Abstract
The etiology of biliary atresia (BA) is unknown, but recent studies suggest a role for rare protein-altering variants (PAVs). Exome sequencing data from the National Birth Defects Prevention Study on 54 child–parent trios, one child–mother duo, and 1513 parents of children with other birth defects were analyzed. Most (91%) cases were isolated BA. We performed (1) a trio-based analysis to identify rare de novo, homozygous, and compound heterozygous PAVs and (2) a case–control analysis using a sequence kernel-based association test to identify genes enriched with rare PAVs. While we replicated previous findings on PKD1L1, our results do not suggest that recurrent de novo PAVs play important roles in BA susceptibility. In fact, our finding in NOTCH2, a disease gene associated with Alagille syndrome, highlights the difficulty in BA diagnosis. Notably, IFRD2 has been implicated in other gastrointestinal conditions and warrants additional study. Overall, our findings strengthen the hypothesis that the etiology of BA is complex.
Keywords: biliary atresia, birth defect, NBDPS, rare variants, whole exome sequencing
1 |. INTRODUCTION
Biliary atresia (BA), a major birth defect with an estimated prevalence of 5–10 per 100,000 births, results in severe liver disease and is the leading indication for pediatric liver transplantation worldwide (Asai et al., 2015; Lakshminarayanan & Davenport, 2016; Sanchez-Valle et al., 2017). Characterized by obstruction of the biliary duct system, children with BA cannot excrete bile from the liver into the intestines to emulsify and help digest fats. Instead, bile is retained in the liver, leading to liver injury, progressive liver fibrosis, and, if untreated, end-stage liver disease by the end of the first year of life (Asai et al., 2015). Approximately 10% of BA cases present as syndromic, for example, with various laterality defects (heterotaxy), including splenic abnormalities and complex cardiac malformations, commonly referred to as the BA splenic malformation syndrome, whereas the remainder of cases present as isolated BA (Berauer et al., 2019; Bezerra et al., 2018; Schwarz et al., 2013).
The genetic architecture of BA, especially isolated BA, remains largely unknown but appears to be complex. Three recent genome-wide association studies revealed that common intronic variants in ADD3, GPC1, ARF6, and EFEMP1 are associated with isolated BA (Chen et al., 2018; Garcia-Barceló et al., 2010; Ningappa et al., 2015). However, some studies have also pointed toward rare variants influencing BA susceptibility. For example, in a study of 67 patients with BA and co-occurring laterality defects, five patients had a rare and potentially deleterious biallelic variant in polycystin-1-like-1 transient receptor potential channel interacting (PKD1L1), a gene associated with ciliary calcium signaling and embryonic laterality determination (Berauer et al., 2019). A recent analysis of exome sequencing (ES) data from 101 children (including 30 child–parent trios) with isolated BA identified 66 rare de novo variants in 66 genes, including potentially deleterious variants in STIP1 and REV1 (Rajagopalan et al., 2020). Furthermore, another study evaluating ES data among nonsyndromic patients from Southeast Asia pointed to the role of rare variants in ciliary genes as underlying BA susceptibility (Lam et al., 2021).
To further elucidate the genetic etiology of BA in children, we sought to identify novel variants associated with isolated BA using ES data from cases, parents, and unrelated controls. We first conducted a family-based analysis using child–parent trios to identify rare de novo, rare homozygous, and rare compound heterozygous protein-altering variants (PAVs). Next, we performed case–control analyses to identify both common and rare PAVs associated with BA.
2 |. MATERIALS AND METHODS
2.1 |. Study population
The National Birth Defects Prevention Study (NBDPS) was a population-based study of over 30 major structural birth defects, which sought to identify environmental and genetic factors associated with these conditions. Details of the study methods and population have been outlined previously (Reefhuis et al., 2015; Yoon et al., 2001). Briefly, sites in 10 U.S. states were included as part of the NBDPS, including Arkansas, California, Georgia, Iowa, Massachusetts, New Jersey, New York, North Carolina, Texas, and Utah. Birth defect surveillance programs from these states ascertained children with eligible defects among pregnancies with estimated dates of delivery between October 1997 and December 2011.
All liveborn children diagnosed with BA were considered for inclusion. First, a board-certified clinical geneticist at each NBDPS site reviewed clinical information abstracted from medical records to verify eligibility (Rasmussen et al., 2003). Consistency across centers was established by a clinical geneticist who performed the final classification of each child diagnosed with BA. Children with known syndromes, chromosomal, or single-gene disorder etiologies were excluded. Next, children with BA were classified as isolated (no additional major birth defects or additional related birth defects only) or multiple (one or more additional major birth defects in an unrelated organ system).
Enrolled mothers completed a computer-assisted telephone interview 6 weeks–2 years after their estimated date of delivery. Following the interview, they were asked to collect buccal cell specimens from themselves, their child (if living), and the child’s father (if available) (Reefhuis et al., 2015). Mothers who participated in the NBDPS with a previous child, those who could not complete the interview in English or Spanish, or those who were incarcerated or otherwise did not have custody of their child at the time of recruitment were excluded.
There were 315 women with eligible pregnancies affected by BA, of whom 216 completed the telephone interview (Reefhuis et al., 2015). Of these, 115 mothers, 105 children, and 95 fathers provided buccal cell specimens. Similarly, 1513 parents from other birth defect groups from the NBDPS with buccal cell specimens for ES were selected as controls (Jenkins et al., 2019). As described previously (Jenkins et al., 2019), two different types of cytobrushes were used to collect specimens during phases of the study: “wet brushes” (1997–2003) (cytobrushes packaged in closed plastic tubes preventing air drying [Cyto-Pak Cytosoft Brushes CP-5B, Medical Packaging Corporation, Camarillo, CA]) and “dry brushes” (2003–2011) (cytobrushes packaged in open paper-backed peel pouches [Cytology Brush Pack CYB-1, Medical Packaging Corporation]). Informed consent was obtained for all participants providing buccal cell specimens, and the study protocol for the NBDPS was approved by the institutional review board at each NBDPS site.
