Abstract
Biomolecular electrostatics has been a subject of computational investigations based on 3D structures. This situation is changing because emerging experimental tools allow us to quantitatively investigate biomolecular electrostatics without any use of structure information. Now, electrostatic potentials around biomolecules can directly be measured for many residues simultaneously by nuclear magnetic resonance (NMR) spectroscopy. This NMR method can be used to study electrostatic aspects of various processes, including macromolecular association and liquid-liquid phase separation. Applications to structurally flexible biomolecules such as intrinsically disordered proteins are particularly useful. The new tools also facilitate examination of theoretical models and methods for biomolecular electrostatics.
Introduction
In life, complex networks of molecular interactions involve electrostatic forces that influence structure and function of biological macromolecules. Electrostatic interactions are crucial for many biomolecular processes such as protein-nucleic acid binding, enzymatic catalysis, and liquid-liquid phase separation [1-6]. Accurate electrostatic information is also key to success in protein engineering [7] and drug design [8]. Thus, electrostatics is important for our fundamental understanding of biomolecular functions as well as for biotechnological development.
Computation of electrostatic potentials in solution from 3D structures using the Poisson-Boltzmann equation solver programs (e.g., APBS [9] and DelPhi [10]) is common in structural biology. However, their validity range should be examined more extensively because the computational method is approximate and uses empirical parameters. Structure-based assessment of electrostatics may also be challenging for structurally flexible biomolecules such as intrinsically disordered proteins (IDPs) and single-stranded RNA.
Recently, it has become possible to directly measure local electrostatic potentials for individual residues of biomolecules by nuclear magnetic resonance (NMR) spectroscopy [11] (Figure 1A). Since this method provides effective near-surface electrostatic potentials (), we referred to it as the method. This NMR method can be used to examine theoretical electrostatic models, because potentials can also be predicted from 3D structures [11-15]. Owing to the de novo nature requiring no structural information, the method also enables electrostatic investigations of conformationally disordered biomolecules [15-19]. In this minireview, we introduce the principle and applications of the method and compare it with other electrostatic methods.
Figure 1.
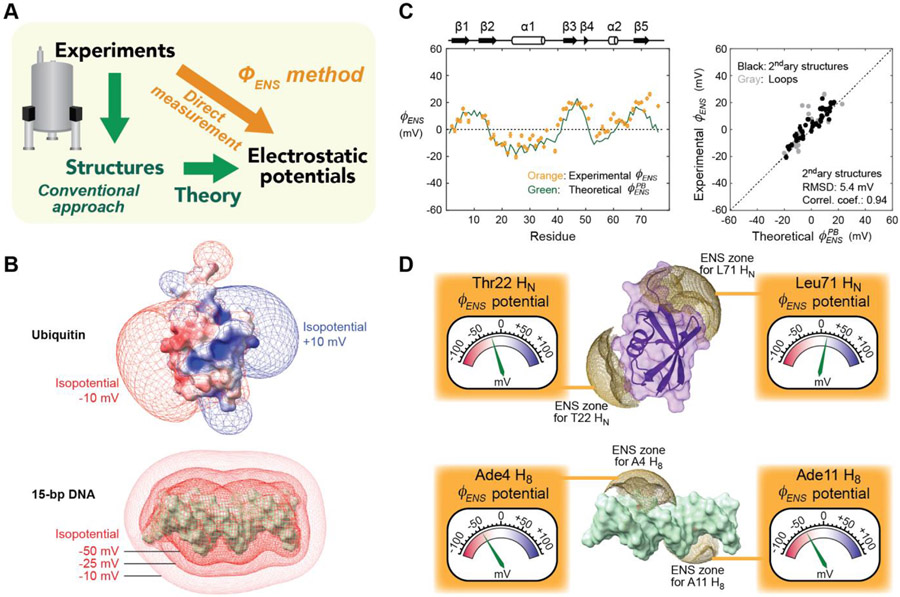
Direct measurement of electrostatic potentials by the method. (A) Conventional (green) vs. new (orange) approaches to analyze electrostatic potentials around biomolecules. (B) Electrostatic potentials around ubiquitin and 15-bp DNA computed with the Adaptive Poisson-Boltzmann Solver (APBS) software. (C) potentials measured for 1HN nuclei of ubiquitin. The experimental data are compared with predictions using Eq. 2. Adapted from Ref. [11]. (D) Physical meanings of potentials. Each potential represents an average electrostatic potential within the effective near-surface (ENS) zone (brown) for the observed 1H nucleus. Some examples for ubiquitin and 15-bp DNA are shown. The structures were drawn with ChimeraX [53]. [Double-column figure]
Challenges in computational assessment of electrostatics
In the past, biomolecular electrostatics has been a subject of computational investigations. Quantitative assessment of biomolecular electrostatics is not as straightforward as it may appear. Biomolecules in solutions are surrounded by mobile ions (e.g., Na+, K+, Cl−), which electrostatically influence the thermodynamic and kinetic properties of macromolecules [20,21]. Distribution of mobile ions around charged biomolecules are nonuniform [22,23]. Electrostatic potentials around biomolecules depend not only on their charged moieties but also on the concentrations and spatial distributions of the surrounding mobile ions. Mobile ions also cause screening, which dampens electric fields [24].
