Abstract
Circadian rhythms and sleep are fundamental biological processes integral to human health. Their disruption is associated with detrimental physiological consequences, including cognitive, metabolic, cardiovascular and immunological dysfunctions. Yet many of the molecular underpinnings of sleep regulation in health and disease have remained elusive. Given the moderate heritability of circadian and sleep traits, genetics offers an opportunity that complements insights from model organism studies to advance our fundamental molecular understanding of human circadian and sleep physiology and linked chronic disease biology. Here, we review recent discoveries of the genetics of circadian and sleep physiology and disorders with a focus on those that reveal causal contributions to complex diseases.
The circadian system coordinates rhythms in physiology and behaviour to the 24-hour light-dark cycle (FIG. 1a). It is composed of a central pacemaker in the hypothalamic suprachiasmatic nucleus (SCN) and peripheral oscillators present in virtually all organs and tissues, ranging from the heart to the liver and even skin cells1,2. Circadian rhythms in the SCN and peripheral oscillators are driven by cell-autonomous transcription-translation feedback loops, referred to as the molecular clock. Whereas the SCN is synchronized to the day-night cycle primarily by light, peripheral oscillators are synchronized by both SCN signals and behaviours, such as eating. The interaction of circadian and homeostatic sleep regulatory processes is known to control the timing and duration of sleep and associated physiology (FIG. 1), but the biological and molecular basis of sleep need, regulation and function in humans is incompletely understood3–5. Disruption of circadian rhythms and sleep is associated with broad negative consequences for health, including cognitive, metabolic, cardiovascular and immunological functions, as well as mood6,7. However, the molecular mechanisms underlying the bidirectional relationships between sleep disorders or circadian disorders and comorbid conditions remain elusive. Insights into fundamental molecular processes and links to other disease systems have come almost entirely from studies in model organisms, reviewed recently for both circadian rhythm8 and sleep9. Given that circadian and sleep traits and disorders are moderately heritable8, human genetic discovery offers a complementary opportunity to advance our fundamental molecular understanding of relevant human biology and its clinical application in the diagnosis and treatment of circadian and sleep disorders and linked chronic diseases.
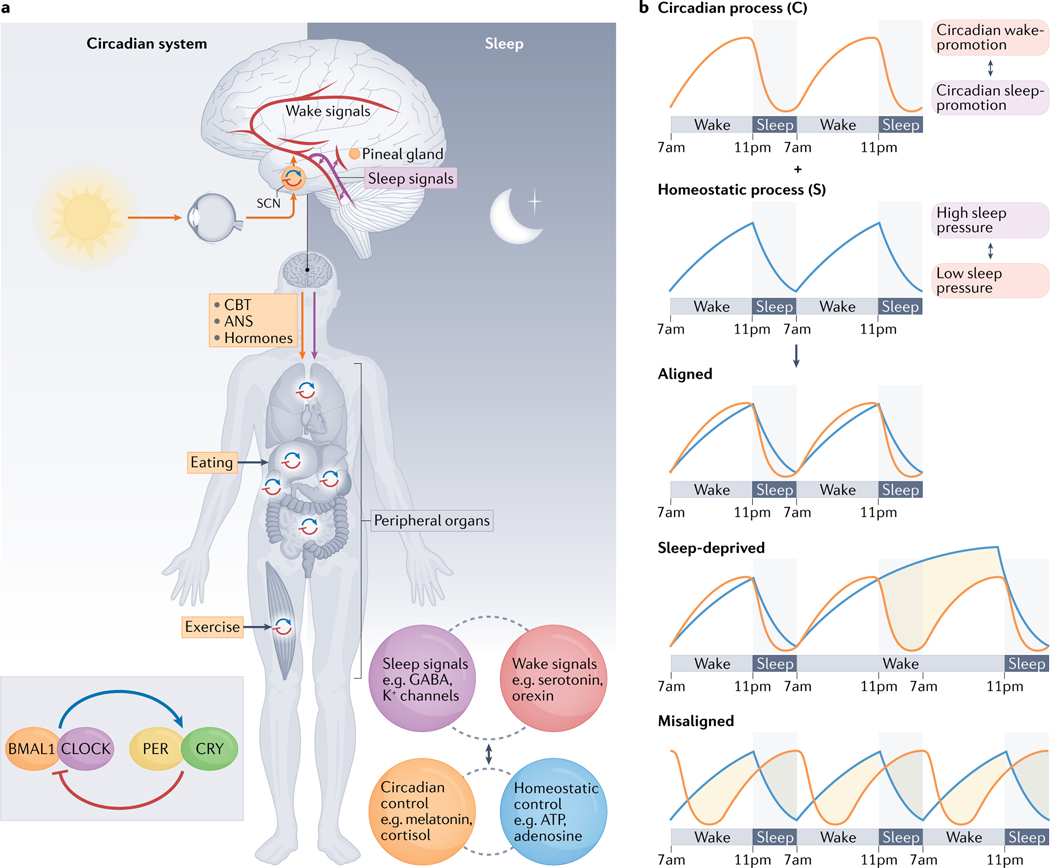
Fig. 1 |. An overview of the circadian and sleep systems.
a | Left: the simplified circadian cell-autonomous transcriptional-translational feedback loop mechanism is depicted together with key anatomic and physiologic features. The master circadian pacemaker, located in the suprachiasmatic nucleus (SCN, orange circle) of the hypothalamus, is entrained primarily by external cues from light. This SCN coordinates 24-hour rhythms in physiology and behaviour through neuronal and hormonal output signals that influence the activity of organs and tissues either directly (for example, the SCN modulating sympathetic tone to the liver or pineal gland, orange arrow) or indirectly by synchronizing peripheral clocks, such that the molecular clock (blue-red circular arrows) in those peripheral organs or tissues then influences their function through the expression of clock-controlled genes. Peripheral oscillators, present in most organs and tissues, can also be synchronized by behaviours such as eating and physical activity. Right: neuroanatomy and sleep-wake circuitry. Many neurotransmitters switch our brains between sleep and wakefulness in a carefully regulated cycle. Key sleep and wake neurotransmitters and known sleep regulators in the brain are listed in the purple and red bubbles. In addition, sleep is regulated both by the circadian process (orange bubble) and the homeostatic process, the drive to sleep that accumulates based on time spent awake (blue bubble). b | The two-process model of sleep3. The timing and structure of sleep are determined by the interaction of a homeostatic (S) and a circadian (C) process. Process S mediates the gradual rise of sleep pressure during wakefulness and its dissipation during sleep. Process C describes the circadian drive for wakefulness. Time courses for processes S and C are shown under aligned sleep-wake cycles, under sleep-deprived conditions and under misaligned sleep-wake cycles. The orange-shaded areas indicate being sleepy or alert at the wrong time. The grey-shaded areas represent sleep episodes except in the sleep-deprived condition, where it represents being awake at the habitual sleep time. ANS, autonomic nervous system; CBT, core body temperature.
Despite the conflation of circadian rhythms and sleep in the perceptions of the general public, the circadian and sleep research fields have evolved mostly separately. Starting more than half a century ago, basic genetic discoveries in circadian biology in Drosophila and mouse occurred rapidly using reverse and forward genetics, as recognized with the 2017 Nobel Prize in Physiology or Medicine. By contrast, genetic insights into sleep regulation have emerged almost exclusively in the past decade (FIG. 2). Conversely, although the clinical importance of sleep disturbances has been well established for over a century, that of circadian disruptions (beyond impact on sleep) have gained wide recognition only recently, as indicated by the budding of ‘circadian clinics’ around the world.
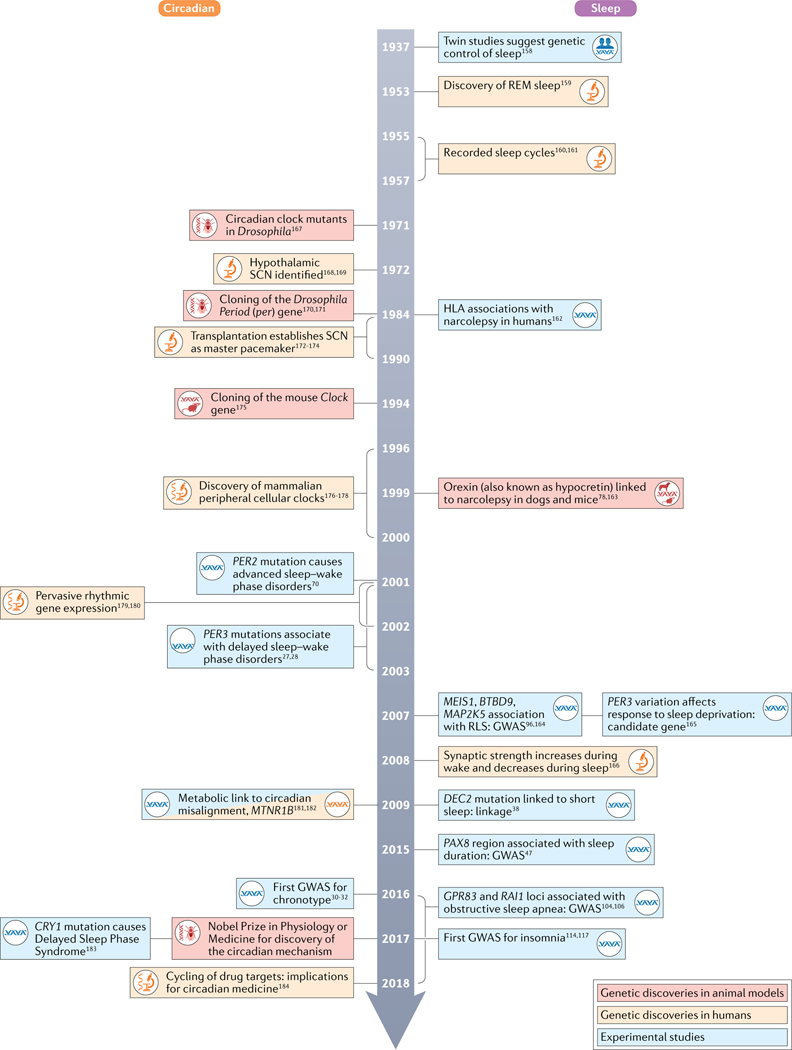
Fig. 2 |. A timeline of circadian and sleep research discoveries with a focus on genetics.
Discoveries are based on publication dates from PubMed (REFS.27,28,30–32,38,47,70,78,96,104,106,114,117,158–184). GWAS, genome-wide association study; REM, rapid eye movement; RLS, restless leg syndrome; SCN, suprachiasmatic nucleus.
Although the development of the instrumental two-process model of sleep by Alexander Borbely and colleagues in the early 1980s formalized the effect of the circadian system on sleep3 (FIG. 1b), only recently have both fields started to move closer together. Their convergence is owing to the recognition that the effects of the circadian timing system and sleep interact on many more levels than previously thought. Examples of their interconnected nature include acute effects of slow wave sleep on neural activity in the SCN10; gating by sleep of the most potent environmental cue (that is, light) to the circadian system11; the effect of circadian timing on REM sleep12; variation in the clock gene PER3 on homeostatic sleep regulation13; and the dramatic impact that sleep deprivation has on circadian rhythm organization14, to name a few.
Here, we focus on recent genetic discoveries primarily using Mendelian family-based approaches and large-scale genome-wide association studies (GWAS) of circadian and sleep phenotypes and disorders, using subjective and objective measurements, supplemented by emerging smaller-scale targeted mechanistic studies. We emphasize the power of the genetics of extreme phenotypes, family-based study designs and of unbiased genome-wide screens of rare and common variation in beginning to define the genetic architecture of sleep and circadian traits and the discovery of novel biological insights. Next, we discuss the relevance of findings from circadian and sleep genetics to human health. Finally, we describe critical gaps in our understanding and obstacles in the path to clinical translation and describe promising future research approaches and methods. Rather than a comprehensive review, we provide a perspective on the fast-evolving field of circadian rhythm and sleep genetics and links to human disease and health.
Genetics of circadian and sleep physiology
Unravelling the genetic mechanisms underlying circadian rhythm generation has laid the foundation for the development of strong experimental tools with which to interrogate the mechanisms and consequences of circadian rhythms and their disruption. Distinct core clock genes have robust roles in determining circadian period15. By contrast, the number of genes that influence sleep-wake regulation seems to be larger, with less defined function and smaller impact16–18.
The success of genetics in studying circadian biology is based on a number of advantages: key phenotypes that can be measured with high precision and stability, such as circadian period (cycle length) and phase (FIG. 3); ease of assessment in animal models, with activity recordings under constant dark conditions (in humans, period assessments especially require complex and demanding circadian protocols); a mechanism not requiring complex neural network or organ functions, but being cell-autonomous, enabling single-cell experiments (although there is clearly additional complexity in the multi-oscillator system of the organism as a whole); and a consistent transcription-translation mechanism across phyla and evolution.
