Abstract
We have investigated whether the identity of the coreceptor (CCR5, CXCR4, or both) used by primary human immunodeficiency virus type 1 (HIV-1) isolates to enter CD4+ cells influences the sensitivity of these isolates to neutralization by monoclonal antibodies and CD4-based agents. Coreceptor usage was not an important determinant of neutralization titer for primary isolates in peripheral blood mononuclear cells. We also studied whether dualtropic primary isolates (able to use both CCR5 and CXCR4) were differentially sensitive to neutralization by the same antibodies when entering U87MG-CD4 cells stably expressing either CCR5 or CXCR4. Again, we found that the coreceptor used by a virus did not greatly affect its neutralization sensitivity. Similar results were obtained for CCR5- or CXCR4-expressing HOS cell lines engineered to express green fluorescent protein as a reporter of HIV-1 entry. Neutralizing antibodies are therefore unlikely to be the major selection pressure which drives the phenotypic evolution (change in coreceptor usage) of HIV-1 that can occur in vivo. In addition, the increase in neutralization sensitivity found when primary isolates adapt to growth in transformed cell lines in vitro has little to do with alterations in coreceptor usage.
Human immunodeficiency virus type 1 (HIV-1) enters CD4+ T cells via an interaction with CD4 and coreceptor molecules, the most important of which yet identified are the chemokine receptors CXCR4 and CCR5 (4, 12, 23, 26, 28, 32). CXCR4 is used by T-cell line-tropic (T-tropic) primary isolates or T-cell line-adapted (TCLA) lab strains, whereas CCR5 is used by primary isolates of the macrophage-tropic (M-tropic) phenotype (4, 12, 23, 26, 28, 32). Most T-tropic isolates and some TCLA strains are actually dualtropic in that they can use both CXCR4 and CCR5 (and often other coreceptors such as CCR3, Bonzo/STRL33, and BOB/gpr15), at least in coreceptor-transfected cells (18, 24, 30, 54, 89). The M-tropic and T-tropic/dualtropic nomenclature has often been used interchangeably with the terms “non-syncytium-inducing” (NSI) and “syncytium-inducing” (SI), although it is semantically imprecise to do so.
M-tropic viruses are those most commonly transmitted sexually (3, 33, 87, 106) and from mother to infant (2, 72, 81). If T-tropic strains are transmitted, or when they emerge, this is associated with a more rapid course of disease in both adults (17, 37, 46, 51, 52, 76, 78, 82, 92, 101) and children (6, 45, 84, 90). However, T-tropic viruses emerge in only about 40% of infected people, usually only several years after infection (76, 78). A well-documented, albeit anecdotal, study found that when a T-tropic strain was transmitted by direct transfer of blood, its replication was rapidly suppressed: the T-tropic virus was eliminated from the body, and M-tropic strains predominated (20). These results suggest that there is a counterselection pressure against the emergence of T-tropic strains during the early stages of HIV-1 infection in most people. But what is this pressure?
Since the M-tropic and T-tropic phenotypes are properties mediated by the envelope glycoproteins whose function is to associate with CD4 and the coreceptors, a selection pressure differentially exerted on M- and T-tropic viruses could, in principle, act at the level of virus entry. In other words, neutralizing antibodies to the envelope glycoproteins, or the chemokine ligands of the coreceptors, could theoretically interfere more potently with the interactions of T-tropic strains with CXCR4 than with M-tropic viruses and CCR5. A differential effect of this nature could suppress the emergence of T-tropic viruses. Consistent with this possibility, neutralizing antibodies are capable of preventing the CD4-dependent association of gp120 with CCR5 (42, 94, 103), and chemokines can also prevent the coreceptor interactions of HIV-1 (8, 13, 23, 28, 70).
Here, we explore whether the efficiency of HIV-1 neutralization is affected by coreceptor usage. Although earlier studies have not found T-tropic strains to be inherently more neutralization sensitive than M-tropic ones (20, 40, 44), previously available reagents and techniques may not have been adequate to fully address this question. One major problem is that even single residue changes can drastically affect both antibody binding to neutralization epitopes and the HIV-1 phenotype (25, 55, 62, 67, 83, 91), and so studies using relatively unrelated viruses and a fixed antibody (polyclonal or monoclonal) preparation have two variables to contend with: the viral phenotype (coreceptor use) and the antigenic structure of the virus and hence the efficiency of the antibody-virion interaction.
We have used a new experimental strategy to explore whether coreceptor usage affects neutralization sensitivity in the absence of other confounding variables: the use of dualtropic viruses able to enter CD4+ cells via either CCR5 or CXCR4. By using a constant HIV-1 isolate or clone and the same monoclonal antibodies (MAbs) or CD4-based reagents as neutralizing agents, we can ensure that the only variable under study in the neutralization reaction is the nature of the coreceptor used for entry. Our major conclusion is that there is no strong association between coreceptor usage and neutralization sensitivity for primary HIV-1 isolates. Independent studies have reached the same conclusion (53a, 59). The emergence of T-tropic (SI) viruses in vivo may be unlikely to be due to escape from antibody-mediated selection pressure.
MATERIALS AND METHODS
Viruses and antibodies.
All virus isolates and clones were from genetic subtype B. The origins and characteristics of each virus are listed in Table 1. All virus stocks, including TCLA isolates, were grown and titrated in mitogen-stimulated peripheral blood mononuclear cells (PBMC) as described previously (96). The 50% tissue culture infective dose (TCID50) refers to the virus titer on PBMC. The phenotypes (NSI or SI) of the test viruses have been described previously (Table 1) or were determined according to the ability to form syncytia in MT-2 cells (36, 82). Isolate P17/H9 was generated by passaging P17 once in H9 cells. Supernatant from this passage was then used to produce virus stocks from PBMC.
TABLE 1.
HIV-1 isolates and clones used in this studya
| Virus | Tropismb | Clonalityc | Replication in:
|
Source | Reference | |
|---|---|---|---|---|---|---|
| U87MG-CD4-CCR5 cells | U87MG-CD4-CXCR4 cells | |||||
| CCR5 primary | ||||||
| SF162 | NSI | Isolate | − | C. Cheng-Mayer | 86 | |
| JR-FL | NSI | Biol. clone | + | − | I. S. Y. Chen | 71 |
| Case B2-6-92 | NSI | Isolate | +/− | − | R. I. Connor | 18 |
| ADA (clAD8) | NSI | Mol. clone | + | − | M. A. Martin | 93 |
| CCR5 + CXCR4 primary | ||||||
| Isolate C7/86 | SI | Isolate | ++ | +++ | R. I. Connor | 18 |
| DH123 | SI | Mol. clone | ++ | ++ | R. Shibata | 85 |
| 2076 cl1 | SI | Biol. clone | ++ | ++ | P. R. Clapham | 89 |
| 2076 cl3 | SI | Biol. clone | ++ | ++ | P. R. Clapham | 89 |
| P17 | SI | Isolate | ++ | +++ | R. A. Fisher | 97 |
| Case B5/86 | SI | Isolate | +/− | ++ | R. I. Connor | 18 |
| 89.6 | SI | Mol. clone | + | ++ | R. G. Collman | 16 |
| CXCR4 primary | ||||||
| HC4 | SI | Biol. clone (p) | − | +++ | S. E. Forte | 34 |
| HC7 | SI | Biol. clone (p) | − | +++ | S. E. Forte | 34 |
| 2075 | SI | Isolate | − | +++ | P. R. Clapham | 89 |
| 2044 | SI | Isolate | − | ++ | P. R. Clapham | 89 |
| 2005 cl1 | SI | Biol. clone | − | +/− | P. R. Clapham | 89 |
| ACH-P | SI | Isolate | +/− | +++ | T. Wrin | 102 |
| CCR5 + CXCR4 TCLA | ||||||
| SF2 | SI | Mol. clone | ++ | + | C. Cheng Mayer | 86 |
| P17/H9 | SI | Isolate | +++ | +++ | A. Trkola and R. A. Fisher | 97 |
| C17 | SI | Isolate | ++ | +++ | R. A. Fisher | 97 |
| CXCR4 TCLA | ||||||
| NL4-3 | SI | Mol. clone | − | +++ | W. O’Brien | 1 |
| ACH-H9 | SI | Isolate | +/− | +++ | T. Wrin | 102 |
| ACH-p/H9 | SI | Isolate | + | +++ | T. Wrin | 102 |
Grouped according to coreceptor usage pattern.
