raising the question of what the molecular mechanics of meiotic HORMAD remodeling look like in this system.
10. A unified model of PCH-2’s role in meiotic prophase
How to integrate this complex and disparate data into an integrated model of PCH-2 function in meiotic prophase? The theme that emerges is an important meiotic role for PCH-2 and its orthologs with common features and species-specific embellishments. For example, a common feature to all systems is that PCH-2 remodels meiotic HORMADs in solution (to make them available for loading onto meiotic chromosomes and/or shuttle them into the nucleus) (see #1 in Fig. 2) and/or on chromosomes to regulate the role of meiotic HORMADs in inter-homolog recombination, affecting the number and distribution of crossovers.
Fig. 2.
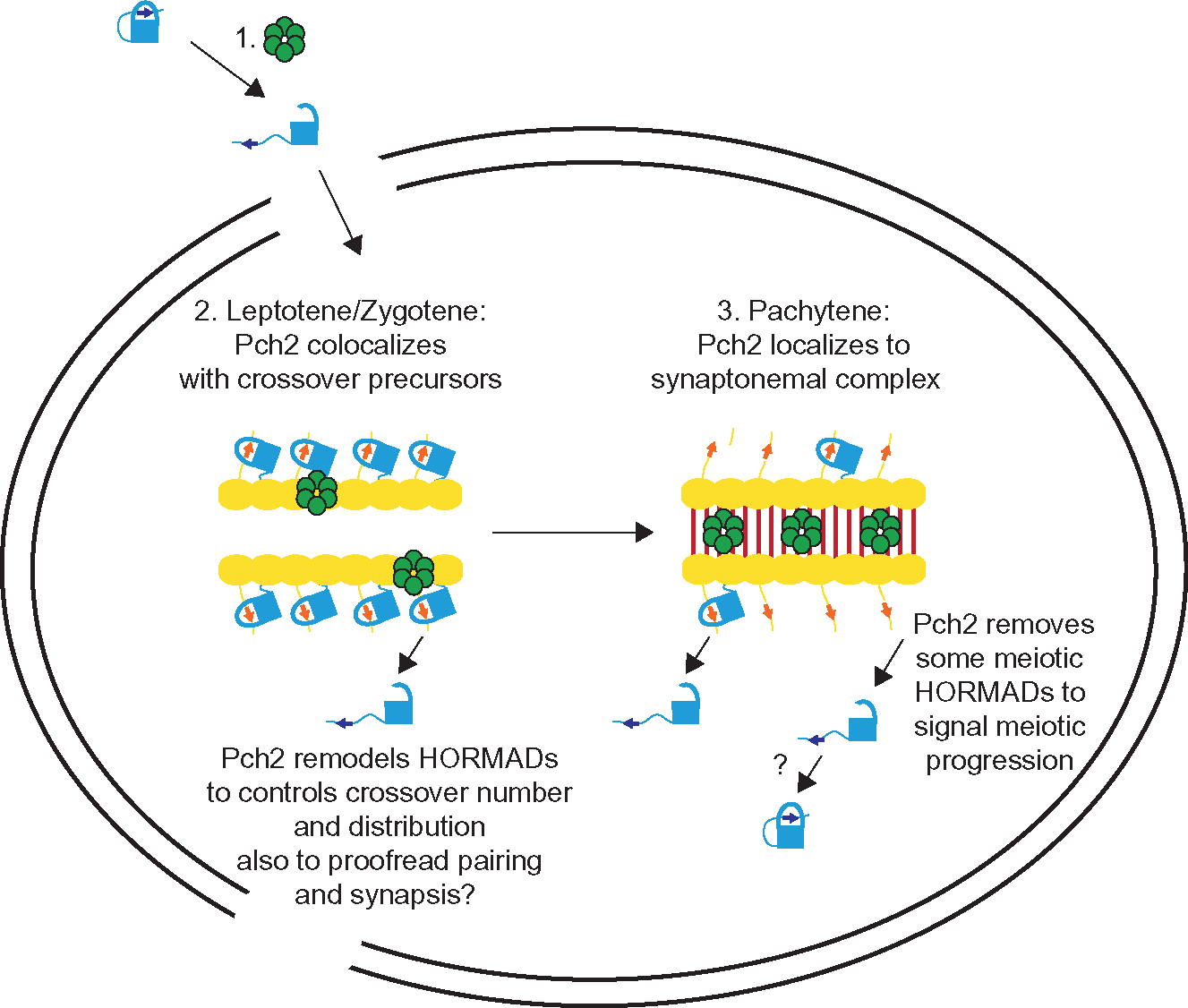
Model for Pch2/TRIP13/PCH2 function in systems in which recombination precedes synapsis, such as budding yeast, plants and mice. 1. Pch2 (green hexamer) remodels meiotic HORMADs from their closed conformation, bound to its own closure motif, to the extended conformation to enable its entry into meiotic nuclei and assembly on meiotic chromosomes. 2. During leptotene/zygotene, Pch2 forms puncta on meiotic chromosomes, colocalizing with crossover precursors, remodeling meiotic HORMADs from closed to extended to contribute to the gradual implementation of crossover number and distribution and possibly proofread homolog pairing and synapsis. Yellow ovals represent axis proteins that contain closure motifs and recruit meiotic HORMADs. 3. During pachytene, Pch2 localizes to the synaptonemal complex between synapsed homologous chromosomes, depleting meiotic HORMADs to limit meiotic recombination, drive meiotic progression and participate in the last stages of implementing crossover number and distribution through the remodeling of a reduced pool of meiotic HORMADs on meiotic chromosomes.