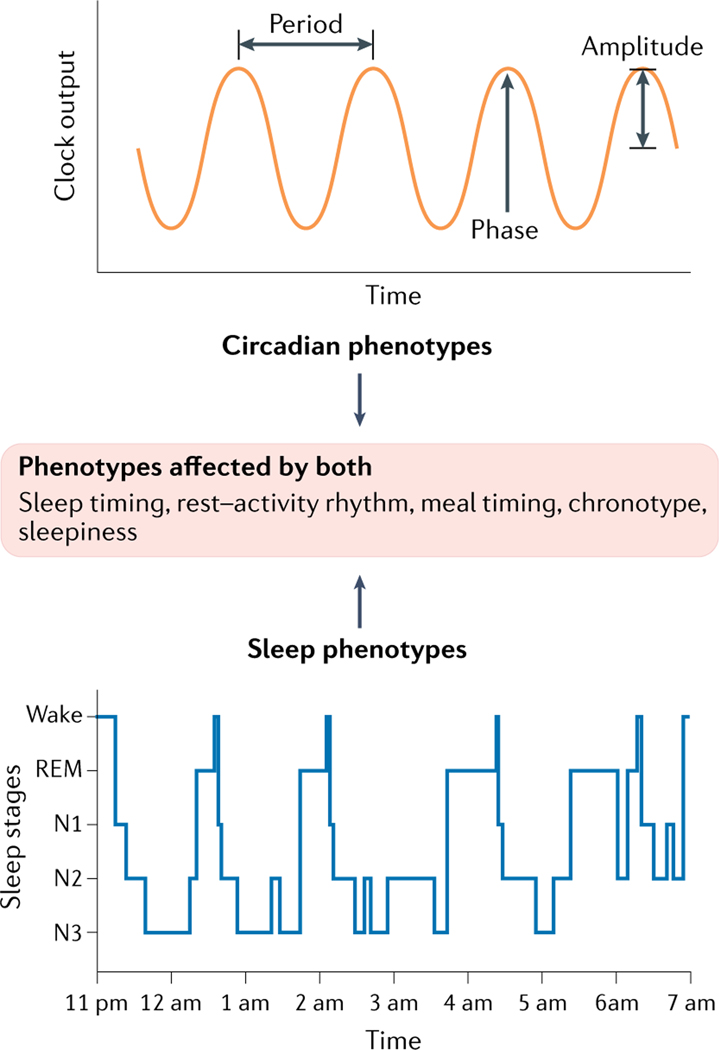
Fig. 3 |. Measurable approximates of circadian rhythm and sleep physiologic phenotypes.
Properties of circadian rhythms include phase, amplitude and period (top panel), whereas sleep phenotypes include sleep latency, duration and quality, sleep stage duration, intensity and depth, as well as temporal and spatial organization of complex sleep brain waves, and associated physiologic and behavioural changes (some aspects shown in bottom panel). Phenotypes affected by both systems are highlighted in the middle panel. Sleep progresses through the individual stages of sleep during the night in 4–6 cycles lasting 70–120 minutes each. Cycles consist of three stages of non-REM sleep (NREM, N1-N3) and the rapid eye movement (REM) sleep stage. N1 involves the transition from wakefulness to sleep with brain waves beginning to slow. N2 involves the further slowing of heart rate and breathing together with a drop in body temperature. N2 is marked by brief bursts of brain activity including spindles and K-complexes. N3 is deep sleep marked by the slowest brain waves. Awakening is difficult during N3. Top part of figure (circadian phenotypes) reprinted with permission from REF.185, OUP. Bottom part of figure (sleep phenotypes) adapted from REF.186, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
By contrast, sleep is a multidimensional phenotype. It can include, for example, sleep duration and timing, but also the spectral power of electroencephalograph (EEG) signals at particular frequency bands, rapid eye movement, muscle atonia, and complex EEG waveforms, such as spindles and K-complexes). Assessment of multiple physiological signals (such as brain activity, eye movements, muscle tone and, depending on the goals, other signals including respiratory signals and leg movements) concurrently (for example, with polysomnography as the gold standard assessment of sleep) is required. Moreover, to observe sleep behaviours, the organism must have complex multicellular mechanisms, and sleep behaviours can reflect varying phenotypic definitions depending on species and ontogeny (for example, the assessment of sleep by classic EEG is not possible in organisms that lack a central nervous system).
Phenotyping approaches
Circadian phenotypes and disorders are approximated by self-reporting using questionnaires and diaries, objectively approximated or estimated using wearable devices (for example, wrist accelerometers), melatonin or body temperature profiles in controlled conditions, blood-based omics biomarkers, such as monocyte transcriptome-based internal circadian timing predictions, and primary human cell culture, or through physician diagnosis and phenotypes derived from electronic health records. However, masking is an important concept in circadian assessments, where an environmental or behavioural factor acutely influences a biological measure intended to assess circadian control and thereby obscures or confounds this circadian measure and renders it invalid or imprecise. For example, light has a masking effect on the release of melatonin, resulting in acute suppression of melatonin concentrations (for example, as assessed in blood, saliva or urine). As another example, the direct effects of sleep, eating and physical activity on body temperature obscure the ability to measure circadian phase based on this measure if these factors are not tightly controlled for. This concept applies to virtually all measures that can be used for circadian phase assessment, although the sensitivity of the measure to masking effects depends on the measure used. For example, although the rise of melatonin concentrations can be assessed relatively accurately merely by ensuring dim light conditions, most other measures are more severely masked by the sleep-wake, rest-activity, postural and/or fasting-eating cycles19,20.
Normal sleep patterns and sleep disorders are also assessed by self-reported questionnaires, sleep diaries, wearable devices such as accelerometers and mobile polysomnography, and the gold-standard polysomnography (see Supplementary Table 1 for the relative strengths and limitations of these assessment approaches).
For genetic studies, an important consideration is the trade-off between phenotypes derived from self-report, from objective measurements and from electronic health records, particularly in light of the rise of wearable devices, which allow real-time phenotyping that is increasingly scalable. There is still much to learn about the heterogeneity of phenotypes and the validity of cruder outcome measures or diagnostic codes relative to gold-standard phenotypes. Indeed, the comparison of the genetics of different measures of sleep and circadian rhythms can serve as an approach to determine whether comparable or diverse phenotypes are being measured21.
Circadian and sleep phenotypes
Circadian-related and sleep-associated factors such as chronotype (for example, morningness or eveningness), sleep timing, sleep duration and sleep quality are quantitative traits that vary among individuals in the general population. Twin and family studies suggest a substantial genetic component, with heritability estimated at 10–45% for self-reported behaviours and up to 96% for EEG-measured characteristics of sleep architecture22,23. Although molecular determinants have been poorly understood, both familial sequencing studies and large-scale GWAS have started to identify genetic loci for circadian and sleep behaviours as summarized below (FIG. 4a), although sample sizes in accelerometry GWAS still lag behind self-reported traits, leading to differences in statistical power. Moving forwards, scaling up studies with objectively measured sleep traits paired with genetic information will lead to increased biological insights.
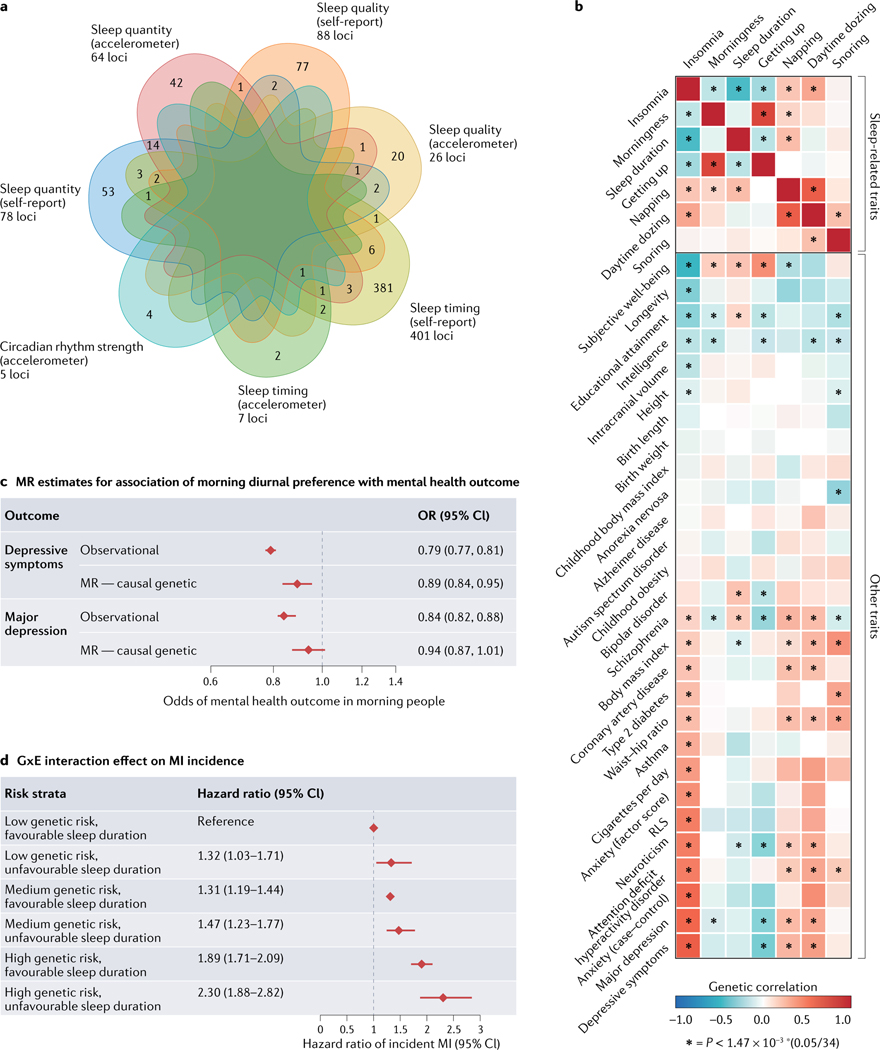
Fig. 4 |. Sleep and circadian rhythms link to health using genetics.
a | Number of genetic loci identified for circadian and sleep phenotypes in the general population shows domain-specific overlap primarily for measures of sleep timing (summarized from published genome-wide association studies (GWAS) discussed in the main text16–18,33,34,49,87,91,101,106,116). b | Evidence of genetic links between sleep/circadian traits/disease and neuropsychiatric and metabolic traits showing a heatmap of genetic correlations between insomnia symptoms GWAS and sleep, neuropsychiatric and metabolic trait GWAS. Positive genetic correlation is shown in red and negative genetic correlation is shown in blue. Significant correlations are shown with an asterisk116. c | Utilizing GWAS results to probe the causal relationship between diurnal preference (chronotype) and major depression, using Mendelian randomization (MR) to demonstrate a protective association of earlier diurnal preference with lower odds of depressive symptoms134. d | Sleep duration interacts with genetic risk of myocardial infarction (MI) to affect the risk of incident MI187. Bars represent 95% confidence intervals (CIs). GxE, gene-by-environment interaction; OR, odds ratio; RLS, restless legs syndrome. Part b reprinted from REF.116, Springer Nature Limited. Part c adapted from REF.134, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Part d adapted with permission from REF.187, Elsevier.
Chronotype and sleep timing.
Genetic studies in model organisms have established that circadian rhythms are generated by a cell-autonomous molecular clock mechanism comprising transcriptional-translational feedback loops24,25. Chronotype, or diurnal preference for morning or evening behaviour, varies in the population as a quantitative trait and is in part driven by variation in environment sensing, circadian period or sleep homeostatic processes26. Identification of clock gene mutations in families with extremely advanced or delayed sleep timing demonstrated the cross-species conservation of the clock mechanism in influencing daily human behaviour. Furthermore, modelling these naturally occurring human mutations in mice led to mechanistic insights into clock regulation by phosphorylation and protein degradation mechanisms. This family-based gene discovery approach has been successful in identifying and characterizing human clock gene mutations that contribute to Mendelian circadian rhythm disorders that advance or delay circadian phase.