As judged by syncytium formation in MT-2 cells.
Whether virus is an uncloned isolate, a biological (biol.) clone, or a molecular (mol.) clone and whether it was of pediatric origin (p).
Human MAbs 2F5, 2G12, 15e, 19b, 447-52D, and IgG1b12, and the CD4-based molecules soluble CD4 (sCD4) and CD4-immunoglobulin G2 (IgG2), have been described previously (5, 29, 39, 43, 66, 69, 95, 96). MAb 447-52D was purchased from Cellular Products Inc. (Buffalo, N.Y.). The ability of each MAb (other than 2F5) to bind to gp120 from each virus was checked by enzyme-linked immunosorbent assay, using nonionic detergent-treated supernatants from virus-infected cultures as the source of gp120 (61, 63). Nonreactive MAb-virus combinations were excluded from the neutralization analyses in Fig. 1 and 2. These combinations were MAb 15e with viruses Case B5/86; and MAb 19b with viruses 2076, P17, P17/H9, C17, Case B5/86, 2075, 2044, and 2005 clone 1 (cl1). We were not able to check for isolate 2005 cl1 whether its resistance to neutralization by MAb 2F5 was due to a change in the epitope sequence. Isolate DH123 has a change in the 2F5 epitope sequence from ELDKWAS to ALDKWAS. However, we have shown that this change does not interfere with binding or neutralization by this MAb (69, 96).
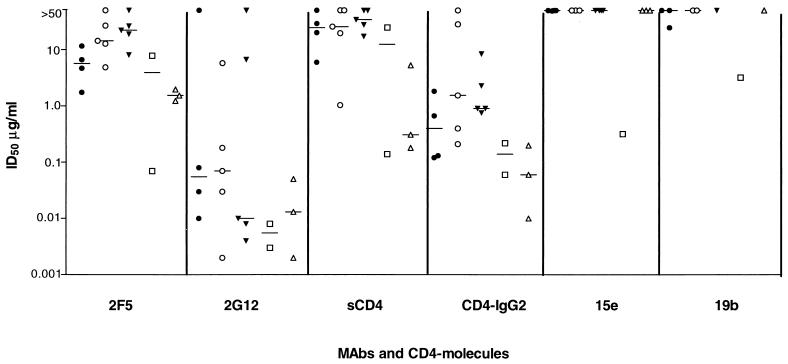
FIG. 1.
Neutralization of HIV-1 by MAbs and CD4-based molecules in PBMC. Viruses are divided into five subgroups: CCR5 primary (•); CCR5 + CXCR4 primary (○); CXCR4 primary (▾); CCR5 + CXCR4 TCLA (□); and CXCR4 TCLA (▵). Values shown are derived from one representative experiment out of two to five performed. When no neutralization at 50 μg/ml was observed, a value >50 μg/ml is reported. For each neutralizing agent and virus category, the median values are depicted by a bar. Data are shown for all viruses listed in Table 1 except for 2076 cl1, HC4, and P17/H9. These viruses are closely related to isolates 2076 cl3, HC7, and C17, respectively, and their neutralization properties were also comparable. Data for only one of each pair are therefore presented. Isolate C7/86 was not tested in PBMC.
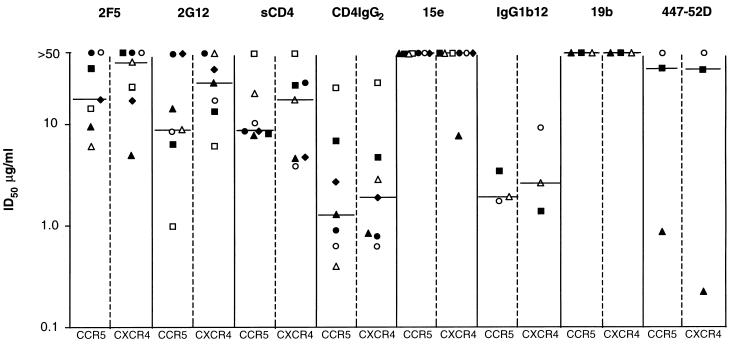
FIG. 2.
Neutralization of dualtropic HIV-1 isolates in U87MG-CD4 cells. The dualtropic isolates used were SF2 (▴), C7/86 (▪), DH123 (▵), 2076 cl3 (□), P17 (•), P17/H9 (○), and C17 (⧫). Values shown are derived from one representative experiment out of two to five performed. When no neutralization at 50 μg/ml was observed, a value >50 μg/ml is reported. For each neutralizing agent, ID50 values obtained on the CCR5-expressing (CCR5) and the CXCR4-expressing (CXCR4) U87MG-CD4 cells are shown. The median values for each reagent on the two cell lines are depicted by a bar.
Cell lines.
Human neuronal U87MG-CD4 cells stably transfected with CCR5 or CXCR4 (U87MG-CD4-CCR5 and U87MG-CD4-CXCR4 cells) were maintained in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal calf serum (FCS), glutamine, antibiotics, puromycin (1 μg/ml; Sigma Chemical), and neomycin (G418; 300 μg/ml; Sigma) and were split twice a week (24, 42).
Human osteosarcoma HOS-CD4 cells stably transfected with CCR5 or CXCR4 and the green fluorescent protein (GFP) reporter gene under the control of the HIV-2 long terminal repeat (designated GHOST-CCR5 and GHOST-CXCR4 cells) were maintained in DMEM containing 10% FCS, glutamine, antibiotics, G418 (500 μg/ml), hygromycin (100 μg/ml), and puromycin (1 μg/ml) and were split twice a week (48).
Determination of coreceptor use by HIV-1.
U87MG-CD4-CCR5 and U87MG-CD4-CXCR4 cells (5 × 104 per well) were seeded into a 24-well plate for 1 day and then incubated with HIV-1 for 16 h before removal of unbound virus by two washes in culture medium. Extracellular p24 antigen was measured 3 and 6 days after infection (96).
Neutralization assay using human PBMC.