For systems such as budding yeast, plants and mice, in which crossover recombination is initiated prior to synapsis, Pch2/PCH2/TRIP13’s role in regulating crossover number and distribution is central and occurs prior to synapsis, possibly to ensure a wider distribution of crossovers beyond DSB “hot spots.” However, since crossover precursors are essential for robust pairing and synapsis in these systems (Baudat et al., 2000; Giroux et al., 1989; Grelon et al., 2001; Romanienko & Camerini-Otero, 2000) and Pch2/PCH2 colocalizes with crossover precursors in budding yeast and plants (Lambing et al., 2015; Joshi et al., 2009), this remodeling on chromosomes is also likely relevant for pairing and synapsis (see #2 in Fig. 2), possibly proofreading them for accuracy.
Once synapsis initiates in these systems, PCH-2 localizes to SC (see #3 in Fig. 2) to deplete or redistribute meiotic HORMADS and signal successful synapsis, the progression of most crossover precursors into appropriate DNA repair pathways and that meiosis should progress. This depletion also restores the sister chromatid as a viable DNA repair template, allowing for the completion of DNA repair, and in some systems, downregulates DSB formation. Further, this depletion may be selective: work in budding yeast has shown that its meiotic HORMAD, Hop1, persists at chromosome ends, leading to an increase in DSBs on shorter chromosomes (Subramanian et al., 2019), thus guaranteeing they get crossovers. Finally, although meiotic HORMADs are depleted from chromosomes during synapsis, they are not completely removed (Cuacos et al., 2021; Lambing et al., 2015; Wojtasz et al., 2009), suggesting roles for both meiotic HORMADs and Pch2/PCH2/TRIP13 even when chromosomes are synapsed, such as contributing to the gradual implementation of crossovers and their control and distribution during synapsis.
In systems in which synapsis precedes recombination, such as C. elegans, the data for PCH-2’s role in pairing and synapsis is clearest (Deshong et al., 2014; Giacopazzi et al., 2020) (Fig. 3A) and is likely accomplished by PCH-2 remodeling meiotic HORMADs on chromosomes directly involved in driving pairing and synapsis. Once chromosomes synapse, PCH-2 localizes to the SC and remodels meiotic HORMADs on these synapsed chromosomes to coordinate control of meiotic recombination so that it is more widely distributed than simply at the first places to pair, synapse or experience DSBs. This remodeling also helps implement crossover assurance and interference (Fig. 3).
