SUMMARY
Throughout development, cell fate decisions are converted into epigenetic information that determines cellular identity. Covalent histone modifications are heritable epigenetic marks and are hypothesized to play a central role in this process. In this report, we assess the concordance of histone H3 lysine 4 dimethylation (H3K4me2) and trimethylation (H3K4me3) on a genome-wide scale in erythroid development by analyzing pluripotent, multipotent, and unipotent cell types. Although H3K4me2 and H3K4me3 are concordant at most genes, multipotential hematopoietic cells have a subset of genes that are differentially methylated (H3K4me2+/me3−). These genes are transcriptionally silent, highly enriched in lineage-specific hematopoietic genes, and uniquely susceptible to differentiation-induced H3K4 demethylation. Self-renewing embryonic stem cells, which restrict H3K4 methylation to genes that contain CpG islands (CGIs), lack H3K4me2+/me3− genes. These data reveal distinct epigenetic regulation of CGI and non-CGI genes during development and indicate an interactive relationship between DNA sequence and differential H3K4 methylation in lineage-specific differentiation.
INTRODUCTION
Metazoan development can be viewed as a series of cell fate choices that results in the progressive restriction of cellular differentiation potential. This process begins with the determination of the three germ layers at gastrulation and results in the production of adult tissues composed almost entirely of fully committed unipotential cells. During development, each cell must retain an epigenetic memory of the cell fate decisions by which it was produced, and this memory determines the cell’s developmental potential (Fisher, 2002). A central question in developmental biology is how the progressive restriction in developmental potential is encoded in the epigenetic memory of developing cells.
It is clear that cells encode epigenetic information in part through alterations in chromatin state (Fisher, 2002; Margueron et al., 2005). The regulation of chromatin state is accomplished by a number of interrelated mechanisms, including covalent modifications of histone tails, DNA methylation at CpG residues, nucleosomal remodeling, and the incorporation of histone variants. The mechanisms by which histone modifications function, and how they are related to other chromatin regulating activities, are now being elucidated. For example, a variety of covalent histone modifications have been correlated with the euchromatic state, including dimethylation and trimethylation of histone H3 lysine 4 (H3K4me2 and H3K4me3, respectively), dimethylation of histone H3 lysine 36 (H3K36me2), acetylation of H3 lysine 9 and H4 lysine 16 (H3K9ac and H4K16, respectively), and hyperacetylation of both H3 and H4 (Fischle et al., 2003; Margueron et al., 2005; Nightingale et al., 2006). However, the relationships between these marks are only beginning to be defined, and there is growing evidence that the various euchromatic histone modifications are not functionally equivalent (Bernstein et al., 2006; Santos-Rosa et al., 2002; Wysocka et al., 2005). In particular, the relationship between H3K4me2 and H3K4me3 is unclear.
The manner in which histone modifications are targeted to specific genomic loci is also largely obscure but appears to include mechanisms that function both in trans and in cis. Transcriptional coregulators have well-described histone-directed enzymatic activities, and these coregulators are often targeted through their interaction with sequence-specific DNA-binding transcription factors (Torchia et al., 1998). The presence of these activities is a direct regulator of gene activation. In addition, correlations between H3K4me3 and CpG islands (CGIs) as well as between H3K27me3 and highly conserved noncoding regions of the genome have been described, implying that features of the primary genomic sequence play an important role in defining the epigenetic state of the cell (Bernstein et al., 2006; Tanay et al., 2007).
In this study, we sought to determine the genome-wide distribution of the euchromatic H3K4me2 and H3K4me3 histone modifications at the promoter regions of genes in multipotential and erythroid-differentiated hematopoietic cells, as well as pluripotential murine embryonic stem (mES) cells, in order to gain insight into how H3K4 methylation may regulate developmental potential. Three major findings are reported here. First, we demonstrate that a multipotential hematopoietic cell line maintains a population of genes in a unique epigenetic state defined by the presence of dimethylation, but not trimethylation, of histone H3 (H3K4me2+/H3K4me3−). These genes are transcriptionally poised, reflect the differentiation potential of the cells, and are specifically regulated in a lineage-specific manner upon erythroid differentiation. Second, we demonstrate that H3K4me2 is targeted to poised (H3K4me2+/me3−) genes in a transcription start site (TSS)-independent manner, in contrast to the TSS targeted enrichment observed at active genes (H3K4me2+/me3+). We implicate critical hematopoietic transcription factors in the TSS-independent targeting of H3K4me2 to these poised genes. Finally, we demonstrate that the methylation state of H3K4 is highly correlated with the presence of CGIs in these genes and is regulated during development, from embryonic stem cells through terminal hematopoietic differentiation. In the multipotential hematopoietic cell line, H3K4me2+ non-CGI genes are highly enriched for lineage-specific hematopoietic genes, while H3K4me3− CGI genes are largely developmental regulatory genes. In ES cells, the distribution of H3K4me2 and H3K4me3 is still highly correlated with the CGI status, but the population of H3K4me2+/me3 genes is absent. We propose that the epigenetic regulation of CGI and non-CGI genes is profoundly different throughout development of the hematopoietic system and reflects the various hierarchical functions of these genes during hematopoiesis.