The Poisson-Boltzmann equation-based computations take mobile ions into consideration and calculate electrostatic potentials. Programs such as APBS and DelPhi compute electrostatic potentials on grid points in a sufficiently large 3D space containing a biomolecular structure [9,10] (see Figure 1B for example). The computation utilizes a continuum dielectric approximation and requires a point charge for each atom of the biomolecule. Typically, the charge of each atom in titratable groups is calculated from a specified pH and a pKa predicted from the structure. However, even advanced pKa prediction methods give root mean square errors as large as 0.8 [25,26], which may cause considerable errors in electric charges. Structural flexibility of biomolecules adds another layer of complexity in assessment of electrostatics [27].
Experimental measurement of near-surface electrostatic potentials
The method allows us to directly measure local electrostatic potentials for many residues of biomolecule (Figure 1C). This NMR method provides the effective near-surface electrostatic potentials () that represent average electrostatics in exterior space near the molecular surface close to the observed 1H nuclei. The method is useful to validate theoretical electrostatic models [11-14] and to investigate electrostatic impacts of inter- and intra-molecular interactions [11,15-17]. Table I summarizes applications of the methods for biophysical research.
Table I.
Biophysical applications of the method.
| Applications | References |
|---|---|
| • Examination of theoretical electrostatic models | 11-15 |
| • Electrostatic aspects of molecular binding | 11, 16, 17 |
| • Impact of salt on biomolecular electrostatics | 11, 13, 17 |
| • Role of electrostatics in LLPS | 16, 17 |
| • Electrostatics of disordered proteins | 15-19 |
| • Electrostatics of nucleic acids | 14 |
| • Structural ensemble | 11,15 |
Paramagnetic relaxation enhancement (PRE) arising from two analogous paramagnetic cosolutes with different charges are used to determine potentials. PRE arising from paramagnetic cosolutes has been referred to as solvent PRE [28,29]. Solvent PRE rates () for 1H transverse magnetizations can readily be measured with various methods using paramagnetic and corresponding non-paramagnetic samples [18,29]. The potential is defined as [11]:
| [1] |
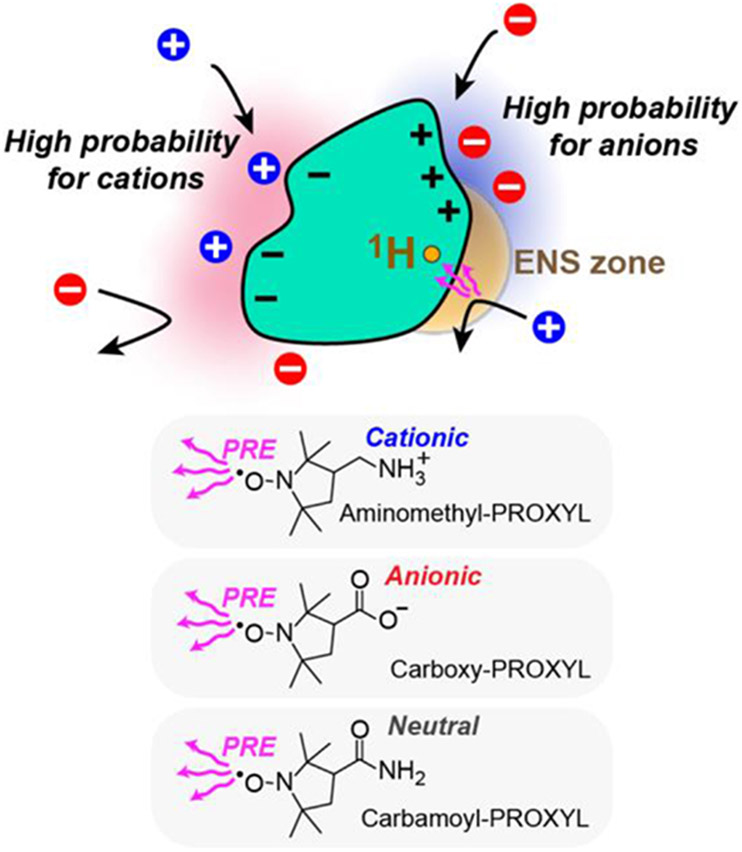
where and are the solvent PRE rates for the two paramagnetic cosolutes a and b at an identical concentration; and are their charge valence; is the elementary charge; is the Boltzmann constant; and is temperature. The spatial distributions of the charged cosolutes around a biomolecule are related to local electrostatic potentials. This allows one to obtain electrostatic information. The PRE rates are governed by the paramagnetic cosolutes within a zone proximal to the observed 1H nucleus, which we refer to as the effective near-surface (ENS) zone. The potential represents an average electrostatic potential within the ENS zone [11] (Figure 1D).