PBMC were isolated from healthy blood donors by Ficoll-Hypaque centrifugation and then stimulated for 2 to 3 days with phytohemagglutinin (5 μg/ml) and interleukin-2 (100 U/ml) (a gift of Hoffmann-La Roche, Nutley, N.J.). Neutralization was assessed as described previously (96). The virus inoculum was adjusted to 400 to 1,000 TCID50/ml in assay medium (RPMI 1640, 10% FCS, 100 U of interleukin-2 per ml, glutamine, and antibiotics), and 50-μl aliquots were incubated with serial dilutions of the MAbs or CD4-based molecules (50 μl) for 1 h at 37°C. The calculated 50% inhibitory doses (ID50s) refer to the concentrations of these agents in this preincubation mixture. PBMC (4 × 105 in 100 μl of medium) were then added. The final concentration of virus in the cultures was 20 to 50 TCID50/well, corresponding to 100 to 250 TCID50/ml. The supernatant medium (50 μl) was assayed for p24 antigen at least once between days 4 and 12 postinfection, depending on the growth kinetics of the isolate (96). As the virus inoculum was not washed out at any stage of the experiment, the residual input p24 concentration was also measured and subtracted from all test results. If virus production in the cultures had not reached its peak on day 6, the cultures were fed with 100 μl of medium without adding fresh MAbs and then reanalyzed for p24 production on subsequent days. The production of p24 antigen in the absence of MAb was designated 100%, and the ratios of p24 production in MAb-containing cultures were calculated relative to this value. The MAb concentrations causing 50 and 90% reduction in p24 production were determined by linear regression analysis. If the appropriate degree of inhibition was not achieved at the highest MAb concentration, a value of >50 μg/ml was recorded.
Neutralization assay on U87MG-CD4-CCR5 and U87MG-CD4-CXCR4 cells.
Cells (5 × 104) were seeded into wells of a 24-well plate 1 day prior to the experiment. The virus inoculum was adjusted to 1,000 to 4,000 TCID50/ml in assay medium (DMEM, 10% FCS, glutamine, and antibiotics), and 50-μl aliquots were incubated with serial dilutions of MAbs or CD4-based molecules (50 μl) for 1 h at 37°C. The calculated inhibitory doses refer to the concentration of these agents in this preincubation mixture. After 1 h, 100 μl of the mixture was added to the cells for 16 h, giving a final virus concentration of 50 to 200 TCID50 per well. Unbound virus was removed by two washes in culture medium. On days 4 to 9 postinfection, the cultures were examined microscopically for syncytium formation and the supernatant was analyzed for the presence of p24 antigen. If virus production in the cultures had not reached its peak on day 5, the cultures were fed with fresh medium and then reanalyzed for p24 on subsequent days. The extent of neutralization was determined as for the PBMC-based assay.
Neutralization assay on GHOST-CCR5 or GHOST-CXCR4 cells.
The procedure was similar to that used with the U87MG-CD4 cells except that on days 4 to 9 postinfection, the cultures were examined microscopically for the presence of fluorescent cells. When sufficient fluorescent cells (>20 per well) were visible, the cultures were harvested. The cells were washed once with phosphate-buffered saline (PBS), incubated with 100 μl of PBS containing 2 mM EDTA for 5 min, and detached from the plates by vigorous pipetting. PBS (100 μl) and 10% formaldehyde (50 μl) were added for 2 h at 4°C to inactivate HIV-1. The cells (10,000 per sample) were then analyzed by using a FACScalibur machine (Becton Dickinson). The number of GFP-positive cells counted in the absence of MAb was designated 100%, and the ratios of GFP-positive cells in MAb-containing cultures were calculated relative to this value. The extent of neutralization was otherwise determined as for the PBMC-based assay.
RESULTS
Neutralization sensitivity of HIV-1 primary isolates in PBMC is independent of CCR5 or CXCR4 usage.
We first measured the sensitivity of a panel of primary and TCLA isolates to neutralization by MAbs and CD4-based reagents in a standard PBMC-blast assay of virus replication (Fig. 1; Table 2). The neutralizing reagents used were human MAbs 2F5 (69, 96), 2G12 (95, 96), 15e (43), 19b (66), and 447-52D (29, 39), sCD4, and the tetrameric CD4-IgG2 molecule (5). These were selected on the grounds that their binding sites were strongly conserved within the B subtype (63, 96) and, whenever possible, that they had proven efficacy at neutralizing primary isolates (29, 96). We tested by enzyme-linked immunosorbent assay whether these agents (other than 2F5) were able to recognize monomeric gp120 from each virus (61, 63); the failure of a MAb to bind monomeric gp120 indicates the outright absence of its epitope from the virus, and so no neutralization would be expected for this trivial reason (61). Nonreactive MAb-virus combinations (listed in Materials and Methods) were excluded from the neutralization analyses.
TABLE 2.
Neutralization of HIV-1 by MAbs and CD4-based molecules in PBMC
| Cells | ID50 (μg/ml)a
|
||||||
|---|---|---|---|---|---|---|---|
| 2F5 | 2G12 | sCD4 | CD4-IgG2 | 15e | 447-52D | 19b | |
| CCR5 primary | |||||||
| SF162 | 11.6 | 0.03 | 5.99 | 0.67 | 49 | 8.1 | 24.6 |
| JR-FL | 1.74 | 0.01 | 20.1 | 0.13 | >50 | ND | >50 |
| Case B2-6-92 | 4.7 | >50 | >50 | 1.83 | >50 | ND | >50 |
| ADA | 6.6 | <0.08 | 29.1 | 0.12 | >50 | ND | ND |
| CCR5 + CXCR4 primary | |||||||
| DH123 | >50 | 5.8 | 25.6 | 0.4 | >50 | ND | >50 |
| 2076 cl1 | 26.7 | 0.18 | >50 | 28.3 | >50 | ND | NB |
| P17 | 14.4 | 0.07 | 19.6 | 1.55 | ND | ND | NB |
| Case B5/86 | 4.88 | 0.03 | >50 | >50 | NB | >50 | NB |
| 89.6 | 12.7 | 0.002 | 1.04 | 0.21 | >50 | ND | >50 |
| CXCR4 primary | |||||||
| HC7 | 22.2 | <0.004 | 27.8 | 0.76 | ND | ND | >50 |
| 2075 | 18.9 | 6.7 | 17.3 | 0.91 | >50 | ND | NB |
| 2044 | 26.3 | >50 | >50 | 8.45 | >50 | >50 | NB |
| 2005 cl1 | >50 | 0.01 | >50 | 0.91 | >50 | ND | NB |
| ACH-P | 8.09 | 0.008 | 34.4 | 2.3 | >50 | 18.1 | ND |
| CCR5 + CXCR4 TCLA | |||||||
| SF2 | 0.07 | 0.003 | 0.14 | 0.06 | 0.32 | <0.08 | 3.21 |
| C17 | 7.8 | 0.008 | 24.9 | 0.22 | ND | ND | NB |
| CXCR4 TCLA | |||||||
| NL4-3 | 1.98 | 0.002 | 0.18 | 0.06 | >50 | ND | >50 |
| ACH-H9 | 1.54 | 0.05 | 0.31 | 0.01 | >50 | ND | ND |
| ACH-P/H9 | 1.23 | 0.013 | 5.3 | 0.2 | >50 | 30.7 | ND |
Derived from one representative experiment out of two to five performed. When no neutralization at 50 μg/ml was observed, a value >50 μg/ml is reported. ND, not done; NB, no binding (i.e., neutralization activity could not be evaluated because the MAb epitope is absent from the viral glycoprotein).
The viruses tested included biological and molecular clones of adult and pediatric origin (Table 1). The abilities of the different viruses to use CCR5 or CXCR4 for entry were assessed by determining the extent of their replication in U87MG-CD4 cells stably expressing these coreceptors (24, 42) (Table 1). This information was used to divide the virus panel into the following subgroups: CCR5 primary, CCR5 + CXCR4 primary, CXCR4 primary, CCR5 + CXCR4 TCLA, and CXCR4 TCLA (Tables 1 and 2; Fig. 1). The neutralization titer (ID50) was plotted for each virus against each MAb or CD4-based molecule according to these subgroups (Fig. 1).