Fig. 3.
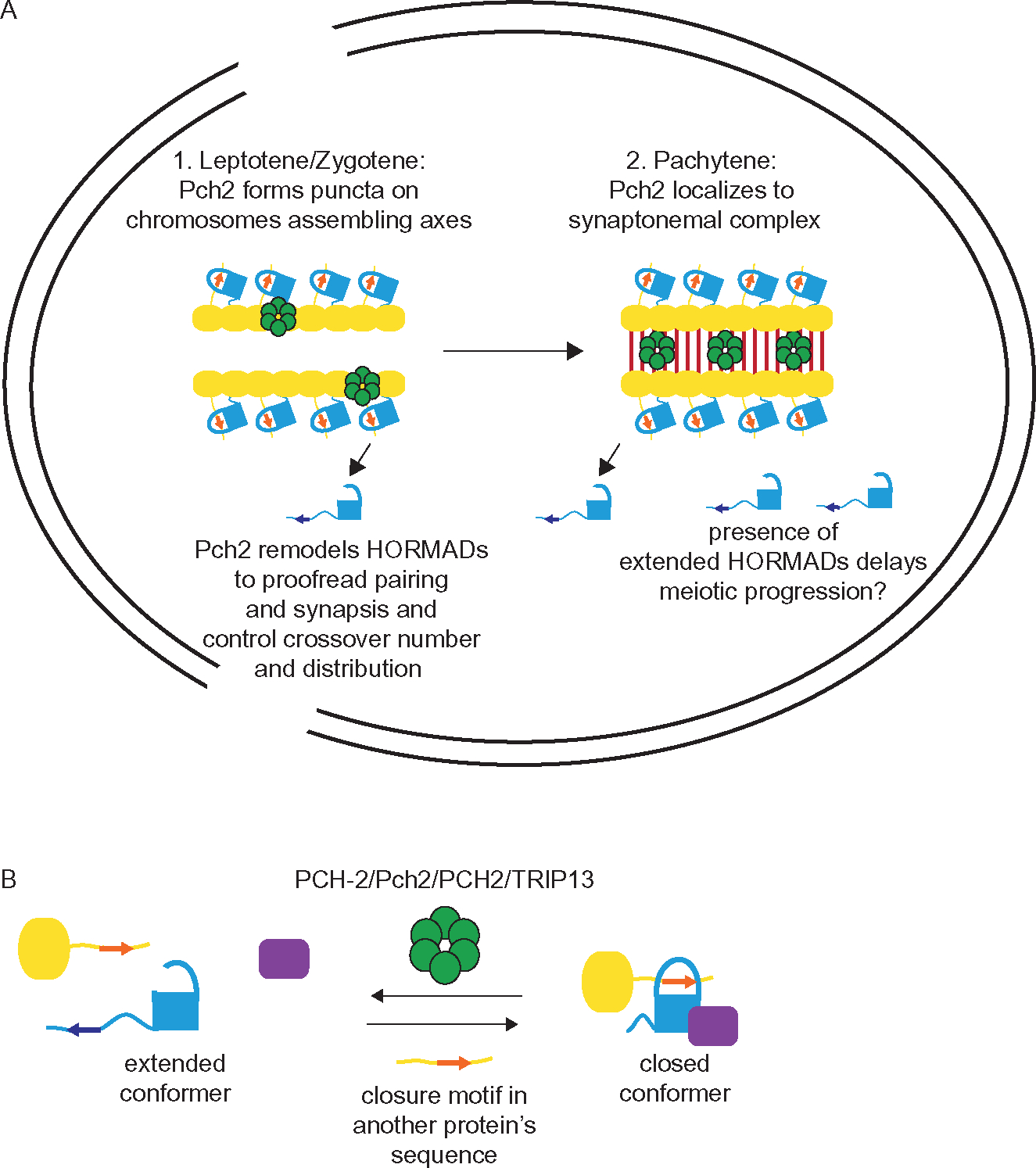
Model for PCH-2 function in systems in which synapsis precedes recombination and PCH-2 localizes to the synaptonemal complex, such as C. elegans. (A) 1. During leptotene/zygotene, Pch2 forms puncta on meiotic chromosomes, remodeling meiotic HORMADs from closed to extended to control to proofread homolog pairing and synapsis. Yellow ovals represent axis proteins that contain closure motifs and recruit meiotic HORMADs. In this system, the assembly of meiotic HORMADs on chromosomes drives meiotic progression. 2. During pachytene, PCH-2 localizes to the synaptonemal complex between synapsed homologous chromosomes, meiotic HORMADs remain on chromosomes and remodeling meiotic HORMADs from closed to extended to contribute to the gradual implementation of crossover number and distribution. The presence of soluble extended conformers of at least one meiotic HORMAD, HTP-1, delays meiotic progression. (B) A model for how PCH-2’s remodeling of meiotic HORMADs could destabilize interhomolog intermediates involved in pairing, synapsis and/or recombination on meiotic chromosomes, contributing to their proofreading and regulation.
