Abstract
Purpose.
HOXB13 is an androgen receptor (AR) co-regulator specifically expressed in cells of prostatic lineage. We sought to associate circulating tumor cell (CTC) HOXB13 expression with outcomes in men with mCRPC treated with abiraterone or enzalutamide.
Methods.
We conducted a retrospective analysis of the multicenter prospective PROPHECY trial of mCRPC men (NCT02269982, n=118) treated with abiraterone/enzalutamide. CTC detection and HOXB13 complementary DNA (cDNA) expression was measured using a modified Adnatest, grouping patients into 3 categories: CTC 0 (undetectable); CTC+ HOXB13 CTC low (<4 copies) or CTC+ HOXB13 CTC high. The HOXB13 threshold was determined by maximally selected rank statistics for prognostic associations with overall survival (OS) and progression-free survival (PFS).
Results.
We included 102 men with sufficient CTC HOXB13 cDNA, identifying 25%, 31%, and 44% of patients who were CTC 0, CTC+ HOXB13 low, and CTC+ HOXB13 high, respectively. Median OS were 25.7, 27.8, and 12.1 months while the median PFS were 9.0, 7.7 and 3.8 months, respectively. In subgroup analysis among men with CellSearch CTCs ≥5 copies/ml and adjusting for prior abi/enza treatment and Halabi clinical risk score, the multivariate HR for HOXB13 CTC detection was 2.39 (95% CI 1.06-5.40) for OS and 2.78 (95% CI 1.38-5.59) for PFS, respectively. Low HOXB13 CTC detection was associated with lower CTC PSA, PSMA, AR-FL, and AR-V7 detection, and more liver/lung metastases (41% vs 25%).
Conclusion.
Higher CTC HOXB13 expression is associated with AR-dependent biomarkers in CTCs and is adversely prognostic in the context of potent AR inhibition in men with mCRPC.
Introduction
The androgen receptor (AR) has been a primary target for systemic therapy in men with advanced prostate cancer, and potent AR inhibitors have improved the survival of men with metastatic hormone sensitive and castration-resistant prostate cancer (mCRPC)1-5. However, the AR is commonly expressed in normal tissues, leading to on-target toxicities due to whole body AR inhibition, such as cardiovascular risk, fatigue, falls, fractures, and hot flashes. In addition, AR inhibition leads to acquired resistance and further pathway activation through cancer-specific AR upregulation, mutation, and paracrine/autocrine androgen synthesis6-10 as well as constitutive activation through the expression of AR isoforms lacking the androgen ligand-binding domain11-15, particularly AR-V716-19. Thus, novel prostate cancer specific targets for therapy are needed.
HOXB13 is a transcription factor and member of the homeobox gene family, and is critical for embryonic and prostate development20-23. Familial mutations in HOXB13 such as G84E on chromosome 17q21-22 are found in men with hereditary prostate cancer, increasing the risk of the disease by more than ten-fold24-27, and recent studies suggest the X285K mutation in HOXB13 may be enriched in men of African ancestry and aggressive disease28. In addition, HOXB13 plays a key role in reprogramming the prostate cancer epigenome where it cooperates with AR to drive the transcription of genes whose products drive the pathobiology of cancer, such as MYC and E2F28,29. HOXB13 can facilitate AR and AR-V7 binding to DNA and leads to cell survival in the castrate state and in the setting of potent AR inhibition30,31. Thus, HOXB13 represents a novel prostate cancer specific target and the therapeutic exploitation of which may have particular utility in prostate cancer.
We retrospectively analyzed the prospective multicenter PROPHECY trial32 to test associations between circulating tumor cell (CTC) HOXB13 detection with clinical outcomes in the setting of abiraterone or enzalutamide therapy in men with mCRPC. We hypothesized that HOXB13 high mCRPC patients would have adverse progression free and overall survival. We found that most men had HOXB13 detection, and that men with more than 4 copies of HOXB13 had a higher risk of progression or death, particularly in those men with elevated CTCs, and independent prior therapy clinical prognostic factors. We also identified a small subgroup (~20%) of men with mCRPC and no detectable HOXB13 who were enriched in visceral metastases and had lower AR-FL and AR-V7 detection.
Methods
Patients
At five clinical sites, we prospectively enrolled men with progressive, high-risk mCRPC initiating standard-of-care treatment with enzalutamide or abiraterone. Prior exposure to enzalutamide or abiraterone was permitted for men who were planning to receive the alternative agent. Inclusion and exclusion criteria have been published previously, and required ≥2 poor prognosis clinical factors32-34 (Table 1). Patient demographics, including age, race, Gleason score, Karnofsky score, PSA, and other labs are included in Table 1. All patients provided written informed consent under institutional review board approval at all participating centers within the Department of Defense (DOD)-funded Prostate Cancer Clinical Trial Consortium (PCCTC)35. This study was approved by local institutional review boards at participating sites per institutional policy and the Declaration of Helsinki.
Table 1.
Baseline characteristics of the patients enrolled according to CTC (by Adnatest) and CTC HOXB13 detection.