As expected, the primary HIV-1 isolates had a wide range of sensitivity to neutralization by the different test reagents, with ID50s ranging from <0.002 to >50 μg/ml for 2G12. Consistent with previous results, 2F5, 2G12, and CD4-IgG2 neutralized most of the primary isolates, whereas sCD4, 15e (to a CD4-binding-site-related epitope), and 19b (to a V3 loop epitope) did not (5, 29, 66, 96). However, among the primary isolates, there was no consistent segregation of neutralization sensitivity or insensitivity with coreceptor usage (Fig. 1). Some CCR5-using viruses were sensitive to neutralization and some were insensitive, and a similar pattern of variation was found among CXCR4-using viruses. Grouped together, the CXCR4-using primary isolates were slightly more sensitive to 2G12, but less sensitive to 2F5 and CD4-IgG2, than the CCR5-using isolates, but the differences were small and the overlap was considerable (Fig. 1). Similar results were observed when 90% neutralization titers (ID90s) were compared, although 90% neutralization was very rarely achieved with sCD4, 15e, and 19b (data not shown). The TCLA viruses were, as a group, only a little more sensitive than the primary isolates to the reagents used in this study, but again there was no segregation between neutralization sensitivity and coreceptor usage (Fig. 1; Table 2). MAbs such as 15e and 19b, known to have activity against TCLA (T-tropic) viruses (43, 66), were not able to neutralize primary T-tropic viruses any better than primary M-tropic viruses; a primary T-tropic virus is not the same as a TCLA virus (62).
Neutralization of dualtropic HIV-1 isolates in U87MG-CD4 cells is independent of CCR5 or CXCR4 usage.
To confirm these observations in a different experimental system, we studied whether dualtropic primary viruses were differentially sensitive to neutralization by the same MAb or CD4-based reagent when they used CCR5 or CXCR4 for entry (Fig. 2; Table 3). By using the same virus and neutralizing agent, we reduce the number of variables in the experiment to one: the identity of the coreceptor. For this experiment, we used U87MG-CD4-CCR5 and U87-CD4-CXCR4 cells (24, 42).
TABLE 3.
Neutralization of dualtropic HIV-1 isolates in U87MG-CD4 cells
| Virus | ID50 (μg/ml)a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2F5
|
2G12
|
sCD4
|
CD4-IgG2
|
15e
|
IgG1b12
|
19b
|
447-52D
|
|||||||||
| CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | CCR5 | CXCR4 | |
| SF2 | 9.6 | 4.8 | 14.1 | 25.6 | 7.81 | 4.6 | 1.27 | 0.84 | >50 | 7.7 | ND | ND | >50 | >50 | 0.86 | 0.22 |
| Isolate C7/86 | 35.6 | >50 | 6.34 | 13.3 | 8.02 | 24.4 | 6.8 | 4.71 | >50 | >50 | 3.4 | 1.4 | >50 | >50 | 34.7 | 34.3 |
| DH123 | 6 | 40.3 | 8.8 | >50 | 20.3 | 17.5 | <0.4 | 2.9 | >50 | >50 | 1.9 | 2.6 | >50 | >50 | ND | ND |
| 2076 cl3 | 14.2 | 23.2 | 0.99 | 6.12 | >50 | >50 | 23 | 25.8 | >50 | >50 | ND | ND | NB | NB | ND | ND |
| P17 | >50 | >50 | >50 | >50 | 8.7 | 25.8 | 0.91 | 0.78 | >50 | >50 | ND | ND | NB | NB | ND | ND |
| P17/H9 | >50 | >50 | 8.4 | 17.2 | 10.5 | 3.9 | 0.62 | 0.62 | >50 | >50 | 1.7 | 9.0 | NB | NB | >50 | >50 |
| C17 | 17.5 | 17.1 | >50 | 34.8 | 8.69 | 4.8 | 2.67 | 1.9 | >50 | >50 | ND | ND | NB | NB | ND | ND |
Derived from one representative experiment out of two to five performed. When no neutralization at 50 μg/ml was observed, a value >50 μg/ml is reported. For each neutralizing agent, ID50 values obtained on the CCR5-expressing (CCR5) and the CXCR4-expressing (CXCR4) U87MG-CD4 cells are shown. ND, not done; NB, no binding.
The results of this experiment were broadly similar to what was observed with the PBMC-based neutralization assay. Neutralization titers (ID50 or ID90) varied over a considerable range, and between different viruses and neutralizing agents, but there was no consistent indication that neutralization sensitivity was a function of coreceptor usage (Fig. 2 and data not shown). The same conclusion was reached whether a MAb effective (IgG1b12) or ineffective (15e) against primary isolates was used, despite both recognizing overlapping epitopes near the CD4-binding site on gp120. Similar results were observed irrespective of whether the test virus was an isolate, a biological clone, or a molecular clone (Table 1; Fig. 2).
Neutralization of dualtropic HIV-1 isolates in GFP-expressing HOS cells is independent of CCR5 or CXCR4 usage.
HOS-CD4 cells that stably expressed transfected CCR5 or CXCR4 and the GFP reporter under the control of the HIV-2 long terminal repeat were created (48). These cells, designated GHOST-CXCR4 and GHOST-CCR5, fluoresce when infected with HIV-1 because the GFP reporter gene is transactivated, allowing quantitation of HIV-1 entry by fluorescence-activated cell sorting. Broadly similar cell lines have been described by others recently (27, 37, 98); the principle is the same as in the well-known MAGI assay, which has a β-galactosidase endpoint (49). We used the GHOST-CXCR4 and GHOST-CCR5 cells to compare how the entry of HIV-1 primary and TCLA isolates was neutralized by sCD4 and CD4-IgG2. The procedure is not a single-cycle assay of HIV-1 neutralization since, in principle, subsequent rounds of HIV-1 infection could occur and be detected. The endpoint is measured after 4 to 9 days (depending on the isolate), when sufficient GFP has accumulated in the infected cells to be quantitated.
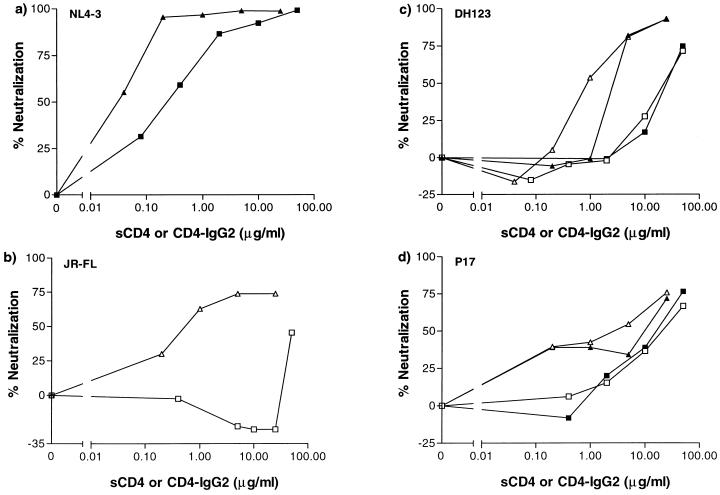
The clonal TCLA virus NL4-3 was sensitive to neutralization by both sCD4 and its more potent derivative, CD4-IgG2, in the GHOST-CXCR4 cells (Fig. 3a). In contrast, the primary M-tropic clone JR-FL was insensitive to neutralization by sCD4 on the GHOST-CCR5 cells, although it was neutralized effectively by CD4-IgG2 (Fig. 3b). These results are consistent with previous reports on the relative neutralization sensitivities of primary and TCLA viruses to CD4-based reagents (5, 21, 53, 60, 61, 64, 100) and establish the validity of using the GHOST cells in neutralization assays. As expected from prior knowledge of their coreceptor usage, NL4-3 did not replicate in the GHOST-CCR5 cells, nor did JR-FL in the GHOST-CXCR4 cells (data not shown). However, two dualtropic viruses entered both GHOST-CCR5 and GHOST-CXCR4 cells. We therefore compared the sensitivities of the dualtropic viruses to sCD4 and CD4-IgG2 on the two lines. As observed with the U87MG-CD4 cells, the potency with which sCD4 and CD4-IgG2 neutralized P17 (60, 97) and DH123 (85) on the GHOST cells was independent of which coreceptor (CCR5 or CXCR4) was expressed on these cells (Fig. 3c and d). This finding confirms that the identity of the coreceptor used by a primary HIV-1 isolate to enter a CD4+ cell is not a major influence on the efficiency with which it is neutralized.
