Abstract
Two regions of the EBNA-3A protein of Epstein-Barr virus were shown to be capable of binding to the cell protein RBP-Jk (also known as CBF-1), a component of the Notch signaling pathway. Consistent with this binding, EBNA-3A inhibited reporter gene expression from plasmids containing RBP-Jk DNA binding sites within their promoters, including the Cp promoter. When EBNA-3A was linked to a GAL4 DNA binding domain, it repressed the activity of a promoter containing GAL4 binding sites at all plasmid concentrations tested. However, a deletion mutant of EBNA-3A lacking amino acids 100 to 364 showed a biphasic response in the GAL4 assay: it inhibited transcription at low DNA concentrations but activated it at high DNA concentrations. There appears to be a gene activation function within EBNA-3A that is masked in the full-length protein in this assay. Current models for EBNA-3 function have stressed transcription repression through binding to RBP-Jk, but we consider an alternative scheme in which the role of the binding of EBNA-3A, -3B, and -3C to RBP-Jk is to buffer the levels of active EBNA-3 protein. We have also found that the behavior of EBNA-3A in a cell fractionation procedure that distinguishes insoluble matrix from soluble cell fractions is modified by EBNA-LP, indicating a further novel level of interplay between the EBNA proteins.
Six viral genes have been shown to be required for efficient transformation of B lymphocytes by Epstein-Barr virus (EBV). These produce the nuclear proteins EBNA-1, -2, -3A, -3C, and -LP and the membrane protein LMP-1 (reviewed in reference 25). The timing of expression of the transforming genes and the levels of the viral proteins are coordinated; this fact is apparent both in the organization of the viral genome and in the strategy used to express the viral genes. This coordination is thought to reflect the complexity of driving infected resting B cells into the cell cycle and subsequently maintaining their growth without causing apoptosis. Also, some of the viral proteins (e.g., LMP-1) are toxic if they are expressed at too high a level (14). The interactions and coordination of activity of the transforming genes are thus important issues in EBV transformation, along with the biochemical functions of their protein products.
EBNA-3A is essential for EBV transformation of B lymphocytes (50) but may only be required during the establishment of the immortalized cell line (23). It is a nuclear phosphoprotein with a calculated protein molecular mass of 103 kDa but migrates on sodium dodecyl sulfate (SDS) gels at about 145 kDa (16, 22, 38). The coding exons of the EBNA-3A, -3B, and -3C genes are adjacent on the viral genome, and the three genes are similar in their exon structure and in the N-terminal parts of their coding sequences (10).
The mechanism of action of EBNA-3A in viral transformation is not known, but the discovery that the EBNA-3A, -3B, and -3C proteins can all bind to the cell DNA binding protein RBP-Jk, also known as CBF1, has greatly advanced analysis of their functions (28, 39, 40, 52, 54). Of the three EBNA-3 proteins, EBNA-3A has been reported to show the weakest binding to RBP-Jk in vitro (40, 52, 54). The region of EBNA-3A that binds to RBP-Jk has been mapped in two different studies (40, 54). There is a discordance in the published data, since one study concluded that amino acids 1 to 138 of EBNA-3A are sufficient (40) and the other mapped the binding site to amino acids 1 to 223, with residues between positions 172 and 223 being essential (54).
RBP-Jk is also bound by EBNA-2, and part of the activation of gene expression caused by EBNA-2 is mediated through its interaction with RBP-Jk, which provides the specific DNA binding function absent in EBNA-2 (reviewed in reference 9). EBNA-3A, -3B, and -3C binding to RBP-Jk has been reported to prevent the binding of RBP-Jk to its target DNA sequence in vitro (39, 40, 52, 54). The same general model has been proposed for all of the EBNA-3 proteins, in which they negatively modulate the transcription activation caused by EBNA-2; the EBNA-3 proteins are thus seen as antagonistic to the activation of Notch pathway gene expression that is mediated by EBNA-2 through RBP-Jk (20, 29, 40, 52, 54). Measurement of the amounts of RBP-Jk complexed with EBNA-2, -3A, -3B, and -3C has shown that significant proportions of RBP-Jk in cells are complexed with each of the proteins (20). The inhibition of EBNA-2-activated transcription would modulate cell and viral promoters dependent on EBNA-2, including the EBV Cp promoter, thus resulting in a feedback loop of inhibition by EBNA-3 proteins of their own synthesis.
The EBNA-LP protein (also known as EBNA-5) cooperates with EBNA-2 to activate gene expression (15, 34, 45), but the mechanism by which this cooperative activation occurs is not yet understood. Early in infection, EBNA-LP is expressed at a very high level, but as the immortalized cells grow, the EBNA-LP protein level subsides. EBNA-LP is a phosphoprotein (36) whose phosphorylation is altered at different stages of the cell cycle (26). In newly infected cells, EBNA-LP is diffusely present in the nucleoplasm (48, 49), but in established lymphoblastoid cell lines (LCLs), the EBNA-LP protein is located at discrete sites in the nucleus (36) which are identical to the sites at which the PML protein is found, now known as ND10 domains (48). These represent a nuclear compartment in which several proteins have been found to be located, including PML and a specific fraction of the Rb protein in the cell (18, 19). In transfected cells, EBNA-LP is present both diffusely in the nucleoplasm and in nuclear bodies, but these sites are not the ND10 domains (48). EBNA-LP has also been found to associate with Hsp70 (27, 32) and has been shown to be attached to the nuclear matrix in LCLs (36). EBNA-3A is also located at specific sites in the nucleus, but these seem to be different from the ND10 domains at which EBNA-LP is found in established LCLs (36).
In this paper, we analyze further the binding of EBNA-3A to RBP-Jk and show that two independent segments of the EBNA-3A protein are both able to bind in vitro to RBP-Jk. We also show that cotransfection of EBNA-LP is able to modulate the distribution of EBNA-3A, altering its association with the nuclear matrix cell fraction. Deletion analysis of EBNA-3A with GAL4 fusion proteins has revealed a novel transcription activation function for EBNA-3A, perhaps analogous to that reported previously for EBNA-3C (33), although there is no substantial sequence homology between the two proteins in the parts involved in transcription activation. Consideration of the transcriptional properties of the EBNA-3A protein has led us to consider an alternative interpretation of the significance of the binding of EBNA-3 proteins to RBP-Jk.
MATERIALS AND METHODS
Cell cultures.