| Baseline characteristics |
CTC=0 N=25 |
CTC+ & HOXB13 low (<4) N=32 |
CTC+ & HOXB13 high (>=4) N=45 |
|---|---|---|---|
| Age, median years (range) | 78 (57-92) | 72 (45-92) | 74 (48-87) |
| Race: white/black/other (%) | 88/4/8 | 84.4/12.5/3.1 | 82.2/8.9/8.9 |
| Gleason Sum 8-10 (%) | 52 | 69 | 53 |
| Karnofsky Score >=90 (%) | 64 | 75 | 62 |
| CIN Positive (>=1 CTCs/mL) (%) | 9.5 | 21.4 | 68.2 |
| NE Positive (>= 2 CTCs/mL) (%) | 0 | 0 | 18.2 |
| Median CTC PSA copies (Range) | 0 | 5.3 (0-389.9) | 124.9 (0-29520.5) |
| Serum PSA, median ng/mL (range) | 5.1 (0.1-256.3) | 14.2 (0.3,319.0) | 49.1 (0.3,2021.4) |
| Median CTC PSMA copies (Range) | 0 | 5.6 (0-214.7) | 133.9 (0-36743.5) |
| JHU CTC AR-V7 positive (%) | 0 | 9.4 | 51.1 |
| Epic CTC AR-V7 positive (%) | 0 | 0 | 22.7 |
| CTC AR-FL >0 (%) | 8 | 53.1 | 95.6 |
| Hemoglobin <12 g/dl (%) | 44 | 28.1 | 48.9 |
| Hemoglobin, median g/dl (range) | 12.5 (9.3-14.9) | 13.1 (10.0-15.9) | 12.0 (8.7-15.4) |
| Alkaline phosphatase, median U/L (range) | 81.0 (30.0-357.0) | 88.0 (41.0-233.0) | 146.0 (53.0-1430.0) |
| Elevated alkaline phosphatase (%) | 24.0 | 25.0 | 62.2 |
| Serum LDH, median U/L (range) | 163.0 (121.0-523.0) | 173.0 (110.0-332.0) | 237.0 (140.0-2538.0) |
| Elevated serum LDH (%) | 24.0 | 18.8 | 51.1 |
| Presence of liver or lung metastasis (%) | 28.0 | 37.5 | 22.2 |
| CellSearch CTC (%) | |||
| 0 | 56.0 | 18.8 | 4.4 |
| 1-4 | 20.0 | 31.3 | 15.6 |
| >=5 | 12.0 | 40.6 | 77.8 |
| Missing | 12.0 | 9.4 | 2.2 |
| CellSearch CTC median (range) | 0 (0-11) | 3 (0-109) | 26 (0-12972) |
Study Design and Assessments
The present study is a retrospective analysis of the PROPHECY trial, a prospective multicenter study evaluating the ability of baseline (pre-treatment) AR-V7 status in CTCs to predict treatment outcomes with abiraterone/enzalutamide as well as subsequent taxane chemotherapy upon disease progression. Patients treated with prior enzalutamide were offered abiraterone and vice versa, and patients without prior abiraterone or enzalutamide exposure were treated according to physician’s choice. Upon disease progression, patients were offered standard-of-care taxane chemotherapy with either docetaxel or cabazitaxel. All authors vouch for the completeness and data integrity and for the fidelity of the study to the clinical protocol (available online). Peripheral-blood samples for analysis of CTC number and CTC AR-V7 status were obtained from eligible patients at pre-specified time points: baseline prior to abiraterone or enzalutamide initiation; at clinical, radiographic, or biochemical progression on abiraterone or enzalutamide; and at progression on taxane-based chemotherapy. CellSearch CTC enumeration was performed at these timepoints on all patients and processed in a CAP/CLIA-approved central laboratory at MSKCC36,37.
Treatment selection was at the discretion of the treating physician without the knowledge of AR-V7 status. Laboratory investigators were blinded to clinical outcomes. All data sets were separately sent to the study statistician (SH) who unblinded the data after database lock.
Analysis of Circulating Tumor Cells
CTCs were analyzed in two central laboratories, each blinded to the results of the other. CTC identification by the Epic Sciences CTC nuclear AR-V7 assay and CTC heterogeneity evaluations were performed as described previously18,32,38,39. The Johns Hopkins AR-V7 modified AdnaTest was performed as previously described using validated methods16,40-43. Established standard operating procedures for sample collection, overnight shipping, processing, and analysis were followed by study sites and the central laboratory at the Johns Hopkins University16,40-43.
Clinical Outcomes
The primary efficacy endpoint in the trial was PFS on abiraterone or enzalutamide therapy, defined from date of registration to clinical/radiographic progression or death, whichever occurred first. Radiographic progression was assessed at each center using PCWG3-modified RECIST 1.1 soft tissue and bone scan criteria44. Clinical progression was defined as a composite endpoint including death, escalating pain or other symptomatic progression, initiation of new systemic therapy, or a skeletal-related event.
Data Analysis
CTC detection and specific HOXB13 cDNA detection and expression was measured using a modified Adnatest using appropriate controls. For CTC HOXB13 cDNA quantification, quantitative PCR (forward primer 5’-TTACCAGTCTTGGGCTCTCG-3’, and reverse primer 5’-TGTACGGAATGCGTTTCTTG-3’) was performed and the absolute HOXB13 copy numbers were quantified using the standard dilution curve generated from the 167bp HOXB13 amplicon. CTC HOXB13 status was categorized into 3 groups: undetectable CTCs (CTC = 0 based on CTC PSMA & PSA = 0); CTC+ but HOXB13 CTC low (< 4 copies) or high (≥4 copies) where the HOXB13 cutoff point was determined by the maximally selected rank statistic45 for prognostic associations based on the OS endpoint. Detection of CTCs was defined as the presence of positive control mRNA for PSMA or PSA above negative controls and performed in the Jun Luo laboratory at Johns Hopkins.
The primary objective of PROPHECY and the present analysis was to validate that patients with pre-treatment HOXB13 high CTCs have shortened PFS and OS with abiraterone/enzalutamide compared with HOXB13 low patients. We described the distribution of HOXB13 detection across patients and compared the baseline characteristics of patients according to HOXB13 detection. Details regarding the overall study design have been published elsewhere32.
The proportional hazards model was used for assessing the prognostic value of HOXB13 CTC status for PFS and OS adjusting for prior abiraterone/enzalutamide treatment and Halabi et al. prognostic factors (risk score), including PSA level, alkaline phosphatase, LDH, opioid analgesic use, ECOG performance status, albumin, hemoglobin, and metastatic site (visceral, bone, node only)46 and CTC enumeration. The Kaplan-Meier product-limit approach was used to estimate the median PFS and OS distributions by CTC HOXB13 status.
Data Availability
Data can be available upon request for investigators interested and who contact the corresponding author (Armstrong) with a valid proposal but will not be publicly posted given the confidentiality of the clinical data.