FIG. 3.
Neutralization of dualtropic HIV-1 isolates in GHOST-CCR5 and GHOST-CXCR4 cells. HIV-1 replication was assessed by determination of fluorescence intensity in GHOST-CXCR4 cells (▴, ▪) or GHOST-CCR5 cells (□, ▵) in the presence of the indicated concentrations of sCD4 (▪, □) or CD4-IgG2 (▴, ▵). The extent of neutralization at each concentration was determined relative to the fluorescence intensity in the absence of added sCD4 or CD4-IgG2. (a) TCLA molecular clone NL4-3; (b) molecularly cloned M-tropic primary virus JR-FL; (c) dualtropic, molecularly cloned primary virus DH123; (d) dualtropic primary isolate P17.
DISCUSSION
We were unable to obtain any evidence that M- and T-tropic primary HIV-1 isolates are differentially sensitive to neutralization by MAbs or CD4-based reagents, or that coreceptor usage (CCR5 versus CXCR4) was a significant variable affecting neutralization sensitivity. Similar results have been obtained in independent studies (53a, 59). There is, however, one important caveat to our conclusion: we could obtain only limited information on whether the V3 loop is of differential importance to the neutralization of M- and T-tropic primary viruses (as opposed to primary and TCLA strains). The reason for this is that V3 loop MAbs which are able to neutralize more than just a few primary viruses do not exist. Many V3 MAbs neutralize TCLA strains very potently (9, 61, 62, 65, 66), but only a few of these have been reported to neutralize primary isolates (39), and their breadth and potency against these viruses are very limited (29, 61). We could find no V3 MAb that was able to neutralize efficiently more than a few of the dualtropic primary viruses that we tested.
Another factor affecting analysis of V3 loop-mediated neutralization is sequence variation. The most common sequence change that occurs during the phenotypic switch from an M-tropic (NSI) to a T-tropic (SI) strain (i.e., an alteration in coreceptor usage) is an increase in the positive charge of the V3 loop (22, 62). More specifically, residues 306 and 315 tend to be negatively charged or neutral in NSI strains but positively charged in SI viruses (11, 22, 35, 51, 56). Models of ligand interactions with seven-transmembrane receptors suggest that an important (but not the only) component of the binding reaction is a charge-based one involving anionic residues on the N-terminal domain of the receptor and cationic ones on the ligand (41, 57, 68, 88, 99). Thus, it is feasible that the charge change in the V3 loop associated with the phenotypic switch helps create a binding site for CXCR4 on T-tropic viruses and that the V3 loop plays a greater role in the interactions of HIV-1 with CXCR4 than with CCR5 (12, 14). If this is the case, antibodies to the V3 loop could interfering more effectively with the CXCR4 interactions of HIV-1 than with CCR5 interactions and so be better able to neutralize T-tropic primary viruses. Unfortunately, amino acid substitutions affecting the ability of HIV-1 to interact with different coreceptors are also likely to alter the affinity of an antibody for the same region of the protein, especially in the context of native, oligomeric envelope glycoproteins. Antigenic variation in the V3 loop affecting MAb-gp120 interactions and sequence variation affecting coreceptor-gp120 interactions are interlocking parameters that complicate analyses. However, the limited data that we were able to obtain indicated that neutralization via the V3 loop is independent of coreceptor usage, as it is for other epitopes (Tables 2 and 3). A similar conclusion has been reached by others (53a).
Only a very modest difference in replication competence can have a profound impact on the emergence of viral variants in vivo (15). Hence, we cannot exclude the possibility that small differences in the neutralizability of M- and T-tropic viruses that are undetectable in vitro can have more significant effects in vivo. But these notes of caution notwithstanding, our main conclusion is that differences in neutralizing antibody-mediated selection pressures are unlikely to account for the emergence of T-tropic (SI) viruses in vivo. What then could, bearing in mind that the pressure must be exerted on the envelope glycoproteins? One attractive theory is that the CXCR4 ligand stroma-derived factor 1 is produced at relatively high concentrations by cells in the stroma of lymphoid tissues and so is able to specifically prevent HIV-1 interactions with CXCR4 (31). It will be important to verify this experimentally. Another possibility involves alterations in the range of CD4+ T cells able to support HIV-1 replication, because of differential expression of CXCR4 and CCR5 on T-cell subsets (104).
Clearly, TCLA strains of HIV-1 are neutralization sensitive compared to T-tropic primary isolates that have the ability to enter a T-cell line. A virus that has become adapted to growth on a T-cell line is not, therefore, the same after passage as it was before it entered the cell line in the first place (9, 62). The acquisition of full neutralization sensitivity usually takes several passages in cell lines (58, 80, 102). Thus, genetic changes in the envelope glycoproteins, rather than epigenetic factors such as glycosylation or differences in virion-associated adhesion factors, are likely to dominate the transition between the neutralization-resistant (T-tropic primary) and the neutralization-sensitive (TCLA) state. Adhesion factor-dependent variables may, however, also contribute to the phenomenon (38, 77). What are these genetic changes, what do they do, and why do they do it? The answers to all these questions are not entirely clear, but some evidence is emerging.
As discussed above, changes in the V3 loop are important for the creation of the T-tropic phenotype in primary isolates. And they are also likely to be important for the transition between an M-tropic virus and a TCLA one. However, the passage of a T-tropic primary virus through a cell line to create a TCLA isolate is not always associated with V3 loop sequence changes (53, 60, 73–75). This is, perhaps, understandable now it is known that both T-tropic primary isolates and TCLA viruses tend to be able to use both CCR5 and CXCR4 (18, 89); there is more to HIV-1 adaptation to growth on cell lines than a change in coreceptor use (53, 75). During this adaptation, the neutralization-sensitive phenotype is created by changes in the HIV-1 envelope glycoproteins. As this also occurs with other lentiviruses (7, 10, 19, 105), a general mechanism may be involved. We have suggested previously that the neutralization-sensitive phenotype might be created in vitro because antibody selection pressure is absent in cell culture (61, 62). If the price that HIV-1 and other lentiviruses pay to be able to resist the binding of neutralizing antibodies in vivo is a modest reduction in the rate at which they bind to and penetrate CD4+ T cells, then the relaxation of antibody selection pressure in vitro might permit the evolution of variants able to replicate more rapidly in cell lines. There are no reports that neutralization-sensitive viruses evolve when primary isolates are passaged continuously in primary T-cells in vitro, although this has not been studied systematically. It can, however, happen occasionally with simian immunodeficiency virus during passage in primary rhesus PBMC (58).