Candidate gene association studies for chronotype that focused on core clock genes identified common coding variants in PER327–29, suggesting a genetic basis for human chronotype. Recent large-scale GWAS using self-reported diurnal preference in the UK Biobank and 23andMe cohorts have found over 350 genetic loci surpassing genome-wide significance, comprising genes enriched in circadian rhythm, cyclic adenosine monophosphate (cAMP), glutamate and insulin signalling pathways, and those expressed in the retina, hindbrain, hypothalamus and pituitary gland30–33. Variants implicate regions at clock genes (PER1, PER2, PER3, CRY1 and ARNTL), retinal and hypothalamic genes that may reflect inputs to the clock or integration in the SCN (for example, VIP and missense single-nucleotide polymorphisms (SNPs) at RGS16 and PATJ) and those involved in sleep-wake processes, for example, a missense SNP in the orexin receptor gene HCRTR2 (also known as OXR2); however, specific molecular mechanisms, cell types and tissue types linking genotype to phenotype remain to be experimentally elucidated.
Large-scale GWAS of timing of maximal or lowest activity and midpoint of sleep using measures from 7-day accelerometry show a strong association of chronotype with objective estimates of sleep and activity timing but not objectively measured sleep duration or quality16. However, the absence of concurrent sleep diaries in current large-scale wrist accelerometry datasets limits precise measurement of sleep onset or offset. GWAS of relative circadian amplitude derived from daily rest-activity rhythms in the UK Biobank identified two associations at the NFASC and SLC25A17 gene regions, with genetic links to mood instability34.
A few studies have examined the mechanistic or physiologic effects of newly discovered genetic variants. Carriers of a PER2 missense variant that confers eveningness were found to have a longer circadian period, or circadian cycle length, as measured in highly controlled in-laboratory experiments35. Investigation of sleep-wake regulation in zebrafish and integration with human genetic evidence for sleep traits showed that EGFR signalling and its downstream pathways have a central and conserved role in regulating sleep36. A GWAS of cellular circadian rhythm function measured through circadian gene expression in cultured cord blood fibroblasts identified protein catabolism and the COP9 signalosome (which stabilizes the essential circadian protein BMAL1) as essential for human circadian variation37. These studies highlight interindividual differences in circadian timing are partially innate and point towards the core molecular clock as one mechanistic pathway.
Sleep duration and sleep quality
The first human genetic studies of sleep duration targeted monogenic conditions such as rare familial natural short sleep (FNSS), where affected family members regularly exhibit unusually short sleep with no apparent adverse consequences. Linkage and candidate gene sequencing highlighted a rare missense mutation in DEC2, a regulator of both the molecular clock38 and of orexin signalling39. Subsequently, other DEC2 missense mutations were found to be associated with short sleep and enhanced resistance to sleep deprivation, confirming an important role for this gene in human sleep39,40. Exome sequencing in families with natural short sleep with Mendelian inheritance identified mutations in GRM1 (REF.41), ADRB1 (REF.42) and NPSR (REF.43) that led to increased neuronal activity and shorter sleep when modelled in mice. These studies suggest that increased global or local brain cellular excitability may be a mechanism for natural short sleep. This experimental follow-up has been critical to establish functional significance44–46. These studies have been complemented by recent GWAS of self-reported habitual sleep duration17,31,47,48, daytime sleepiness49 and frequency of daytime napping50 in adults. The strongest and most reproducible association for sleep duration has been at the PAX8 locus — PAX8 encodes a transcription factor involved in development, particularly of the kidney and thyroid gland — with an effect size estimated at 2.44 minutes per allele for self-reported sleep duration17,47 and 3.24 minutes per allele estimated by objective 7-day activity monitoring16. Genes at self-reported sleep duration loci are enriched in neuronal pathways including striatum and subpallium development, mechanosensory response, dopamine binding, synaptic neurotransmission and plasticity17.
GWAS of self-reported daytime sleepiness and napping, reflecting deficits in sleep quality and quantity, arousal pathways or natural preferences, have identified 42 and 123 loci, respectively49,50. As daytime sleepiness and frequent napping may result from insufficient sleep, underlying sleep disorders or genetic variation in sleep/wake or circadian regulatory processes, a cross-trait mapping and clustering approach may prove particularly helpful in defining distinct contributing biological mechanisms, as for other complex traits51. Indeed, clustering of genetic loci that lead to daytime sleepiness based on their association with other subjective and objective sleep measures identified two distinct daytime sleepiness genetic mechanisms that reflect either a higher propensity for sleep (longer and more consolidated sleep) or higher sleep fragmentation49. GWAS of objective sleep quality measures, such as sleep efficiency and number of sleep episodes from actigraphy, found enrichment for serotonin-processing genes16. Overall, the genetic architectures of sleep timing traits and chronotype differ substantially from that of sleep quality and quantity traits, as evidenced by low genetic correlation (rg) at the genome-wide level (for example, rg = −0.06 between morningness and sleep duration). However, several genetic variants are associated with multiple sleep timing, duration and quality phenotypes, suggesting a fundamental role in sleep regulation with multiple phenotypic effects. For example, a missense variant in orexin receptor 2 (HCRTR2), a gene implicated in REM sleep regulation and narcolepsy and a known drug target for insomnia, increases daytime sleepiness and associates with higher napping frequency, earlier sleep timing and morningness chronotype50.
In sum, GWAS in adults have implicated many genomic regions of association with very small individual effects, suggesting that circadian and sleep behaviours in adults reflect multiple underlying processes and are highly polygenic, with individual common genetic variants conferring small effects, as has been observed for other complex traits and diseases52. These findings in humans are also consistent with the polygenicity of sleep phenotypes and behaviours in mice53 and in flies54,55. Given that circadian rhythms and sleep behaviour change dramatically throughout the lifespan56,57, genetic studies in children and adolescents, at puberty and menopause, and in the elderly are needed. The largest genetic study of sleep duration in children suggests that there is little genetic overlap with adult sleep duration, but no robust genetic findings have been reported58. By contrast, a polygenic score of 351 morningness preference loci from adults predicted morningness and earlier sleep timing in adolescents59. More genetic studies of common and rare variation in children and adults from more diverse populations are needed, and leveraging well phenotyped families and individuals with extreme phenotypes will continue to be a powerful strategy in uncovering the biological basis of sleep and circadian rhythm phenotypes.
Circadian and sleep disorders
Circadian and sleep disorders can affect the quality, timing and amount of sleep and lead to daytime impairment. Circadian rhythm and sleep disorders can be categorized into dyssomnias and parasomnias60,61, with dyssomnias characterized by trouble falling or staying asleep, often associated with excessive daytime sleepiness (also known as hypersomnolence), and parasomnias characterized by abnormal activities during sleep (TABLE 1). Genetic discoveries for circadian and sleep disorders so far illustrate their complex genetic architecture and integral nature.
Table 1 |.
Known primary sleep disorders
| Sleep disorder | Definition and examples |
|---|---|
| Insomnias | Self-reported difficulty falling or staying asleep, poor sleep quality |
| Parasomnias | Disruptive sleep-related disorders, which includes non-REM sleep-related parasomnias, such as sleep terrors, and REM-related parasomnias, such as nightmare disorder and REM sleep behaviour disorder |
| Central disorders of hypersomnolence | Disorders of daytime sleepiness despite normal night-time sleep and aligned circadian rhythm, such as narcolepsy types 1 and 2, and idiopathic hypersomnia |
| Circadian rhythm sleep-wake disorders | Sleep disruptions owing to misalignment between environment and/or circadian system and sleep-wake cycle, such as jet lag/shift work disorder, advanced or delayed sleep-wake phase disorder, and non-24-hour rhythm disorder |
| Sleep-related movement disorders | Restless leg syndrome |
| Sleep-related breathing disorders | Central sleep apnea, obstructive sleep apnea |
Circadian rhythm sleep-wake disorders
Extreme circadian rhythm disorders, collectively called circadian rhythm sleep-wake phase disorders (SWPDs), include disorders with biological and environmentally driven atypical circadian rhythms, including irregular circadian rhythms, non-24-hour sleep-wake rhythm disorder (non-entrained), which is most often associated with blindness, greatly advanced sleep timing (advanced SWPD), and greatly delayed sleep timing (delayed SWPD)5. Of these, advanced SWPD, as defined by International Classification of Sleep Disorders (ICSD) criteria is prevalent in up to 0.21% of the population62 and delayed SWPD is reported in about 3%5,63,64, with circadian and non-circadian subtypes65. Mendelian forms of advanced SWPD and delayed SWPD have been identified, and linkage or sequencing studies have implicated rare variants in the genes PER2, PER3, CRY1, CRY2, CSNK1D and TIMELESS, confirming the involvement of molecular clock genes previously identified in model organism studies27,28,66–71. Variants have been functionally linked to physiological changes in circadian rhythm cycle length (period) or entrainment to light, as well as molecular changes in phosphorylation in mice, demonstrating the power of human gene discovery followed by functional and mechanistic studies in animal models. However, so far only a limited number of families with circadian disorders have been studied72. In designing future family-based studies, it is important to report a new gene to be confidently implicated only when variants in the same gene appear in multiple unrelated affected individuals (two or more families), and also to report statistical evidence for segregation72. Furthermore, implicated variants may not be fully penetrant, and for circadian variants, effects may be masked by environmental factors, so recall-by-genotype studies with deeper phenotyping in controlled conditions may also be important to reveal relevant human biology. Notably, some circadian forms of advanced SWPD and delayed SWPD likely represent extremes at the end of the chronotype distribution; for example, rare variants (minor allele frequency = 0.4%) in PER3 contribute both to diurnal preference and associate with clinically defined advanced SWPD and delayed SWPD28,33,73.
Narcolepsy
Narcolepsy is a rare teen-onset dyssomnia that affects the sleep-wake cycle, characterized by an intense urge to sleep at unusual times, vivid hallucinations at night and REM sleep dysregulation. Narcolepsy can also occur with cataplexy, a physical collapse upon strong emotional stimulus (type 1) or without cataplexy (type 2) and affects approximately 1 in 2,000 people74. Although narcolepsy is mostly sporadic, there is familial enrichment, with a 20–40-fold increased risk compared to the general population75,76. Through human genetic studies of narcoleptic cases came the ground-breaking first evidence that narcolepsy is an autoimmune disorder, with a striking enrichment of a specific HLA antigen serotype in cases77. These studies were complemented by canine genetic studies of narcolepsy mapping the genetic defect to the orexin receptor type 2 gene (HCRTR2)78, pinpointing the orexin/hypocretin system as specifically affected by this autoimmune disorder. An autoimmune reaction to orexin and molecular mimicry to influenza in narcolepsy type 1 results in selective loss of neurons in the lateral hypothalamus that produce orexin79. GWAS and rare variant exome sequencing studies have identified 8 regions of the genome and 3 genes associated with narcolepsy type 1, in addition to 9 HLA serotypes, with most loci and genes supporting the inflammation and autoimmune hypotheses75,80–83. Nearly all patients with narcolepsy type 1 carry HLA-DQB1 *0602 on at least one chromosome, but this allele is also relatively common, demonstrating that it is not sufficient for disease development84,85; however, HLA-DQB1*0602 serotype testing may be advantageous in diagnosis of narcolepsy86. The interplay between genetics and environment has been demonstrated, as infections and other environmental triggers interact with a susceptible immune background82. Studying narcolepsy has led to an understanding of the critical part the orexin system plays in sleep and wakefulness and the autoimmune underpinnings of narcolepsy (reviewed in REFS.75,87). The genetics of narcolepsy provides a successful example of how to advance from genetics to biological mechanisms and towards the clinic (TABLE 2).
Table 2 |.