Results
Between May 2015 and January 2017, we enrolled 118 men with high-risk mCRPC from 5 academic medical centers. Of these, 15 patients were excluded due to insufficient CTC cDNA for HOXB13 analysis, and one patient was excluded due to a lack of follow up data, leaving 102 patients included for the present analysis (CONSORT diagram in Supplementary Figure 1). Baseline characteristics of the cohort are described in Table 1, according to CTC and HOXB13 status, and in Supplementary Table 1, according to CTC and Cellsearch CTC.
Overall in the PROPHECY study (n=102), 22% of men had no CTCs detectable (n=22) and had no HOXB13 cDNA detection. Only 2.9% (n=3) had HOXB13 detection and no PSMA or PSA CTC detection. Of those men with detectable CTCs (n=77), 25% (n=19) had no detectable HOXB13, while the majority of men at 75% (n=58) had both CTC and HOXB13 detection. The median HOXB13 copy numbers was 7 among those with positive CTC HOXB13 detection. Using the gold standard FDA cleared CellSearch assay (n=95), we also found that the majority of men with detectable CTCs (n=73) using this platform had detectable HOXB13 expression (n=56, 77%), and only 23% (n=17) of men with detectable CTCs had no HOXB13 cDNA detection. HOXB13 was moderately correlated with AR-FL copy number (Spearman coefficient of 0.80) and AR-V7 copy number (Spearman 0.60), indicating that most patients had AR driven, HOXB13 expressing CTCs in this mCRPC-adenocarcinoma cohort prior to the receipt of abiraterone or enzalutamide.
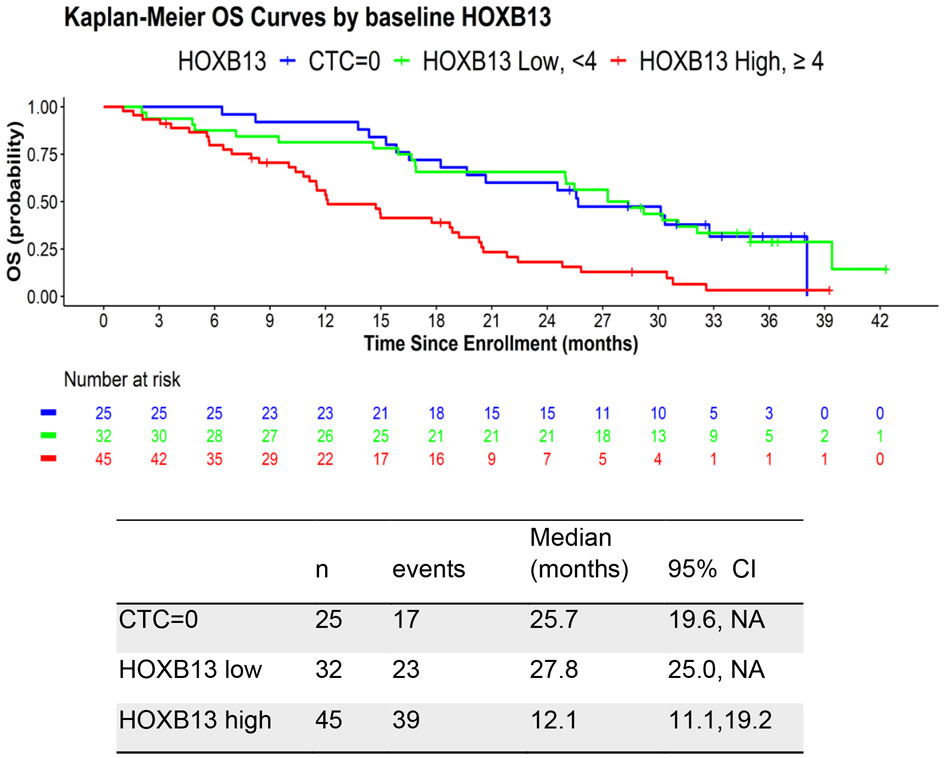
In order to examine the prognostic significance of HOXB13 CTC detection and levels, we analyzed and developed an optimal cutoff for HOXB13 for predicting overall survival from the time of initiation of potent AR inhibitor therapy. We identified an optimal cut-off of 4 copies of HOXB13, in which 44% of PROPHECY patients were CTC+ HOXB13 high, 31% were CTC+ HOXB13 low, and 25% were CTC negative. Using this optimal threshold, the median overall survival in these 3 groups was 12.1, 27.8, and 25.7 months, respectively (Figure 1). The univariate HR for OS for positive HOXB13 CTC detection was 2.76 (95% CI 1.54-4.93), compared to negative CTC detection (Table 2). After adjusting for prior treatment (abi/enza), Halabi clinical risk score, the multivariable HR for OS for positive HOXB13 CTC detection, using the optimal threshold, was 1.99 (95% CI 1.02-3.89) compared to negative CTC detection.
Figure 1.
Kaplan Meier plot of OS in men with mCRPC based on CTC and HOXB13 CTC detection.
Table 2.
Association of CTC HOXB13 expression with overall survival after treatment with abiraterone or enzalutamide, univariate and multivariate analysis.