RESULTS
The Distribution of the H3K4me2 among Lineage-Specific Promoters Reflects the Differentiation Potential of Bone-Marrow-Derived Cell Populations and Hematopoietic Cell Lines
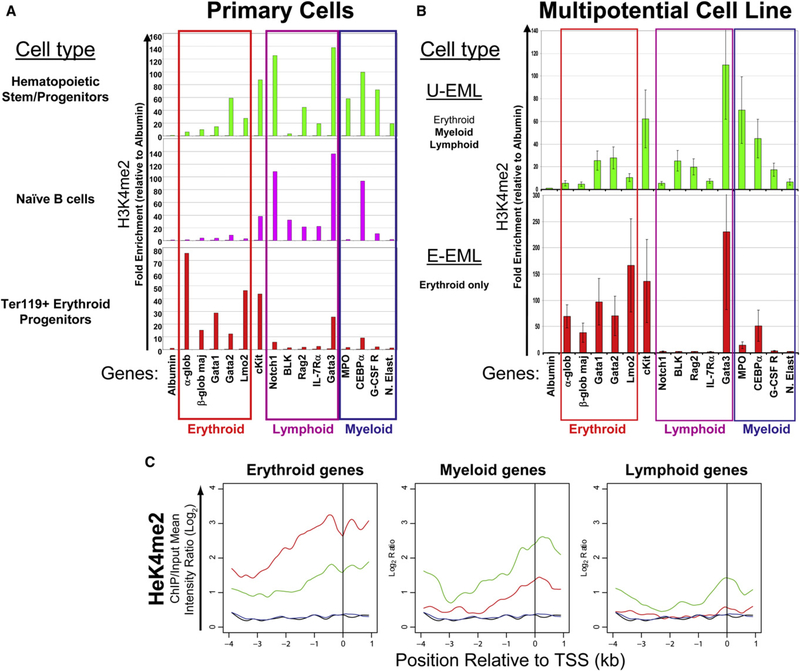
To determine the distribution of the H3K4me2 mark in vivo, ChIP assays were performed on multipotential stem/progenitor (Sca+/Kit+/Lin−), erythroid-restricted (Ter119+), and B lymphoidrestricted (B220+/IgMLo/CD23+) primary cell populations purified from mouse bone marrow and spleen. H3K4me2 enrichment was determined by qPCR at a panel of lineage-restricted (erythroid, myeloid, and lymphoid) promoter regions. In general, the distribution of H3K4me2 at these promoters reflected the developmental potential of the cell population assayed (Figure 1A and see Figure S1 available online), with broad enrichment in the multipotential cells and more restricted enrichment in the erythroid and lymphoid cell populations.
Figure 1. The Distribution of H3K4me2 Enrichment at Lineage-Specific Hematopoietic Promoters Reflects the Differentiation Potential of Hematopoietic Cell Lines.
(A) Primary cells were purified from mouse bone marrow (multipotential and erythroid cells) and spleen (B lymphoid cells), and anti-H3K4me2 ChIP assays were performed. All hematopoietic genes are enriched with H3K4me2 in the multipotential cell population, whereas the distribution was restricted in erythroid and lymphoid cells in a lineage-specific manner. ChIP enrichment was measured by qPCR at 15 hematopoietic promoters that are organized according to their lineage affiliation. The liver-specific promoter albumin was included as a negative control. The experiment was performed in duplicate, and the pattern of H3K4me2 enrichment was reproducible in the replicate (Figure S1).