A major contribution to understanding the overall phenomenon of cell line adaptation is provided by the work of Kabat and colleagues, who showed that the relevant selection pressure on primary T-tropic viruses is for variants which have a higher affinity for CD4 (53, 75). This gives them the ability to infect cell lines expressing relatively low CD4 concentrations compared to the amount on primary CD4+ T cells (53, 75). This would also explain why TCLA viruses are so sensitive to neutralization by sCD4 (21, 40, 64, 100), which we have argued is merely coincidental to their sensitivity to neutralizing antibodies against related epitopes (61, 62). No such selection pressure occurs when primary viruses are passaged in primary CD4+ T cells, perhaps explaining why the primary isolate phenotype is usually maintained under these conditions.
A meld of these arguments is that in some as yet undefined way, the tertiary and quaternary structure of the envelope glycoproteins alters during passage on cell lines to better expose receptor-binding sites (CD4 and/or coreceptor sites), increase the efficiency of virus-cell attachment and entry, and (coincidentally) make the virus more sensitive to the actions of neutralizing antibodies. It remains to be determined whether neutralization sensitivity occurs solely because antibodies can attach more rapidly to key sites on the envelope glycoproteins of TCLA viruses, or whether the destabilization of their gp120-gp41 linkage also contributes (21, 47, 50, 64, 79, 100).
As we learn more of how the HIV-1 envelope glycoproteins function, and of the mechanisms by which they resist neutralizing antibodies, it becomes apparent how difficult it will be to induce antibodies by vaccination that will be able to counter this virus. Viral evolution has created a pathogen that is necessarily insensitive to humoral immunity. Yet HIV-1 does have its weak links, for it can be potently neutralized by a limited subset of human antibodies (29, 96). An emphasis of vaccine design must, therefore, be to find ways to create immunogens able to induce antibodies of these specificities efficiently.
ACKNOWLEDGMENTS
We are very grateful to David Montefiori for helpful discussions. We thank Cecilia Cheng-Mayer, Paul Clapham, Ron Collman, Ruth Connor, Serene Forte, Mal Martin, Riri Shibata, John Sullivan, Jon Weber, and Ullrich Desselberger for HIV-1 isolates and clones. We also thank Paul Maddon and Graham Allaway for the gifts of soluble CD4 and CD4-IgG2 and Dennis Burton for MAb IgG1b12. We appreciate the technical assistance of Simon Monard, Jeremy Segal, and Jamie Matthews.
This study was supported by NIH grants AI36082 and AI41420 and by the Pediatric AIDS Foundation. A.T. is a Fellow of the Austrian Program for Advanced Research and Technology; V.N.K. is supported by a postdoctoral fellowship from the Damon Runyon/Walter Winchell Foundation; D.R.L. is an Investigator of the Howard Hughes Medical Institute; J.P.M. is an Elizabeth Glaser Scientist of the Pediatric AIDS Foundation.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad N, Baroudy B M, Baker R C, Chappey C. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J Virol. 1995;69:1001–1012. doi: 10.1128/jvi.69.2.1001-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert J, Fiore J, Fenyö E M, Pedersen C, Lundgren J D, Gerstoft J, Nielsen C. Biological phenotype of HIV-1 and transmission. AIDS. 1995;9:822–823. doi: 10.1097/00002030-199507000-00029. [DOI] [PubMed] [Google Scholar]
- 4.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 5.Allaway G P, Davis-Bruno K L, Beaudry G A, Garcia E B, Wong E L, Ryder A M, Hasel K W, Gauduin M-C, Koup R A, McDougal J S, Maddon P J. Expression and characterization of CD4IgG2, a novel heterotetramer which neutralizes primary HIV-1 isolates. AIDS Res Hum Retroviruses. 1995;11:533–540. doi: 10.1089/aid.1995.11.533. [DOI] [PubMed] [Google Scholar]
- 6.Balotta C, Viganò A, Riva C, Colombo M C, Salvaggio A, de Pasquale M P, Crupi L, Papagno L, Galli M, Moroni M, Principi N. HIV type 1 phenotype correlates with the stage of infection in vertically infected children. AIDS Res Hum Retroviruses. 1996;12:1247–1253. doi: 10.1089/aid.1996.12.1247. [DOI] [PubMed] [Google Scholar]
- 7.Baldinotti F, Matteucci D, Mazzetti P, Giannelli C, Bandecchi P, Tozzini F, Bendinelli M. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J Virol. 1994;68:4572–4579. doi: 10.1128/jvi.68.7.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R., and D. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11(Suppl. A):587–598. [PubMed]
- 10.Cammarota G, Matteucci D, Kistello M, Nicoletti E, Giannecehini S, Bendinelli M. Reduced sensitivity to strain-specific neutralization of laboratory-adapted feline immunodeficiency virus after one passage in vivo: association with amino acid substitutions in the V4 region of the surface glycoprotein. AIDS Res Hum Retroviruses. 1996;12:173–175. doi: 10.1089/aid.1996.12.173. [DOI] [PubMed] [Google Scholar]
- 11.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard G, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;86:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Cocchi F, DeVico A L, Garzino-Derno A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha and MIP-1 beta as the major HIV suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Derno A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 15.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 16.Collman R, Balliet J W, Gregory S A, Friedman H, Kolson D L, Nathanson N, Srinivasan A. An infectious molecular clone of an unusual macrophage-tropic and highly cytopathic strain of human immunodeficiency virus type 1. J Virol. 1992;66:7517–7521. doi: 10.1128/jvi.66.12.7517-7521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replication capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1 infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cook R F, Berger S L, Rushlow K E, McManus J E, Cook S J, Harrold S, Raabe M L, Montelaro R C, Issel C J. Enhanced sensitivity to neutralizing antibodies in a variant of equine infectious anemia virus is linked to amino acid substitutions in the surface unit envelope glycoprotein. J Virol. 1995;69:1493–1499. doi: 10.1128/jvi.69.3.1493-1499.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornellisen M, Mulder-Mampinga G, Veenstra J, Zorgdrager F, Kuiken C, Hartman S, Dekker J, van der Hoek L, Sol C, Coutinho R, Goudsmit J. Syncytium-inducing (SI) phenotype suppression at seroconversion after intramuscular inoculation of a non-syncytium-inducing/SI phenotypically mixed human immunodeficiency virus population. J Virol. 1995;69:1810–1818. doi: 10.1128/jvi.69.3.1810-1818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daar E S, Li X L, Moudgil T, Ho D D. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc Natl Acad Sci USA. 1990;87:6574–6578. doi: 10.1073/pnas.87.17.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirement for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 24.Deng H, Unutmaz D, Kewal-Ramani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 25.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 26.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S, Parmentier M, Collman R G, Doms R W. A dual-tropic, primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;86:1149–1159. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 27.Dorsky D I, Wells M, Harrington R D. Detection of HIV-1 infection with a green fluorescent protein reporter system. J Acquired Immune Defic Syndr. 1996;13:308–313. doi: 10.1097/00042560-199612010-00002. [DOI] [PubMed] [Google Scholar]