The EBV-negative human Burkitt lymphoma-derived cell line DG75 (4) was grown at 37°C in RPMI 1640 medium supplemented with 15% (vol/vol) heat-inactivated fetal calf serum, 2 mM glutamine, and 100 U each of penicillin and streptomycin per ml.
Human HeLa and 293 epithelial cell lines were grown at 37°C in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) heat-inactivated fetal calf serum, 2 mM glutamine, and 100 U each of penicillin and streptomycin per ml.
Reporter plasmids.
pBLCAT2 contains the herpes simplex virus thymidine kinase promoter upstream of the chloramphenicol acetyltransferase (CAT) gene (30), and pUASCAT contains five copies of the GAL4 DNA binding sequence (upstream activating sequence) upstream of the thymidine kinase promoter in the pBLCAT2 vector (53). The plasmid used for normalization of transfection efficiency, pCMV19βGal, has the β-galactosidase gene under the control of the cytomegalovirus (CMV) immediate-early promoter.
p1.4Cp-CAT encodes the CAT gene under the control of the B95-8 Cp region (B95-8 EBV nucleotides 9911 to 11336) (41), and p1.4Cp-CAT E2RE mut is similar but has a 5-nucleotide mutation of the RBP-Jk binding site (7).
The reporter plasmids 4Jk-TK-CAT wt and E2RE mut were previously described (identical to pTK-CAT-Cp4x and pTK-CAT-CpM4x, respectively, of Waltzer et al. [51]). They contain four wild-type or mutant copies, respectively, of a double-stranded oligonucleotide containing the RBP-Jk binding site of the Cp promoter cloned into pBLCAT2. They were kind gifts from Evelyne Manet and Alain Sergeant.
GAL4 plasmids.
pGAL4-E3A(1-944) was constructed by cloning into a GAL4 vector (see below) the EBNA-3A gene isolated by Pfu PCR from cosmid p23 (13) with primers TTTGTTGGATCCACCATGGACAAGGACAGGCCG and CACACTTCTAGACCTTTGCTTAGGCCTCA. The GAL4 vector used for this procedure was derived from pBS(RSV)GAL4-E2F1(368-437) (21) by removal of its E2F1 fragment by BamHI-XbaI digestion, leaving sites into which the PCR fragment could be cloned. pGAL4-E3A(Δ100-364) was prepared by elimination of a BglII fragment (B95-8 EBV nucleotides 92539 to 93422) in pGAL4-E3A(1-944). Control vector pGAL4(1-147) has been described elsewhere (3).
EBNA expression plasmids.
pSNOC-LP, in which the expression of the EBNA-LP gene is driven by the CMV immediate-early promoter in the pSNOC vector, was described previously (1). pSNOC-E3A was constructed by Pfu PCR from B95-8 cosmid p23 with primers GCCAGAGTCGACATGGACAAGGACAGGCCG and CACACTTCTAGACCTTTGCTTAGGCCTCA and cloned as a SalI-XbaI fragment into pSNOC. The p294-EBNA-1 plasmid, in which EBNA-1 is expressed from the CMV immediate-early promoter, was a kind gift from Wolfgang Hammerschmidt. The pSV-EBNA-2 expression plasmid (8) contains a BglII-NotI fragment of the BamHI WYH region (B95-8 EBV nucleotides 44664 to 50628) cloned into pSV2-gpt and was a kind gift from Lars Rymo.
EBNA-3A mutant plasmids and glutathione S-transferase (GST) fusion plasmids.
pSP72-E3A(1-944) was constructed by subcloning of the EBNA-3A T2 cDNA clone (5) between the SalI and EcoRI sites of pSP72. pSP72-E3A(Δ100-364) was prepared by elimination of a BglII fragment (B95-8 EBV nucleotides 92539 to 93422) from pSP72-E3A(1-944), and pSP72-E3A(1-524) was similarly prepared by deletion of a StuI fragment (B95-8 EBV nucleotides 93900 to 95160).
Further deletion mutants were prepared by truncation of pSP72-E3A(1-944) with restriction enzymes and recircularization of the blunt-ended fragments: pSP72-E3A(1-277) with EcoRI (B95-8 EBV nucleotide 93162), pSP72-E3A(1-224) with SmaI (B95-8 EBV nucleotide 93002), pSP72-E2A(1-172) with BstEII (B95-8 EBV nucleotide 92847), pSP72-E3A(1-138) with NcoI (B95-8 EBV nucleotide 92743), pSP72-E3A(1-124) with BamHI (B95-8 EBV nucleotide 92703), and pSP72-E3A(1-99) with BglII (B95-8 EBV nucleotide 92539).
EBNA-3A mutants lacking the N terminus of EBNA-3A were cloned in vector cDNA3HA, a kind gift from Eric Lam. cDNA3HA-E3A(124-364) was prepared by cloning the BamHI-BglII fragment (B95-8 EBV nucleotides 92703 to 93422) from pSP72-E3A into cDNA3HA at the BamHI site. Similarly, cDNA3HA-E3A(622-826) and cDNA3HA-E3A(500-944) were prepared by cloning a HincII fragment (B95-8 EBV nucleotides 94195 to 94808) and ApaLI-EcoRV fragments (B95-8 EBV nucleotides 93826 to 95239), respectively, from pSP72-E3A into the EcoRV site of cDNA3HA. cDNA3HA-E3A(224-566) was prepared by cloning the SmaI fragment (B95-8 EBV nucleotides 93002 to 94028) from pSP72-E3A into the blunted XhoI site of cDNA3HA.
The GST–RBP-Jk plasmid was kindly supplied by Diane Hayward, and the empty vector pGEX-3X was purchased from Pharmacia. All plasmids were purified by anion-exchange chromatography (Qiagen, Inc.).
Transient transfection assays.
DG75 cells were transfected by electroporation (3). HeLa and 293 cells were transfected by calcium phosphate coprecipitation essentially as described previously (12). In all cases, the total amount of effector DNA in each transfection remained constant by making up the difference with the appropriate empty vector, pGAL4(1-147) or pSNOC. A constant amount of pCMV19-βGal was cotransfected as an internal control (2 μg in DG75 and HeLa cells and 0.5 μg in 293 cells).
Cells were harvested 40 to 48 h after transfection, and lysates were prepared by three cycles of freezing-thawing. The β-galactosidase activity of each sample was determined by use of chlorophenol red–β-d-galactopyranoside as a substrate (Boehringer). For CAT assays, a sample of lysate was incubated with [14C]acetyl coenzyme A and chloramphenicol, the reaction mixture was extracted with ethyl acetate, and the organic phase was added to scintillation fluid prior to counting (46). All counts were normalized to β-galactosidase activity. All transfections were performed at least three times with different batches of plasmid DNA.