PROXYL or TEMPO derivatives have been used in the methods [11-19]. For example, aminomethyl-PROXYL, carboxy-PROXYL, and carbamoyl-PROXYL (, −1, and 0, respectively, at neutral pH) were used (Figure 2). Combination of positively and negatively charged paramagnetic probes is ideal for accurate measurements [19]. For strongly charged systems (e.g., nucleic acids), PRE arising from one of the charged probes may be too small for determination. In such a case, a pair of analogous neutral and charged probes can be used [14,15,18]. Tetramethyl nitroxide is known to interact with hydrophobic surfaces of proteins [30-32]. If the hydrophobic or other non-electrostatic interactions are the same for the two analogous cosolutes, the contributions of such interactions are canceled in the ratio [11,13]. Consequently, in Eq. 1 illuminates electrostatic components.
Figure 2.
Examples of paramagnetic cosolutes for the method. It is crucial that electrostatic interactions between the biomolecule and the cosolutes governs differences in the spatial distributions of the cosolutes around the biomolecule. Hydrophobic interactions at the tetramethyl nitroxide moiety common to the cosolutes does not affect the method [11,13]. [Single-column figure]
Prediction of potentials from 3D structure
A convenient feature of potential data is that they can be predicted from 3D structures. Owing to this feature, data are useful for evaluation of theoretical models and methods (see Figure 1C for example). A simple way to predict potential is to use the following equation together with electrostatic potentials at individual grid points around the molecule [11,12]:
| [2] |
in which is the distance from the 1H nucleus to the grid point ; and is a factor that represents the accessibility of the grid point and is either 1 (accessible) or 0 (inaccessible). Predictions with Eq. 2 typically yield the root mean squared difference of ~5 mV compared with the experimental potentials [11,12,14].
Eq. 2 assumes the same spatial distribution for the charge and the unpaired electron. This assumption may be simplistic because the charged moiety and the unpaired electron of the PROXYL derivatives are on the opposite sides of the molecules. Angular correlations for the paramagnetic probes with respective to the biomolecule are not taken into account in Eq. 2 either. Chen et al. demonstrated that potentials can be predicted more accurately by using atomistic models of the paramagnetic cosolutes along with appropriate treatment of the angular correlations [13], which is computationally more expensive.