PCH2’s role is clearly different in Drosophila, where PCH2 appears to regulate meiotic progression and when chromosomes are competent for crossover formation from the nuclear periphery ( Joyce & McKim, 2010), raising the obvious question: what is its meiotic substrate(s)? One possible candidate is Mad2 which controls cell cycle progression (Rodriguez-Bravo et al., 2014) and has been shown to regulate meiotic synapsis in C. elegans through its localization at the nuclear periphery (Devigne & Bhalla, 2021). Another compelling candidate is Rev7, either through its capacity in regulating recombination (Clairmont et al., 2020) or cell cycle progression (Listovsky & Sale, 2013; Pfleger, Salic, Lee, & Kirschner, 2001).
Pch2/PCH2’s remodeling of HORMADs in the cytoplasm to make them available for their roles in meiotic nuclei is clear in both budding yeast and plants (Herruzo et al., 2021; Yang et al., 2020). What does PCH-2’s remodeling of meiotic HORMADs directly on chromosomes accomplish, aside from their depletion to signal meiotic progression in some systems? Aside from their adoption of multiple conformers to accomplish different functions, another hallmark of HORMADs is a dimerization interface that enables interactions with other factors (Rosenberg & Corbett, 2015). One possibility is that PCH-2’s remodeling of meiotic HORMADs on chromosomes destabilizes interactions at this interface that are required for pairing, synapsis and/or interhomolog recombination (Fig. 3B). This model could explain PCH-2’s proofreading function during pairing and synapsis and potentially its role in controlling the number and distribution of crossovers. In particular, PCH-2’s ability to destabilize some crossover intermediates to favor their distribution elsewhere in the genome during the progressive implementation of crossover recombination could account for its observed involvement in crossover assurance, interference and homeostasis in almost all systems to date.
A final set of species-specific embellishments are the variations in molecular mechanisms that produce the checkpoint responses that rely on HORMADs and are regulated by PCH2 in response to meiotic defects. It seems clear that, when confronted with meiotic defects, the PCH-2/HORMAD module often exploits meiotic processes specific to species, such as MSCI during mammalian spermatogenesis, to implement its effect on meiotic progression. These species-specific mechanisms could also explain the variation in stringency of pachytene checkpoint responses among different species, even among sexes in the same species. Since these differences in checkpoint stringency have been used to explain the ability of some organisms to tolerate genetic incompatibilities and changes in ploidy (Li et al., 2009), it is possible that future studies of the PCH-2/HORMAD module will expand this discussion to our understanding of speciation and hybrid sterility.
11. Conclusion
The role of PCH-2 in meiosis has long been enigmatic and confusing, defying an easy and accessible characterization of its conserved function among the variety of systems in which meiosis is currently studied. However, its conservation, even in systems lacking meiotic HORMADs, suggests an important role in meiotic regulation. When assessed in the context of the multiple, different ways that essential events in meiosis are themselves regulated in these systems, a conserved function for PCH-2 becomes clearer and differences can be interpreted as species-specific embellishments on this conserved function. Indeed, given that sexual reproduction is known for evolutionary innovation, these differences may provide as much insight into how organisms coordinate recombination with synapsis as conservation and similarities do.