GST fusion pull-down assays.
GST and the fusion protein GST–RBP-Jk were purified as described elsewhere (3). Coupled transcription-translation reactions were carried out with the TNT system (Promega); labelling was done with [35S]methionine. In vitro-synthesized proteins were adsorbed to GST-coated beads for 1 h by rotation at 4°C in 100 μl of EBC buffer (140 mM NaCl, 0.5% Nonidet P-40 [NP-40], 50 mM Tris-Cl [pH 8.0], 0.5 mM phenylmethylsulfonyl fluoride, 2 mM dithiothreitol, 5 μg of aprotinin per ml, 1 mg of bovine serum albumin per ml). The beads were sedimented by centrifugation, and the cleared supernatant was incubated with GST or GST–RBP-Jk protein on beads in a total volume of 200 μl at 4 or 30°C for 30 min. The beads were washed three times with 1 ml of NETN buffer (300 mM NaCl, 1 mM EDTA, 0.5% NP-40, 20 mM Tris-Cl [pH 8.0]). The beads were heated at 95°C for 5 min in SDS sample buffer (45), and bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography. Results were quantified by densitometry of autoradiographs or PhosphorImager analysis.
Fractionation and immunoblotting.
DG75 cells were harvested 48 h after transfection, washed twice with cold phosphate-buffered saline, and resuspended in RIPA buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, 100 μg of phenylmethylsulfonyl fluoride per ml, 5 μg of aprotinin per ml). They were left on ice for 30 min. The cells were further disrupted by sonication as needed. The lysates were centrifugated for 5 min at 4°C. The pellets were resuspended in SDS sample buffer and heated at 95°C for 5 min. The supernatants were diluted in SDS sample buffer and heated at 95°C for 5 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to Protan nitrocellulose (Schleicher & Schuell), and blocked with 5% dried milk in TBST (150 mM NaCl, 10 mM Tris-Cl [pH 7.4], 0.05% Tween 20) for 1 h at room temperature. Antibodies were applied overnight at 4°C with 5% dried milk in TBST. The antibodies used were mouse monoclonal antibody JF186 to EBNA-LP (11) diluted 1:10, mouse monoclonal antibody T2.78 to EBNA-3A (42) diluted 1:10, mouse monoclonal antibody sc-510 to GAL4 (Santa Cruz Biotechnology) diluted 1:500, mouse monoclonal antibody PE-2 to EBNA-2 (Dako) diluted 1:500, and mouse monoclonal antibody EBNA-OT1x to EBNA-1 (6) diluted 1:1,500. After extensive washing, the secondary antibody, a horseradish peroxidase-conjugated sheep anti-mouse antibody (Amersham) diluted 1:2,500, was applied for 30 min at 4°C. Detection was performed with enhanced chemiluminescence analysis (Amersham).
RESULTS
Two regions of EBNA-3A bind to RBP-Jk.
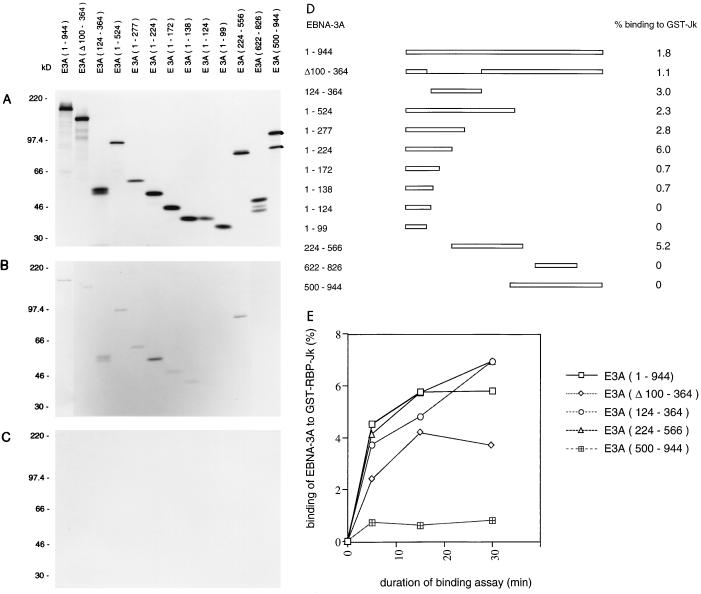
The previously reported deletion analyses of EBNA-3A binding to RBP-Jk (40, 54) used sequential deletion mutations from the C terminus of the protein. They did not consider the possibility that there might be more than one RBP-Jk binding site within EBNA-3A. There is also a discordance in the published data on the boundaries of the RBP-Jk binding segment of EBNA-3A (40, 54). We independently prepared a series of deletion mutants of EBNA-3A and assayed their ability to associate with RBP-Jk. In these experiments, the EBNA-3A protein was labelled with [35S]methionine by in vitro translation, and RBP-Jk was isolated as a fusion protein with GST. The proportion of input EBNA-3A that bound to GST–RBP-Jk was estimated with the binding of the same EBNA-3A protein to GST as a negative control (Fig. 1A to C). Consistent with earlier reports, the results (Fig. 1D) showed that truncation of EBNA-3A from the C terminus increased the proportion of the protein which bound to a fixed amount of GST–RBP-Jk (for example, mutant 1-277 bound better than 1-944). Deletion analysis of the remaining part of EBNA-3A showed that two regions of EBNA-3A were competent to bind GST-RBP-Jk (mutants 1-224 and 224-566 both bound efficiently). Further truncation of each of these regions indicated that the more N-terminal binding region, which lies within residues 1 to 224, suffered a partial loss of activity as it was truncated to 172 and 138 and then a complete loss of activity when it was further shortened to 124. Deletion of sequences between 100 and 364 (the main region of sequence homology among EBNA-3A, -3B, and -3C) in the context of the whole protein only slightly reduced GST–RBP-Jk binding. The N-terminal GST–RBP-Jk binding site should be inactivated by this deletion, so it appears that part of the more C-terminal binding site lies between residues 364 and 500. This conclusion would be altered if there were complementation between a residual part of the N-terminal site and the C-terminal site in mutant Δ100-364. Similar relative affinities of the key fragments were observed at 4 and 30°C (data not shown) and over a time course of binding at 30°C (Fig. 1E). The results showed that there are at least two sites for the association of EBNA-3A and RBP-Jk in vitro, only one of which was detected in earlier studies.