Applications to intrinsically disordered proteins
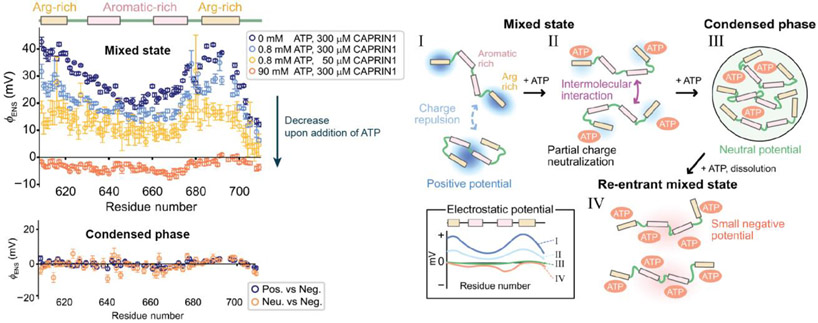
Because it does not require structural information, the method is particularly useful for electrostatic investigations of IDPs. Liquid-liquid phase separation (LLPS) play various regulatory roles in biology [33]. Recently, Kay and coworkers applied the method to map per-residue surface electrostatic potential of the CAPRIN1 protein along its trajectory of LLPS [16,17] (Figure 3). In the absence of ATP, where this IDP was not phase-separated, potentials around the Arg-rich regions were positive. However, as ATP was added and the protein transitioned into phase separation, potentials of the region decreased and became close to zero [16]. The decrease in potentials reflect electrostatic interactions between the positively charged Arg-rich region and the negatively charged ATP molecules. When the ATP concentration was as high as 90 mM, the potentials became even negative, leading to re-entrance into a mixed state. The studies on CAPRIN1 demonstrated the effectiveness of the method for investigations of LLPS. Electrostatics are crucial for LLPS of some proteins. For example, co-partitioning of transcription regulators through LLPS is governed by patterned charge blocks in IDRs [34]. We anticipate that the method will reveal more about the electrostatic aspects of LLPS.
Figure 3.
Mapping of potentials along the adenosine triphosphate (ATP)-induced phase-separation trajectory for CAPRIN1 [16]. As ATP was added, the potentials around CAPRIN1 progressively decreased. Upon phase separation, the potentials became close to zero, with ~5 ATP molecules associated with each CAPRIN1 chain. Increasing the ATP concentration further inverted the potentials, leading to re-entrance into a mixed phase. Adapted from Ref. 16. [Double-column figure]
Clore and coworkers applied the method to both the folded and unfolded states of the drkN SH3 domain [15]. The variation of potentials among different residues was smaller in the unfolded state, presumably due to averaging of various structures. Using snapshots of a replica-exchange molecular dynamics (MD) trajectory on the unfolded state of this protein [35], potentials were predicted and found to agree well with experimental potentials. This seems to support the validity of the MD ensemble for the unfolded state. In conjunction with other experimental data (e.g., NMR chemical shifts, residual dipolar couplings), potential data may facilitate experimental assessment of structural ensembles of IDPs/IDRs.
Advantages over other methods for electrostatic potentials
Electron-electron double resonance (ELDOR) can also provide near-surface electrostatic potentials using an extrinsic probe attached to a biomolecule in the presence of charged or neutral paramagnetic cosolutes [36]. Cysteine modification kinetics or diffusion-enhanced fluorescence energy transfer data have also been used to estimate electrostatic potentials [37,38]. However, these methods provide only one electrostatic potential for each sample, requiring preparation of multiple samples to measure electrostatic potentials at multiple sites. In contrast, the method provides electrostatic potentials at many sites simultaneously. For example, potentials were measured for > 150 different sites for ubiquitin, a 76-residue protein [11,12]. Furthermore, the method does not require any chemical modification of biomolecules.
In principle, cryogenic electron microscopy (cryo-EM) can directly provide electrostatic potential relevant to atomic scattering factor [39,40]. However, cryo-EM electrostatic potential maps do not provide quantitative information of electrostatic potentials in the exterior space around the biomolecules. Radiation damage also makes it practically difficult to quantitatively analyze cryo-EM electrostatic potential maps [39]. In contrast, the method provides quantitative information of electrostatic potentials in exterior space around the biomolecules in solutions under physiological conditions. Thus, the method is currently the most powerful experimental method for quantitative investigations of electrostatic potentials.
Comparison with vibrational spectroscopy methods for electric fields
Vibrational Stark effect (VSE) spectroscopy has been used to investigate external electric fields that perturb the energies for vibrational transitions of covalent bonds [41]. Since covalent bonds that exhibit unique infrared (IR) signals are desirable for VSE spectroscopy, nitrile C≡N or aldehyde/ketone C=O bonds, which are absent in natural proteins, are often introduced through chemical modifications, amber suppression, or ligand binding. Site-specific 13C-labeling and 12C ─ 13C difference spectra were also used to analyze the VSE for specific sites [42,43]. For aldehyde groups, electric field orientations can be extracted using two directional vibrational probes by exploiting the VSE of C=O and C-D bonds [44]. IR signal intensities of nitrile C≡N bonds were also found to be useful for analyzing electric fields [45]. Using VSE spectroscopy along with enzyme kinetics for wild-type and mutant enzymes, Boxer and coworkers demonstrated the role of electrostatics in enzymatic catalysis [43,46,47].