From genetics to therapy for an example sleep disorder and future directions in genetics of sleep and circadian disorders
| Level | Narcolepsy | Future directions |
|---|---|---|
| Gene | Canine narcolepsy gene HCRTR2 identified Genome-wide association study identifies HLA and other immune-related genes |
Common and rare variant discovery In silico and experimental analyses to identify causal variants and effector genes Multi-ancestry analyses Leveraging biobanks and electronic health records Homozygous loss-of-function mutation carriers |
| Tissue and pathway | Autoimmune mechanism based on molecular mimicry Loss of orexin neurons in hypothalamus T cell-mediated killing of orexin neurons |
Epigenetics Spatiotemporal transcriptomics Integrative multi-omics In vitro cell models Timing of cellular function |
| Circuits | Orexin neurons integrate sleep-wake, circadian, motivation and visceral signals to promote arousal, lower REM sleep, increase reward and learning, locomotion and sympathetic tone | Imaging and brain function Multi-oscillator network Animal models Organoid models |
| Physiology | Loss of orexin causes instability of sleep-wake state Key role of orexin in flipflop switch During wakefulness, orexin stimulates wake-promoting areas, solidifying wakefulness During sleep, sleep-promoting areas inhibit orexin as well as wake-promoting areas, solidifying sleep |
Objective, scalable measures Recall-by-genotype human mechanistic trials Sleep deprivation and circadian misalignment effects |
| Disorder | Narcolepsy Poor wake maintenance Sleep fragmentation Effects on metabolism and feeding Effects on reward behaviour Triggered by environment |
Population-specific polygenic risk scores Subtyping of disease based on genetic signatures Gene-by-environment effects |
| Therapy | Symptoms-based Orexin receptor agonists in development Preventive immune-based therapies in development Stem cell replacement therapy for hypocretin neurons |
New therapies based on causal mechanisms Behavioural interventions Precision chronotherapy |
Restless legs syndrome
Restless legs syndrome (RLS) is a dyssomnia and a neurological disorder affecting 5–15% of the adult population, characterized by an urge to move the legs that worsens at night, disrupts sleep and is improved by walking or stretching88. There is a strong genetic component to RLS, with 60% of cases having affected relatives, with familial RLS onset typically prior to age 45 years. Brain regional iron deficiency has been implicated in RLS pathophysiology by post-mortem, neuroimaging and cerebrospinal fluid studies, and dopamine dysfunction is implicated because of the clinical efficacy of dopamine agonists89,90. Genetic factors identified to date explain about 12% of the observed disease variance; through GWAS91, there are now 19 loci associated with RLS, with another six implicated through a transcriptome-wide association study92. The findings of these genetic studies, reviewed recently93, and functional follow-up studies in zebrafish and mice94,95 implicate neural development pathways in the pathophysiology of RLS and further underscore the importance of iron metabolism and homeostasis and dopamine dysfunction in RLS. Lead signals implicate the genes MEIS1, involved in neurogenesis and neuronal specification; BTBD9, important in metamorphosis and limb pattern formation in Drosophila and linked to both periodic limb movements in sleep and iron deficiency96; and SKOR1, a regulator of cell fate of spinal cord dorsal horn interneurons that relay pain and touch, and a repressor of genes enriched in neurodevelopmental processes97. These observations link to previous neurophysiological studies that have described changes in sensorimotor networks and increased excitability in cortical neurons, particularly in the motor cortex, and that identified opioid receptors in the pathogenesis of RLS88.
Sleep-disordered breathing
Sleep-disordered breathing encompasses a spectrum of disorders that includes central sleep apnea, sleep-related hypoventilation and obstructive sleep apnea (OSA), the latter characterized by recurrent episodes of upper airway obstruction (apneas and hypopneas). OSA, the most prevalent of these disorders, is found in more than 15% of adults, with higher prevalence associated with older age, male sex and Asian or Hispanic ancestries. Obstructed breathing causes intermittent hypoxaemia, sleep fragmentation and sympathetic nervous system surges, increasing the risk of a wide variety of chronic health problems. Genetic studies have examined OSA using multiple indices: self-reported snoring (a hallmark symptom), International Classification of Diseases, Ninth and Tenth Revisions (ICD) codes for OSA diagnosis from electronic health records, and quantitative measures from overnight sleep studies, including the Apnea Hypopnea Index (AHI), respiratory event length and oxygen saturation levels and patterns. Heritability for the AHI is estimated to vary from 6% to 30%, while respiratory event length heritability is as high as 60%98. Obesity is associated with a two- to sixfold increased prevalence of OSA, and OSA and obesity have a high genetic correlation (rg = 0.72). Adjustment of OSA genetic associations for obesity generally has decreased the magnitude of signals in obesity pathways, although signals have often become stronger in pathways associated with respiratory, inflammatory and neurological disorders. Recent large-scale studies within biobanks of snoring99 or ICD-coded OSA100 have identified genetic associations for individuals with European ancestries, with smaller multi-ancestry cohort studies leveraging admixture101 (for example, identifying FECH) or linkage102,103 signals to improve power, including rare variant associations (for example, CAV1 and DLC1 ). The latter studies have directed research of OSA to pathways that modulate inflammation, iron and steroid metabolism, lung function and hypoxia responses, which are all in the early stages102,103. Genetic associations of sleep apnea are heterogeneous, with some ancestry-specific104, some stage-specific (for example, REM versus non-REM sleep)105 and some sex-specific (for example, RAI1 in men)106, indicating the value of employing trans-ancestry, stage-specific and sex-stratified analysis approaches for complex phenotypes such as OSA as well as other sleep and circadian traits107.
Insomnia
Insomnia is characterized by chronic difficulties in falling asleep and/or maintaining sleep with associated daytime impairment. Insomnia disorder, defined as significant insomnia at least three nights a week for at least three months, is seen in approximately 10% of the population and is linked to adverse long-term health outcomes108, whereas occasional insomnia symptoms are seen in approximately 30% of the population. Early studies of insomnia heritability have come mainly from twin studies (heritability of 22% to 59% in adults, 14% to 71% in children), with a longitudinal twin study demonstrating stability over time and differential heritability by sex (around 59% in women and around 38% in men109. Candidate gene studies and early small GWAS (n <10,000) paved the way for large-scale studies of insomnia genetics by combining validated questionnaires of insomnia symptoms and actigraphy-measured phenotypes with genetic data110–113. Large-scale GWAS based on questionnaires assessing insomnia symptoms, self-report of physician diagnosis and use of sleep prescription aids (n >100,000) have identified many loci (554 risk loci as of the most recent GWAS preprint; 202 published loci from meta-analysis of 23andMe and UK Biobank) implicating axons, striatum, medium spiny neurons and the hypothalamus in insomnia pathophysiology114–117. These studies estimate SNP-based heritability to be about 8% and suggest a highly polygenic architecture, but few specific genes have been implicated so far118. Insomnia GWAS identified associations with RLS loci, with follow-up analyses pointing towards undiagnosed RLS cases reporting insomnia symptoms115,117. Importantly, the genetic associations with insomnia are robust even when considering several possible confounders, such as caffeine consumption, socioeconomic status and existing comorbidities. Polygenic risk scores for insomnia associate with physician-diagnosed or ICD-10 diagnosed insomnia disorder in smaller cohorts115, but large-scale GWAS of physician-diagnosed insomnia disorder are still lacking. Comprehensive reviews of insomnia genetics119 and sleep disorders in children have recently been reported120.
Lessons learned from disorders
GWAS studies of circadian and sleep phenotypes highlight important lessons. The first is the challenge of heterogeneity and misclassification. For example, GWAS of insomnia symptoms revealed associations with validated genetic variants for RLS121, and further analyses demonstrated that this association is most probably owing to the inclusion of individuals with RLS reporting insomnia symptoms115,117,121. Insomnia, particularly occasional insomnia, is probably not a singular condition but a cluster of symptoms with a range of underlying causes. Future genetic studies need to dissect this heterogeneity. Another lesson is in the importance of phenotype definitions, as phenotype definitions range from symptoms, and subjective and objective sleep quality, to clinical disorder diagnosis. The scalability of questionnaire-based phenotypes and ICD codes has enabled increased sample size and genetic power to detect associations, but clinical specificity will be necessary to understand molecular specificity and expand the utility of genetic findings for prediction and prevention. For example, in the latest meta-analysis of insomnia genetics122, despite a sample size increase, the newly discovered genetic loci explain less of the variation than a prior study116, perhaps owing to poorer phenotyping and characteristics of the additional individuals sampled (r2 = 2.0% in the largest study of 2.3 million versus r2 = 2.6% in the 2019 study of 1.3 million). Therefore, there is a need for both standardized and easy-to-use phenotypic instruments. Phenotypic instruments assessed at scale will advance gene discovery from large-scale cohorts, and precise, detailed phenotyping in smaller, well designed cohorts will enable gene characterization. Nonetheless, data from currently available studies, for example, for sleep apnea, emphasize that analysis of even simple phenotypes (from medical records) can identify promising signals and genetic correlations with other disorders, whereas more detailed analysis (from polysomnography) can elucidate potential mechanisms related to disease severity (oxygen desaturation), subtype (state specificity), sexual dimorphism and ancestry-driven variants. Lastly, more studies with rigorous unbiased genetic approaches are necessary to capture the full spectrum of genetic mutations. An increase in exome and genome sequencing of rare disorder cases and families together with population outlier extremes is necessary. Genetic studies must also push towards more robust trans-ancestry genetic approaches combined with high-throughput downstream functional assays to advance biology and pinpoint new therapeutic targets for circadian and sleep disorders.
Genetic insights for therapy
Within both the field of circadian rhythms and sleep, there is plenty of need for new therapeutics and for personalization. Particularly in sleep therapeutics, pharmacological treatments are often aimed at symptoms rather than underlying causes. Human genetic studies of circadian rhythms and sleep open the door to potential novel targets for drug development, as well as tailoring of current therapeutics123. Genetic studies also have the unique ability to identify drug targets in causal pathways.
In narcolepsy type 1, active drug development is shifting to mechanism-based therapeutics, including targeting the immune and orexin systems124. Current narcolepsy type 1 treatment consists primarily of psychostimulants for excessive daytime sleepiness and sodium oxybate or antidepressants for cataplexy and REM sleep deregulation. Under development are agonists acting at the orexin-2 receptor and orexin replacement therapy via implanted intrathecal catheter or intranasal administration. Furthermore, given that the genetic background of over 98% of all patients with narcolepsy type 1 (versus 25% of controls) carry the HLA class II HLA-DQB1 *0602 allele, immune-based therapies are being investigated for the treatment of those at risk of developing narcolepsy and preventing the destruction of orexin neurons, with an active search for reliable biomarkers. In addition to pharmacological treatments, genetic revelation of the mechanisms of narcolepsy type 1 have led to both immune-targeted diagnosis methods and stem cell replacement therapy for orexin neurons125 (TABLE 2). Therefore, it is critical to continue genetic investigations of the underlying mechanisms of circadian and sleep disorders to enable new targeted therapeutics, diagnostics and preventative interventions.
Genetic links to other complex diseases
Over the years, many epidemiologic and experimental studies have linked circadian and sleep traits and disorders with facets of human health and disease, including cardiometabolic and psychiatric disorders4,6,8, but it is unclear to what degree sleep and circadian disorders are markers, consequences and/or causal in disease pathology. Human genetics could play an important part in resolving the directionality of associations126. This work is crucial in demonstrating the central role of circadian rhythms and sleep in human health and arguing for the inclusion of circadian and sleep health as a central component of preventative and diagnostic healthcare. Recent circadian and sleep GWAS have enabled both studies of genetic relationships to health and disease and causal interrogation of those genetic connections (FIGS. 4,5).
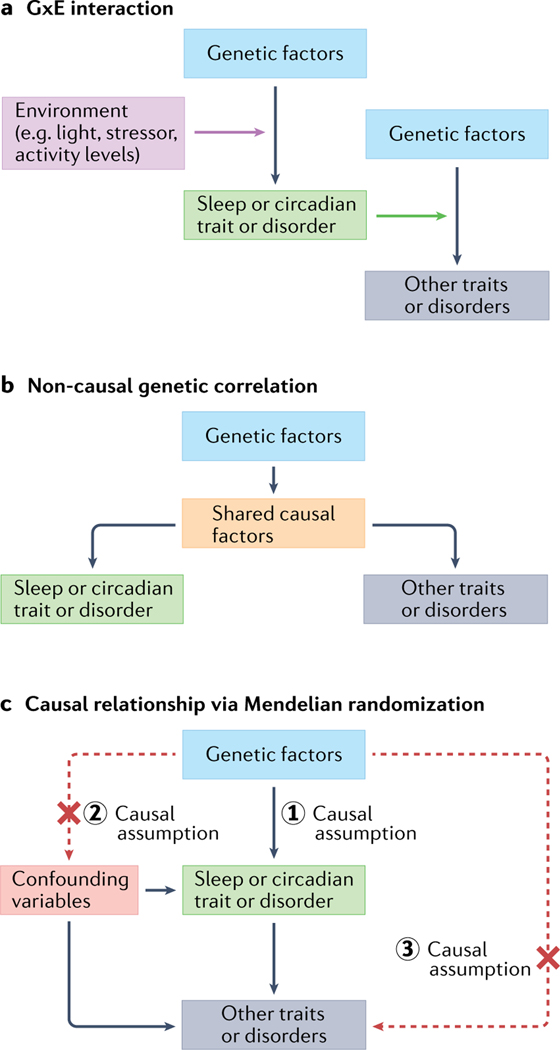
Fig. 5 |. Potential sources of sleep and circadian genetic relationships to environmental factors and other traits and disorders.
a | Gene by environment (GxE) interactions of sleep and circadian traits and disorders where sleep and circadian rhythms can act both as outcomes and environmental modifiers of disease. b | Similar to part a with shared causal factors (pleiotropy). Non-causal relationships between sleep/circadian traits or disorders and other traits or disorders can occur when genetic factors are independently associated with both outcomes. c | Graphical representation of a causal genetic relationship between sleep/circadian traits and disorders with downstream traits/disorders interrogated by Mendelian randomization analysis. For causality testing using Mendelian randomization, the three causal assumptions highlighted must be satisfied: (1) genetic factors (for example, associated SNPs) are strongly and clearly linked to sleep/circadian traits/disorders (‘the exposure’); (2) the genetic factors are not linked to any confounders; and (3) the genetic factors should be linked to the outcomes through the sleep/circadian traits/disorders only. Black arrows indicate causal links and red-dashed lines indicate potential violations of Mendelian randomization assumptions.