References
- Alfieri C, Chang L, & Barford D (2018). Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13. Nature, 559, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis JM, Smith AV, & Roeder GS (2000). Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Molecular and Cellular Biology, 20, 4838–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboni M, Yang C, Komaki S, Brun J, & Schnittger A (2020). COMET Functions as a PCH2 cofactor in regulating the HORMA domain protein ASY1. Current Biology, 30(4113–4127), e4116. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, & Keeney S (2000). Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Molecular Cell, 6, 989–998. [DOI] [PubMed] [Google Scholar]
- Bhalla N, & Dernburg AF (2005). A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science, 310, 1683–1686. [DOI] [PubMed] [Google Scholar]
- Bhalla N, & Dernburg AF (2008). Prelude to a division. Annual Review of Cell and Developmental Biology, 24, 397–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GV, Barot A, & Kleckner N (2008). Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proceedings of the National Academy of Sciences of the United States of America, 105, 3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, & Hunter N (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell, 117, 29–45. [DOI] [PubMed] [Google Scholar]
- Brown SD, Jarosinska OD, & Lorenz A (2018). Genetic interactions between the chromosome axis-associated protein Hop1 and homologous recombination determinants in Schizosaccharomyces pombe. Current Genetics, 64, 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Zhang D, Schaffer DE, Iyer LM, & Aravind L (2015). Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Research, 43, 10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capilla-Perez L, Durand S, Hurel A, Lian Q, Chambon A, Taochy C, et al. (2021). The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, & Cha RS (2008). Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell, 132, 758–770. [DOI] [PubMed] [Google Scholar]
- Caryl AP, Armstrong SJ, Jones GH, & Franklin FC (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma, 109, 62–71. [DOI] [PubMed] [Google Scholar]
- Chakraborty P, Pankajam AV, Lin G, Dutta A, Krishnaprasad GN, Tekkedil MM, et al. (2017). Modulating crossover frequency and interference for obligate crossovers in Saccharomyces cerevisiae meiosis. G3 (Bethesda), 7, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gaglione R, Leong T, Bednor L, de Los Santos T, Luk E, et al. (2018). Mek1 coordinates meiotic progression with DNA break repair by directly phosphorylating and inhibiting the yeast pachytene exit regulator Ndt80. PLoS Genetics, 14, e1007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clairmont CS, Sarangi P, Ponnienselvan K, Galli LD, Csete I, Moreau L, et al. (2020). TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nature Cell Biology, 22, 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, et al. (2012). Homeostatic control of recombination is implemented progressively in mouse meiosis. Nature Cell Biology, 14, 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Nabeshima K, Villeneuve A, & Zetka M (2004). A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Current Biology, 14, 585–592. [DOI] [PubMed] [Google Scholar]
- Couteau F, & Zetka M (2005). HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes & Development, 19, 2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, & Zetka M (2011). DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Developmental Cell, 20, 353–363. [DOI] [PubMed] [Google Scholar]
- Cuacos M, Lambing C, Pachon-Penalba M, Osman K, Armstrong SJ, Henderson IR, et al. (2021). Meiotic chromosome axis remodelling is critical for meiotic recombination in Brassica rapa. Journal of Experimental Botany, 72, 3012–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, et al. (2011). Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nature Cell Biology, 13, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, & Villeneuve AM (1998). Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell, 94, 387–398. [DOI] [PubMed] [Google Scholar]
- Deshong AJ, Ye AL, Lamelza P, & Bhalla N (2014). A quality control mechanism coordinates meiotic prophase events to promote crossover assurance. PLoS Genetics, 10, e1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devigne A, & Bhalla N (2021). Mad1’s ability to interact with Mad2 is essential to regulate and monitor meiotic synapsis in C. elegans. PLoS Genetics, 17, e1009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacopazzi S, Vong D, Devigne A, & Bhalla N (2020). PCH-2 collaborates with CMT-1 to proofread meiotic homolog interactions. PLoS Genetics, 16, e1008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux CN, Dresser ME, & Tiano HF (1989). Genetic control of chromosome synapsis in yeast meiosis. Genome, 31, 88–94. [DOI] [PubMed] [Google Scholar]
- Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, & Zetka M (2008). HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Developmental Cell, 14, 263–274. [DOI] [PubMed] [Google Scholar]
- Gray S, & Cohen PE (2016). Control of meiotic crossovers: From double-strand break formation to designation. Annual Review of Genetics, 50, 175–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, & Pelletier G (2001). AtSPO11–1 is necessary for efficient meiotic recombination in plants. The EMBO Journal, 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Chin GM, & Villeneuve AM (2007). C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genetics, 3, e191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruzo E, Lago-Maciel A, Baztan S, Santos B, Carballo JA, & San-Segundo PA (2021). Pch2 orchestrates the meiotic recombination checkpoint from the cytoplasm. PLoS Genetics, 17, e1009560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruzo E, Ontoso D, Gonzalez-Arranz S, Cavero S, Lechuga A, & San-Segundo PA (2016). The Pch2 AAA+ ATPase promotes phosphorylation of the Hop1 meiotic checkpoint adaptor in response to synaptonemal complex defects. Nucleic Acids Research, 44, 7722–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruzo E, Santos B, Freire R, Carballo JA, & San-Segundo PA (2019). Characterization of Pch2 localization determinants reveals a nucleolar-independent role in the meiotic recombination checkpoint. Chromosoma, 128, 297–316. [DOI] [PubMed] [Google Scholar]
- Ho HC, & Burgess SM (2011). Pch2 acts through Xrs2 and Tel1/ATM to modulate interhomolog bias and checkpoint function during meiosis. PLoS Genetics, 7, e1002351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochwagen A, Tham WH, Brar GA, & Amon A (2005). The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell, 122, 861–873. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, & Byers B (1990). The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell, 61, 73–84. [DOI] [PubMed] [Google Scholar]
- Hollister JD, Arnold BJ, Svedin E, Xue KS, Dilkes BP, & Bomblies K (2012). Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genetics, 8, e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J, Tang D, Shen Y, Xue Z, Wang H, Shi W, et al. (2016). P31comet, a member of the synaptonemal complex, participates in meiotic DSB formation in rice. Proceedings of the National Academy of Sciences of the United States of America, 113, 10577–10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, & Borner GV (2009). Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genetics, 5, e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Brown MS, Bishop DK, & Borner GV (2015). Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Molecular Cell, 57, 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, & McKim KS (2009). Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics, 181, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, & McKim KS (2010). Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLoS. Genet 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kostow N, & Dernburg AF (2015). The chromosome axis mediates feedback control of CHK-2 to ensure crossover formation in C. elegans. Developmental Cell, 35, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Rosenberg SC, Kugel CL, Kostow N, Rog O, Davydov V, et al. (2014). The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Developmental Cell, 31, 487–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogo H, Tsutsumi M, Inagaki H, Ohye T, Kiyonari H, & Kurahashi H (2012). HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes to Cells, 17, 897–912. [DOI] [PubMed] [Google Scholar]
- Kogo H, Tsutsumi M, Ohye T, Inagaki H, Abe T, & Kurahashi H (2012). HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes to Cells, 17, 439–454. [DOI] [PubMed] [Google Scholar]
- Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, et al. (2015). Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genetics, 11, e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, & Taylor SS (2012). The spindle assembly checkpoint. Current Biology, 22, R966–R980. [DOI] [PubMed] [Google Scholar]
- Latypov V, Rothenberg M, Lorenz A, Octobre G, Csutak O, Lehmann E, et al. (2010). Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Molecular and Cellular Biology, 30, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Barringer BC, & Barbash DA (2009). The pachytene checkpoint and its relationship to evolutionary patterns of polyploidization and hybrid sterility. Heredity (Edinburgh), 102, 24–30. [DOI] [PubMed] [Google Scholar]
- Li XC, & Schimenti JC (2007). Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLoS Genetics, 3, e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listovsky T, & Sale JE (2013). Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. The Journal of Cell Biology, 203, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F, McFarlane RJ, et al. (2004). S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. Journal of Cell Science, 117, 3343–3351. [DOI] [PubMed] [Google Scholar]
- Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, et al. (2002). Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science, 296, 2222–2225. [DOI] [PubMed] [Google Scholar]
- MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, & Dernburg AF (2005). Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell, 123, 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Bourc’his D, de Rooij DG, Bestor TH, Turner JM, & Burgoyne PS (2008). Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. The Journal of Cell Biology, 182, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik R, Kopylov M, Gomez-Llorente Y, Jain R, Johnson RE, Prakash L, et al. (2020). Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis. Nature Structural & Molecular Biology, 27, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, & Malone RE (1996). Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics, 144, 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, & Villeneuve AM (2005). HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes & Development, 19, 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, et al. (1998). Meiotic synapsis in the absence of recombination. Science, 279, 876–878. [DOI] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, et al. (2013). Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. Plant Cell, 25, 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Fozard JA, Hartley M, Henderson IR, Bomblies K, & Howard M (2021). Diffusion-mediated HEI10 coarsening can explain meiotic crossover positioning in Arabidopsis. Nature Communications, 12, 4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S, Altendorfer E, Saito TT, Martinez-Garcia M, & Colaiacovo MP (2021). HIM-17 regulates the position of recombination events and GSP-1/2 localization to establish short arm identity on bivalents in meiosis. Proceedings of the National Academy of Sciences of the United States of America, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, & Hollingsworth NM (2005). Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Molecular Biology of the Cell, 16, 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, et al. (2011). Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell, 146, 372–383. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Salic A, Lee E, & Kirschner MW (2001). Inhibition of Cdh1-APC by the MAD2-related protein MAD2L2: a novel mechanism for regulating Cdh1. Genes & Development, 15, 1759–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugar E, Burnett C, Chen X, & Hollingsworth NM (2017). Coordination of double strand break repair and meiotic progression in yeast by a Mek1-Ndt80 negative feedback loop. Genetics, 206, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina VB, & Vader G (2020). Homeostatic control of meiotic prophase checkpoint function by Pch2 and Hop1. Current Biology, 30(22), 4413–4424.e5. [DOI] [PubMed] [Google Scholar]
- Rinaldi VD, Bolcun-Filas E, Kogo H, Kurahashi H, & Schimenti JC (2017). The DNA damage checkpoint eliminates mouse oocytes with chromosome synapsis failure. Molecular Cell, 67(1026–1036), e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, et al. (2014). Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell, 156, 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelens B, Barroso C, Montoya A, Cutillas P, Zhang W, Woglar A, et al. (2019). Spatial regulation of polo-like kinase activity during Caenorhabditis elegans meiosis by the nucleoplasmic HAL-2/HAL-3 complex. Genetics, 213, 79–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, & Keeney S (2010). Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLoS Genet, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, & Camerini-Otero RD (2000). The mouse Spo11 gene is required for meiotic chromosome synapsis. Molecular Cell, 6, 975–987. [DOI] [PubMed] [Google Scholar]
- Rosenberg SC, & Corbett KD (2015). The multifaceted roles of the HORMA domain in cellular signaling. The Journal of Cell Biology, 211, 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera A, Vozdova M, Fernandez J, Sebestova H, Capilla L, Frohlich J, et al. (2017). Recombination correlates with synaptonemal complex length and chromatin loop size in bovids-insights into mammalian meiotic chromosomal organization. Chromosoma, 126, 615–631. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, & Franklin FC (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & Development, 21, 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo PA, & Roeder GS (1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell, 97, 313–324. [DOI] [PubMed] [Google Scholar]
- Schwacha A, & Kleckner N (1994). Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell, 76, 51–63. [DOI] [PubMed] [Google Scholar]