(B) Anti-H3K4me2 ChIP assays were performed on FACS-purified undifferentiated and erythroid-differentiated EML cells. All hematopoietic genes are enriched with H3K4me2 in the undifferentiated cells. However, the enrichment of nonerythroid promoters is substantially reduced following differentiation, while enrichment of the erythroid promoters is augmented. Experiments performed in triplicate; error bars depict SEM.
(c) Genome-wide location analysis. Three sets of lineage-specific genes (21 erythroid, 16 myeloid, and 23 lymphoid) were identified from the literature. The spatial distribution of mean H3K4me2 enrichment in U-EMLs (green) and E-EMLs (red) is plotted for the three sets of lineage-specific genes, as well as a set on nonhematopoietic lineage-specific genes (blue in U-EML and black in E-EML), across the 5 kb promoter region on the microarrays. All three sets of lineage-specific genes display H3K4me2 enrichment in the U-EML cells. Following erythroid differentiation, the enrichment in myeloid and lymphoid genes is lost, while the enrichment of the erythroid genes is augmented.
To perform more detailed studies, we assessed the consistency of this relationship in hematopoietic cell lines where cell numbers were not limited. Studies in four well-characterized hematopoietic cell lines recapitulated the findings in the primary cell populations, with enrichment of H3K4me2 at lineage-restricted promoters reflecting each cell line’s developmental potential (Figure S2). To confirm that these patterns were a direct result of cellular differentiation potential, the enrichment of H3K4me2 and hyperacetylated histone H4 (acH4) was also determined in the multipotential EML cell line before and after differentiation into the erythroid lineage. After demonstrating that the Sca+ population of EML cells are homogeneously multipotent by clonal analysis (Figure S3), ChIP assays performed on FACS-purified undifferentiated and erythroid-differentiated EML cells (U-EML and E-EML, respectively; Figure S4) demonstrated that all of the hematopoietic promoters were enriched with H3K4me2 and acH4 in the multipotential cell populations, whereas this mark was largely confined to the erythroid genes in the erythroid cells (Figure 1B and Figure S5). Importantly, the enrichment of H3K4me2 at the promoter regions was not merely a reflection of the expression of these genes, as its presence did not always correlate with gene expression (Figure S6). On the whole, these studies demonstrated that the EML cell line is a valuable model for studying the regulation of covalent histone modifications during hematopoietic differentiation. However, due to the limited scope of the ChIP-qPCR method and the presence of outlier genes (gata3), it was important to perform an unbiased, genome-wide analysis in order to confirm the differentiation-induced effect.
Genome-wide Analysis of H3K4me2 in EML Cells
Because the ChIP-qPCR approach is limited in scope, genome-wide location analysis (ChIP-chip) was performed in U-EML and E-EML cells to map differentiation-induced changes. As a confirmation of the reliability of the ChIP-chip technique, the data generated from these experiments were highly reproducible and reflected our ChIP-qPCR results with high fidelity (Figure S7). The spatial profile of H3K4me2 distribution revealed that this modification tends to be localized in the region immediately surrounding transcription start sites (TSS) of genes, with a localized dip in enrichment at the TSS, a pattern that has been reported previously (Figure S8; Mito et al., 2005).
Analysis of H3K4me2 enrichment at promoters (see Experimental Procedures) identified over 7000 enriched (H3K4me2+) promoters in U-EML cells. Following erythroid differentiation, this number was reduced by over 5%, with 796 genes losing H3K4me2 enrichment and 375 genes gaining H3K4me2. To classify the genes that lost H3K4me2, gene ontology (GO) analysis was performed. Of the 60+ GO categories that were overrepresented in those genes, half of them were directly related to hematopoiesis, specifically nonerythroid (mainly myeloid and lymphoid) hematopoietic categories (Table S1). In contrast, only one hematopoietic GO category (O2 transport) was included among those that gained the H3K4me2 mark.
In a more focused analysis, the enrichment of H3K4me2 was compared between 60 hematopoietic lineage-specific (myeloid, erythroid, and lymphoid) genes and 71 nonhematopoietic lineage-specific genes, as identified from the literature. This analysis revealed that three lineage-specific sets were H3K4me2 enriched in the undifferentiated EML cells (Figure 1C, green tracings) when compared to nonhematopoietic genes (black tracings; Wilcoxon test p value < 8.7 × 1012). Upon erythroid differentiation, the degree of enrichment (Figure 1C, red tracings) was significantly reduced in the nonerythroid promoters (p value < 2.6 × 10−4), whereas it was augmented at the erythroid promoters (p value < 3.36 × 10−3). This phenomenon can be further visualized at the level of individual genes (Figure S9A). To account for any potential bias in the curation of the genes, the identical analysis was performed on a set of “differentiation-induced” genes selected based on expression data (unexpressed prior to differentiation and induced by at least 4-fold upon differentiation). The results of both the promoter-wide mean intensity ratio profiles (Figure S9B) and the undifferentiated versus differentiated enrichment scatter plots (Figure S9C) were very similar to the erythroid genes in the previous analysis.