Shared genetic architecture
Examination of patterns of genetic correlations, or shared genetic influences are beginning to provide insights into relationships between circadian or sleep and cardio-metabolic and psychiatric traits90,98,127 (FIGS. 4a,5). For example, genetic correlations between insomnia and psychiatric disorders such as anxiety and depression are higher (rg = 0.45–0.59) than between insomnia and most other sleep traits, suggesting that the aetiology of insomnia shares biological processes more with psychiatric disorders than with sleep-wake regulatory processes115,116,128. Furthermore, genetic sharing with different sleep phenotypes differs between disorders, suggesting that diseases may be distinguishable by distinct patterns of sleep disruption.
Genetic evidence for causality
Mendelian randomization is a genetic method that utilizes human genetic variants associated with a risk factor as natural experiments to test whether the risk factor may reflect a cause of the disease, thus reducing influences from confounders and reverse causality that are limitations of other epidemiologic approaches in humans129,130 (FIGS. 4b,5). For example, in the field of lipid and cholesterol biology, Mendelian randomization helped to eliminate high-density lipoprotein cholesterol from the causal pathway for heart attack, enabling the redirection of drug targets131. For circadian rhythms, prior observational prospective epidemiologic studies have linked chronotype with depression132,133, reporting effects in both directions: early depression associated with increasing eveningness preference in youth, and chronotype associated with incident depression in youth and adult women. Mendelian randomization results from several studies confirm a causal effect of the ‘lifelong intervention’ of genetic variation for morning chronotype as protective for psychiatric disorders, especially major depressive disorder31–33,134,135. A recent randomized control trial that targeted sleep timing, meal timing, light exposure, caffeine intake, napping behaviour and exercise behaviour provided evidence for the feasibility of interventions to produce clinically relevant shifts in circadian rhythms136. In recent studies of insomnia and short sleep, a causal link to coronary artery disease and myocardial infarction was demonstrated114–117, further supporting numerous observational studies linking insomnia with cardiac outcomes137,138. This connection has emphasized the need for randomized control trials of treatment of sleep disruption as a preventive therapy for heart disease and advocacy to reduce the health disparities around sleep disruption. Taken together, these two examples make a strong case for the use of human genetics to understand the causal pathways of circadian rhythms and sleep in health and disease. Mouse models with naturally occurring or knockout mutations in clock genes have been found to have metabolic disorders such as obesity and type 2 diabetes mellitus, infertility, increased tumour development in response to stress, premature ageing, impaired cognition, memory or mood phenotypes (reviewed in REF.15), suggesting that clock genes have key roles in disease. How these causal links translate to humans remains an important question to answer. Genetic studies also support bidirectional causal links between sleep disorders and disease, such as insomnia and depression, consistent with models suggesting that sleep disturbances contribute to a spiraling exacerbation of disease139.
Sleep disorder markers of degeneration
Genetic studies suggest that some sleep disorders may be early markers of progressive neurodegenerative disease pathology. REM behaviour disorder (RBD) is a rare parasomnia, affecting 0.3–1.1% of adults, in which people act out their dreams and lack atonia during REM sleep, which may reflect a prodromal neurodegenerative synucleinopathy140. Indeed, more than 90% of patients with RBD progress to Parkinson disease or other dementias within 15 years of diagnosis. Overall, genetic characterization suggests that RBD-associated Parkinson disease is a subtype of Parkinson disease, with contributions from some Parkinson disease-predisposing alleles, for example, in GBA, but not from others, for example, in LRRK2 (REF.141). Deeper understanding of the genetic architecture of RBD, and discovery of genetic modifiers of rate of progression from RBD to neurodegenerative disease, will have clinical value for precision diagnosis, management and treatment. Although circadian and sleep dysfunction is present in patients with Parkinson disease, the underlying genetic mechanisms and links to RBD remain unknown142. In addition, recent studies suggest genetic liability for Alzheimer disease may predict short sleep, lending support to investigation of sleep oscillations as an early marker143,144. Similarly, fatal familial insomnia is a progressive neurodegenerative disease caused by dominant mutations in the prion protein PRNP that presents with sleep disturbances and insomnia (which progressively worsen as toxic prion proteins accumulate in the brain)145.
Circadian and sleep behaviour as a modifier
Circadian and sleep disruption are known modifiable risk factors for many common, complex diseases. Given their increasing prevalence in modern society, an important question is whether circadian and sleep behaviours modify the genetic risk of other diseases (FIGS. 4c,5). While highly relevant, studies focusing on circadian or sleep behaviour as modifiers of genetic risk of disease are just emerging. A twin study, as a classical method of disentangling the contributions of genetic and environmental factors for a trait of interest, first found that shorter sleep duration increased genetic influences on body mass index (BMI)146. Recent studies in the CHARGE consortium have successfully used genome-wide gene-sleep duration interaction testing to identify novel loci for blood pressure147 and lipid traits148. However, heterogeneity between different study cohorts, in both measurements of lifestyle or environmental and genetic variables, and cultural heterogeneity, can introduce more variability compared with a similarly sized single-cohort study. Thus, the recent development in large-scale prospective cohorts with rich information on lifestyle and environments has provided a unique opportunity to circumvent the aforementioned limitations. Indeed, recent studies utilizing UK Biobank data have identified that poor sleep characteristics (for example, short sleep duration, daytime nap/sleepiness, insomnia and snoring) increase the genetic risk of obesity-related measures149,150 and the risks of coronary heart disease and stroke151.
As an emerging field of investigation, there are still many gaps. For example, most gene-circadian/sleep behaviour studies were performed in adults with European ancestries. It is important to learn whether population groups of different ancestries or paediatric populations share the same interactions. The multi-ancestry sleep-by-SNP interaction study148 and another study in Chinese children and adolescents152 are beginning to address this gap.
In addition, almost all previous studies used lifestyle questionnaires to obtain circadian or sleep behaviour, which are prone to recall bias and limited to a few easily attainable phenotypes (Supplementary Table 1). As more cohorts are starting to implement accelerometer-based movement monitors or wearables that measure sleeping EEG and/or heart rate, objectively assessing sleep behaviour may provide more accurate estimates of time taken to fall asleep, regularity and fragmentation of sleep, depth of sleep as well as rest-activity rhythms. Moreover, further longitudinal studies are needed that address the limited ‘life course’ assessment of lifestyles and behaviours or ‘exposomes’ over time and capitalize on the opportunity to use low burdensome data from wearables and ‘nearables’, as well as public databases on pollution. The All of Us cohort is a large initiative aimed at collecting comprehensive environmental and genetics data, with sleep data potentially available via wearables (for example, Fitbit). Importantly, as growing evidence has shown that meal timing can be a critical diurnal behaviour to affect metabolic health153,154 (BOX 1 ), future studies should consider including its assessment for gene-behaviour analyses.
Box 1 |. Food timing and MTNR1B diabetes risk allele-mediated glucose intolerance.
Genetic links between sleep, circadian and metabolic factors have been well established in mice and flies188. Genetic evidence for this link in humans has included the discovery of variants in the clock gene CRY2 and melatonin receptor MTNR1B in glucose regulation and type 2 diabetes mellitus risk189 (FIG. 2), association at the body mass index (BMI)-associated FTO/IRX3/IRX5 and PATJ loci with natural variation in sleep traits (reviewed in REF.190), and genetic links between daytime napping and obesity traits50. Here, we focus on one example: MTNR1B.
GWAS for glycaemic traits identified variants in the melatonin receptor 2 gene (MTNR1B) associated with an increased risk of type 2 diabetes mellitus182. among these variants, the MTNR1B rs10830963 SNP confers the strongest effects on disposition index (the product of both insulin secretion and insulin sensitivity), fasting glucose level191,192 and first-phase insulin secretion193. Importantly, the significant associations between the MTNR1B rs10830963 SNP and glycaemic traits were reported in multiethnic populations. The risk allele of this locus is common across ethnic groups (allele frequency of 7.1% in the african/african american (‘afr’ in gnomaD) population, 20.7% in the american admixed/Latino (‘amr’ in gnomaD) population, 28.9% in people of non-Finnish European (‘nfe’ in gnomaD) ancestries, and 43% in East asians (‘eas’ in gnomaD)194), indicating broad relevance. The mechanism underlying this association remains an active area of investigation. The prevailing hypothesis is based on the inhibitory effect of melatonin signalling on insulin release from the pancreatic islets192,195. a key finding came from functional studies in which upregulation of MTNR1B mRNa expression was found in the islets from the carriers of the common rs10830963 variant (the risk G-allele)196,197, with the risk allele increasing FoXa2-bound enhancer activity in islet- and liver-derived cells and allele-specific differences in NEuRoD1 binding in islet-derived cells. Indeed, a small placebo-controlled trial observed that exogenous melatonin worsens glucose tolerance, particularly in MTNR1B common risk allele carriers as opposed to non-carriers198. Such an effect also applies to endogenous physiological melatonin: in a randomized crossover study, the MTNR1B risk allele, but not the non-risk allele, impaired glucose tolerance in response to late night versus early dinners (that is, in the presence of elevated endogenous melatonin concentrations)199,200. Interestingly, carriers of the rs10830963 risk allele show extended melatonin secretion in the morning201, which suggests that eating too early may also impair postprandial glucose tolerance in risk-allele carriers. These data reflect the potential interaction effects of certain behaviours (for example, late-night dinner, early breakfast) and the MTNR1B common variant, highlighting the importance of personalized recommendations in vulnerable populations. Rare variants in MTNR1B have also been reported to be associated with diabetes risk and glucose control202; however, replication studies are needed.
Conclusions and future perspectives
The fields of circadian rhythms and sleep are poised to make significant advances in human biology and improve human health (TABLE 2). Notably, gene discovery at a large scale has become possible primarily through large biobanks with coarse questionnaire-based and wrist-activity monitor phenotyping and limited demographic composition, and few follow-up mechanistic studies are available. Thus, systems-level insights are yet to be gained for most sleep and circadian traits and disorders.
To enable further genetic insights, new methods of scalable phenotyping will need to be balanced with measurement specificity. New apps and devices for scalable dynamic monitoring of circadian and sleep behaviour are emerging that take advantage of changes in accelerometry, temperature, oxygen saturation, heart rate variability and brain electrical activity during sleep as well as molecular phenotyping to capture internal body timing or circadian phase. Integration of these technologies paired together with biobanks and biorepositories of genetic information including whole-exome and whole-genome sequencing will enable novel gene discovery. Increasingly large biobanks with circadian rhythm and sleep measures are necessary to understand the phenotypic consequences of rare loss-of-function mutations. Although genetic studies using physiologically defined phenotypes have been limited by sample size, and studies utilizing codes based on electronic health records or questionnaires have been limited by phenotypes that do not characterize disease severity or subtypes of disease (for example, REM-predominant OSA) and have misclassification bias, there is further opportunity to triangulate these approaches to leverage the power of the large, simple phenotype study with smaller, deep-phenotyping studies to identify variants that drive susceptibility for specific circadian and sleep disorder subtypes, and potentially uncover novel disease mechanisms. In parallel, it will continue to be important to integrate insights from lessons learned from genetic efforts in other diseases and electronic health-care settings to catalyse large-scale genomic analyses and translation into a clinical setting for sleep and circadian rhythm traits and disorders155.
The next challenge is the synthesis of discoveries into biological mechanisms and insights that enable clinical translation. To turn the recent wave of GWAS into advances in human biology, large-scale functional follow-up studies will need to be conducted in tandem with deep dives into cell-type- and developmental-stage-specific effects. The challenge of understanding the effect of genetic variants on biological function is common to all fields of human genetics, and insights and resources generated in other fields will prove useful in deriving insights into biological mechanisms for sleep and circadian rhythm human genetics156. Cellular models of large-scale gene perturbation with cell-autonomous readouts of clock gene cycling or sleep-related neuronal activity are also likely to be powerful resources in understanding gene function. Although in vitro functional studies are harder for systemic processes such as sleep, studies that evaluate function at a large scale, such as sequence-based omics analyses may help towards biological interpretation at scale. Importantly, genotype-targeted physiologic studies that interrogate how genetic variants for sleep and sleep disorders influence EEG measures, or for circadian traits influence circadian physiology are expected to provide important insights into biological mechanisms.