Information from VSE data differs from that from data. VSE data provide electric fields at the observed bonds inside a molecule of interest, whereas data provide average electrostatic potentials in a local exterior space close to the observed 1H nuclei. The electric field F is the gradient (∂/∂x, ∂/∂y, ∂/∂z) of the electrostatic potential multiplied by −1. With a charge valence z, the electric field gives the electric force zeF, whereas the electrostatic potential gives the electrostatic energy . The VSE and methods are complementary regarding biomolecular electrostatics.
Conclusions
Recent methodological advances have enabled direct electrostatic measurements for many sites on biological macromolecules through experiments. Direct measurements of electrostatic potentials facilitate investigations of inter- or intra-molecular electrostatic interactions, particularly for those involving IDPs/IDRs. Electrostatic interactions involving IDRs impact thermodynamic and kinetic properties of some proteins [48-52]. Direct measurements of electrostatics may greatly facilitate quantitative investigations of such electrostatic interactions. The emerging experimental tools for electrostatics can be applied to a wide variety of biomolecular processes such as electrostatic steering, post-translational modifications, and co-partitioning in LLPS. Experimental measurements of electrostatic potentials also allow for examination of theoretical electrostatic methods.
Acknowledgements
This work was supported by Grants R35-GM130326 (to J.I.) and R01-GM037657 (to B.M.P.) from the National Institutes of Health and by Grants H-2104-20220331 (to J.I.) and H-013 (to B.M.P.) from the Welch Foundation. We thank Drs. Chuanying Chen, Michael Sherman, and Razie Yousefi for useful discussions.
References
- 1.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM: Electrostatic Basis for Enzyme Catalysis. Chemical Reviews 2006, 106:3210–3235. [DOI] [PubMed] [Google Scholar]
- 2.Fried SD, Boxer SG: Electric Fields and Enzyme Catalysis. Annu Rev Biochem 2017, 86:387–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu B, Pettitt BM, Iwahara J: Dynamics of ionic interactions at protein–nucleic acid interfaces. Acc Chem Res 2020, 53:1802–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Privalov PL, Dragan AI, Crane-Robinson C: Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res 2011, 39:2483–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou HX, Pang X: Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem Rev 2018, 118:1691–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honig B, Nicholls A: Classical electrostatics in biology and chemistry. Science 1995, 268:1144–1149. [DOI] [PubMed] [Google Scholar]
- 7.Vizcarra CL, Mayo SL: Electrostatics in computational protein design. Curr Opin Chem Biol 2005, 9:622–626. [DOI] [PubMed] [Google Scholar]
- 8.Anderson AC: The process of structure-based drug design. Chem Biol 2003, 10:787–797. [DOI] [PubMed] [Google Scholar]
- 9.Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, Brookes DH, Wilson L, Chen J, Liles K, et al. : Improvements to the APBS biomolecular solvation software suite. Protein Sci 2018, 27:112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Jia Z, Chakravorty A, Pahari S, Peng Y, Basu S, Koirala M, Panday SK, Petukh M, Li L, et al. : DelPhi Suite: New Developments and Review of Functionalities. J Comput Chem 2019, 40:2502–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••11. Yu B, Pletka CC, Pettitt BM, Iwahara J: De novo determination of near-surface electrostatic potentials by NMR. Proc Natl Acad Sci U S A 2021, 118:e2104020118. This is the first paper on the method. Experimental potential data for ubiquitin and a protein-DNA complex were used to examine the validity of the Poisson-Boltzmann theory.
- 12.Yu B, Pletka CC, Iwahara J: Protein Electrostatics Investigated through Paramagnetic NMR for Nonpolar Groups. J Phys Chem B 2022, 126:2196–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •13. Chen C, Yu B, Yousefi R, Iwahara J, Pettitt BM: Assessment of the Components of the Electrostatic Potential of Proteins in Solution: Comparing Experiment and Theory. J Phys Chem B 2022, 126:4543–4554. This paper demonstrates that structure-based prediction of potentials can be improved by considering angular correlation between the paramagnetic probes and the biomolecule. The validity range was examined for a wide range of ionic strength (up to 735 mM).