These results demonstrate that multipotential hematopoietic cells maintain hematopoietic lineage-specific gene promoters marked with euchromatic histone modifications and, upon the specification and differentiation down one lineage, these marks are lost from promoters associated with the alternative hematopoietic lineages.
Differentiation-Induced Loss of H3K4me2 Is Restricted to the Set of Transcriptionally Poised H3K4me2+/me3 Genes
While H3K4me2 and H3K4me3 are highly correlated in terms of localization at active genes, there is some evidence in yeast and higher eukaryotes that there are differences in their function (Bernstein et al., 2005; Santos-Rosa et al., 2002; Wysocka et al., 2005). To compare the roles of these two marks in multipotential and differentiated hematopoietic cells, we compared their genome-wide distribution by ChIP-chip in U-EML and E-EML cells.
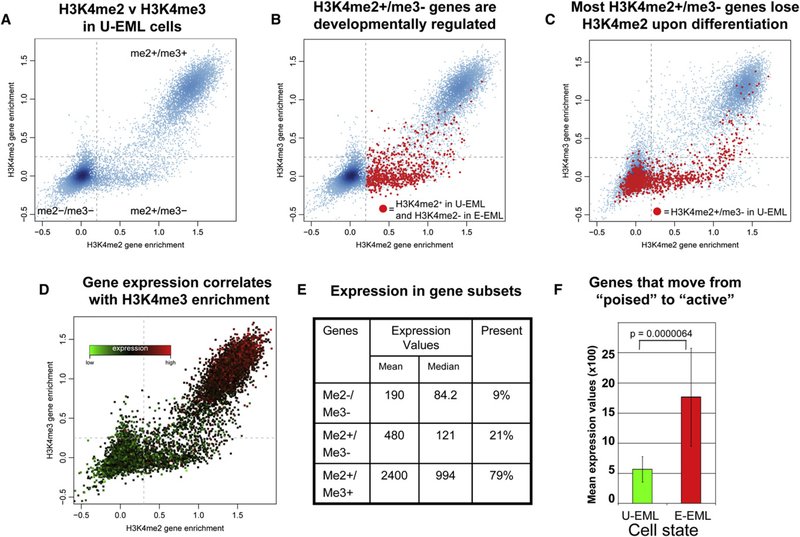
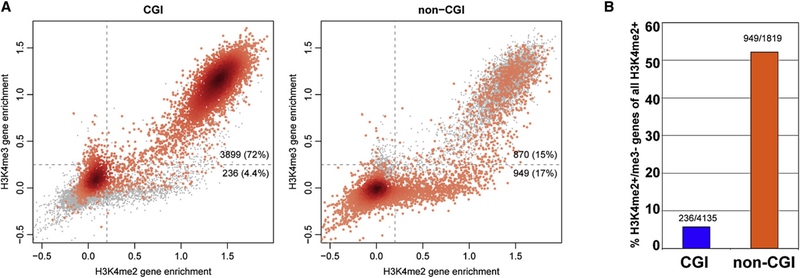
As expected, the H3K4me2 and H3K4me3 state was identical (me2+/me3+ or me2/me3) for most genes (Figure 2A). However, a subset of genes revealed a discordant pattern of H3K4 methylation in which H3K4me2 was present but H3K4me3 was absent (H3K4me2+/me3−). Interestingly, the reciprocal population (H3K4me2−/me3+) was essentially absent in both U-EML and E-EML cells.
Figure 2. H3K4me2+/me3 Genes Are Developmentally Regulated and Transcriptionally Inactive.
(A) H3K4me2 (x axis) and H3K4me3 (y axis) enrichment values in U-EML were determined by ChIP-chip and displayed as a scatterplot. H3K4me2+/me3+ andH3K4me2−/me3+ genes comprise the two dominant gene populations. There is an additional population of H3K4me2+/me3 (bottom right) genes but very few of the reciprocal population (H3K4me2−/me3+; upper left).