2.2 |. Specimen processing, sequencing, and alignment
Specimen processing, sequencing, and alignment were performed in collaboration with the National Institutes of Health Intramural Sequencing Center (NISC) at the National Human Genome Research Institute (NHGRI) and the University of Washington Center for Mendelian Genomics. Detailed procedures have been previously described (Jenkins et al., 2019). Specifically, due to DNA quality and quantity considerations, 64 child–parent trio specimens from only dry brushes were subjected to ES. Buccal specimens with adequate DNA amounts (≥200 ng assessed by quantitative real-time polymerase chain reaction [PCR] targeting the RNaseP gene) were sent to the NISC at the NHGRI for processing and sequencing. The ES capture kit used at NISC was a standard commercially available kit, the NimbleGen Seq-Cap EZ Exome+UTR Library (Version 3.0) and covered 96 Mb (Roche NimbleGen, 2013). The DNA was sheared mechanically, and targeted fragments were captured by probe hybridization and amplified before sequencing (Jenkins et al., 2019). NISC generated read lengths of 126 bases on an Illumina HiSeq 2500 instrument. Paired-end reads generated approximately 250 base pairs (bp) of sequence from each fragment in the library. A total of 38 million paired-end 126 bp reads were targeted and as many as 48 libraries were pooled and sequenced across as many lanes as needed to achieve the targeted number of reads (938 million read pairs or 76 million reads pre-library); thus, 5–6 libraries were run per lane. Image analysis and base calling were performed using the Illumina Genome Analyzer Pipeline software (version 1.18.64.0) with default parameters. In preparation for ES, ten trios and one father failed during sequencing library preparation due to bad reagents from contaminated library kits, and there was insufficient DNA quantity available for a second library rebuild. The final sample of BA cases sequenced comprised 54 child–parent trios and one child–mother duo. Cases were predominantly isolated BA (n = 50), whereas five had additional major birth defects.
After ES at NISC, Binary Alignment Map files were sent to the University of Washington Center for Mendelian Genomics for reprocessing. Reads were aligned to human reference (hg19hs37d5) using BWA-MEM (Burrows-Wheeler Aligner v0.7.10) (Li et al., 2009). Read data from a flow-cell lane were treated independently for alignment and quality control (QC) purposes in instances where merging of data from multiple lanes was required (e.g., for DNA sample multiplexing). Read-pairs not mapped within ±2 standard deviations (SD) of the average library size (~150 ± 15 bp for exomes) were removed. All aligned read data were subject to the following steps: (a) “duplicate removal” (Picard MarkDuplicates v1.111); (b) indel realignment (the Genome-Analysis-Toolkit (GATK) IndelRealigner v3.2–2); and (c) base quality recalibration (GATK BaseRecalibrator v3.2–2). Variant detection and genotyping were performed using the HaplotypeCaller tool from GATK v3.2. Following GATK best practices, variant quality score recalibration was performed. Variants flagged as low quality or potential false positives (quality score ≤ 50, long homopolymer run >4, quality by depth <5, or within a cluster of single nucleotide polymorphisms) were excluded.
2.3 |. Variant and sample quality control
Following specimen sequencing and alignment, we performed all subsequent ES data QC. In particular, all variants underwent additional genotype, variant, and sample QC prior to case–control analyses. We first removed variants with mean genotype depth < 10 reads. Variants were removed if they were multi-allelic, failed the Hardy–Weinberg equilibrium (HWE) check at p < 10−6 or had a call rate <0.99. GATK best practices (McKenna et al., 2010) for variant prioritization were applied, whereby variants with heterozygosity values >54.69 were removed. Next, the variant quality score recalibration pipeline in GATK v4.1.2 was implemented, utilizing seven informative annotation profiles (Quality by Depth, Mapping Quality, MQRankSum, ReadPos-RankSum, Fisher strand, Strand odds ratio, and InbreedingCoeff) to quantify the quality of all variants. Single nucleotide variants (SNVs) and insertions/deletions (INDELs) were evaluated independently using unique annotation profiles as recommended by GATK. Top-quality SNVs and INDELs were selected at the 99.7 and 99.0 percentile, respectively. Lastly, only uniquely mapped variants with a 100-mers mappability score of one were evaluated (Karimzadeh et al., 2018).
Sample QC involved filtering on mean sample’s genotype depth, number of variants, number of singletons, inbreeding coefficient, heterozygous-to-homozygous ratio, transition-to-transversion ratio, and missingness. Specifically, individuals were excluded if any of the evaluated metrics fell beyond ±6 SD from the sample mean. Sample kinship for genetic relatedness and genetic ancestry estimation were evaluated with KING v2.2 (Manichaikul et al., 2010) and PRIMUS v1.9 (Staples et al., 2013), respectively. Individuals identified to be duplicates, related at second degree or closer with cases, or parents of children with BA were excluded from the final sample for the case–control association analysis.
2.4 |. Child–parent trio analysis
ES data was available for 54 child–parent trios. In each trio, variants were identified using Platypus v0.8.1 (Rimmer et al., 2014), and variant annotation was conducted using ANNOVAR (Wang et al., 2010) for information on variant type, alternate allele frequency (AAF) in the gnomAD v2.1.1 population database (Karczewski et al., 2020), and multiple in silico predictions of variant deleteriousness that included rare exome variant ensemble learner (REVEL) (Ioannidis et al., 2016) and phred-scaled combined annotation dependent depletion (CADD) (Kircher et al., 2014) scores. To identify potential pathogenic variants, we prioritized rare de novo (novel or gnomAD AAF <0.0001), homozygous (novel or gnomAD AAF <0.001), and compound heterozygous (novel or gnomAD AAF <0.001) variants.
2.5 |. Case–control analysis
Differences in case and control groups by demographic characteristics were compared using the Pearson χ2 test. Common and rare variants were evaluated separately. Specifically, we utilized the single variant score test in Rvtests (Zhan et al., 2016) to identify common variants (minor allele frequency [MAF] ≥0.05) associated with BA. The association of rare variants (MAF <0.05) was evaluated using a gene-based approach. To prioritize for PAVs, we evaluated rare missense and rare synonymous variants independently since we expected synonymous variants to be unassociated. Furthermore, in the gene-based association analyses, we only evaluated genes with (1) at least two variants in the overall study sample and (2) at least one variant present in children with BA. Principal components (PCs) were calculated using PLINK v1.9 (Purcell et al., 2007) to capture unmeasured ancestry structure in the study population. We conducted the sequence kernel-based association test (SKAT) for a combined effect of rare variants using the SKAT v2.0.1 package in R v4.1.1 (Ionita-Laza et al., 2013; Lee et al., 2012). Briefly, SKAT is a region-based test for the joint effects of the individual variant score test statistic. Within a prespecified genomic region of multiple rare variants, SKAT performs a multiple regression approach directly regressing a phenotype on genetic variants and covariates, and SKAT p-values for the association are computed analytically (Lee et al., 2012; Wu et al., 2011). All analyses were applied using the efficient resampling method for the inclusion of extremely rare variants (Lee et al., 2016). Assuming an additive genetic model, all statistical models for common and rare variants were adjusted for sex and the first five PCs to account for population stratification. Quantile–quantile plots and genomic inflation factors were evaluated for signs of genomic inflation. Raw association p-values were corrected for multiple testing using the Bonferroni correction approach, and statistically significant findings were defined at the corrected p < 0.05.