- 14.Yu B, Wang X, Iwahara J: Measuring Local Electrostatic Potentials Around Nucleic Acids by Paramagnetic NMR Spectroscopy. J Phys Chem Lett 2022, 13:10025–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •15. Okuno Y, Schwieters CD, Yang Z, Clore GM: Theory and Applications of Nitroxide-based Paramagnetic Cosolutes for Probing Intermolecular and Electrostatic Interactions on Protein Surfaces. J Am Chem Soc 2022, 144:21371–21388. potentials were analyzed for the folded and unfolded states of the drkN SH3 domain.
- ••16. Toyama Y, Rangadurai AK, Forman-Kay JD, Kay LE: Mapping the per-residue surface electrostatic potential of CAPRIN1 along its phase-separation trajectory. Proc Natl Acad Sci U S A 2022, 119:e2210492119. The authors demonstrate elegant applications of the method to phase separation of an IDP.
- 17.Toyama Y, Rangadurai AK, Forman-Kay JD, Kay LE: Surface electrostatics dictate RNA-binding protein CAPRIN1 condensate concentration and hydrodynamic properties. J Biol Chem 2022, 299:102776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyama Y, Rangadurai AK, Kay LE: Measurement of 1Hα transverse relaxation rates in proteins: application to solvent PREs. J Biomol NMR 2022, 76:137–152. [DOI] [PubMed] [Google Scholar]
- •19. Kaushik Rangadurai A, Toyama Y, Kay LE: Practical considerations for the measurement of near-surface electrostatics based on solvent paramagnetic relaxation enhancements. J Magn Reson 2023, 349:107400. Some different combinations of paramagnetic probes were tested for the method.
- 20.Lipfert J, Doniach S, Das R, Herschlag D: Understanding nucleic acid-ion interactions. Annu Rev Biochem 2014, 83:813–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu B, Iwahara J: Experimental approaches for investigating ion atmospheres around nucleic acids and proteins. Comput Struct Biotechnol J 2021, 19:2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He W, Chen YL, Pollack L, Kirmizialtin S: The structural plasticity of nucleic acid duplexes revealed by WAXS and MD. Sci Adv 2021, 7:eabf6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu B, Pletka CC, Iwahara J: Quantifying and visualizing weak interactions between anions and proteins. Proc Natl Acad Sci U S A 2021, 118:e2015879118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chazalviel JN: Coulomb screening by mobile charges: Application to materials science, chemistry, and biology. New York: Springer; 1999. [Google Scholar]
- 25.Coskun D, Chen W, Clark AJ, Lu C, Harder ED, Wang L, Friesner RA, Miller EB: Reliable and Accurate Prediction of Single-Residue pKa Values through Free Energy Perturbation Calculations. J Chem Theory Comput 2022, 18:7193–7204. [DOI] [PubMed] [Google Scholar]
- 26.Stanton CL, Houk KN: Benchmarking pKa Prediction Methods for Residues in Proteins. J Chem Theory Comput 2008, 4:951–966. [DOI] [PubMed] [Google Scholar]
- 27.Georgescu RE, Alexov EG, Gunner MR: Combining Conformational Flexibility and Continuum Electrostatics for Calculating pKas in Proteins. Biophys J 2002, 83:1731–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenard AJ, Mulder FAA, Madl T: Solvent paramagnetic relaxation enhancement as a versatile method for studying structure and dynamics of biomolecular systems. Prog Nucl Magn Reson Spectrosc 2022, 132-133:113–139. [DOI] [PubMed] [Google Scholar]
- 29.Clore GM, Iwahara J: Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 2009, 109:4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••30. Okuno Y, Szabo A, Clore GM: Quantitative Interpretation of Solvent Paramagnetic Relaxation for Probing Protein-Cosolute Interactions. J Am Chem Soc 2020, 142:8281–8290. The theory described in this paper laid the foundation for the method.