(B) The set of genes that loses H3K4me2 upon erythroid differentiation (red) was marked on the plot from (A), revealing that the set of developmentally regulated genes is strongly overrepresented in the me2+/me3− subset.
(C) The set of genes that were H3K4me2+/me3 (within dashed red box) in U-EML cells are highlighted in red in this scatterplot depicting the H3K4me2 versus H3K4me3 state of genes following erythroid differentiation (in E-EML). The large majority of genes are demethylated at H3K4, becoming H3K4me2−/me3−. A minority of genes remain H3K4me2+/me3−, and even fewer become H3K4me2+/me3+. Those that become me2+/me3+ are transcriptionally activated (see [F]).
(D) The U-EML H3K4me2 versus H3K4me3 scatterplot in (A) was color coded with each gene’s Affymetrix expression value, with green reflecting the lowest values and red the highest.
(E) The mean and median expression values and the percentage of “Present” genes are depicted for the H3K4me2−/me3−, H3K4me2+/me3−, and H3K4me2+/ me3+ gene sets.
(F) The mean Affymetrix expression values in U-EML and E-EML are provided for the set of poised genes (H3K4me2+/me3− in U-EML) that gain H3K4me3 following erythroid differentiation (H3K4me2+/me3+ in E-EML). Error bars depict SEM.
The number of H3K4me2+/H3K4me3− genes was reduced following erythroid differentiation, from 14% in U-EML cells to 9.8% in E-EML cells (p < 10−14). In Figure 2B, those genes that will lose H3K4me2 upon erythroid differentiation are highlighted in the U-EML scatter plot. This figure clearly shows that the majority of these genes are H3K4me2+/me3− prior to differentiation. Furthermore, the majority of H3K4me2+/me3− genes in U-EML cells become H3K4me2+/me3− upon differentiation, while some remain me2+/me3− and fewer become me2+/ me3+ (Figure 2C). Given that the large majority of H3K4me2+ genes are also H3K4me3+, this distribution of developmentally labile genes is highly nonrandom and demonstrates that residence in the H3K4me2+/H3K4me3− population of genes is highly dependent on the differentiation state of the cell.
It has been shown in yeast that genes enriched with H3K4me2 and lacking H3K4me3 at their promoter regions are not actively expressed but are uniquely poised for the activation of expression (Santos-Rosa et al., 2002). Similarly, gene expression profiling of U-EML cells revealed that only 21% of H3K4me2+/ H3K4me3− genes were called Present by the Affymetrix analysis, whereas 79% of H3K4me2+/me3+ genes were Present (Figures 2D and 2E). The H3K4me2+/me3− genes do show some low-level expression, but remain substantially below the me2+/ me3+ genes (Figure 2E). Additionally, the expression of genes that transition from H3K4me2+/me3− to H3K4me2+/me3+ upon erythroid differentiation are substantially upregulated upon differentiation (Wilcoxon test p value < 6.4 × 10−6; Figure 2F), providing further evidence that the acquisition of H3K4me3 enrichment correlates with transcriptional activation and that H3K4me2+/me3− genes are truly poised. In our subsequent analyses, we refer to the H3K4me2−/me3−, H3K4me2+/ me3−, and H3K4me2+/me3+ gene subsets as inactive, poised, and active, respectively, recognizing that these titles reflect the general behavior of each group as a whole (not every individual gene within each group). Taken together, these data demonstrate that the set of K4me2+/me3− genes in U-EML cells are developmentally labile and poised for transcription or further silencing in response to differentiation.