2.6 |. Pathogenic variant validation
Potential pathogenic variants identified in the child–parent trio analysis and their inheritance patterns were further validated by an orthogonal DNA-sequencing method. Target amplicons were amplified from genomic DNA using conventional PCR (HotStarTaqDNA polymerase, QIAGEN), and PCR amplification products were analyzed by Sanger sequencing using established methods.
3 |. RESULTS
3.1 |. Study characteristics
Our initial population included 1568 individuals, including 50 isolated BA cases, 5 cases with multiple defects, and 1513 controls, that underwent ES with 754,935 variants available prior to QC. Following sample QC, 17 controls did not pass the inclusion threshold, and 32 controls failed relatedness checks, which included two duplicated controls and 30 controls related at second–degree or closer to children with BA. Similarly, following variant QC, we excluded 536,571 (71.1%) variants–34,999 (4.6%) variants failed the HWE threshold p < 10−6; 426,983 (56.6%) had a call rate <0.99; 71,289 (9.4%) failed GATK best practices; and 3300 (0.4%) had a 100-mers mappability score less than one. The final dataset for the child–par ent trio analysis included 54 child–parent trios. For the case–control analysis following QC, ES data on 55 cases, 1481 unrelated controls, and 218,364 SNVs and INDELs were selected.
Overall, half of the children with BA (n = 28, 50.9%) and controls (n = 740, 50.0%) were male (Table 1). Among those with BA, 54.5% were of European ancestry (n = 30), 20.0% of African ancestry (n = 11), and 10.9% of Asian ancestry (n = 6) based on PRIMUS v1.9 genetic ancestry estimation. In comparison, while the majority (n = 1023, 69.1%) of the controls were also of European ancestry, 7.8% (n = 115) and 5.5% (n = 82) were of African and Asian ancestry, respectively.
TABLE 1.
Demographic characteristics of children with biliary atresia and unrelated parents of children without biliary atresia with exome sequencing data and enrolled in the National Birth Defects Prevention Study, 1997–2011.
| Demographic characteristics, n (%) | BA children (n = 55) | Non-BA parents (n = 1481) | p-Value* |
|---|---|---|---|
| Sex | |||
| Male | 28 (50.9) | 740 (50.0) | 0.89 |
| Female | 27 (49.1) | 741 (50.0) | |
| Genetic ancestrya | |||
| European | 30 (54.5) | 1023 (69.1) | 0.01 |
| Native American | 4 (7.3) | 113 (7.6) | |
| African | 11 (20.0) | 115 (7.8) | |
| Asian | 6 (10.9) | 82 (5.5) | |
| Admixed | 4 (7.3) | 148 (10.0) |
Abbreviation: BA, biliary atresia.
Genetic ancestry was estimated with PRIMUS v1.9.
p-Value based on the Pearson χ2 test.
3.2 |. Child–parent trio analysis
Rare de novo, homozygous, and compound heterozygous PAVs were prioritized in 54 BA child–parent trios. Overall, a total of 42 rare de novo PAVs were identified in 27 (50.0%) children with BA, of which five (11.9%) were loss-of-function variants (Table S1). However, no de novo PAVs were recurrent across more than one trio. A novel de novo stop-gain variant in the Notch receptor 2 (NOTCH2) gene (NM_024408.4:c.5194C > T,p.Gln1732Ter) was confirmed by Sanger sequencing (Table 2). Moreover, we identified two children with BA with compound heterozygous variants in the polycystic kidney disease 1 like 1 gene (PKD1L1), NM_138295.5:c.8485G > C(p.Glu2829Gln) / NM_138295.5:c.7552G > A(p.Ala2518Thr) and NM_138295.5: c.6473 + 2_6473 + 3del / NM_138295.5:c.731C > T(p.Pro244Leu) (Table 2). All variants in NOTCH2 and PDK1L1 were identified among children with isolated BA and orthogonally confirmed with Sanger sequencing. Additional homozygous (n = 1 in two children with BA) and compound heterozygous (n = 4 in six children with BA) variants are outlined in Tables S2 and S3, respectively.
TABLE 2.
Rare protein-altering variants identified from exome sequencing data of children with biliary atresia and their parents enrolled in the National Birth Defects Prevention Study, 1997–2011.
| Inheritance pattern | Child | Gene | Variant | Variant type | AAFa | REVEL | CADD | ClinVar variation ID |
|---|---|---|---|---|---|---|---|---|
| De novo | ||||||||
| 9 | NOTCH2 | NM_024408.4:c.5194C > T (p.Gln1732Ter) | Stop gain | 0 | – | 40.0 | – | |
| Autosomal recessive | ||||||||
| 23 | PKD1L1 | NM_138295.5:c.8485G > C (p.Glu2829Gln) | Missense | 0 | 0.1 | 14.4 | – | |
| NM_138295.5:c.7552G > A (p.Ala2518Thr) | Missense | 2.5 × 10−5 | 0.14 | 9.64 | – | |||
| 32 | PKD1L1 | NM_138295.5:c.6473 + 2_6473 + 3del | Deletion | 4.0 × 10−4 | – | – | 235796 | |
| NM_138295.5:c.731C > T (p.Pro244Leu) | Missense | 4.2 × 10−3 | 0.05 | 7.61 | 787669 |
Abbreviations: AAF, alternate allele frequency; REVEL, rare exome variant ensemble learner score; CADD, phred-scaled combined annotation-dependent depletion score.
Average alternate allele frequency based on gnomAD v2.1.1 database.