FIG. 1.
GST pull-down assay identifying two regions within EBNA-3A capable of binding to RBP-Jk. (A) In vitro-translated proteins analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. This film was exposed for one-seventh the time used in panels B and C. (B) In vitro-translated EBNA-3A proteins that bound to GST-RBP-Jk in a binding assay performed at 4°C. (C) Negative control binding of in vitro-translated EBNA-3A proteins to GST at 4°C. Positions of protein size markers are shown in kilodaltons. (D) EBNA-3A and deletion mutants. Numbers are the amino acids of the 944-amino-acid EBNA-3A protein present, except for Δ100-364, in which residues 100 to 364 are absent. The percentage of input protein that bound to GST-RBP-Jk in panel B was quantified by densitometry and is shown on the right. (E) Time course of GST-RBP-Jk binding with EBNA-3A mutants at 30°C. The bound EBNA-3A protein was quantified by PhosphorImager analysis and expressed as a percentage of the input protein.
EBNA-3A can repress transcription from the Cp promoter.
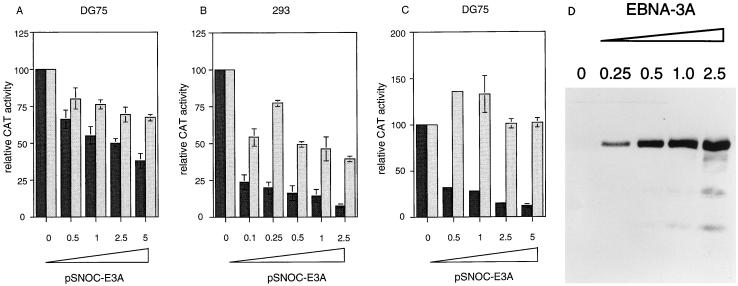
No cell or viral promoter has been reported to be regulated by wild-type EBNA-3A. Since the EBV Cp promoter is known to be inhibited by EBNA-3C expression (37), we also tested the ability of EBNA-3A to inhibit Cp. As shown in Fig. 2A, transfection of increasing amounts of an EBNA-3A plasmid in DG75 cells progressively inhibited Cp reporter activity, although the effect was small (about twofold). A larger repression (about 10-fold) was observed in 293 cells (Fig. 2B). Even 0.1 μg of the pSNOC-E3A plasmid caused a fourfold inhibition of Cp reporter activity in 293 cells. The progressively increasing inhibition of Cp activity correlated with increased EBNA-3A protein expression in the transfected cells (Fig. 2D). The ability of EBNA-3A to inhibit Cp is likely to be mediated through an interaction of EBNA-3A with RBP-Jk, since EBNA-3A also inhibits the activity of a model promoter dependent on the activity of four RBP-Jk sites (Fig. 2C). This finding was specific to the RBP-Jk sites, since EBNA-3A failed to inhibit the residual activity of a corresponding promoter construct in which the RBP-Jk binding sites had been mutated. There may thus be feedback inhibition from EBNA-3A of its own promoter, Cp, through the RBP-Jk site, independent of EBNA-2. The same situation has been proposed for EBNA-3C (37), but the effect with EBNA-3A is smaller than that observed with EBNA-3C.
FIG. 2.
Repression of promoters containing RBP-Jk binding sites by EBNA-3A. The results in panels A to C are expressed as relative CAT activity, the CAT activity with the empty vector pSNOC being set at 100. (A) EBNA-3A inhibits Cp in DG75 cells. pSNOC-E3A (0.5 to 5 μg) was titrated on p1.4Cp-CAT (black bars) or p1.4Cp-CAT E2RE mut (grey bars). (B) EBNA-3A inhibits Cp in 293 cells. pSNOC-E3A (0.1 to 2.5 μg) was titrated as in panel A. (C) EBNA-3A inhibits a promoter containing four RBP-Jk binding sites. pSNOC-E3A (0.5 to 5 μg) was titrated in DG75 cells on 4Jk-TK-CAT (black bars) or 4Jk-TK-CAT E2RE mut (grey bars). (D) Western immunoblot analysis of EBNA-3A expression. Samples from panel B were analyzed with antibody T2.78. EBNA-3A concentrations are given in micrograms.
Although mutation of the RBP-Jk sites in the 4Jk-TK-CAT construct resulted in a promoter whose residual activity was unaffected by EBNA-3A, there was a small repressive effect of EBNA-3A on the Cp promoter in which the RBP-Jk site had been mutated (Fig. 2A). This result may imply that EBNA-3A is also able to affect the RBP-Jk-independent activity of Cp or alternatively that the mutation introduced into Cp does not completely prevent RBP-Jk binding in cells, even though binding to the mutated site is abolished in vitro (7).
Induction of gene expression by an EBNA-3A deletion mutant targeted to a promoter.
Much of the EBNA-3A protein sequence does not appear to be required for binding to RBP-Jk, so we investigated other possible transcriptional effects of EBNA-3A. Since it is not known whether EBNA-3A can bind directly to DNA, we analyzed the effects of EBNA-3A on gene expression using a fusion of the GAL4 DNA binding domain to EBNA-3A on a model promoter containing a GAL4 binding site. Consistent with a recently reported analysis of EBNA-3A in HeLa cells with the GAL4 system (52), GAL4-E3A was found to repress transcription in the human B-cell line DG75 (Fig. 3A). A similar result has been reported with GAL4 fusions to EBNA-3C (3, 52). These phenomena have been discussed in the context of RBP-Jk binding exhibited by both proteins.
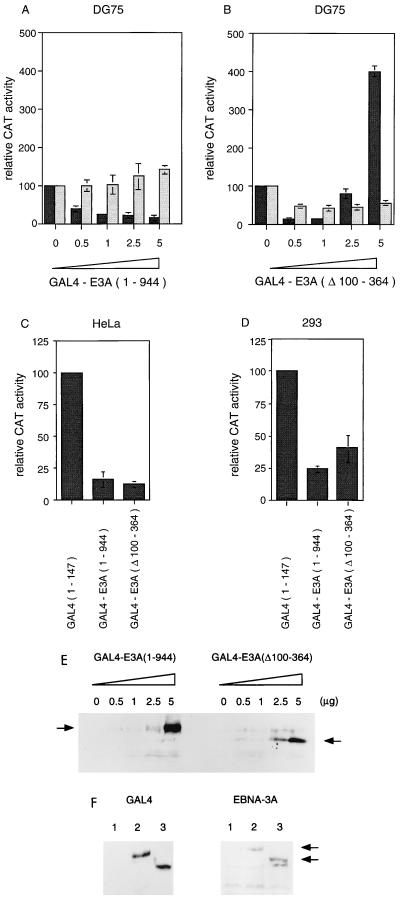
FIG. 3.