- 28.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 29.D’Souza M P, Livnat D, Bradac J A, Bridges S H the AIDS Clinical Trials Group Antibody Selection Working Group; Collaborating Investigators. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J Infect Dis. 1997;175:1075–1062. doi: 10.1086/516443. [DOI] [PubMed] [Google Scholar]
- 30.Farzan M, Choe M, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 32.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 33.Fiore J R, Björndal A, Peipke K A, Di Stefano M, Angarano G, Pastore G, Gaines H, Fenyö E M, Albert J. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology. 1994;204:297–303. doi: 10.1006/viro.1994.1534. [DOI] [PubMed] [Google Scholar]
- 34.Forte S E, Sullivan J L, Somasundaran M. In vitro characterization of HIV type 1 biological clones from asymptomatic and symptomatic pediatric patients. AIDS Res Hum Retroviruses. 1996;12:1585–1593. doi: 10.1089/aid.1996.12.1585. [DOI] [PubMed] [Google Scholar]
- 35.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouchier R A M, Meyaard L, Bruwer M, Hovenkamp E, Schuitemaker H. Broader tropism and higher cytopathicity for CD4+ T cells of a syncytium-inducing compared to a non syncytium-inducing HIV-1 isolate as a mechanism for accelerated CD4+ T cell decline in vivo. Virology. 1996;219:87–95. doi: 10.1006/viro.1996.0225. [DOI] [PubMed] [Google Scholar]
- 37.Gervaix A, West D, Leoni L M, Richman D D, Wong-Staal F. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci USA. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez M B, Hildreth J E K. Antibody to adhesion molecule LFA-1 enhances plasma neutralization of human immunodeficiency virus type 1. J Virol. 1995;69:4628–4632. doi: 10.1128/jvi.69.8.4628-4632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorny M K, Conley A J, Karwowska S, Buchbinder A, Xu J-Y, Emini E A, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J Virol. 1992;66:7358–7342. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groenink M, Moore J P, Broersen S, Schuitemaker H. Equal gp120 retention and neutralization resistance of phenotypically distinct primary human immunodeficiency virus type-1 variants upon soluble CD4 treatment. J Virol. 1995;69:523–527. doi: 10.1128/jvi.69.1.523-527.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hébert C A, Chuntharapai A, Smith M, Colby T, Kim J, Horuk R. Partial functional mapping of the human interleukin-8 type A receptor. J Biol Chem. 1993;268:18549–18553. [PubMed] [Google Scholar]
- 42.Hill C M, Deng H-K, Unutmaz D, Kewal-Ramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency viruses types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho D D, McKeating J A, Li X L, Moudgil T, Daar E S, Sun N-C, Robinson J E. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J Virol. 1991;65:489–493. doi: 10.1128/jvi.65.1.489-493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hogervorst E, de Jong J, van Wijk A, Bakker M, Valk M, Nara P L, Goudsmit J. Insertion of primary syncytium-inducing (SI) and non-SI envelopes in human immunodeficiency virus type 1 (HIV-1) LAI reduces neutralization sensitivity to autologous, but not heterologous, HIV 1 antibodies. J Virol. 1995;69:6342–6351. doi: 10.1128/jvi.69.10.6342-6351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutto C, Zhou Y, He J, Geffin R, Hill M, Scott W, Wood C. Longitudinal studies of viral sequence, viral phenotype, and immunological parameters of human immunodeficiency virus type 1 infection in perinatally infected twins with discordant disease courses. J Virol. 1996;70:3589–3598. doi: 10.1128/jvi.70.6.3589-3598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlsson A, Parsmyr K, Aperia K, Sandström E, Fenyö E M, Albert J. MT-2 cell tropism of human immunodeficiency virus type 1 isolates as a marker for response to treatment and development of drug resistance. J Infect Dis. 1994;170:1367–1375. doi: 10.1093/infdis/170.6.1367. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson G B, Gao F, Robinson J, Hahn B, Sodroski J. Increased envelope spike density and stability are not required for the neutralization resistance of primary human immunodeficiency viruses. J Virol. 1996;70:6136–6142. doi: 10.1128/jvi.70.9.6136-6142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.KewalRamani, V. N., and D. R. Littman. Unpublished data.
- 49.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:3026–3031. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klasse P J, Moore J P. Quantitative model of the sensitivity to antibody- and soluble CD4-mediated neutralization of primary isolates and T-cell line-adapted strains of human immunodeficiency virus type 1. J Virol. 1996;70:3668–3677. doi: 10.1128/jvi.70.6.3668-3677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koot M, Keet I P M, Vos A H V, De Goede R E Y, Roos M T L, Coutinho R A, Miedema F, Schellekens P T A, Tersmette M. Prognostic value of HIV 1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 52.Koot M, van’t Wout A B, Kootstra N A, de Goede R E Y, Tersmette M, Schuitemaker H. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:349–354. doi: 10.1093/infdis/173.2.349. [DOI] [PubMed] [Google Scholar]
- 53.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infection by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.LaCasse R A, Follis K E, Moudgil T, Trahey M, Binley J M, Planelles V, Zolla-Pazner S, Nunberg J H. Coreceptor utilization by human immunodeficiency virus type 1 is not a primary determinant of neutralization sensitivity. J Virol. 1998;72:2491–2495. doi: 10.1128/jvi.72.3.2491-2495.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao F, Alkhatib G, Peden K C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKnight A, Weiss R A, Shotton C, Takeuchi Y, Hoshino H, Clapham P R. Change in tropism upon immune escape by human immunodeficiency virus. J Virol. 1995;69:3167–3170. doi: 10.1128/jvi.69.5.3167-3170.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milich L, Margolin B, Swanstrom R. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol. 1993;67:5623–5634. doi: 10.1128/jvi.67.9.5623-5634.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monteclaro F S, Charo I F. The amino-terminal extracellular domain of the MCP-1 receptor, but not the RANTES/MIP-1α receptor, confers chemokine sensitivity. Evidence for a two-step mechanism for MCP-1 receptor activation. J Biol Chem. 1996;271:19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 58.Montefiori, D. C. Personal communication.