3.3 |. Case–control association analysis
Overall, 78,316 rare PAVs in 6919 genes and 48,642 rare synonymous variants in 6206 genes passed QC. The gene-based testing identified a significant association between BA and IFRD2 (p = 3.75 × 10−6; Bonferroni corrected p = 0.03) (Table 3). The IFRD2 gene-based association test was based on 21 rare PAVs, of which three variants–NM_006764.5: c.1016C > T(p.Ser339Phe), NM_006764.5:c.427G > A(p.Gly143Ser), and NM_006764.5:c.791G > A(p.Arg264Gln)–had a p < 0.05 based on the single variant Score test (Table S4). Among cases that carried PAVs in IFRD2, 50.0%, 12.5%, 25.0%, 12.5%, and 0.0% were of White, Hispanic, Black, Asian, and Mixed ancestry, respectively, while 27.2%, 18.4%, 18.4%, 8.8%, and 27.2% of controls were of White, Hispanic, Black, Asian, and Mixed ancestry, respectively (Fisher exact test p = 0.3). No other genes met the gene-based Bonferroni corrected p < 0.05 threshold. The gene-based genomic inflation factor in the missense rare PAVs analysis was observed at 1.13 (Figure S1). Additionally, no significant associations were identified among rare synonymous variants (Figure S2).
TABLE 3.
Gene identified from the sequence kernel-based association analysis of rare missense protein-altering variants in exome sequencing data of children with biliary atresia enrolled in the National Birth Defects Prevention Study, 1997–2011.
| Gene | No. rare variants | Case cumulative AAFa | Control cumulative AAFa | SKAT p-value (adjusted*) |
|---|---|---|---|---|
| IFRD2 | 21 | 0.004 | 0.002 | 3.75 × 10−6 (0.03) |
Abbreviations: AAF, alternate allele frequency; SKAT, sequence-based kernel association test.
Cumulative AAF was calculated as the total alternate allele count over the total allele number across all variants in a gene within each cohort.
p-Values adjusted for multiple testing using Bonferroni correction.
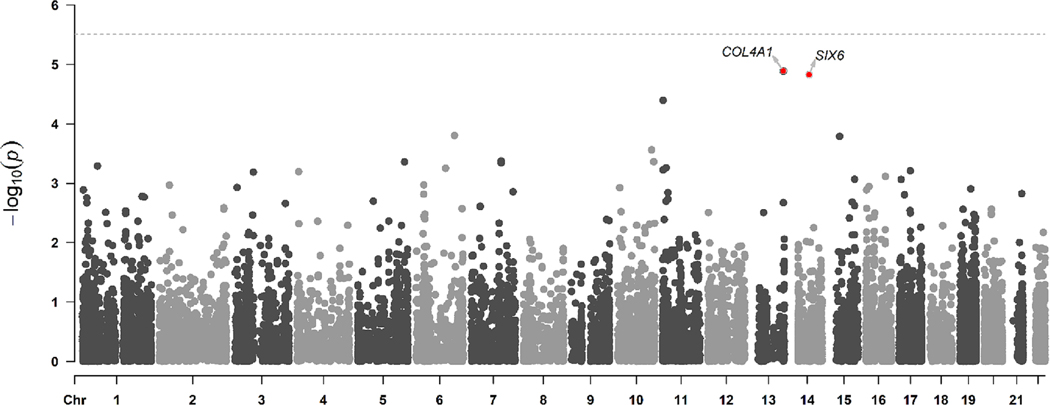
In the analysis of common variants, 15,944 SNVs and INDELs at MAF ≥0.05 were evaluated (Figure 1, Figure S3). While no variants were significant at the Bonferroni corrected threshold (p < 3.14 × 10−5), the top three hits included two synonymous variants–NM_001845.6:c.3189A > T(p.Arg1063=) and NM_001845.6: c.3183G > A(p.Gly1061=)–in COL4A1 and a missense variant–NM_007374.3:c.421C > A(p.His141Asn)–in SIX6 (Supplementary Table 5). In the gene-based association analysis, there was no statistical association for COL4A1 (p = 0.3). No rare PAVs were present for SIX6.
FIGURE 1.
Manhattan plot for common variants evaluated in a genome-wide association analysis from exome sequencing data of children with biliary atresia and unrelated parents of children without biliary atresia enrolled in the National Birth Defects Prevention Study, 1997–2011. Horizontal threshold line indicates the Bonferroni corrected p-value.
4 |. DISCUSSION
Overall, our study adds to emerging evidence on the role of the genetic underpinnings of isolated BA. Specifically, among children with isolated BA, we observed variants in PDK1L1, a gene similarly described among children with syndromic BA (Berauer et al., 2019) and identified susceptibility PAVs in IFRD2. Our assessment did not suggest that recurrent de novo PAVs account for a sizeable proportion of cases. However, while we did not identify any recurrent de novo PAVs across trios, our finding related to NOTCH2, a disease gene found in children with Alagille syndrome, may point to the unique challenges of diagnosing BA.
Variants in the Notch signaling pathway, including in NOTCH2, underlie Alagille syndrome, which can mimic BA in early infancy with presentations of cholestasis and bile duct paucity (Gilbert et al., 2019; Kamath et al., 2012; ShenTu et al., 2021). Hence, it is possible that the infant identified with a variant in NOTCH2 was misdiagnosed with BA. This is plausible because distinguishing BA and Alagille syndrome can be a clinical challenge. Both phenotypes may present similarities in the first weeks of life, and occasionally infants with Alagille syndrome will undergo a Kasai hepatoportoenterostomy, a surgical procedure that is the first line of treatment for BA (Hartley et al., 2009; Lee et al., 2015). Furthermore, the infant with the variant in NOTCH2 had a Kasai procedure reported in the medical record, but information on long term follow-up was not available. Alternatively, we cannot rule out the possibility that NOTCH2 variants and the associated Notch signaling pathway might have a role in the development of BA, and the lack of NOTCH2 variants in additional study cases may point to BA having multiple etiologies (Mao et al., 2018; Zagory et al., 2017). Future larger studies could help inform if variants in Notch pathway genes are found in a subset of BA cases.