Comparison of transcription regulation by EBNA-3A and EBNA-3A(Δ100-364) in the GAL4 system. The results in panels A to D are expressed as relative CAT activity, the CAT activity with vector pGAL4(1-147) being set at 100. (A) GAL4-E3A inhibits transcription from a GAL4-dependent promoter in transfected DG75 cells. GAL4-E3A (0.5 to 5 μg) was titrated on pUASCAT (5 μg; black bars) or pBLCAT2 (5 μg; grey bars). (B) Effect of GAL4-E3A(Δ100-364) on transcription in transfected DG75 cells. Symbols are as in panel A. (C) GAL4-E3A and GAL4-E3A(Δ100-364) both inhibit transcription from pUASCAT in transfected HeLa cells (5 μg of pUASCAT and 5 μg of GAL effector DNA were transfected). (D) GAL4-E3A and GAL4-E3A(Δ100-364) both inhibit transcription from pUASCAT in transfected 293 cells (5 μg of pUASCAT and 5 μg of GAL effector DNA were transfected). (E) Western blot analysis of GAL4-E3A and GAL4-E3A(Δ100-364) expression in transfected DG75 cells. Samples from panels A and B were assayed with GAL4 monoclonal antibody sc-510. The arrows indicate the GAL4-E3A and GAL4-E3A(Δ100-364) proteins. (F) Western blot of extracts of transfected 293 cells with sc-510 anti-GAL4 (left panel) or T2.78 anti-EBNA-3A (right panel) antibodies. Lanes: 1, GAL4(1-147); 2, GAL4-E3A; 3, GAL4-E3A(Δ100-364). Only the high-molecular-weight EBNA-3A region of the gel is shown, so the small GAL4(1-147) protein is not seen in lane 1 of the left panel.
By analyzing additional mutants of EBNA-3A, we have found evidence for a further novel function of EBNA-3A. In DG75 cells at low levels of input DNA, the deletion mutant GAL4-E3A(Δ100-364) repressed like wild-type GAL4-E3A, but at higher levels of transfected DNA, GAL4-E3A(Δ100-364) was found to be an activator of transcription (Fig. 3B). This effect was dependent on the presence of a GAL4 DNA binding site in the target promoter. Since the activation phenomenon was not observed with a fusion of full-length EBNA-3A to GAL4, it appears that the part of EBNA-3A that includes some of the sequences involved in RBP-Jk binding acts to mask this transcription-activating property of EBNA-3A. Western blotting analysis of the GAL4 fusion proteins in DG75 cells (Fig. 3E) showed approximately similar levels of expression of the GAL4-E3A and GAL4-E3A(Δ100-364) proteins and the expected increasing expression of the GAL4-E3A(Δ100-364) protein with increasing input DNA. So, the biphasic response in CAT activity is not due to biphasic expression of the GAL4-E3A(Δ100-364) protein.
In the assays shown here, activation by GAL4-E3A(Δ100-364) depended on the cell type used for transfection. Although activation by GAL4-E3A(Δ100-364) was reproducibly observed in DG75 lymphoid cells (in 10 different experiments with three different preparations of plasmid DNA), no activation occurred in HeLa or 293 cells (Fig. 3C and D). In fact, in these cells, GAL4-E3A(Δ100-364) repressed transcription to a similar degree as full-length GAL4-E3A(1-944), about three- to fivefold. Similar levels of GAL4-EBNA-3A expression from the wild-type and GAL4-E3A(Δ100-364) fusion protein constructs were found by Western blotting of extracts of transfected cells with antibodies to both GAL4 and EBNA-3A (Fig. 3F).
To exclude the possibility that the lack of transcription activation in 293 and HeLa cells by GAL4-E3A(Δ100-364) could be a consequence of lower levels of expression of the transfected GAL4 constructs in these cells, the levels of protein expression in 293 and DG75 cells were compared by Western blotting, correcting for transfection efficiency. The transfection efficiencies (20% for 293 and 0.75% for DG75) were measured by immunofluorescence and fluorescence-activated cell sorter analyses (data not shown). The results indicated that with equivalent levels of input DNA, about 30 times as much fusion protein was expressed per cell in 293 cells as in DG75 cells, so the lack of activation was not a consequence of lower levels of protein expression. Titration of the GAL4-E3A(Δ100-364) plasmid to levels of input DNA 10-fold lower than that shown in Fig. 3D also failed to reveal any activation.
The binding of RBP-Jk to EBNA-2, -3A, -3B, and -3C links and coordinates the functions of these proteins in the cell. Since the levels of LMP-1 are partly determined by the activity of EBNA-2 (24), RBP-Jk would play a key role in modulating the activity or level of five of the six transforming genes of EBV. The remaining EBV gene required for efficient transformation, EBNA-LP, does not appear to associate with RBP-Jk (20), but we showed (see below) that its expression appears to modulate the biochemical distribution of EBNA-3A in transfected cells.
Effects of EBNA-LP on EBNA-3A distribution in the cell.
EBNA-LP and EBNA-3A are both nuclear proteins. EBNA-3A is observed under the immunofluorescence microscope as a pattern of dots within the nucleus of EBV-infected cells, presumably as a consequence of association with some uncharacterized structure within the nucleus. EBNA-LP is also present as dots, which have been shown to be ND10 domains, where PML, Rb, and Hsp70 may be located under some conditions. In biochemical extraction procedures, most of the EBNA-LP in LCLs behaves as though it is attached to the nuclear matrix (36, 47).