| N | HR | 95% CI | |
|---|---|---|---|
| Univariate analysis | |||
| CTC=0 | 25 | Ref | |
| HOXB13<4 | 32 | 0.98 | 0.52, 1.84 |
| HOXB13≥4 | 45 | 2.76 | 1.54, 4.93 |
| Adjusting for prior abi/enza treatment, Halabi risk score * | |||
| CTC=0 | 20 | Ref | |
| HOXB13<4 | 27 | 0.91 | 0.44, 1.88 |
| HOXB13≥4 | 43 | 1.99 | 1.02, 3.89 |
| Adjusting for prior abi/enza treatment, Halabi risk score*, CellSearch CTC cutoff at 5 ** | |||
| CTC=0 | 18 | Ref | |
| HOXB13<4 | 25 | 0.76 | 0.35, 1.62 |
| HOXB13≥4 | 42 | 1.39 | 0.64, 3.01 |
| Adjusting for prior abi/enza treatment, Halabi risk score*, CellSearch CTC cutoff at 1, 5 ** | |||
| CellSearch CTC=0 & HOXB13=0 | 15 | Ref | |
| CellSearch CTC=0 & HOXB13>0 | 3 | 2.09 | 0.56, 7.81 |
| CellSearch 1<=CTC<=4 & HOXB13=0 | 11 | 0.80 | 0.29, 2.19 |
| CellSearch 1<=CTC<=4 & HOXB13>0 | 11 | 2.00 | 0.75, 5.33 |
| CellSearch CTC>=5 & HOXB13=0 | 5 | 2.45 | 0.76, 7.89 |
| CellSearch CTC>=5 & HOXB13>0 | 40 | 2.39 | 1.06, 5.40 |
12 patients did not have risk scores
7 patients did not have CellSearch CTC
However, the multivariable HR for OS for positive HOXB13 CTC detection decreased to 1.39 (95% CI 0.64-3.01) after adjusting for prior treatment, Halabi clinical risk score, and CellSearch CTC criteria (5 or more CTCs). This indicates that HOXB13 CTC detection is strongly associated with CTC burden. To further examine the value of HOXB13 CTC detection, we categorized patients into 6 groups based on both HOXB13 and CellSearch CTC where any detection of HOXB13 cDNA (>0) was used as a threshold and 1 and 5 were used as cutoff points for CellSearch CTC. The combination of detectable HOXB13 and CellSearch ≥ 5 copies/ml increases the multivariable HR for OS (2.39, 95% CI 1.06-5.40). After adjusting for JHU CTC AR-V7 detection, the multivariate HR for OS decreased to 1.57 (95% CI 0.67-3.68; Supplementary Table 2) and after adjusting for EPIC CTC nuclear AR-V7 detection, the multivariate HR for OS decreased to 2.19 (95% CI 0.95-5.04; Supplementary Table 3), which are consistent with the primary results.
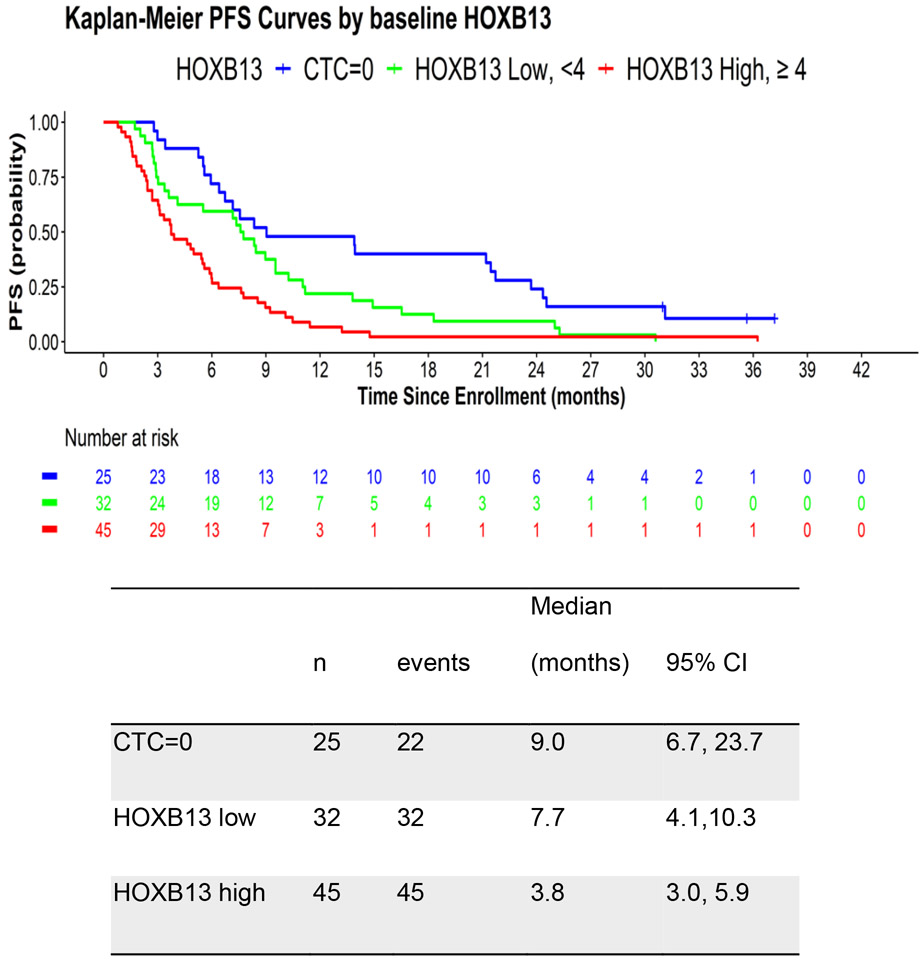
The median progression free survival (PFS) in the CTC=0, HOXB13<4 and HOXB13≥4 groups are 9.03, 7.67 and 3.75 months, respectively (Figure 2). In univariate analysis, HOXB13 CTC detection was associated with worse PFS (HR 3.17, 95% CI 1.86-5.40) using the optimal threshold of 4 HOXB13 copies (Table 3). After adjusting for prior treatment and Halabi clinical risk score, the multivariable HR for PFS for HOXB13 CTC detection using the optimal threshold was 2.31 (95% CI 1.24-4.30). In addition, the multivariable HR for PFS for HOXB13 CTC detection was 2.14 (95% CI 1.04-4.42) after adjusting for prior treatment, Halabi clinical risk score, and CellSearch CTC criteria (5 or more CTCs). Similar to OS, the combination of detectable HOXB13 and CellSearch ≥ 5 copies/ml increases the multivariate HR for PFS to 2.78 (95% CI 1.38-5.59). After adjusting for JHU CTC AR-V7 detection, the multivariate HR for OS decreased to 2.32 (95% CI 1.11-4.84; Supplementary Table 4) and after adjusting for EPIC CTC nuclear AR-V7 detection, the multivariate HR for OS decreased to 2.40 (95% CI 1.17-4.89; Supplementary Table 5), which remain consistent with the primary results and suggest independent negative prognostic associations for abiraterone/enzalutamide treatment outcomes.