Our finding of compound heterozygous PAVs in PKD1L1 among two children with BA supports a prior report of 67 patients with BA and co-occurring laterality defects, in which five children were identified with bi-allelic variants in this gene (Berauer et al., 2019). Unlike the previous assessment where all patients had co-occurring laterality defects, individuals in our study were predominantly isolated cases, suggesting these groups could have overlapping etiologies. Of interest, the variant NM_138295.5:c.6473 + 2_6473 + 3del has been reported independently by three clinical groups to be likely pathogenic (https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000235796.4) and was identified to be associated with laterality defects in humans (Vetrini et al., 2016). PKD1L1 is a member of the polycystic kidney disease family of large membrane proteins called polycystin proteins. Located in the primary cilia of the renal epithelium, PKD1L1 along with PKD2 form a Ca2+ channel complex that regulates the ciliary motility and extracellular fluid flow, two processes required for left–right axis formation in vertebrates (Delling et al., 2013; Grimes et al., 2016; Hojo et al., 2007; Kamura et al., 2011). Given the function of cilia in modulating biliary flow and its contribution to cholangiopathies (Mansini et al., 2018; Masyuk et al., 2008), it is biologically plausible that abnormal cholangiocyte ciliary structure and function contribute to cholangiopathy development observed in BA. However, the specific mechanistic role of PKD1L1 in the pathogenesis of cholangiopathy and BA remains largely unknown.
In one of the only other large-scale trio-based sequencing assessments of isolated BA, Rajagopalan et al. prioritized 66 de novo variants in 66 genes including potentially deleterious variants in STIP1 and REV1 in an analysis of 30 child–parent trios (Rajagopalan et al., 2020). However, in our evaluation of 54 child–parent trios, we did not observe de novo PAVs in STIP1 or REV1. This absence in replication of previously reported de novo variants could be attributed to the genetic ancestry of the study populations or the genetic heterogeneity of BA. For example, two recent family-based sequencing assessments of BA among Asian children identified variants in novel genes including AMER1, INVS, OCRL, PCNT, KIF3B, and TTC17 suggesting a genetic heterogeneity of BA (Lam et al., 2021; Tran et al., 2021). The lack of replication and identification of de novo variants across multiple studies highlight the complexity of the etiology of BA and support the hypothesis that isolated BA is multifactorial.
In an independent analysis of child–parent trios from Southeast Asia (n = 89), investigators concluded that variants in ciliary genes may play a role in susceptibility to nonsyndromic BA (Lam et al., 2021). To further explore these findings in our population, we evaluated variants in these genes using a similar strategy. Specifically, Lam et al. noted that 37.5% of protein truncating de novo variants identified in trios were in ciliary genes, whereas in our population, none of the five protein-truncating de novo variants were in ciliary genes. Additionally, Lam et al. reported 31.5% of individuals with BA carried at least one rare damaging variant in a ciliary gene, while we observed that 7.4% of cases carried these variants. Moreover, we did not detect an increased burden of rare variants among ciliary or liver expressed ciliary gene sets (SKAT p = 0.4 in both gene sets). As with the assessment by Rajagopalan et al., differences in findings could be due to the etiologic complexity of BA, as well as differences in genetic ancestry across populations.
A notable finding in our case–control analysis was the identification of IFRD2 among children with BA harboring rare PAVs. Reports on the role of IFDR2 on BA etiology are limited; however, there is evidence suggesting IFRD2 plays an important role in gastrointestinal development. For example, IFRD2 was highly expressed in the hepatic primordium in the initial stages of embryogenesis in a murine model (Buanne et al., 1998). IFRD2, along with its paralogue IFRD1, are thought to be involved in fat metabolism and adipogenesis where Wnt signaling, an important negative regulator of adipocyte differentiation, was highly upregulated in IFRD2 knockout mice (Vietor et al., 2020). More recently, IFRD2 variants have been identified in relation to sporadic colorectal cancer and high light scatter reticulocyte count in human studies (Barton et al., 2021; Yu et al., 2018). Additionally, IFRD2 has been described to be associated with interferon (IFN) activities, a cytokine with involvement in immunomodulatory responses, which may further support its potential implication in BA development (Cheluvappa et al., 2015; Stark et al., 1998). For example, studies involving human BA livers have observed affected hepatic microenvironments to be pro-inflammatory and pro-fibrotic with overexpression of activation markers including IFN-γ (Asai et al., 2015; Mack et al., 2004). While the mechanisms underlying the association between IFRD2 and BA are unclear, exploring this finding in independent populations is warranted.
In our assessment of common variants, we did not observe any of the previously reported BA associated loci in ADD3, GPC1, ARF6, or EFEMP1 (Chen et al., 2018; Garcia-Barcelo et al., 2010; Ningappa et al., 2015). However, this is not unexpected as the reported variants were intronic and not captured in our sequencing platform. However, we did observe some other interesting variants. Specifically, while not statistically significant after correcting for multiple testing, we identified two synonymous variants in COL4A1 and one missense variant in SIX6. While the variants identified in COL4A1 were reported to be likely benign in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000258250.19, https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000258251.19), an up-regulated transcription of several inflammatory and fibrosis genes, including COL4A1, was observed in studies using murine models with chronic cholangitis (Nakken et al., 2007, 2009). In human tissues with hepatocellular carcinoma, COL4A1 has been hypothesized to promote cell proliferation and metastasis (Wang et al., 2020; Zhang et al., 2021). As with IFRD2, the role of these variants and genes on BA susceptibility is not clear and should be considered in future assessments.
Our study should be considered in light of certain limitations. As with previous assessments of BA, our sample size was relatively small, allowing us to identify only highly penetrant rare variants. This limitation is partly a function of the rarity and low prevalence of BA, and future studies leveraging multiple data sources to increase the sample size would improve statistical power to detect variants with small or moderate effects on BA risk. In addition, the use of parents from other birth defect groups as controls may bias our results; however, we did not observe significant genomic inflation from the common variants or gene-based analyses. Nonetheless, further studies may benefit from including unaffected children as controls for association testing.
Our study also has several strengths. A primary strength is the use of a trio-based design to discern between inherited versus de novo PAVs. Another strength is that children were systematically ascertained for the NBDPS using population-based birth defect surveillance programs. NBDPS is a multisite study with active surveillance methods that ascertain ethnically diverse population-based cases rather than hospital or clinic-based cases, minimizing potential selection bias. Finally, medical records for each child were reviewed by clinical geneticists, producing a well-characterized population.