Given the number of loci that influence circadian and sleep disorders, a systematic, integrative approach that takes advantage of external biological resources and information on functional and tissue-specific enrichments across risk loci may serve as a complementary approach to prioritize causal genes and identify underlying causal biological processes. As the number of signals for a sleep disorder increases, large-scale functional genomic analyses can uncover biological clues into the molecular and cellular processes that play a core part in pathogenesis, and the spatial and temporal context of their disruption. Enrichment of GWAS signals in regulatory elements active in specific tissue types, or in elements regulated by the molecular clock, suggest that, as more disease loci are found, the biology will converge to specific molecular, cellular and systemic processes. This level of global analyses is only beginning for sleep genetic studies, but GWAS has revealed novel biology for other complex traits, for example uncovering the role of the complement pathway in age-related macular degeneration and autophagy in inflammatory bowel disease.
Convergence of loci and pathways implicated by both rare Mendelian disorders and common variant associations can provide further insight into phenotypic consequences of dose-dependent perturbation of a key circadian or sleep gene. For example, rare, highly penetrant alleles in PER3 contribute to extremes of chronotype (advanced or delayed sleep phase syndromes), and common alleles in PER3 contribute to chronotype and sleep timing in the general population33. Comprehensive exome sequencing of PER3 variation in large biobanks suggests that a continuous spectrum of morningness alleles exists across frequencies and effect sizes, with the potential to illuminate genotype-phenotype relationships157. Extrapolating from this example to other circadian and sleep genes, this allelic spectrum can inform on the direction of effect (that is, whether gain or loss of gene function leads to the phenotype), and genotype-driven analysis can show whether an allele has variable penetrance in the population. One immediate goal of the field should be to test current polygenic scores, with special sensitivity to their applicability across diverse-ancestry groups, for predictive utility to integrate genetic risk of circadian and sleep disruption into the clinical framework for severe complications such as anesthaesia-induced delirium or suicide risk in individuals with depression.
The genetics of circadian biology has benefited from strong effects of a limited set of clock genes, its cell-autonomous basis and high precision of phenotypes, whereas challenges in genetics of sleep physiology and disorders include heterogeneous mechanisms, systemic manifestation and complex phenotypes. Genetic discoveries are expected to reveal novel biology and connections to disease that may offer promising avenues for precision medicine. Thus, genetics and genomics will remain powerful approaches to enhance biological understanding of circadian and sleep disorders and linked diseases in humans.
Supplementary Material
Acknowledgements
J.M.L. is supported by NIH grant number K01 HL136884. J.Q. is supported by NIH grant number K99HL148500. S.R. is supported by NIH grants on genetics of sleep apnea. E.M. is supported by NIH grants. F.A.J.L.S. is supported by NIH grants R01 DK105072, R01 HL140574, R01 HL153969 and R01 DK102696. R.S. is supported by NIH grants R01 DK105 072, R01 DK102696, R01 DK107859, R01 HL14 6751 and R2 1 AG068890, DOD grant W81XWH2010776, a Dreem Jury prize (in-kind support) and a Phyllis and Jerome Lyle Rappaport MGH Research Scholar Award.
S.R. reports receipt of NIH grants that include studies of the genetics of sleep apnea. F.A.J.L.S. served on the Board of Directors for the Sleep Research Society and has received consulting fees from the University of Alabama at Birmingham. The interests of F.A.J.L.S. were reviewed and managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. The consultancies of F.A.J.L.S. are not related to the current work. R.S. is a founder and shareholder of Magnet Biomedicine, not related to the current work. R.S. has received in-kind support from Dreem.
Glossary
- Suprachiasmatic nucleus (SCN)
Located in the hypothalamus, the SCN is the master circadian oscillator and coordinates circadian rhythms throughout the body
- Circadian rhythm
An approximately 24-hour rhythm in cellular, physiological or behavioural processes that are driven by the internal circadian timekeeping system; this rhythm is sustained in the absence of environmental and behavioural rhythms and can be entrained to solar day/night cycle by light
- Molecular clock
Molecular mechanism driving circadian rhythms, consisting of transcriptional-translational feedback loops of core clock genes
- Homeostatic
The sleep-wake homeostat balances the need for sleep and wakefulness and regulates sleep intensity. The longer you are awake the greater your body’s need for sleep (‘homeostatic sleep pressure’), which subsides during sleep (‘sleep dissipation’). The sleep-wake homeostat works in concert with the circadian rhythm
- REM sleep
REM sleep, which occurs primarily during the latter half of a sleep cycle (primarily owing to the influence of process C), is associated with the highest brain activity during sleep and with dreaming. During REM sleep, most of the body’s skeletal muscles are paralysed
- Clock genes
Core clock genes are directly involved in the primary transcriptional-translational feedback loops. By contrast, clock-controlled genes are those genes whose expression is driven by the transcriptional-translational feedback loops within cells and tissues, resulting in circadian oscillations in their function
- Circadian period
Duration of time for completion of a full cycle, that is, the time it takes to move from a reference point in a rhythmic process (a particular phase) to return to the same point (for example, time difference between two sequential dim light melatonin onsets). in humans, the endogenous circadian period is roughly 24.15 hours
- Phase
Reference time within the circadian cycle, for example, at which a particular event occurs such as the minimum or maximum of a circadian measure
- Electroencephalograph (EEG)
A measurement of electrical brain activity that uses sensors and can be used to identify stages of sleep
- Polysomnography (PSG)
A measurement of multiple parallel channels of data during sleep, typically including EEG and measures of muscle activity (electromyography), eye movements (electrooculography), and heart rate (electrocardiography). For clinical PSGs, the following measures are typically measured in addition: oxygen levels (pulse oximetry), breathing (respiratory belts, nasal pressure sensor, thermistor), and leg movements (leg electromyography). PSG is frequently used to diagnose a variety of sleep disorders, such as sleep apnea
- Wearable devices
Also known as wearables, these devices record real-time environmental, behavioural and/or physiological parameters such as accelerometry, skin temperature and heart rate
- Melatonin
A hormone produced by the pineal gland during the circadian night, and which can be suppressed by retinal light in mammals. Synthesis and secretion of melatonin into the circulation is controlled by the SCN, and therefore melatonin is used as a marker of the phase of internal circadian time
- Actigraphy
A non-invasive wrist-worn accelerometer-based objective measurement of average motor activity over a period of time
- Polygenic score
Also known as polygenic risk score. A score that summarizes genetic liability to a trait or disease and is typically calculated by aggregating the weighted effect of many trait-associated genetic variants
- Excessive daytime sleepiness
Difficulty staying awake or alert during the day, which is often a symptom of sleep problems and/or disorders
- Entrainment
The synchronization of a (here, circadian) rhythm to an environmental, behavioural, or other circadian cycle, such as the light/dark cycle, eating/fasting or temperature cycles
- Sleep–wake cycle
The daily pattern of alternating wakefulness and sleep with a periodicity typically approximating the 24-hour day–night cycle
- Nearables
Devices worn not on but near to the body, with a variety of sensors that can measure and record signals, such as snoring sounds, body movement or heart rate, as physiological proxies to estimate (for example) sleep duration, timing and/or quality
Footnotes
Competing interests
The other authors report no competing interests.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41576-022-00519-z.
References
- 1.Mohawk JA, Green CB & Takahashi JS Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci 35, 445–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibner C, Schibler U. & Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol 72, 517–549 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Borbély AA A two process model of sleep regulation. Hum. Neurobiol 1, 195–204 (1982). [PubMed] [Google Scholar]
- 4.Buysse DJ Sleep health: can we define it? Does it matter? Sleep 37, 9–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy JF et al. Circadian rhythm sleep-wake disorders: gaps and opportunities. Sleep 44, zsaa281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chellappa SL, Vujovic N, Williams JS & Scheer FAJL Impact of circadian disruption on cardiovascular function and disease. Trends Endocrinol. Metab 30, 767–779 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kecklund G. & Axelsson J. Health consequences of shift work and insufficient sleep. BMJ 355, i5210 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Rijo-Ferreira F. & Takahashi JS Genomics of circadian rhythms in health and disease. Genome Med. 11, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allada R, Cirelli C. & Sehgal A. Molecular mechanisms of sleep homeostasis in flies and mammals. Cold Spring Harb. Perspect. Biol 9, a027730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deboer T, Vansteensel MJ, Détári L. & Meijer JH Sleep states alter activity of suprachiasmatic nucleus neurons. Nat. Neurosci 6, 1086–1090 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Khalsa SBS, Jewett ME, Cajochen C. & Czeisler CA A phase response curve to single bright light pulses in human subjects. J. Physiol 549, 945–952 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijk DJ & Czeisler CA Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans. J. Neurosci 15, 3526–3538 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasan S. et al. A human sleep homeostasis phenotype in mice expressing a primate-specific PER3 variable-number tandem-repeat coding-region polymorphism. FASEB J. 28, 2441–2454 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Möller-Levet CS et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl Acad. Sci. USA 110, E1132–E1141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi JS, Hong H-K, Ko CH & McDearmon EL The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet 9, 764–775 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones SE et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat. Commun 10, 1585 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dashti HS et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat. Commun 10, 1100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doherty A. et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat. Commun 9, 5257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duffy JF & Dijk DJ Getting through to circadian oscillators: why use constant routines? J. Biol. Rhythms 17, 4–13 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Wang W. et al. Using Kleitman’s Forced Desynchrony protocol to assess the intrinsic period of circadian oscillators and estimate the contributions of the circadian pacemaker and the sleep-wake homeostat to physiology and behavior in clinical research. Nat. Protoc (In Press, 2022).
- 21.Perez-Pozuelo I. et al. The future of sleep health: a data-driven revolution in sleep science and medicine. npj Digit. Med 3, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ambrosius U. et al. Heritability of sleep electroencephalogram. Biol. Psychiat 64, 344–348 (2008). [DOI] [PubMed] [Google Scholar]
- 23.De Gennaro L. et al. The electroencephalographic fingerprint of sleep is genetically determined: a twin study. Ann. Neurol 64, 455–460 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Vitaterna MH, Shimomura K. & Jiang P. Genetics of circadian rhythms. Neurol. Clin 37, 487–504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreani TS, Itoh TQ, Yildirim E, Hwangbo D-S & Allada R. Genetics of circadian rhythms. Sleep. Med. Clin 10, 413–421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roenneberg T. & Merrow M. Entrainment of the human circadian clock. Cold Spring Harb. Symp. Quant. Biol 72, 293–299 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Archer SN et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep 26, 413–415 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Ebisawa T. et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2, 342–346 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons MJ et al. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNβ3 with sleep measures. J. Sleep. Res 23, 595–604 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y. et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat. Commun 7, 10448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones SE et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 12, e1006125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane JM et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat. Commun 7, 10889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones SE et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat. Commun 10, 343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson A. et al. Genome-wide association study of circadian rhythmicity in 71,500 UK biobank participants and polygenic association with mood instability. eBioMedicine 35, 279–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang A-M et al. Chronotype genetic variant in PER2 is associated with intrinsic circadian period in humans. Sci. Rep 9, 5350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DA et al. Evolutionarily conserved regulation of sleep by epidermal growth factor receptor signaling. Sci. Adv 5, eaax4249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaspar L. et al. The genomic landscape of human cellular circadian variation points to a novel role for the signalosome. eLife 6, e24994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He Y. et al. The transcriptional repressor DEC2 regulates sleep length in mammals. Science 325, 866–870 (2009). This paper described the first gene for natural short sleep discovered using a human genetic family-based approach.