Figure 2.
Kaplan Meier plot of PFS in men with mCRPC based on CTC and HOXB13 CTC detection.
Table 3.
Association of CTC HOXB13 expression with progression-free survival on abiraterone/enzalutamide, univariate and multivariate analysis
| N | HR | 95% CI | |
|---|---|---|---|
| Univariate analysis | |||
| CTC=0 | 25 | Ref | |
| HOXB13<4 | 32 | 1.80 | 1.03, 3.12 |
| HOXB13≥4 | 45 | 3.17 | 1.86, 5.40 |
| Adjusting for prior abi/enza treatment, Halabi risk score * | |||
| CTC=0 | 20 | Ref | |
| HOXB13<4 | 27 | 1.78 | 0.97, 3.28 |
| HOXB13≥4 | 43 | 2.31 | 1.24, 4.30 |
| Adjusting for prior abi/enza treatment, Halabi risk score*, CellSearch CTC cutoff at 5 ** | |||
| CTC=0 | 18 | Ref | |
| HOXB13<4 | 25 | 1.74 | 0.88, 3.43 |
| HOXB13≥4 | 42 | 2.14 | 1.04, 4.42 |
| Adjusting for prior abi/enza treatment, Halabi risk score*, CellSearch CTC cutoff at 1, 5 ** | |||
| CellSearch CTC=0 & HOXB13=0 | 15 | Ref | |
| CellSearch CTC=0 & HOXB13>0 | 3 | 1.69 | 0.47, 6.04 |
| CellSearch 1<=CTC<=4 & HOXB13=0 | 11 | 1.62 | 0.71, 3.70 |
| CellSearch 1<=CTC<=4 & HOXB13>0 | 11 | 2.48 | 1.05, 5.81 |
| CellSearch CTC>=5 & HOXB13=0 | 5 | 1.53 | 0.52, 4.46 |
| CellSearch CTC>=5 & HOXB13>0 | 40 | 2.78 | 1.38, 5.59 |
12 patients did not have risk scores
7 patients did not have CellSearch CTC
Finally, we explored if the baseline clinical characteristics of men with mCRPC were associated with HOXB13 CTC detection or absence using CellSearch CTC, hypothesizing that men predisposed to lineage plasticity and AR independence may have lower HOXB13 detection as well. HOXB13 low but CTC+ men did not differ from HOXB13 high CTC+ men based on age, race, Gleason sum, or functional status. However, the CTCs of these patients did differ, as HOXB13 low CTCs were more likely to have low CTC PSA (median 3 copies vs 79 copies), low CTC PSMA (0 copies vs 99 copies), and low CTC AR-FL (47 vs. 89 copies) and CTC AR-V7 (6 vs. 42 copies). HOXB13 CTC low patients were less likely to have Epic nuclear AR-V7 protein detection as well (0% vs. 17%). In addition, HOXB13 low men with mCRPC were more likely to have liver or lung metastases (41% vs 25%) but less likely to have a high serum LDH (12 vs 48%). Interestingly, a subset of HOXB13 negative and Adnatest CTC 0 patients with detectable CTCs by Cellsearch (n=8, Supplementary Table 1) had the highest rate of visceral metastases at 50%, suggesting AR/HOXB13 indifferent differentiation. These data suggest that HOXB13 low mCRPC may have a different AR low and viscerotropic phenotype, but an intermediate prognosis as compared to the poor prognosis associated with the more AR and AR-variant positive HOXB13/CTC positive cohort of men with mCRPC. In comparison, men without CTCs have an overall better prognosis.
Discussion
In the present analysis, we have evaluated the detection of HOXB13 mRNA in CTC enriched blood from men with mCRPC and identified an independent prognostic association for HOXB13 high CTCs with both progression-free and overall survival in the context of potent AR inhibition. Men with HOXB13 high CTCs had adverse prognoses and short times to progression and survival even after adjusting for CTC enumeration and validated clinical prognostic factors. We find that the majority of men with mCRPC (75%) were HOXB13 CTC high. We also identified a subgroup of 25% of patients that lack HOXB13 detection, despite having evaluable CTCs, and these men lacked AR-V7 detection, had lower PSMA and PSA CTC expression, and were enriched in liver and lung metastases. While this small subset did not have an adverse prognosis as compared to CTC positive patients, this data suggests our assay may detect patients that exhibit early lineage plasticity and AR independence.
The present study builds on a number of prior liquid biopsy assays of prognostic and potentially predictive clinical relevance and utility in men with mCRPC. Our prior work from the multicenter, blinded, and prospective PROPHECY trial demonstrates that AR-V7 detection in CTCs by either the JHU mRNA or Epic Sciences nuclear AR-V7 protein assay was associated with a very low probability of confirmed PSA decline and short OS and PFS, and thus have little evidence of clinical benefit from abiraterone or enzalutamide32. HOXB13 detection strongly associated with AR and AR-V7, suggesting co-expression in the majority of such patients. Should an anti-HOXB13 therapeutic be developed, we would anticipate a greater therapeutic index than potent AR inhibitors in such patients, due to the lack of likely toxicities in normal tissues, which lack HOXB13 detection.
Some men with mCRPC have CTCs with absent or very heterogenous HOXB13 expression, similar to what we have reported previously for neuroendocrine and lineage plasticity biomarkers in CTCs in this setting. In this same PROPHECY trial, we have shown that additional biomarkers of AR therapy resistance and poor outcome include chromosomal instability, CTC heterogeneity, CTC lineage plasticity and the neuroendocrine phenotype47, genomic alterations such as CHD1 loss or MYCN gain48, and overexpression of CTC PSMA49. The current study suggests that lack of HOXB13 detection is associated with the lack of CTC PSMA, AR, and PSA expression, and thus novel biomarker approaches are needed for such poor prognosis patients. HOXB13 may also be important in regulating AR therapy resistance through AR reprogramming, and given its tumor specific expression, may thus be a suitable biomarker and precision medicine target.