In conclusion, our assessment adds to our growing understanding of the genetic etiologies underlying isolated BA and the potential complexity and heterogeneity of this phenotype. Future assessments would benefit from larger sample sizes, as our assessment does not suggest that a large proportion of cases are due to highly penetrant rare variants. Additionally, our data do not support that recurrent de novo variants play an important role in BA susceptibility. While our findings support the role of PKD1L1 in the developmental origins of BA, our findings related to NOTCH2 and IFRD2 warrant additional study.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by R01HD093660 to A.J. Agopian and Philip J. Lupo, U01DD001285 to Wendy N. Nembhard, and through Centers for Disease Control and Prevention cooperative agreements under PA #96043, PA #02081, FOA #DD09-001, FOA #DD13-003, and NOFO #DD18-001 to the Centers for Birth Defects Research and Prevention participating in the National Birth Defects Prevention Study (NBDPS) and/or the Birth Defects Study To Evaluate Pregnancy exposureS (BD-STEPS). This work was additionally supported by the Division of Intramural Research of the National Human Genome Research Institute, National Institutes of Health. Sequencing data were reprocessed and analyzed by the University of Washington Center for Mendelian Genomics and was funded by the National Human Genome Research Institute and the National Heart, Blood, and Lung Institute grants UM1 HG006493 and U24 HG008956. We also would like to thank all NBDPS participants, scientists, staff, the Genetics Collaborative Working Group, and the California Department of Public Health, Maternal Child and Adolescent Health Division for providing surveillance data from California for this study. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention, the National Institutes of Health, or the California Department of Public Health.
Funding information
Birth Defects Study To Evaluate Pregnancy exposureS; Centers for Disease Control and Prevention; National Birth Defects Prevention Study; National Heart, Lung, and Blood Institute; National Human Genome Research Institute; National Institutes of Health
Footnotes
CONFLICT OF INTEREST STATEMENT
Sanjiv Harpavat participates in a Data Safety Monitoring Board (DSMB) for a therapeutic clinical trial for biliary atresia. The DSMB is sponsored by Syneos Health. No other conflict of interest to declare for the rest of the authors.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
Data from the NBDPS are not released to the public. Qualified researchers can be granted access to NBDPS data for analysis through collaboration with one of the Centers for Birth Defects Research and Prevention.
REFERENCES
- Asai A, Miethke A, & Bezerra JA (2015). Pathogenesis of biliary atresia: Defining biology to understand clinical phenotypes. Nature Reviews. Gastroenterology & Hepatology, 12(6), 342–352. 10.1038/nrgastro.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AR, Sherman MA, Mukamel RE, & Loh P-R (2021). Whole-exome imputation within UK biobank powers rare coding variant association and fine-mapping analyses. Nature Genetics, 53(8), 1260–1269. 10.1038/s41588-021-00892-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berauer J-P, Mezina AI, Okou DT, Sabo A, Muzny DM, Gibbs RA, Hegde MR, Chopra P, Cutler DJ, Perlmutter DH, Bull LN, Thompson RJ, Loomes KM, Spinner NB, Rajagopalan R, Guthery SL, Moore B, Yandell M, Harpavat S, … Childhood Liver Disease Research Network (ChiLDReN). (2019). Identification of polycystic kidney disease 1 like 1 gene variants in children with biliary atresia splenic malformation syndrome. Hepatology, 70(3), 899–910. 10.1002/hep.30515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra JA, Wells RG, Mack CL, Karpen SJ, Hoofnagle JH, Doo E, & Sokol RJ (2018). Biliary atresia: Clinical and research challenges for the twenty-first century. Hepatology, 68(3), 1163–1173. 10.1002/hep.29905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buanne P, Incerti B, Guardavaccaro D, Avvantaggiato V, Simeone A, & Tirone F. (1998). Cloning of the human interferon-related developmental regulator (IFRD1) gene coding for the PC4 protein, a member of a novel family of developmentally regulated genes. Genomics, 51(2), 233–242. 10.1006/geno.1998.5260 [DOI] [PubMed] [Google Scholar]
- Cheluvappa R, Eri R, Luo AS, & Grimm MC (2015). Modulation of interferon activity-associated soluble molecules by appendicitis and appendectomy limits colitis-identification of novel anti-colitic targets. Journal of Interferon and Cytokine Research, 35(2), 108–115. 10.1089/jir.2014.0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gilbert MA, Grochowski CM, McEldrew D, Llewellyn J, Waisbourd-Zinman O, Hakonarson H, Bailey-Wilson JE, Russo P, Wells RG, Loomes KM, Spinner NB, & Devoto M. (2018). A genome-wide association study identifies a susceptibility locus for biliary atresia on 2p16.1 within the gene EFEMP1. PLoS Genetics, 14(8), e1007532. 10.1371/journal.pgen.1007532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delling M, DeCaen PG, Doerner JF, Febvay S, & Clapham DE (2013). Primary cilia are specialized calcium signalling organelles. Nature, 504(7479), 311–314. 10.1038/nature12833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Barceló M-M, Yeung M-Y, Miao X-P, Tang CS-M, Cheng G, Chen G, So M-T, Ngan ES-W, Lui VC-H, Chen Y, Liu X-L, Hui K-JWS, Li L, Guo W-H, Sun X-B, Tou J-F, Chan K-W, Wu X-Z, Song Y-Q, … Tam PK-H (2010). Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Human Molecular Genetics, 19(14), 2917–2925. 10.1093/hmg/ddq196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MA, Bauer RC, Rajagopalan R, Grochowski CM, Chao G, McEldrew D, Nassur JA, Rand EB, Krock BL, Kamath BM, Krantz ID, Piccoli DA, Loomes KM, & Spinner NB (2019). Alagille syndrome mutation update: Comprehensive overview of JAG1 and NOTCH2 mutation frequencies and insight into missense variant classification. Human Mutation, 40(12), 2197–2220. 10.1002/humu.23879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes DT, Keynton JL, Buenavista MT, Jin X, Patel SH, Kyosuke S, Vibert J, Williams DJ, Hamada H, Hussain R, Nauli SM, & Norris DP (2016). Genetic analysis reveals a hierarchy of interactions between Polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genetics, 12(6), e1006070. 10.1371/journal.pgen.1006070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Davenport M, & Kelly DA (2009). Biliary atresia. Lancet, 374(9702), 1704–1713. 10.1016/S0140-6736(09)60946-6 [DOI] [PubMed] [Google Scholar]