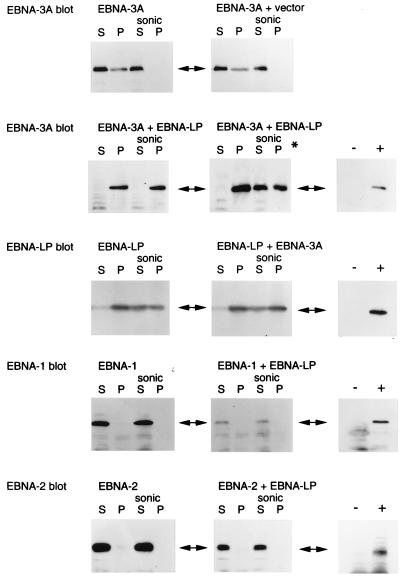
We tested whether the presence of EBNA-LP might affect the behavior of EBNA-3A in cells by transfecting expression vectors for the two proteins into DG75 cells. The transfected cells were lysed with RIPA buffer, and the lysates were centrifuged to separate the soluble material from the insoluble matrix. The matrix was then extracted with SDS sample buffer, which almost completely dissolved it. Aliquots of the RIPA buffer-soluble material and the insoluble matrix were analyzed by Western blotting for EBNA-3A. When a plasmid expressing EBNA-3A was transfected alone or with a control (empty) vector, the EBNA-3A was distributed between the soluble and insoluble matrix fractions (Fig. 4); sonication of the cells in RIPA buffer before centrifugation made all of the EBNA-3A RIPA buffer soluble. In contrast, when a plasmid expressing EBNA-LP was cotransfected with the EBNA-3A plasmid, the EBNA-3A was almost entirely found in the RIPA buffer pellet under our standard extraction conditions (Fig. 4). Only when the extraction time in RIPA buffer was extended to 60 min did sonication render some of the EBNA-3A RIPA buffer soluble in the EBNA-LP-cotransfected cells. When an EBNA-LP expression vector was transfected alone, the EBNA-LP was mostly RIPA buffer insoluble, and cotransfection with the EBNA-3A plasmid had no effect on the distribution of EBNA-LP. As a control to check that EBNA-LP did not cause all proteins expressed from cotransfected plasmids to become RIPA buffer insoluble, the EBNA-LP expression plasmid was cotransfected with expression vectors for EBNA-2 or EBNA-1. EBNA-LP made no difference in the distribution of these proteins in the RIPA buffer extracts; EBNA-2 and EBNA-1 were always in the RIPA buffer-soluble fractions.
FIG. 4.
EBNA-3A, a nuclear protein, is relocated to the nuclear matrix fraction by transfection of EBNA-LP. Western immunoblotting for the indicated proteins was done with extracts of DG75 cells transfected with combinations of EBNA expression plasmids. RIPA buffer extracts of transfected cells were centrifuged, yielding a soluble fraction (S) and a pellet (P). Where indicated (sonic), the RIPA buffer extracts were sonicated before centrifugation. Standard RIPA buffer extractions were done for 30 min, but in the panel marked with an asterisk, the extractions were done for 60 min. In each case (right-hand panels), expression of the transfected EBNA protein was demonstrated by comparison of cells transfected with the EBNA plasmid (+) and cells transfected with the corresponding empty vector (−).
DISCUSSION
Localization effects of EBNA-LP.
Cotransfection of an expression vector for EBNA-LP caused a relocation of EBNA-3A expressed from a transfected plasmid into a RIPA buffer-insoluble form. This effect was specific for EBNA-3A; EBNA-1 or EBNA-2 was unaffected by EBNA-LP. Because the endogenous levels of EBNA-3A are very low in LCLs, we have not been able to test whether EBNA-3A and EBNA-LP bind to each other in LCLs, but the fact that they do not seem to be localized in the same spots (36, 47) argues against this idea. When the proteins are newly made, however, they may be accessible to each other. It is notable that early in infection the distribution of EBNA-LP is more diffuse (47) and the protein is expressed at a higher level than in established LCLs. Our transient transfection results indicate that the expression of EBNA-LP may alter the nuclear structure such that EBNA-3A, which is already partially in the RIPA buffer-insoluble fraction, becomes almost entirely insoluble. This idea suggests a further novel level of interplay between the EBNA proteins.
Multiple functions of EBNA-3A.
Using in vitro binding assays, we have shown that more than one region of EBNA-3A is able to bind to RBP-Jk. Although we have attempted transfection experiments with a panel of GAL4-E3A deletion mutants (data not shown) to assess the relative significance of these binding sites in gene regulation, interpretation of those experiments has been limited by the instability of many of the mutated proteins in transfected cells. We have therefore focused on the relative activities of GAL4 fusions of wild-type EBNA-3A and the GAL4-E3A(Δ100-364) mutant, both of which are similarly well expressed upon transfection. The properties of GAL4-E3A(Δ100-364) indicate that EBNA-3A contains both repression and activation functions, which can be revealed under different conditions and in different cell types. A region which can act as an activation domain has also been identified (33) in EBNA-3C (amino acids 724 to 826), and both activation and repression effects of EBNA-3C on the promoter for LMP-1 have been reported (33). There is, however, no obvious primary sequence homology between EBNA-3A and -3C in the C-terminal parts of the proteins, even though both contain repeat sequences (2).
The ability of EBNA-3A to act as both an activator and a repressor of transcription, depending on conditions, has precedents in some cell transcription factors. For example, the Sp3 protein functions as a repressor when it is bound to a promoter through multiple DNA binding sites but as an activator when it is targeted to a promoter through a single binding site (31). It is a bifunctional regulator whose predominant effect depends on the context of the Sp3 DNA binding sites and on the nature of corepressors present in the cell background. Another example is the Acanthamoeba TATA-binding protein (TBP) promoter binding factor, which can regulate TBP in a dose-related manner (17). At low concentrations, this factor binds to a cis element located upstream of the TATA box and functions as an activator. When its concentration increases, it can also bind to a lower-affinity sequence located between the TATA box and the transcription initiation site, and then it acts as a repressor. The TBP promoter binding factor is thus a transcription factor whose action is determined by both its concentration and the sequence context of its binding sites.
The way in which the Drosophila Krüppel protein acts as a dual-function regulator may be of particular relevance to our observations with the GAL4-E3A(Δ100-364) protein. When assayed on promoters containing a single Krüppel binding site, the Krüppel protein is converted from an activator to a repressor, depending on its concentration (44). At low concentrations, it is a monomer that activates transcription, whereas at higher concentrations, it is a homodimer that binds to the same DNA sequence as the monomer but acts as a repressor. The two forms of the Krüppel protein specifically contact distinct general transcription factors, the monomers binding transcription factor IIB and the dimers interacting with transcription factor IIEβ (43).
It would be interesting to determine whether the repression-activation transition observed with GAL4-E3A(Δ100-364) is due to multimerization of the protein, which might affect its ability to interact with other transcription factors, or is due to titration of a cell cofactor that is limiting in DG75 cells. The properties of a previously reported (50) truncated EBNA-3A protein which appeared to act in a dominant negative manner, preventing the function of wild-type EBNA-3A, may also be consistent with EBNA-3A complexing with other proteins or itself when it is active in cells. The cell context is probably also important since in our experiments, no activation was detected in HeLa or 293 cells, perhaps suggesting a cell type-specific cofactor for EBNA-3A.