- Shin YH, McGuire MM, & Rajkovic A (2013). Mouse HORMAD1 is a meiosis i checkpoint protein that modulates DNA double- strand break repair during female meiosis. Biology of Reproduction, 89, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva N, Ferrandiz N, Barroso C, Tognetti S, Lightfoot J, Telecan O, et al. (2014). The fidelity of synaptonemal complex assembly is regulated by a signaling mechanism that controls early meiotic progression. Developmental Cell, 31, 503–511. [DOI] [PubMed] [Google Scholar]
- Stanzione M, Baumann M, Papanikos F, Dereli I, Lange J, Ramlal A, et al. (2016). Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nature Cell Biology, 18, 1208–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, MacQueen AJ, Vader G, Shinohara M, Sanchez A, Borde V, et al. (2016). Chromosome synapsis alleviates Mek1-dependent suppression of meiotic DNA repair. PLoS Biology, 14, e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, Zhu X, Markowitz TE, Vale-Silva LA, San-Segundo PA, Hollingsworth NM, et al. (2019). Persistent DNA-break potential near telomeres increases initiation of meiotic recombination on short chromosomes. Nature Communications, 10, 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, & Vacquier VD (2002). The rapid evolution of reproductive proteins. Nature Reviews. Genetics, 3, 137–144. [DOI] [PubMed] [Google Scholar]
- Tromer EC, van Hooff JJE, Kops G, & Snel B (2019). Mosaic origin of the eukaryotic kinetochore. Proceedings of the National Academy of Sciences of the United States of America, 116, 12873–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM (2007). Meiotic sex chromosome inactivation. Development, 134, 1823–1831. [DOI] [PubMed] [Google Scholar]
- Ur SN., & Corbett KD. (2021). Architecture and dynamics of meiotic chromosomes. Annual Review of Genetics, 55, 497–526. [DOI] [PubMed] [Google Scholar]
- Vader G (2015). Pch2(TRIP13): Controlling cell division through regulation of HORMA domains. Chromosoma, 124, 333–339. [DOI] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, & Hochwagen A (2011). Protection of repetitive DNA borders from self-induced meiotic instability. Nature, 477, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hooff JJ, Tromer E, van Wijk LM, Snel B, & Kops GJ (2017). Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Reports, 18(9), 1559–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AMV, Komives EA, & Corbett KD (2017). Conformational dynamics of the Hop1 HORMA domain reveal a common mechanism with the spindle checkpoint protein Mad2. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AS, & Holliday R (2009). The evolution of meiosis from mitosis. Genetics, 181, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, et al. (2012). Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes & Development, 26, 958–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, Boonsanay V, et al. (2009). Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. -, 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HY, & Burgess SM (2006). Two distinct surveillance mechanisms monitor meiotic chromosome metabolism in budding yeast. Current Biology, 16, 2473–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hu B, Portheine SM, Chuenban P, & Schnittger A (2020). State changes of the HORMA protein ASY1 are mediated by an interplay between its closure motif and PCH2. Nucleic Acids Research, 48, 11521–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, et al. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Current Biology, 23, 2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Kim DH, Dereli I, Rosenberg SC, Hagemann G, Herzog F, et al. (2017). The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding. The EMBO Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Lau RK, Mathews IT, Birkholz EA, Watrous JD, Azimi CS, et al. (2020). HORMA domain proteins and a Trip13-like ATPase regulate bacterial cGAS-like enzymes to mediate bacteriophage immunity. Molecular Cell, 77(709–722), e707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Rosenberg SC, Moeller A, Speir JA, Su TY, & Corbett KD (2015). TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, & Villeneuve AM (2012). COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell, 149, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Kim Y, & Dernburg AF (2016). Meiotic recombination and the crossover assurance checkpoint in Caenorhabditis elegans. Seminars in Cell & Developmental Biology, 54, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, & Alani E (2009). The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genetics, 5, e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, Sonntag Brown M, Chen C, & Alani E (2011). Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics, 188, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, & Muller F (1999). Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Development, 13, 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]