- 31.Petros AM, Mueller L, Kopple KD: NMR identification of protein surfaces using paramagnetic probes. Biochemistry 1990, 29:10041–10048. [DOI] [PubMed] [Google Scholar]
- 32.Pintacuda G, Otting G: Identification of protein surfaces by NMR measurements with a pramagnetic Gd(III) chelate. J Am Chem Soc 2002, 124:372–373. [DOI] [PubMed] [Google Scholar]
- 33.Alberti S, Gladfelter A, Mittag T: Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons H, Veettil RT, Pradhan P, Fornero C, De La Cruz N, Ito K, Eppert M, Roeder RG, Sabari BR: Functional partitioning of transcriptional regulators by patterned charge blocks. Cell 2023, 186:327–345.e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuno Y, Yoo J, Schwieters CD, Best RB, Chung HS, Clore GM: Atomic view of cosolute-induced protein denaturation probed by NMR solvent paramagnetic relaxation enhancement. Proc Natl Acad Sci U S A 2021, 118:e2112021118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin YK, Hubbell WL: Determination of electrostatic potentials at biological interfaces using electron-electron double resonance. Biophys J 1992, 61:1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu J, Kobertz WR, Deutsch C: Mapping the Electrostatic Potential within the Ribosomal Exit Tunnel. J Mol Biol 2007, 371:1378–1391. [DOI] [PubMed] [Google Scholar]
- 38.Meltzer RH, Lurtz MM, Wensel TG, Pedersen SE: Nicotinic acetylcholine receptor channel electrostatics determined by diffusion-enhanced luminescence energy transfer. Biophys J 2006, 91:1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marques MA, Purdy MD, Yeager M: CryoEM maps are full of potential. Curr Opin Struct Biol 2019, 58:214–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J: Experimental charge density from electron microscopic maps. Protein Sci 2017, 26:1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fried SD, Boxer SG: Measuring Electric Fields and Noncovalent Interactions Using the Vibrational Stark Effect. Acc Chem Res 2015, 48:998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider SH, Kozuch J, Boxer SG: The Interplay of Electrostatics and Chemical Positioning in the Evolution of Antibiotic Resistance in TEM beta-Lactamases. ACS Cent Sci 2021, 7:1996–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43. Ji Z, Kozuch J, Mathews II, Diercks CS, Shamsudin Y, Schulz MA, Boxer SG: Protein Electric Fields Enable Faster and Longer-Lasting Covalent Inhibition of beta-Lactamases. J Am Chem Soc 2022, 144:20947–20954. The vibrational Stark effect was analyzed using 12C-13C difference spectra to reveal the role of electric fields in covalent inhibition of β-lactamase.
- ••44. Zheng C, Mao Y, Kozuch J, Atsango AO, Ji Z, Markland TE, Boxer SG: A two-directional vibrational probe reveals different electric field orientations in solution and an enzyme active site. Nat Chem 2022, 14:891–897. The first demonstration of experimental determination of electric field orientations using aldehyde C=O and C-D bonds.
- 45.Weaver JB, Kozuch J, Kirsh JM, Boxer SG: Nitrile Infrared Intensities Characterize Electric Fields and Hydrogen Bonding in Protic, Aprotic, and Protein Environments. J Am Chem Soc 2022, 144:7562–7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fried SD, Bagchi S, Boxer SG: Extreme electric fields power catalysis in the active site of ketosteroid isomerase. Science 2014, 346:1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ji Z, Boxer SG: beta-Lactamases Evolve against Antibiotics by Acquiring Large Active-Site Electric Fields. J Am Chem Soc 2022, 144:22289–22294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, et al. : Extreme disorder in an ultrahigh-affinity protein complex. Nature 2018, 555:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Greenblatt HM, Bigman LS, Yu B, Pletka CC, Levy Y, Iwahara J: Dynamic Autoinhibition of the HMGB1 Protein via Electrostatic Fuzzy Interactions of Intrinsically Disordered Regions. J Mol Biol 2021, 433:167122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zaharias S, Zhang Z, Davis K, Fargason T, Cashman D, Yu T, Zhang J: Intrinsically disordered electronegative clusters improve stability and binding specificity of RNA-binding proteins. J Biol Chem 2021, 297:100945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schütz S, Bergsdorf C, Goretzki B, Lingel A, Renatus M, Gossert AD, Jahnke W: The Disordered MAX N-terminus Modulates DNA Binding of the Transcription Factor MYC:MAX. J Mol Biol 2022, 434:167833. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Bigman LS, Greenblatt HM, Yu B, Levy Y, Iwahara J: Negatively charged, intrinsically disordered regions can accelerate target search by DNA-binding proteins. Nucleic Acids Res 2023, 51:4701–4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE: UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 2018, 27:14–25. [DOI] [PMC free article] [PubMed] [Google Scholar]