Significance of the interaction of EBNA-3 proteins with RBP-Jk: coordinating role of RBP-Jk.
EBV is exceptionally efficient in B-cell immortalization, and this fact is reflected in the large number of viral genes required for the process. These genes are organized in both time and level of expression. The order of expression of the EBNA proteins is apparently determined by the locations of the EBNA genes in the genome and by sequential promoter usage, the Wp promoter being active upon infection, followed by the Cp promoter, which normally takes over expression of the EBNA genes in established LCLs. The layout of the EBNA genes in the genome is consistent with the timing of expression, EBNA-LP being the first detectable protein, closely followed by EBNA-2, the EBNA-3 family, and EBNA-1. The amounts of the EBNA proteins are controlled to some extent at the transcriptional level, the Cp promoter being under feedback control from the EBNA proteins in both positive and negative ways. Because the EBNA proteins appear to have low turnover rates, coordination of their activity most likely occurs at the protein level. One of the simplest ways to coordinate the activities of the proteins would be for them to be buffered through interaction with a single common factor.
RBP-Jk has been shown to bind to EBNA-2 and to EBNA-3A, -3B, and -3C. The binding of EBNA-2 to RBP-Jk has been well characterized and shown to be important in the role of EBNA-2 in EBV transformation. Measurement of the amounts of RBP-Jk bound to EBNA-2 and to EBNA-3A, -3B, and -3C has shown that the association with the EBNA-3 proteins is quantitatively significant compared to the amount of RBP-Jk bound to EBNA-2 (20), and the binding of the EBNA-3 proteins has been interpreted as counterbalancing the activating effects of EBNA-2 mediated through RBP-Jk (20, 40, 52, 54), for example, on the Cp promoter. However, Cp is in fact only partially dependent on EBNA-2–RBP-Jk in LCLs, since the binding site for RBP-Jk can be mutated without a loss of Cp function (7). It is also remarkable that EBV should require four large viral proteins comprising 79% of the sequence complexity of the six viral proteins required for transformation to control signalling through RBP-Jk. The conventional explanation of the role of EBNA-3A, 3B, and 3C is negative modulation of the activation by EBNA-2 of signalling through RBP-Jk, the parts of the EBNA-3 proteins not required for RBP-Jk binding presumably integrating other signals into this pathway. However, we have considered an alternative interpretation of the significance of the binding of EBNA-3 proteins to RBP-Jk which might be equally relevant.
EBNA-3A, -3B, and -3C are clearly related genes and probably arose through gene multiplication events, but the relationship of their exon structures and protein sequences is only significant in the N-terminal parts of the proteins. The parts of the proteins that are related in primary sequence lie within the region that is involved in binding to RBP-Jk. Although all three proteins have repetitive sequence elements in the C-terminal regions, these are unrelated at the sequence level. In this alternative model, EBNA-3A, -3B, and -3C might have different roles in EBV biology; the significance of the binding to RBP-Jk would be to buffer the levels of the active protein, the EBNA-3–RBP-Jk complexes representing inactive forms of the EBNA-3 proteins in this respect. This model supposes that there are novel functions unrelated to RBP-Jk binding for EBNA-3A and EBNA-3C in EBV transformation. Clues to these functions might be available in the gene activation that we observed in a deletion mutant of EBNA-3A (see above), in the activation domain reported for EBNA-3C (33), and in the cooperation of EBNA-3C with activated Ha-ras in the transformation of primary murine fibroblasts (35). There is considerable evidence that the levels of EBV transforming proteins have to be tightly regulated in LCLs, and this buffering of EBNA-3 proteins on RBP-Jk would be an effective way of coordinating the level of EBNA-3 proteins with EBNA-2–RBP-Jk activity.
ACKNOWLEDGMENTS
We thank Alan Rickinson and Elliott Kieff for sending copies of their manuscripts (15, 34) prior to publication and Roger Watson, Evelyne Manet, Alain Sergeant, Lars Rymo, Eric Lam, Diane Hayward, and Wolfgang Hammerschmidt for providing plasmids.
This work was supported by the Ludwig Institute for Cancer Research.
REFERENCES
- 1.Allan G J, Inman G J, Parker B D, Rowe D T, Farrell P J. Cell growth effects of Epstein-Barr virus leader protein. J Gen Virol. 1992;73:1547–1551. doi: 10.1099/0022-1317-73-6-1547. [DOI] [PubMed] [Google Scholar]
- 2.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 3.Bain M, Watson R J, Farrell P J, Allday M J. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J Virol. 1996;70:2481–2489. doi: 10.1128/jvi.70.4.2481-2489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Bassat H, Goldblum N, Mitrani S, Goldblum T, Yoffey J M, Cohen M M, Bentwich Z, Ramot B, Klein E, Klein G. Establishment in continuous culture of a new type of lymphocyte from a “Burkitt like” malignant lymphoma (line D.G.-75) Int J Cancer. 1977;19:27–33. doi: 10.1002/ijc.2910190105. [DOI] [PubMed] [Google Scholar]
- 5.Bodescot M, Brison O, Perricaudet M. An Epstein-Barr virus transcription unit is at least 84 kilobases long. Nucleic Acids Res. 1986;14:2611–2620. doi: 10.1093/nar/14.6.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen M R, Middeldorp J M, Hayward S D. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans T J, Farrell P J, Swaminathan S. Molecular genetic analysis of Epstein-Barr virus Cp promoter function. J Virol. 1996;70:1695–1705. doi: 10.1128/jvi.70.3.1695-1705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fahraeus R, Jansson A, Sjoblom A, Nilsson T, Klein G, Rymo L. Cell phenotype-dependent control of Epstein-Barr virus latent membrane protein 1 gene regulatory sequences. Virology. 1993;195:71–80. doi: 10.1006/viro.1993.1347. [DOI] [PubMed] [Google Scholar]
- 9.Farrell P. Epstein-Barr virus immortalizing genes. Trends Microbiol. 1995;3:105–109. doi: 10.1016/s0966-842x(00)88891-5. [DOI] [PubMed] [Google Scholar]
- 10.Farrell P J. The Epstein-Barr virus genome. In: Klein I G, editor. Advances in viral oncology. New York, N.Y: Raven Press; 1989. pp. 103–132. [Google Scholar]