HOXB13 low CTCs indicate two possibilities: they either reflect low CTC burden (CTC 0) or low HOXB13 expression (CTC+). The HOXB13 low CTC 0 group is a good prognosis low volume disease subset, while the HOXB13 low CTC+ group likely contains neuroendocrine prostate cancer (NEPC) or AR/HOXB13 indifferent cases. The increased rate of visceral metastases despite a lower CTC PSMA, AR/AR-V7, and PSA suggests that some HOXB13 low patients may be enriched for NEPC or AR indifferent double negative phenotypes. HOXB13 expression is retained in >80% of NEPCs or AR-negative CRPCs, although the expression levels are typically lower.50 Decreased HOXB13 expression has been associated with epigenetic silencing and HOXB13 gene body CpG methylation.50
Acknowledging all methods have inherent pros and cons, the Adnatest was the method of choice because it is a standardized and CE-marked test designed to detect CTCs based on the expression of prostate specific transcripts. Similar to many other CTC methods, it requires an enrichment step and there are remaining leukocytes in the sample even after enrichment. However, in the case of Adnatest, its dependence on prostate specific transcripts is expected to alleviate the concern of false positive detection caused by post-enrichment leukocytes.
In conclusion, we have demonstrated that CTC HOXB13 is an independent prognosticator of poor progression-free and overall survival in men with mCRPC treated with abiraterone or enzalutamide. Given the retrospective nature of this study, prospective clinical validation is required. Ideally, HOXB13 biomarker detection using CTCs will be paired with specific HOXB13 molecularly targeted therapies, and in such a context, measuring the detection and heterogeneity of such HOXB13 detection may provide value for identifying the patients with mCRPC most likely to benefit from HOXB13 targeting clinically.
Supplementary Material
Translational Relevance.
HOXB13 is a transcription factor and androgen receptor (AR) co-regulator that can promote prostate cancer development and progression in the setting of castration and potent AR inhibition. In this study, we retrospectively analyzed circulating tumor cells (CTCs) that had been collected during the multicenter prospective PROPHECY trial involving men with metastatic castration-resistant prostate cancer (mCRPC) who received potent AR inhibition with abiraterone or enzalutamide. CTC detection of HOXB13 was associated with worse overall survival and progression free survival. Higher CTC HOXB13 was associated with AR-dependent biomarkers and poor prognosis in men with mCRPC who received potent AR inhibition. Given its prostate cancer specific expression, HOXB13 is a potential liquid biomarker and precision medicine target. HOXB13 detection on a CTC assay could be prognostically useful in selecting patients for AR inhibition therapy or ideally for future HOXB13 targeted therapies.
Acknowledgements
We wish to thank the Prostate Cancer Foundation and Movember for their financial support of this Global Treatment Sciences Challenge Award (PI D McDonnell for present study, and for PROPHECY, PI A Armstrong, Duke), and the US Department of Defense Prostate Cancer Clinical Trial Consortium for infrastructural support for this multicenter study. A. Armstrong was supported by a Prostate Cancer Foundation grant, NIH R01s (1R01CA233585-01 and 5R01CA233585-05), and the DCI P30 CA014236 as well as Duke Cancer Institute shared resources for biostatistics, Flow Cytometry, and Sequencing and Genomic Technologies. This work was partially funded by Department of Defense grants W81XWH-13-PCRP-CCA, W81XWH-17-2-0021 and W81XWH-14-2-0179 (D George, A Armstrong, Duke), W81XWH-15-1-0467 (S Halabi, Duke), W81XWH-18-1-0278 (S Halabi, Duke), W81XWH-14-2-0159 (D Nanus, Weill Cornell), W81XWH-15-2-0018 (R Szmulewitz, Chicago), and W81XWH-16-PCRP-CCRSA (E Antonarakis, Johns Hopkins). D Danila was also supported in part by National Cancer Institute Grants P30CA008748 and the MSKCC Sidney Kimmel Center for Prostate and Urologic Cancers. We wish to thank the study coordinators at Weill Cornell, Duke University, Johns Hopkins, University of Chicago, and Memorial Sloan Kettering Cancer Center. We wish to acknowledge the dedication of our patients to provide blood samples at no clear benefit to them but for the benefits of all patients with prostate cancer.