- 39.Hirano A. et al. DEC2 modulates orexin expression and regulates sleep. Proc. Natl Acad. Sci. USA 115, 3434–3439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pellegrino R. et al. A novel BHLHE41 variant is associated with short sleep and resistance to sleep deprivation in humans. Sleep 37, 1327–1336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi G. et al. Mutations in metabotropic glutamate receptor 1 contribute to natural short sleep trait. Curr. Biol 31, 13–24.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi G. et al. A rare mutation of β 1-adrenergic receptor affects sleep/wake behaviors. Neuron 10.1016/j.neuron.2019.07.026 (2019). [DOI] [PMC free article] [PubMed]
- 43.Xing L. et al. Mutant neuropeptide S receptor reduces sleep duration with preserved memory consolidation. Sci. Transl. Med 11, eaax2014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toda H, Shi M, Williams JA & Sehgal A. Genetic mechanisms underlying sleep. Cold Spring Harb. Symp. Quant. Biol 10.1101/sqb.2018.83.037705 (2019). [DOI] [PubMed]
- 45.Summa KC & Turek FW The genetics of sleep: insight from rodent models. Sleep. Med. Clin 6, 141–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cirelli C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat. Rev. Neurosci 549–560 (2009). [DOI] [PMC free article] [PubMed]
- 47.Gottlieb DJ et al. Novel loci associated with usual sleep duration: the CHARGE consortium genome-wide association study. Mol. Psychiat 20, 1232–1239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishiyama T. et al. Genome-wide association meta-analysis and Mendelian randomization analysis confirm the influence of ALDH2 on sleep duration in the Japanese population. Sleep 10.1093/sleep/zsz046 (2019). [DOI] [PubMed]
- 49.Wang H. et al. Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat. Commun 10, 3503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dashti HS et al. Genetic determinants of daytime napping and effects on cardiometabolic health. Nat. Commun 12, 900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Udler MS et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med. 15, e1002654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Visscher PM, Yengo L, Cox NJ & Wray NR Discovery and implications of polygenicity of common diseases. Science 373, 1468–1473 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diessler S. et al. A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biol. 16, e2005750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harbison ST, McCoy LJ & Mackay TFC Genome-wide association study of sleep in Drosophila melanogaster. BMC Genom. 14, 281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Tunc I, Tansey TR, Pirooznia M. & Harbison ST Identification of genes contributing to a long circadian period in Drosophila melanogaster. J. Biol. Rhythms 36, 239–253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duffy JF, Zitting K-M & Chinoy ED Aging and circadian rhythms. Sleep. Med. Clin 10, 423–434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohayon MM, Carskadon MA, Guilleminault C. & Vitiello MV Meta-analysis of quantitative sleep parameters from childhood to old age in healthy individuals: developing normative sleep values across the human lifespan. Sleep 27, 1255–1273 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Marinelli M. et al. Heritability and genome-wide association analyses of sleep duration in children: the EAGLE consortium. Sleep 39, 1859–1869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merikanto I. et al. Circadian preference and sleep timing from childhood to adolescence in relation to genetic variants from a genome-wide association study. Sleep Med. 50, 36–41 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Sateia MJ International classification of sleep disorders-third edition: highlights and modifications. Chest 146, 1387–1394 (2014). [DOI] [PubMed] [Google Scholar]
- 61.American Psychiatric Association Diagnostic and Statistical Manual of Mental Disorders (DSM-5) 5th edn (American Psychiatric Pub, 2013). [Google Scholar]
- 62.Curtis BJ et al. Extreme morning chronotypes are often familial and not exceedingly rare: the estimated prevalence of advanced sleep phase, familial advanced sleep phase, and advanced sleep-wake phase disorder in a sleep clinic population. Sleep 10.1093/sleep/zsz148 (2019). [DOI] [PMC free article] [PubMed]
- 63.Sivertsen B, Harvey AG, Gradisar M, Pallesen S. & Hysing M. Delayed sleep-wake phase disorder in young adults: prevalence and correlates from a national survey of Norwegian university students. Sleep. Med 77, 184–191 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Paine S-J, Fink J, Gander PH & Warman GR Identifying advanced and delayed sleep phase disorders in the general population: a national survey of New Zealand adults. Chronobiol. Int 31, 627–636 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Murray JM et al. Prevalence of circadian misalignment and its association with depressive symptoms in delayed sleep phase disorder. Sleep 40, zsw002 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Satoh K, Mishima K, Inoue Y, Ebisawa T. & Shimizu T. Two pedigrees of familial advanced sleep phase syndrome in Japan. Sleep 26, 416–417 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Pereira DS et al. Association of the length polymorphism in the human Per3 gene with the delayed sleep-phase syndrome: does latitude have an influence upon it? Sleep 28, 29–32 (2005). [PubMed] [Google Scholar]
- 68.Xu Y. et al. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature 434, 640–644 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Jones CR et al. Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nat. Med 5, 1062–1065 (1999). [DOI] [PubMed] [Google Scholar]
- 70. Toh KL et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291, 1040–1043 (2001). This paper described the first gene for advanced sleep phase syndrome discovered using a human genetic family-based approach.
- 71.Kurien P. et al. TIMELESS mutation alters phase responsiveness and causes advanced sleep phase. Proc. Natl Acad. Sci. USA 10.1073/pnas.1819110116 (2019). [DOI] [PMC free article] [PubMed]
- 72.MacArthur DG et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L. et al. A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proc. Natl Acad. Sci. USA 113, E1536–E1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kornum BR et al. Narcolepsy. Nat. Rev. Dis. Prim 3, 16100 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Ollila HM Narcolepsy type 1: what have we learned from genetics? Sleep 10.1093/sleep/zsaa099 (2020). [DOI] [PMC free article] [PubMed]
- 76.Mignot E. Genetic and familial aspects of narcolepsy. Neurology 50, S16–S22 (1998). [DOI] [PubMed] [Google Scholar]
- 77.Langdon N, Welsh KI, van Dam M, Vaughan RW & Parkes D. Genetic markers in narcolepsy. Lancet 2, 1178–1180 (1984). [DOI] [PubMed] [Google Scholar]
- 78.Lin L. et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98, 365–376 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Luo G. et al. Autoimmunity to hypocretin and molecular mimicry to flu in type 1 narcolepsy. Proc. Natl Acad. Sci. USA 115, E12323–E12332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hor H. et al. Genome-wide association study identifies new HLA class II haplotypes strongly protective against narcolepsy. Nat. Genet 42, 786–789 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Faraco J. et al. ImmunoChip study implicates antigen presentation to T cells in narcolepsy. PLoS Genet. 9, e1003270 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Han F. et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genet. 9, e1003880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Degn M. et al. Rare missense mutations in P2RY11 in narcolepsy with cataplexy. Brain 140, 1657–1668 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Han F. et al. HLA-DQ association and allele competition in Chinese narcolepsy. Tissue Antigens 80, 328–335 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Mignot E, Hayduk R, Black J, Grumet FC & Guilleminault C. HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep 20, 1012–1020 (1997). [PubMed] [Google Scholar]
- 86.Capittini C. et al. Correlation between HLA-DQB1*06:02 and narcolepsy with and without cataplexy: approving a safe and sensitive genetic test in four major ethnic groups. A systematic meta-analysis. Sleep. Med 52, 150–157 (2018). [DOI] [PubMed] [Google Scholar]
- 87.Miyagawa T. & Tokunaga K. Genetics of narcolepsy. Hum. Genome Var 6, 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trenkwalder C. et al. Comorbidities, treatment, and pathophysiology in restless legs syndrome. Lancet Neurol. 10.1016/S1474-4422(18)30311-9 (2018). [DOI] [PubMed]
- 89.Earley CJ et al. Altered brain iron homeostasis and dopaminergic function in restless legs syndrome (Willis-Ekbom disease). Sleep Med. 15, 1288–1301 (2014). [DOI] [PubMed] [Google Scholar]
- 90.Jiménez-Jiménez FJ, Alonso-Navarro H, García-Martín E. & Agúndez JAG Neurochemical features of idiopathic restless legs syndrome. Sleep. Med. Rev 45, 70–87 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Schormair B. et al. Identification of novel risk loci for restless legs syndrome in genome-wide association studies in individuals of European ancestry: a meta-analysis. Lancet Neurol. 16, 898–907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Akçimen F. et al. Transcriptome-wide association study for restless legs syndrome identifies new susceptibility genes. Commun. Biol 3, 373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jiménez-Jiménez FJ, Alonso-Navarro H, García-Martín E. & Agúndez JAG Genetics of restless legs syndrome: an update. Sleep Med. Rev 39, 108–121 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Spieler D. et al. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 24, 592–603 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Drgonova J. et al. Mouse model for protein tyrosine phosphatase D (PTPRD) associations with restless leg syndrome or Willis-Ekbom disease and addiction: reduced expression alters locomotion, sleep behaviors and cocaine-conditioned place preference. Mol. Med 21, 717–725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stefansson H. et al. A genetic risk factor for periodic limb movements in sleep. N. Engl. J. Med 357, 639–647 (2007). [DOI] [PubMed] [Google Scholar]
- 97.Sarayloo F. et al. SKOR1 has a transcriptional regulatory role on genes involved in pathways related to restless legs syndrome. Eur. J. Hum. Genet 28, 1520–1528 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang J. et al. Comparison of heritability estimation and linkage analysis for multiple traits using principal component analyses. Genet. Epidemiol 40, 222–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campos AI et al. Insights into the aetiology of snoring from observational and genetic investigations in the UK Biobank. Nat. Commun 11, 817 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strausz S. et al. Genetic analysis of obstructive sleep apnoea discovers a strong association with cardiometabolic health. Eur. Respir. J 57, 2003091 (2021). [DOI] [PubMed] [Google Scholar]
- 101.Wang H. et al. Admixture mapping identifies novel loci for obstructive sleep apnea in hispanic/latino americans. Hum. Mol. Genet 10.1093/hmg/ddy387 (2018). [DOI] [PMC free article] [PubMed]
- 102.Wang H. et al. Variants in angiopoietin-2 (ANGPT2) contribute to variation in nocturnal oxyhaemoglobin saturation level. Hum. Mol. Genet 25, 5244–5253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liang J. et al. Sequencing analysis at 8p23 identifies multiple rare variants in DLC1 associated with sleep-related oxyhemoglobin saturation level. Am. J. Hum. Genet 105, 1057–1068 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cade BE et al. Genetic associations with obstructive sleep apnea traits in Hispanic/Latino Americans. Am. J. Respir. Crit. Care Med 194, 886–897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cade BE et al. Associations of variants In the hexokinase 1 and interleukin 18 receptor regions with oxyhemoglobin saturation during sleep. PLoS Genet. 15, e1007739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen H. et al. Multiethnic meta-analysis identifies RAI1 as a possible obstructive sleep apnea-related quantitative trait locus in men. Am. J. Respir. Cell Mol. Biol 58, 391–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukherjee S, Saxena R. & Palmer LJ The genetics of obstructive sleep apnoea. Respirology 23, 18–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morin CM et al. Insomnia disorder. Nat. Rev. Dis. Prim 1, 15026 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Lind MJ, Aggen SH, Kirkpatrick RM, Kendler KS & Amstadter AB A longitudinal twin study of insomnia symptoms in adults. Sleep 38, 1423–1430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Amin N. et al. Genetic variants in RBFOX3 are associated with sleep latency. Eur. J. Hum. Genet 24, 1488–1495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ban H-J, Kim SC, Seo J, Kang H-B & Choi JK Genetic and metabolic characterization of insomnia. PLoS ONE 6, e18455 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Spada J. et al. Genome-wide association analysis of actigraphic sleep phenotypes in the LIFE adult study. J. Sleep. Res 25, 690–701 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Parsons MJ et al. Replication of genome-wide association studies (GWAS) loci for sleep in the British G1219 cohort. Am. J. Med. Genet. B 162B, 431–438 (2013). [DOI] [PubMed] [Google Scholar]
- 114.Lane JM et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat. Genet 49, 274–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lane JM et al. Biological and clinical insights from genetics of insomnia symptoms. Nat. Genet 10.1038/s41588-019-0361-7 (2019). [DOI] [PMC free article] [PubMed]
- 116. Jansen PR et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet 10.1038/s41588-018-0333-3 (2019). This largest-to-date GWAS for insomnia identified 202 genetic loci associated with insomnia symptoms and causal links to cardiometabolic traits.