- 11.Finke J, Rowe M, Kallin B, Ernberg I, Rosen A, Dillner J, Klein G. Monoclonal and polyclonal antibodies against Epstein-Barr virus nuclear antigen 5 detect multiple protein species in Burkitt’s lymphoma and lymphoblastoid cell lines. J Virol. 1987;61:3870–3878. doi: 10.1128/jvi.61.12.3870-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham F L, van der Eb A. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 13.Griffin B E, Karran L. Immortalization of monkey epithelial cells by specific fragments of Epstein-Barr virus DNA. Nature. 1984;309:78–82. doi: 10.1038/309078a0. [DOI] [PubMed] [Google Scholar]
- 14.Hammerschmidt W, Sugden B, Baichwal V R. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J Virol. 1989;63:2469–2475. doi: 10.1128/jvi.63.6.2469-2475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hennessy K, Fennewald S, Kieff E. A third viral nuclear protein in lymphoblasts immortalized by Epstein-Barr virus. Proc Natl Acad Sci USA. 1985;82:5944–5948. doi: 10.1073/pnas.82.17.5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Bateman E. Transcription of the Acanthamoeba TATA-binding protein gene. J Biol Chem. 1997;272:3852–3859. doi: 10.1074/jbc.272.6.3852. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W Q, Szekely L, Wendel-Hansen V, Ringertz N, Klein G, Rosen A. Co-localization of the retinoblastoma protein and the Epstein-Barr virus-encoded nuclear antigen EBNA-5. Exp Cell Res. 1991;197:314–318. doi: 10.1016/0014-4827(91)90438-z. [DOI] [PubMed] [Google Scholar]
- 19.Jiang W Q, Wendel-Hansen V, Lundkvist A, Ringertz N, Klein G, Rosen A. Intranuclear distribution of Epstein-Barr virus-encoded nuclear antigens EBNA-1, -2, -3 and -5. J Cell Sci. 1991;99:497–502. doi: 10.1242/jcs.99.3.497. [DOI] [PubMed] [Google Scholar]
- 20.Johannsen E, Miller C L, Grossman S R, Kieff E. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaelin W, Jr, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 22.Kallin B, Dillner J, Ernberg I, Ehlin-Henriksson B, Rosen A, Henle W, Henle G, Klein G. Four virally determined nuclear antigens are expressed in Epstein-Barr virus-transformed cells. Proc Natl Acad Sci USA. 1986;83:1499–1503. doi: 10.1073/pnas.83.5.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart J W, Kremmer E, Delecluse H J, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieff E. Epstein-Barr virus. In: Fields B, Knipe D, Howley P, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 2343–2396. [Google Scholar]
- 26.Kitay M, Rowe D. Cell cycle stage-specific phosphorylation of the Epstein-Barr virus immortalization protein EBNA-LP. J Virol. 1996;70:7885–7893. doi: 10.1128/jvi.70.11.7885-7893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitay M K, Rowe D T. Protein-protein interactions between Epstein-Barr virus nuclear antigen-LP and cellular gene products: binding of 70-kilodalton heat shock proteins. Virology. 1996;220:91–99. doi: 10.1006/viro.1996.0289. [DOI] [PubMed] [Google Scholar]
- 28.Krauer K G, Kienzle N, Young D B, Sculley T B. Epstein-Barr nuclear antigen-3 and -4 interact with RBP-2N, a major isoform of RBP-J kappa in B lymphocytes. Virology. 1996;226:346–353. doi: 10.1006/viro.1996.0662. [DOI] [PubMed] [Google Scholar]
- 29.Le-Roux A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 30.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 32.Mannick J B, Tong X, Hemnes A, Kieff E. The Epstein-Barr virus nuclear antigen leader protein associates with hsp72/hsc73. J Virol. 1995;69:8169–8172. doi: 10.1128/jvi.69.12.8169-8172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitsche F, Bell A, Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker G, Crook T, Bain M, Sara E, Farrell P, Allday M. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene. 1996;13:2541–2549. [PubMed] [Google Scholar]
- 36.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 37.Radkov S, Bain M, Farrell P, West M, Rowe M, Allday M. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J Virol. 1997;71:8552–8562. doi: 10.1128/jvi.71.11.8552-8562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricksten A, Kallin B, Alexander H, Dillner J, Fahraeus R, Klein G, Lerner R, Rymo L. BamHI E region of the Epstein-Barr virus genome encodes three transformation-associated nuclear proteins. Proc Natl Acad Sci USA. 1988;85:995–999. doi: 10.1073/pnas.85.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney C M, Brimmell M, Buschle M, Allan G, Farrell P J, Kolman J L. Host cell and EBNA-2 regulation of Epstein-Barr virus latent-cycle promoter activity in B lymphocytes. J Virol. 1992;66:496–504. doi: 10.1128/jvi.66.1.496-504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe M, Young L S, Cadwallader K, Petti L, Kieff E, Rickinson A B. Distinction between Epstein-Barr virus type A (EBNA 2A) and type B (EBNA 2B) isolates extends to the EBNA 3 family of nuclear proteins. J Virol. 1989;63:1031–1039. doi: 10.1128/jvi.63.3.1031-1039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jackle H. Control of transcription by Krüppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 44.Sauer F, Jackle H. Dimerization and the control of transcription by Krüppel. Nature. 1993;364:454–457. doi: 10.1038/364454a0. [DOI] [PubMed] [Google Scholar]
- 45.Sinclair A J, Palmero I, Peters G, Farrell P J. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 47.Szekely L, Jiang W-Q, Pokrovskaja K, Wiman K G, Klein G, Ringertz N. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA-5 and hsp70 proteins after exposure to heat shock or cell density congestion. J Gen Virol. 1995;76:2423–2432. doi: 10.1099/0022-1317-76-10-2423. [DOI] [PubMed] [Google Scholar]
- 48.Szekely L, Pokrovskaja K, Jiang W Q, de-The H, Ringertz N, Klein G. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J Virol. 1996;70:2562–2568. doi: 10.1128/jvi.70.4.2562-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szekely L, Pokrovskaja K, Jiang W Q, Selivanova G, Lowbeer M, Ringertz N, Wiman K G, Klein G. Resting B-cells, EBV-infected B-blasts and established lymphoblastoid cell lines differ in their Rb, p53 and EBNA-5 expression patterns. Oncogene. 1995;10:1869–1874. [PubMed] [Google Scholar]
- 50.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-Jk repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson R J, Robinson C, Lam E W. Transcription regulation by murine B-myb is distinct from that by c-myb. Nucleic Acids Res. 1993;21:267–272. doi: 10.1093/nar/21.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]