Supported by a grant from the Prostate Cancer Foundation and Movember as well as infrastructure support from the Department of Defense Prostate Cancer Clinical Trials Consortium (DOD PCCTC)
Footnotes
Conflict of interest:
Susan Halabi
Consulting Role from ASCO, AVEO, BMS, Janssen, Sanofi
Research Funding: ASCO, Astellas
David M. Nanus
Advisory Boards: Janssen Oncology, Telix
Research support: Exelixis, Zenith Epigenetics, Plantable, Sagimet Biosciences
Daniel J. George
Advanced Accelerator Applications SA/Novartis – Personal/Financial – 06/2019
American Association for Cancer Research – Sr Editor – Personal/Financial – 11/2013
Astellas – Consultant, Research, Advisory Board – Institutional and Personal/Financial – 04/2010
Astrazeneca – Research, Consultant, Advisory Board, CAPI-281 Steering Committee member - Institutional and Personal/Financial – 10/2018
AVEO Pharmaceuticals – Personal/Financial – 01/2012
Bayer H/C Pharmaceuticals – Consultant, Speaker, Honorarium, Travel accommodations, SC – Personal/Financial – 04/2019
BMS – Research - Institutional
Calithera – Research – Institutional
Eisai – Personal/Financial – 02/2021
Exelixis, Inc – Research, Consultant, Speaker, Honorarium, Travel accommodations - Institutional and Personal/Financial – 11/2015
IdeoOncology (formerly Nexus) – Consultant – 12/2020
Janssen Pharmaceuticals – Research, Consultant, Independent Data Monitoring Committee (IDMC) - Institutional and Personal/Financial – 03/2015
Medscape Education – Personal/Financial – 02/2017
Merck Sharp & Dohme – Consultant – Personal/Financial – 07/2015
Michael J Hennessey Associates – Honorarium, Consultant – Personal/Financial – 05/2017
Millennium Medical Publishing, Clinical Advances in Hematology & Oncology – Co-Editor-in-Chief – Personal/Financial – 11/2013
Myovant Sciences, Inc – Consultant – Personal/Financial – 08/2017
NCI Genitourinary Steering Committee member (Leidos Biomedical Research Inc) – Personal/Financial – 02/2015
Novartis – Research - Institutional
Pfizer – Research, Consultant, Steering Committee, Honorarium - Institutional and Personal/Financial – 10/2016
Propella TX – Consultant (formerly Vizuri) – Personal/Financial – 03/2021
RevHealth, LLC (David Winkler) – Consultant – 01/2021 – Personal/Financial
Sanofi – Research, Consultant, Speaker, Honorarium, Travel accommodations - Institutional and Personal/Financial – 09/2011
Seattle Genetics – Consultant – 10/2020
UroGPO – Honorarium – Personal/Financial – 04/2018
UroToday (Digital Science Press) – Honorarium, Travel accommodations – Personal/Financial – 06/2018
WebMD – Consultant – 05/2011
WilmerHale Attorneys – Personal/Finance – 04/2021
Xcures – Consultant – 8/2021
Emmanuel S. Antonarakis reports grants and personal fees from Janssen, Sanofi, Bayer, Bristol Myers Squibb, Curium, Merck, Pfizer, AstraZeneca, Clovis, Constellation; personal fees from Astellas, Amgen, Blue Earth, Exact Sciences, Invitae, Eli Lilly, and Foundation Medicine; grants from Novartis, Celgene, and grants from Orion outside the submitted work; and has a patent for an AR-V7 biomarker technology that has been licensed to Qiagen.
Daniel Danila
Research support from US Department of Defense, American Society of Clinical Oncology, Prostate Cancer Foundation, Stand Up 2 Cancer, Amgen, Janssen Research & Development, Astellas, Medivation, Agensys, Genentech, CreaTV. Consultant for Angle LLT, Janssen Research & Development, AstraZeneca, BioView LTD, Clovis, Astellas, Medivation, Pfizer, Agensys, Merck.
Russell Zelig Szmulewitz
Honoraria: Astellas Pharma, Pfizer/Astellas
Consulting or Advisory Role: AstraZeneca, Abbvie, Exelixis, Merck, Amgen, Janssen Oncology, Sanofi, Astellas Pharma, Pfizer, Novartis, Eisai
Research Funding (to institution): Abbvie, Astellas Pharma, Macrogenics, Janssen Oncology, Plexxikon, Harpoon therapeutics, Merck, Novartis, Progenics Patent licensed by University of Chicago of which I am co-inventor to Corcept Therapeutics for combination AR/GR inhibition in prostate cancer
Travel, Accommodations, Expenses; Corcept Therapeutics
Jun Luo is the lead inventor of AR-V7 and CTC technologies that are owned by the Johns Hopkins University and licensed to Qiagen and A & G.
Changxue Lu is a co-inventor of AR-V7 and CTC technologies that are owned by the Johns Hopkins University and licensed to Qiagen.
Andrew J. Armstrong
Research support (to Duke) from the NIH/NCI, PCF/Movember, DOD, Astellas, Pfizer, Bayer, Janssen, Dendreon, BMS, AstraZeneca, Merck, Forma, Celgene, Amgen, Novartis
Consulting or advising relationships with Astellas, Epic Sciences, Pfizer, Bayer, Janssen, Dendreon, BMS, AstraZeneca, Merck, Forma, Celgene, Clovis, Exact Sciences, Myovant, Exelixis, Go
The other authors declare no potential conflicts of interest.
Prior presentation: Poster Presented, in part, at the 2023 American Society of Clinical Oncology National Meeting, Chicago IL June 2023
References
- 1.de Bono JS, Logothetis CJ, Molina A, et al. : Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 364:1995–2005, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer TM, Armstrong AJ, Rathkopf DE, et al. : Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 371:424–33, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan CJ, Smith MR, Fizazi K, et al. : Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol 16:152–60, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Armstrong AJ, Lin P, Tombal B, et al. : Five-year Survival Prediction and Safety Outcomes with Enzalutamide in Men with Chemotherapy-naive Metastatic Castration-resistant Prostate Cancer from the PREVAIL Trial. Eur Urol 78:347–357, 2020 [DOI] [PubMed] [Google Scholar]
- 5.Armstrong AJ, Azad AA, Iguchi T, et al. : Improved Survival With Enzalutamide in Patients With Metastatic Hormone-Sensitive Prostate Cancer. J Clin Oncol 40:1616–1622, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CD, Welsbie DS, Tran C, et al. : Molecular determinants of resistance to antiandrogen therapy. Nat.Med 10:33–39, 2004 [DOI] [PubMed] [Google Scholar]
- 7.Stanbrough M, Bubley GJ, Ross K, et al. : Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 66:2815–2825, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Montgomery RB, Mostaghel EA, Vessella R, et al. : Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res 68:4447–54, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora VK, Schenkein E, Murali R, et al. : Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155:1309–22, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson D, Van Allen EM, Wu YM, et al. : Integrative clinical genomics of advanced prostate cancer. Cell 161:1215–1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson PA, Chen YF, Balbas MD, et al. : Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A 107:16759–65, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan SC, Li Y, Dehm SM: Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem 287:19736–49, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu R, Lu C, Mostaghel EA, et al. : Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 72:3457–3462, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Chan SC, Brand LJ, et al. : Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 73:483–489, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo J, Attard G, Balk SP, et al. : Role of Androgen Receptor Variants in Prostate Cancer: Report from the 2017 Mission Androgen Receptor Variants Meeting. Eur Urol 73:715–723, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antonarakis ES, Lu C, Wang H, et al. : AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371:1028–38, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scher HI, Lu D, Schreiber NA, et al. : Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol 2:1441–1449, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher HI, Graf RP, Schreiber NA, et al. : Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp A, Coleman I, Yuan W, et al. : Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J Clin Invest 129:192–208, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeltser L, Desplan C, Heintz N: Hoxb-13: a new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development 122:2475–84, 1996 [DOI] [PubMed] [Google Scholar]