- 117.Hammerschlag AR et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat. Genet 49, 1584–1592 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Khlghatyan J. et al. Fxr1 regulates sleep and synaptic homeostasis. EMBO J. 39, e103864 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lind MJ & Gehrman PR Genetic pathways to insomnia. Brain Sci. 6, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mainieri G. et al. The genetics of sleep disorders in children: a narrative review. Brain Sci. 11, 1245 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.El Gewely M. et al. Reassessing GWAS findings for the shared genetic basis of insomnia and restless legs syndrome. Sleep 10.1093/sleep/zsy164 (2018). [DOI] [PubMed]
- 122.Watanabe K. et al. Genome-wide meta-analysis of insomnia in over 2.3 million individuals implicates involvement of specific biological pathways through gene-prioritization. Preprint at medRxiv 10.1101/2020.12.07.20245209 (2020). [DOI]
- 123.Krystal AD & Prather AA Sleep pharmacogenetics: the promise of precision medicine. Sleep. Med. Clin 14, 317–331 (2019). [DOI] [PubMed] [Google Scholar]
- 124.Barateau L. & Dauvilliers Y. Recent advances in treatment for narcolepsy. Ther. Adv. Neurol. Disord 12, 1756286419875622 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Equihua-Benítez AC, Equihua-Benítez JA, Guzmán-Vásquez K, Prospero-García O. & Drucker-Colín R. Orexin cell transplant reduces behavioral arrest severity in narcoleptic mice. Brain Res. 1745, 146951 (2020). [DOI] [PubMed] [Google Scholar]
- 126.Pingault J-B et al. Using genetic data to strengthen causal inference in observational research. Nat. Rev. Genet 19, 566–580 (2018). [DOI] [PubMed] [Google Scholar]
- 127.Bulik-Sullivan B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Byrne EM The relationship between insomnia and complex diseases-insights from genetic data. Genome Med. 11, 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Emdin CA, Khera AV & Kathiresan S. Mendelian randomization. JAMA 318, 1925–1926 (2017). [DOI] [PubMed] [Google Scholar]
- 130.Smith GD & Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol 32, 1–22 (2003). [DOI] [PubMed] [Google Scholar]
- 131.Voight BF et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet 380, 572–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vetter C. et al. Prospective study of chronotype and incident depression among middle- and older-aged women in the Nurses’ Health Study II. J. Psychiat. Res 103, 156–160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Haraden DA, Mullin BC & Hankin BL The relationship between depression and chronotype: a longitudinal assessment during childhood and adolescence. Depress. Anxiety 34, 967–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.O’Loughlin J. et al. Using Mendelian randomisation methods to understand whether diurnal preference is causally related to mental health. Mol. Psychiat 10.1038/s41380-021-01157-3 (2021). [DOI] [PMC free article] [PubMed]
- 135.Daghlas I, Lane JM, Saxena R. & Vetter C. Genetically proxied diurnal preference, sleep timing, and risk of major depressive disorder. JAMA Psychiat. 10.1001/jamapsychiatry.2021.0959 (2021). [DOI] [PMC free article] [PubMed]
- 136.Facer-Childs ER, Middleton B, Skene DJ & Bagshaw AP Resetting the late timing of ‘night owls’ has a positive impact on mental health and performance. Sleep. Med 60, 236–247 (2019). [DOI] [PubMed] [Google Scholar]
- 137.Javaheri S. & Redline S. Insomnia and risk of cardiovascular disease. Chest 152, 435–444 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zheng B. et al. Insomnia symptoms and risk of cardiovascular diseases among 0.5 million adults: a 10-year cohort. Neurology 10.1212/WNL.0000000000008581 (2019). [DOI] [PMC free article] [PubMed]
- 139.Baranova A, Cao H. & Zhang F. Shared genetic liability and causal effects between major depressive disorder and insomnia. Hum. Mol. Genet 10.1093/hmg/ddab328 (2021). [DOI] [PubMed]
- 140.Högl B, Stefani A. & Videnovic A. Idiopathic REM sleep behaviour disorder and neurodegeneration — an update. Nat. Rev. Neurol 14, 40–55 (2018). [DOI] [PubMed] [Google Scholar]
- 141.Gan-Or Z, Alcalay RN, Rouleau GA & Postuma RB Sleep disorders and Parkinson disease; lessons from genetics. Sleep Med. Rev 41, 101–112 (2018). [DOI] [PubMed] [Google Scholar]
- 142.Gros P. & Videnovic A. Overview of sleep and circadian rhythm disorders in parkinson disease. Clin. Geriatr. Med 36, 119–130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Leng Y, Ackley SF, Glymour MM, Yaffe K. & Brenowitz WD Genetic risk of Alzheimer’s disease and sleep duration in non-demented elders. Ann. Neurol 89, 177–181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lucey BP et al. Sleep and longitudinal cognitive performance in preclinical and early symptomatic Alzheimer’s disease. Brain 10.1093/brain/awab272 (2021). [DOI] [PMC free article] [PubMed]
- 145.Medori R. et al. Fatal familial insomnia, a prion disease with a mutation at codon 178 of the prion protein gene. N. Engl. J. Med 326, 444–449 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Watson NF et al. Sleep duration and body mass index in twins: a gene-environment interaction. Sleep 35, 597–603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang H. et al. Multi-ancestry genome-wide gene-sleep interactions identify novel loci for blood pressure. Mol. Psychiatry 26, 6293–6304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Noordam R. et al. Multi-ancestry sleep-by-SNP interaction analysis in 126,926 individuals reveals lipid loci stratified by sleep duration. Nat. Commun 10, 5121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rask-Andersen M, Karlsson T, Ek WE & Johansson Å Gene-environment interaction study for BMI reveals interactions between genetic factors and physical activity, alcohol consumption and socioeconomic status. PLoS Genet. 13, e1006977 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Celis-Morales C. et al. Sleep characteristics modify the association of genetic predisposition with obesity and anthropometric measurements in 119,679 UK Biobank participants. Am. J. Clin. Nutr 105, 980–990 (2017). This paper demonstrates genetic effects via sleep and chronotype behaviour interactions on obesity-related phenotypes.
- 151.Fan M. et al. Sleep patterns, genetic susceptibility, and incident cardiovascular disease: a prospective study of 385 292 UK biobank participants. Eur. Heart J 41, 1182–1189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fu J. et al. Childhood sleep duration modifies the polygenic risk for obesity in youth through leptin pathway: the Beijing child and adolescent metabolic syndrome cohort study. Int. J. Obes 43, 1556–1567 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Garaulet M. et al. Melatonin effects on glucose metabolism: time to unlock the controversy. Trends Endocrinol. Metab 10.1016/j.tem.2019.11.011 (2020). [DOI] [PMC free article] [PubMed]
- 154.Gill S. & Panda S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 22, 789–798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.eMERGE Consortium Lessons learned from the eMERGE Network: balancing genomics in discovery and practice. HGG Adv. 2, 100018 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lappalainen T. & MacArthur DG From variant to function in human disease genetics. Science 373, 1464–1468 (2021). [DOI] [PubMed] [Google Scholar]
- 157.Weedon MN et al. The impact of Mendelian sleep and circadian genetic variants in a population setting. Preprint at bioRxiv 10.1101/2022.01.04.21268199 (2022). [DOI] [PMC free article] [PubMed]
- 158.Geyer H. Über den Schlaf von Zwillingen. Z. Vererbungslehre 73, 524–527 (1937). [Google Scholar]
- 159.Aserinsky E. & Kleitman N. Regularly occurring periods of eye motility, and concomitant phenomena, during sleep. Science 118, 273–274 (1953). [DOI] [PubMed] [Google Scholar]
- 160.Dement W. & Kleitman N. Incidence of eye motility during sleep in relation to varying EEG pattern. Fed. Proc 14, 216 (1955). [Google Scholar]
- 161.Dement W. & Kleitman N. Cyclic variations in EEG during sleep and their relation to eye movements, body motility, and dreaming. Electroencephalogr. Clin. Neurophysiol 9, 673–690 (1957). [DOI] [PubMed] [Google Scholar]
- 162.Juji T, Satake M, Honda Y. & Doi Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens 24, 316–319 (1984). [DOI] [PubMed] [Google Scholar]
- 163.Chemelli RM et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98, 437–451 (1999). [DOI] [PubMed] [Google Scholar]
- 164.Winkelmann J. et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat. Genet 39, 1000–1006 (2007). [DOI] [PubMed] [Google Scholar]
- 165.Viola AU et al. PER3 polymorphism predicts sleep structure and waking performance. Curr. Biol 17, 613–618 (2007). [DOI] [PubMed] [Google Scholar]
- 166.Vyazovskiy VV, Cirelli C, Pfister-Genskow M, Faraguna U. & Tononi G. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci 11, 200–208 (2008). [DOI] [PubMed] [Google Scholar]
- 167.Konopka RJ & Benzer S. Clock mutants of Drosophila melanogaster. Proc. Natl Acad. Sci. USA 68, 2112–2116 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Moore RY & Eichler VB Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42, 201–206 (1972). [DOI] [PubMed] [Google Scholar]
- 169.Stephan FK & Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl Acad. Sci. USA 69, 1583–1586 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Reddy P. et al. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell 38, 701–710 (1984). [DOI] [PubMed] [Google Scholar]
- 171.Bargiello TA & Young MW Molecular genetics of a biological clock in Drosophila. Proc. Natl Acad. Sci. USA 81, 2142–2146 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Sawaki Y, Nihonmatsu I. & Kawamura H. Transplantation of the neonatal suprachiasmatic nuclei into rats with complete bilateral suprachiasmatic lesions. Neurosci. Res 1, 67–72 (1984). [DOI] [PubMed] [Google Scholar]
- 173.Lehman MN et al. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J. Neurosci 7, 1626–1638 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ralph MR, Foster RG, Davis FC & Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978 (1990). [DOI] [PubMed] [Google Scholar]
- 175.Vitaterna MH et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yamazaki S. et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685 (2000). [DOI] [PubMed] [Google Scholar]
- 177.Tosini G. & Menaker M. Circadian rhythms in cultured mammalian retina. Science 272, 419–421 (1996). [DOI] [PubMed] [Google Scholar]
- 178.Balsalobre A, Damiola F. & Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937 (1998). [DOI] [PubMed] [Google Scholar]
- 179.Grundschober C. et al. Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J. Biol. Chem 276, 46751–46758 (2001). [DOI] [PubMed] [Google Scholar]
- 180.Panda S. et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002). [DOI] [PubMed] [Google Scholar]
- 181.Scheer FAJL, Hilton MF, Mantzoros CS & Shea SA Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl Acad. Sci. USA 106, 4453–4458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Prokopenko I. et al. Variants in MTNR1B influence fasting glucose levels. Nat. Genet 41, 77–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patke A. et al. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169, 203–215.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ruben MD et al. A database of tissue-specific rhythmically expressed human genes has potential applications in circadian medicine. Sci. Transl. Med 10, eaat8806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McClung CR Plant circadian rhythms. Plant Cell 18, 792–803 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Blume C. et al. Across the consciousness continuum-from unresponsive wakefulness to sleep. Front. Hum. Neurosci 10.3389/fnhum.2015.00105 (2015). [DOI] [PMC free article] [PubMed]
- 187. Daghlas I. et al. Sleep duration and myocardial infarction. J. Am. Coll. Cardiol 74, 1304–1314 (2019). This study used a longitudinal study design and Mendelian randomization to investigate the causal relationship between sleep duration and myocardial infarction.
- 188.Panda S. Circadian physiology of metabolism. Science 354, 1008–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dupuis J. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet 42, 105–116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dashti HS & Ordovás JM Genetics of sleep and insights into its relationship with obesity. Annu. Rev. Nutr 10.1146/annurev-nutr-082018-124258 (2021). [DOI] [PubMed]
- 191.Sparsø T. et al. G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release. Diabetes 58, 1450–1456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Lyssenko V. et al. Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat. Genet 41, 82–88 (2009). This study demonstrates a role for a circadian-related gene, MTNR1B, in type 2 diabetes mellitus, highlighting the role of the hormone melatonin in diabetes pathogenesis via a direct inhibitory effect in pancreatic β cells.
- 193.Wood AR et al. A genome-wide association study of IVGTT-based measures of first-phase insulin secretion refines the underlying physiology of type 2 diabetes variants. Diabetes 66, 2296–2309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Karczewski KJ et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Karamitri A. & Jockers R. Melatonin in type 2 diabetes mellitus and obesity. Nat. Rev. Endocrinol 15, 105–125 (2019). [DOI] [PubMed] [Google Scholar]
- 196.Gaulton KJ et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet 47, 1415–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tuomi T. et al. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 23, 1067–1077 (2016). [DOI] [PubMed] [Google Scholar]
- 198.Garaulet M. et al. Common type 2 diabetes risk variant in MTNR1B worsens the deleterious effect of melatonin on glucose tolerance in humans. Metabolism 64, 1650–1657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lopez-Minguez J, Saxena R, Bandín C, Scheer FA & Garaulet M. Late dinner impairs glucose tolerance in MTNR1B risk allele carriers: a randomized, cross-over study. Clin. Nutr 10.1016/j.clnu.2017.04.003 (2017). [DOI] [PMC free article] [PubMed]
- 200.Garaulet M. et al. Interplay of dinner timing and MTNR1B type 2 diabetes risk variant on glucose tolerance and insulin secretion: a randomized crossover trial. Diabetes Care 45, 512–519 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lane JM et al. Impact of common diabetes risk variant in MTNR1B on sleep, circadian, and melatonin physiology. Diabetes 65, 1741–1751 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bonnefond A. et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat. Genet 44, 297–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.