- Hojo M, Takashima S, Kobayashi D, Sumeragi A, Shimada A, Tsukahara T., Yokoi H., Narita T., Jindo T., Kage T., Kitagawa T., Kimur T., Sekimizu K., Miyake A., Setiamarga D., Murakami R., Tsuda S., Ooki S., Kakihar K., … Takeda H. (2007). Right-elevated expression of charon is regulated by fluid flow in medaka Kupffer’s vesicle. Development, Growth & Differentiation, 49(5), 395–405. 10.1111/j.1440-169X.2007.00937.x [DOI] [PubMed] [Google Scholar]
- Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, … Sieh W. (2016). REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. American Journal of Human Genetics, 99(4), 877–885. 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, & Lin X. (2013). Sequence kernel association tests for the combined effect of rare and common variants. American Journal of Human Genetics, 92(6), 841–853. 10.1016/j.ajhg.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MM, Almli LM, Pangilinan F, Chong JX, Blue EE, Shapira SK, White J, McGoldrick D, Smith JD, Mullikin JC, Bean CJ, Nembhard WN, Lou X-Y, Shaw GM, Romitti PA, Keppler-Noreuil K, Yazdy MM, Kay DM, Carter TC, … National Birth Defects Prevention Study. (2019). Exome sequencing of family trios from the National Birth Defects Prevention Study: Tapping into a rich resource of genetic and environmental data. Birth Defects Research, 111(20), 1618–1632. 10.1002/bdr2.1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, & Spinner NB (2012). NOTCH2 mutations in Alagille syndrome. Journal of Medical Genetics, 49(2), 138–144. 10.1136/jmedgenet-2011-100544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura K, Kobayashi D, Uehara Y, Koshida S, Iijima N, Kudo A, Yokoyama T, & Takeda H. (2011). Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development (Cambridge, England), 138(6), 1121–1129. 10.1242/dev.058271 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, … MacArthur DG (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature, 581(7809), 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimzadeh M, Ernst C, Kundaje A, & Hoffman MM (2018). Umap and Bismap: Quantifying genome and methylome mappability. Nucleic Acids Research, 46(20), e120. 10.1093/nar/gky677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics, 46(3), 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayanan B, & Davenport M. (2016). Biliary atresia: A comprehensive review. Journal of Autoimmunity, 73, 1–9. 10.1016/j.jaut.2016.06.005 [DOI] [PubMed] [Google Scholar]
- Lam W-Y, Tang CS-M, So M-T, Yue H, Hsu JS, Chung PH-Y, Nicholls JM, Yeung F, Lee C-WD, Ngo DN, Nguyen PAH, Mitchison HM, Jenkins D, O’Callaghan C, Garcia-Barcelo M-M, Lee S-L, Sham P-C, Lui VC-H, & Tam PK-H (2021). Identification of a wide spectrum of ciliary gene mutations in nonsyndromic biliary atresia patients implicates ciliary dysfunction as a novel disease mechanism. eBioMedicine, 71, 103530. 10.1016/j.ebiom.2021.103530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HP, Kang B, Choi SY, Lee S, Lee S-K, & Choe YH (2015). Outcome of Alagille syndrome patients who had previously received Kasai operation during infancy: A single center study. Pediatric Gastroenterology, Hepatology & Nutrition, 18(3), 175–179. 10.5223/pghn.2015.18.3.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, NHLBI GO Exome Sequencing Project—ESP Lung Project Team, Christiani DC, Wurfel MM, & Lin X. (2012). Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. American Journal of Human Genetics, 91(2), 224–237. 10.1016/j.ajhg.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Fuchsberger C, Kim S, & Scott L. (2016). An efficient resampling method for calibrating single and gene-based rare variant association analysis in case-control studies. Biostatistics, 17(1), 1–15. 10.1093/biostatistics/kxv033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, & 1000 Genome Project Data Processing Subgroup. (2009). The sequence alignment/map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack CL, Tucker RM, Sokol RJ, Karrer FM, Kotzin BL, Whitington PF, & Miller SD (2004). Biliary atresia is associated with CD4+ Th1 cell-mediated portal tract inflammation. Pediatric Research, 56(1), 79–87. 10.1203/01.PDR.0000130480.51066.FB [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, & Chen W-M (2010). Robust relationship inference in genome-wide association studies. Bioinformatics, 26(22), 2867–2873. 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansini AP, Peixoto E, Thelen KM, Gaspari C, Jin S, & Gradilone SA (2018). The cholangiocyte primary cilium in health and disease. Biochimica et Biophysica Acta, Molecular Basis of Disease, 1864(4 Pt B), 1245–1253. 10.1016/j.bbadis.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Tang S, Yang L, & Li K. (2018). Inhibition of the Notch signaling pathway reduces the differentiation of hepatic progenitor cells into Cholangiocytes in biliary atresia. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology, 49(3), 1074–1082. 10.1159/000493290 [DOI] [PubMed] [Google Scholar]
- Masyuk AI, Masyuk TV, & LaRusso NF (2008). Cholangiocyte primary cilia in liver health and disease. Developmental Dynamics, 237(8), 2007–2012. 10.1002/dvdy.21530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, & DePristo MA (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research, 20(9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakken KE, Nygård S, Haaland T, Berge KE, Arnkvaern K, Ødegaard A, Labori KJ, & Raeder MG (2007). Multiple inflammatory-, tissue remodelling- and fibrosis genes are differentially transcribed in the livers of Abcb4 (−/−) mice harbouring chronic cholangitis. Scandinavian Journal of Gastroenterology, 42(10), 1245–1255. 10.1080/00365520701320521 [DOI] [PubMed] [Google Scholar]
- Nakken KE, Nygard S, Haaland TK, Berge KE, Ødegaard A, Labori KJ, & Raeder MG (2009). Gene expression profiles reflect sclerosing cholangitis activity in abcb4 (−/−) mice. Scandinavian Journal of Gastroenterology, 44(2), 211–218. 10.1080/00365520802400867 [DOI] [PubMed] [Google Scholar]