- 21.Economides KD, Capecchi MR: Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 130:2061–9, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Economides KD, Zeltser L, Capecchi MR: Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev Biol 256:317–30, 2003 [DOI] [PubMed] [Google Scholar]
- 23.Norris JD, Chang CY, Wittmann BM, et al. : The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell 36:405–16, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ewing CM, Ray AM, Lange EM, et al. : Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med 366:141–9, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kote-Jarai Z, Mikropoulos C, Leongamornlert DA, et al. : Prevalence of the HOXB13 G84E germline mutation in British men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes. Ann Oncol 26:756–761, 2015 [DOI] [PubMed] [Google Scholar]
- 26.Nyberg T, Govindasami K, Leslie G, et al. : Homeobox B13 G84E Mutation and Prostate Cancer Risk. Eur Urol 75:834–845, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyberg T, Brook MN, Ficorella L, et al. : CanRisk-Prostate: A Comprehensive, Externally Validated Risk Model for the Prediction of Future Prostate Cancer. J Clin Oncol 41:1092–1104, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanayama M, Chen Y, Rabizadeh D, et al. : Clinical and Functional Analyses of an African-ancestry Gain-of-function HOXB13 Variant Implicated in Aggressive Prostate Cancer. Eur Urol Oncol, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pomerantz MM, Li F, Takeda DY, et al. : The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 47:1346–51, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Wu D, Thomas-Ahner JM, et al. : Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc Natl Acad Sci U S A 115:6810–6815, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen DT, Yang W, Renganathan A, et al. : Acetylated HOXB13 Regulated Super Enhancer Genes Define Therapeutic Vulnerabilities of Castration-Resistant Prostate Cancer. Clin Cancer Res 28:4131–4145, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong AJ, Halabi S, Luo J, et al. : Prospective Multicenter Validation of Androgen Receptor Splice Variant 7 and Hormone Therapy Resistance in High-Risk Castration-Resistant Prostate Cancer: The PROPHECY Study. J Clin Oncol 37:1120–1129, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halabi S, Small EJ, Kantoff PW, et al. : Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J.Clin.Oncol 21:1232–1237, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Halabi S, Lin CY, Kelly WK, et al. : Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 32:671–7, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris MJ, Basch EM, Wilding G, et al. : Department of Defense prostate cancer clinical trials consortium: a new instrument for prostate cancer clinical research. Clin Genitourin Cancer 7:51–7, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Bono JS, Scher HI, Montgomery RB, et al. : Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin.Cancer Res. 14:6302–6309, 2008 [DOI] [PubMed] [Google Scholar]
- 37.Scher HI, Heller G, Molina A, et al. : Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 33:1348–55, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scher HI, Lu D, Schreiber NA, et al. : Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scher HI, Armstrong AJ, Schonhoft JD, et al. : Development and validation of circulating tumour cell enumeration (Epic Sciences) as a prognostic biomarker in men with metastatic castration-resistant prostate cancer. Eur J Cancer 150:83–94, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antonarakis ES, Lu C, Luber B, et al. : Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol 1:582–91, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Antonarakis ES, Lu C, Luber B, et al. : Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol 35:2149–2156, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lokhandwala PM, Riel SL, Haley L, et al. : Analytical Validation of Androgen Receptor Splice Variant 7 Detection in a Clinical Laboratory Improvement Amendments (CLIA) Laboratory Setting. J Mol Diagn 19:115–125, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Markowski MC, Silberstein JL, Eshleman JR, et al. : Clinical Utility of CLIA-Grade AR-V7 Testing in Patients With Metastatic Castration-Resistant Prostate Cancer. JCO Precis Oncol 2017, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scher HI, Morris MJ, Stadler WM, et al. : Trial Design and Objectives for Castration-Resistant Prostate Cancer: Updated Recommendations From the Prostate Cancer Clinical Trials Working Group 3. J Clin Oncol 34:1402–18, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hothorn T, Lausen B: On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis 43:121–137, 2003 [Google Scholar]
- 46.Halabi S, Lin CY, Small EJ, et al. : Prognostic model predicting metastatic castration-resistant prostate cancer survival in men treated with second-line chemotherapy. J Natl Cancer Inst 105:1729–37, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown LC, Halabi S, Schonhoft JD, et al. : Circulating Tumor Cell Chromosomal Instability and Neuroendocrine Phenotype by Immunomorphology and Poor Outcomes in Men with mCRPC Treated with Abiraterone or Enzalutamide. Clin Cancer Res 27:4077–4088, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, Halabi S, Kemeny G, et al. : Circulating Tumor Cell Genomic Evolution and Hormone Therapy Outcomes in Men with Metastatic Castration-Resistant Prostate Cancer. Mol Cancer Res 19:1040–1050, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta S, Halabi S, Yang Q, et al. : PSMA-positive Circulating Tumor Cell Detection and Outcomes with Abiraterone or Enzalutamide Treatment in Men with Metastatic Castrate-resistant Prostate Cancer. Clin Cancer Res 29:1929–1937, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel RA, Sayar E, Coleman I, et al. : Characterization of HOXB13 expression patterns in localized and metastatic castration-resistant prostate cancer. J Pathol, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data can be available upon request for investigators interested and who contact the corresponding author (Armstrong) with a valid proposal but will not be publicly posted given the confidentiality of the clinical data.