- 59.Montefiori D C, Collman R G, Fouts T R, Zhou J Y, Bilska M, Hoxie J A, Moore J P, Bolognesi D P. Evidence that antibody-mediated neutralization of human immunodeficiency virus type 1 by sera from infected individuals is independent of coreceptor usage. J Virol. 1998;72:1886–1893. doi: 10.1128/jvi.72.3.1886-1893.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore J P, Burkly L C, Connor R I, Cao Y, Tizard R, Ho D D, Fisher R A. Adaptation of two primary, macrophage-tropic human immunodeficiency virus type 1 isolates to growth in transformed T-cell lines correlates with alterations in the responses of their envelope glycoproteins to soluble CD4. AIDS Res Hum Retroviruses. 1993;9:529–539. doi: 10.1089/aid.1993.9.529. [DOI] [PubMed] [Google Scholar]
- 61.Moore J P, Cao Y, Qing L, Sattentau Q J, Pyati J, Koduri R, Robinson J, Barbas III C F, Burton D R, Ho D D. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore, J. P., and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117–S136. [PubMed]
- 63.Moore J P, McCutchan F E, Poon S-W, Mascola J, Liu J, Cao Y, Ho D D. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J Virol. 1994;68:8350–8364. doi: 10.1128/jvi.68.12.8350-8364.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore J P, McKeating J A, Huang Y, Ashkenazi A, Ho D D. Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol. 1992;66:235–243. doi: 10.1128/jvi.66.1.235-243.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore, J. P., and P. L. Nara. 1991. The role of the V3 domain of gp120 in HIV infection. AIDS 5(Suppl. 2):S21–S33. [DOI] [PubMed]
- 66.Moore J P, Trkola A, Korber B, Boots L J, Kessler II J A, McCutchan F E, Mascola J, Ho D D, Robinson J, Conley A J. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J Virol. 1995;69:122–130. doi: 10.1128/jvi.69.1.122-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moore J P, Yoshiyama H, Ho D D, Robinson J E, Sodroski J. Antigenic variation in gp120s from molecular clones of HIV-1 LAI. AIDS Res Hum Retroviruses. 1993;9:1179–1187. doi: 10.1089/aid.1993.9.1185. [DOI] [PubMed] [Google Scholar]
- 68.Moser R, Dewald B, Barella L, Schumacher C, Baggiolini M, Clark-Lewis I. Interleukin-8 antagonists generated by N-terminal modification. J Biol Chem. 1993;268:7125–7128. [PubMed] [Google Scholar]
- 69.Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H. Cross-neutralizing antibodies against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 71.O’Brien W A, Koyanagi Y, Namazie A, Zhao J-Q, Diagne A, Idler K, Zack J A, Chen I S Y. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 72.Ometto L, Zanotto C, Maccabruni A, Caselli D, Truscià D, Giaquinto E, Ruga E, Chieco Bianchi L, De Rossi A. Viral phenotype and host-cell susceptibility to HIV-1 infection as risk factors for mother-to-child HIV 1 transmission. AIDS. 1995;9:427–434. [PubMed] [Google Scholar]
- 73.Orloff S L, Bandea C I, Kennedy M S, Allaway G P, Maddon P J, McDougal J S. Increase in sensitivity to soluble CD4 by primary HIV type 1 isolates after passage through C8166 cells: association with sequence differences in the first constant (C1) region of glycoprotein 120. AIDS Res Hum Retroviruses. 1995;11:335–342. doi: 10.1089/aid.1995.11.335. [DOI] [PubMed] [Google Scholar]
- 74.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 75.Platt E J, Madiani N, Kozak S L, Kabat D. Infectious properties of human immunodeficiency virus type 1 mutants with distinct affinities for the CD4 receptor. J Virol. 1997;71:883–890. doi: 10.1128/jvi.71.2.883-890.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Richman D D, Bozzette S A. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J Infect Dis. 1994;169:968–974. doi: 10.1093/infdis/169.5.968. [DOI] [PubMed] [Google Scholar]
- 77.Rizzutto C D, Sodroski J G. Contribution of virion ICAM-1 to human immunodeficiency virus infectivity and sensitivity to neutralization. J Virol. 1997;71:4847–4851. doi: 10.1128/jvi.71.6.4847-4851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saag, M. S., S. M. Hammer, and J. M. A. Lange. 1994. Pathogenicity and diversity of HIV and implications for clinical management: a review. J. Acquired Immune Defic. Syndr. 7(Suppl. 2):S2–S11. [PubMed]
- 79.Sattentau Q J, Moore J P. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the glycoprotein gp120 oligomer. J Exp Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sawyer L S W, Wrin M T, Crawford-Miksza L, Potts B, Wu Y, Weber P A, Alfonso R D, Hanson C V. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scarlatti G, Hodara V, Rossi P, Muggiasca L, Bucceri A, Albert J, Fenyö E M. Transmission of human immunodeficiency virus type 1 (HIV-1) from mother to child correlates with viral phenotype. Virology. 1993;197:624–629. doi: 10.1006/viro.1993.1637. [DOI] [PubMed] [Google Scholar]
- 82.Schuitemaker H, Koot M, Kootstra N A, Dergksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Eeftink Schattenkerk J K M, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schutten M, Langedijk J P M, Andeweg A C, Huisman R C, Meloen R H, Osterhaus A D M E. Characterization of a V3 domain-specific neutralizing human monoclonal antibody that preferentially recognizes non-syncytium-inducing human immunodeficiency virus type 1 strains. J Gen Virol. 1995;76:1665–1673. doi: 10.1099/0022-1317-76-7-1665. [DOI] [PubMed] [Google Scholar]
- 84.Sei S, Akiyoshi H, Bernard J, Venzon D J, Fox C H, Schwartzentruber D J, Anderson B D, Kopp J B, Mueller B U, Pizzo P A. Dynamics of virus versus host interactions in children with human immunodeficiency virus type 1 infection. J Infect Dis. 1996;173:1485–1490. doi: 10.1093/infdis/173.6.1485. [DOI] [PubMed] [Google Scholar]
- 85.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, Martin M A. Isolation and characterization of a syncytium-inducing, macrophage/T-cell line-tropic human immunodeficiency virus type 1 isolate that readily infects chimpanzee cells in vitro and in vivo. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 87.Shpaer E G, Delwart E L, Kuiken C L, Goudsmit J, Bachmann M H, Mullins J I. Conserved V3 loop sequences and transmission of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10:1679–1684. doi: 10.1089/aid.1994.10.1679. [DOI] [PubMed] [Google Scholar]
- 88.Siliciano S J, Rollins T E, DeMartino J, Konteatis Z, Malkowitz L, Van Riper G, Bondy S, Rosen H, Springer M S. Two-site binding of C5a by its receptor: an alternative binding paradigm for G protein-coupled receptors. Proc Natl Acad Sci USA. 1994;91:1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spencer L T, Ogino M T, Dankner W M, Spector S A. Clinical significance of human immunodeficiency virus type 1 phenotypes in infected children. J Infect Dis. 1994;169:491–495. doi: 10.1093/infdis/169.3.491. [DOI] [PubMed] [Google Scholar]
- 91.Stamatatos L, Cheng-Mayer C. Structural modulations of the envelope gp120 glycoprotein of HIV-1 upon oligomerization and differential V3 loop-epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J Virol. 1995;69:6191–6198. doi: 10.1128/jvi.69.10.6191-6198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tersmette M, de Goede R E Y, Al B J M, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Theodore T S, Englund G, Buckler-White A, Buckler C E, Martin M A, Peden K W C. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 94.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4 dependent, antibody sensitive interactions between HIV-1 and its co-receptor CCR5. Nature. 1996;384:184–186. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 95.Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore J P, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trkola A, Pomales A B, Yuan H, Korber B, Maddon P J, Allaway G P, Katinger H, Barbas III C F, Burton D R, Ho D D, Moore J P. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol. 1995;69:6609–6617. doi: 10.1128/jvi.69.11.6609-6617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turner S, Tizard R, DeMarinis J, Pepinsky R B, Zullo J, Schooley R, Fisher R. Resistance of primary isolates of human immunodeficiency virus type 1 to neutralization by soluble CD4 is not due to lower affinity with the viral envelope glycoprotein gp120. Proc Natl Acad Sci USA. 1992;89:1335–1339. doi: 10.1073/pnas.89.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 99.Wells T N C, Power C A, Lusti-Narasimhan M, Hoogewerf A J, Cooke R M, Chung C-W, Peitsch M C, Proudfoot A E I. Selectivity and antagonism of chemokine receptors. J Leukocyte Biol. 1996;59:53–60. doi: 10.1002/jlb.59.1.53. [DOI] [PubMed] [Google Scholar]
- 100.Willey R L, Martin M A, Peden K W C. Increase in soluble CD4 binding to and CD4-induced dissociation of gp120 from virions correlates with infectivity of human immunodeficiency virus type 1. J Virol. 1994;68:1029–1039. doi: 10.1128/jvi.68.2.1029-1039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wong M T, Dolan M J, Kozlow E, Doe R, Melcher G P, Burke D S, Boswell R N, Vahey M. Patterns of virus burden and T cell phenotype are established early and are correlated with the rate of disease progression in human immunodeficiency virus type 1-infected persons. J Infect Dis. 1996;173:877–887. doi: 10.1093/infdis/173.4.877. [DOI] [PubMed] [Google Scholar]
- 102.Wrin T, Loh T P, Vennari J C, Schuitemaker H, Nunberg J H. Adaptation to growth in the H9 cell line renders a human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4 induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 104.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C. CCR5 levels and expression pattern correlate with infectability by macrophage tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wyand M S, Manson K H, Garcia-Moll M, Montefiori D, Desrosiers R C. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–3733. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhu T, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]