- Ningappa M, Min J, Higgs BW, Ashokkumar C, Ranganathan S, & Sindhi R. (2015). Genome-wide association studies in biliary atresia. Wiley Interdisciplinary Reviews. Systems Biology and Medicine, 7(5), 267–273. 10.1002/wsbm.1303 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, & Sham PC (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Tsai EA, Grochowski CM, Kelly SM, Loomes KM, Spinner NB, & Devoto M. (2020). Exome sequencing in individuals with isolated biliary atresia. Scientific Reports, 10, 2709. 10.1038/s41598-020-59379-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SA, Olney RS, Holmes LB, Lin AE, Keppler-Noreuil KM, Moore CA, & National Birth Defects Prevention Study. (2003). Guidelines for case classification for the National Birth Defects Prevention Study. Birth Defects Research. Part A, Clinical and Molecular Teratology, 67(3), 193–201. 10.1002/bdra.10012 [DOI] [PubMed] [Google Scholar]
- Reefhuis J, Gilboa SM, Anderka M, Browne ML, Feldkamp ML, Hobbs CA, Jenkins MM, Langlois PH, Newsome KB, Olshan AF, Romitti PA, Shapira SK, Shaw GM, Tinker SC, Honein MA, & National Birth Defects Prevention Study. (2015). The National Birth Defects Prevention Study: A review of the methods. Birth Defects Research. Part A, Clinical and Molecular Teratology, 103(8), 656–669. 10.1002/bdra.23384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmer A, Phan H, Mathieson I, Iqbal Z, Twigg SRF, WGS500 Consortium, Wilkie AOM, McVean G, & Lunter G. (2014). Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nature Genetics, 46(8), 912–918. 10.1038/ng.3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Valle A, Kassira N, Varela VC, Radu SC, Paidas C, & Kirby RS (2017). Biliary atresia: Epidemiology, genetics, clinical update, and public health perspective. Advances in Pediatrics, 64(1), 285–305. 10.1016/j.yapd.2017.03.012 [DOI] [PubMed] [Google Scholar]
- Schwarz KB, Haber BH, Rosenthal P, Mack CL, Moore J, Bove K, Bezerra JA, Karpen SJ, Kerkar N, Shneider BL, Turmelle YP, Whitington PF, Molleston JP, Murray KF, Ng VL, Romero R, Wang KS, Sokol RJ, Magee JC, & Childhood Liver Disease Research and Education Network. (2013). Extrahepatic anomalies in infants with biliary atresia: Results of a large prospective north American multicenter study. Hepatology, 58(5), 1724–1731. 10.1002/hep.26512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ShenTu Y, Mi X, Tang D, Jiang Y, Gao L, Ma X, Zhou B, Yang W, Shi J, Lan D, Chen G, & Gong L. (2021). Alagille syndrome caused by NOTCH2 mutation presented atypical pathological changes. Clinica Chimica Acta, 521, 258–263. 10.1016/j.cca.2021.07.026 [DOI] [PubMed] [Google Scholar]
- Staples J, Nickerson DA, & Below JE (2013). Utilizing graph theory to select the largest set of unrelated individuals for genetic analysis. Genetic Epidemiology, 37(2), 136–141. 10.1002/gepi.21684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, & Schreiber RD (1998). How cells respond to interferons. Annual Review of Biochemistry, 67, 227–264. 10.1146/annurev.biochem.67.1.227 [DOI] [PubMed] [Google Scholar]
- Tran KT, Le VS, Dao LTM, Nguyen HK, Mai AK, Nguyen HT, Ngo MD, Tran QA, & Nguyen LT (2021). Novel findings from family-based exome sequencing for children with biliary atresia. Scientific Reports, 11(1), 21815. 10.1038/s41598-021-01148-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrini F, D’Alessandro LCA, Akdemir ZC, Braxton A, Azamian MS, Eldomery MK, Miller K, Kois C, Sack V, Shur N, Rijhsinghani A, Chandarana J, Ding Y, Holtzman J, Jhangiani SN, Muzny DM, Gibbs RA, Eng CM, Hanchard NA, … Yang Y. (2016). Bi-allelic mutations in PKD1L1 are associated with laterality defects in humans. American Journal of Human Genetics, 99(4), 886–893. 10.1016/j.ajhg.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietor I, Cikes D, Piironen K, Gstir R, Erlacher MD, Hess M, Kuhn V, Degenhart G, Rozman J, Klingenspor M, de Angelis MH, Valovka T, & Huber LA (2020). The negative adipogenesis regulator DLK1 is transcriptionally regulated by TIS7 (IFRD1) and translationally by its orthologue SKMc15 (IFRD2). BioRxiv, 719922. 10.1101/719922 [DOI] [PMC free article] [PubMed]
- Wang K, Li M, & Hakonarson H. (2010). ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research, 38(16), e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Jin H, Hu J, Li X, Ruan H, Xu H, Wei L, Dong W, Teng F, Gu J, Qin W, Luo X, & Hao Y. (2020). COL4A1 promotes the growth and metastasis of hepatocellular carcinoma cells by activating FAK-Src signaling. Journal of Experimental & Clinical Cancer Research, 39(1), 148. 10.1186/s13046-020-01650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC., Lee S., Cai T., Li Y., Boehnke M., & Lin X. (2011). Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics, 89(1), 82–93. 10.1016/j.ajhg.2011.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Rasmussen SA, Lynberg MC, Moore CA, Anderka M, Carmichael SL, Costa P, Druschel C, Hobbs CA, Romitti PA, Langlois PH, & Edmonds LD (2001). The National Birth Defects Prevention Study. Public Health Reports (Washington, D.C.: 1974), 116(Suppl 1), 32–40. 10.1093/phr/116.S1.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Yin B, Qu K, Li J, Jin Q, Liu L, Liu C, Zhu Y, Wang Q, Peng X, Zhou J, Cao P, & Cao K. (2018). Screening for susceptibility genes in hereditary non-polyposis colorectal cancer. Oncology Letters, 15(6), 9413–9419. 10.3892/ol.2018.8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagory JA, Dietz W, Park A, Fenlon M, Xu J, Utley S, Mavila N, & Wang KS (2017). Notch signaling promotes ductular reactions in biliary atresia. The Journal of Surgical Research, 215, 250–256. 10.1016/j.jss.2017.03.051 [DOI] [PubMed] [Google Scholar]
- Zhan X, Hu Y, Li B, Abecasis GR, & Liu DJ (2016). RVTESTS: An efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics, 32(9), 1423–1426. 10.1093/bioinformatics/btw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, & Ding H. (2021). COL4A1, negatively regulated by XPD and miR-29a-3p, promotes cell proliferation, migration, invasion and epithelial-mesenchymal transition in liver cancer cells. Clinical and Translational Oncology, 23(10), 2078–2089. 10.1007/s12094-021-02611-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the NBDPS are not released to the public. Qualified researchers can be granted access to NBDPS data for analysis through collaboration with one of the Centers for Birth Defects Research and Prevention.