Spatial Distribution of H3K4me2 at Active (me2+/me3+) and Poised (me2+/me3−) Genes Demonstrates that the H3K4me2 Mark Is Targeted by TSS-Dependent and TSS-Independent Mechanisms
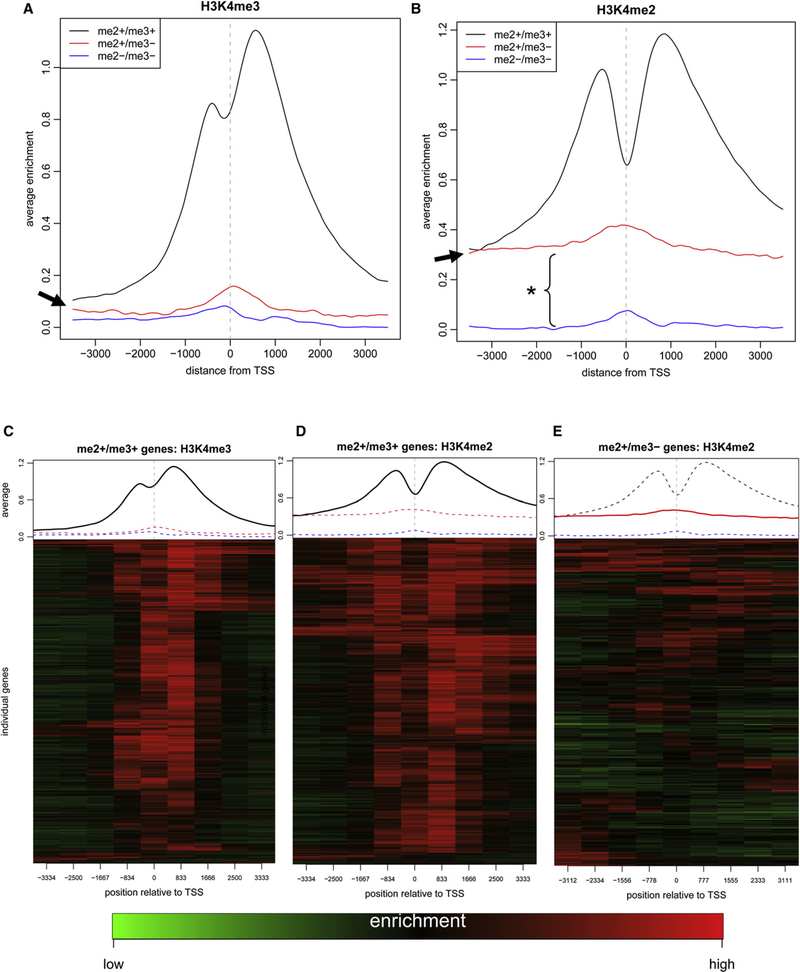
Our data show that both H3K4me2 and H3K4me3 are localized to the 5’ ends of genes in higher eukaryotes. To gain further insight into the regulation of these marks, the mean enrichment profiles for H3K4me2 and H3K4me3 were determined for the newly described inactive (me2−/me3−), poised (me2+/me3−), and active (me2+/me3+) gene sets (Figures 3A and 3B). As expected, the TSSs of active genes were highly enriched for H3K4me2 and H3K4me3, in “double peak” patterns centered at the TSS. H3K4me3 enrichment was strongly biased toward the downstream peak (in the coding region), whereas H3K4me2 was more evenly distributed. Interestingly, H3K4me3 enrichment distant from the TSS in active genes returned almost down to the level seen in inactive genes (arrowhead in Figure 3A), whereas substantial H3K4me2 enrichment remained in active genes away from the TSS (Figure 3B). In contrast to the active genes, on average, the H3K4me2 mark was distributed more uniformly throughout the 7 kb region for the poised genes. Given that the degree of H3K4me2 enrichment in the poised and active genes was equivalent at the most upstream region, it appears that the TSS-targeted H3K4me2 enrichment of active genes accumulates “on top of” the baseline level present in poised genes. We hypothesize that this pattern is the result of two separate mechanisms targeting H3K4me2 to this set of genes prior to and following transcriptional activation.
Figure 3. H3K4me2 Is TSS Independent in Poised Genes and Largely TSS Dependent in Active Genes.
(A) The spatial distribution of H3K4me3 enrichment in U-EMLs is plotted for the three subsets of genes across the 7 kb region on the microarrays. The TSS is located in the center of the 7 kb region (vertical dashed line). While the enrichment at active genes (black line) is localized within the immediate vicinity of the TSS, there is little enrichment at poised and inactive genes. As the distance from the TSS increases, the enrichment of active genes drops to the enrichment level of the inactive genes (arrow).
(B) The spatial distribution of the average H3K4me2 enrichment is plotted as for H3K4me3 in (A). H3K4me2 enrichment (red line) is TSS independent at poised genes (H3K4me2+/me3−). Active genes (H3K4me2+/me3+, black line) show increased H3K4me2 enrichment in the proximity of the TSS. Interestingly, at the 5’ and 3’ extremes, H3K4me2 enrichment drops down to the level seen in poised genes. Note the region between the poised genes and the inactive genes (blue line) that comprises the “baseline” enrichment (*).
(C) The H3K4me3 profile was generated for each active (me2+/me3+) gene by averaging the values in 700 bp windows across the 7 kb region, and the resulting profiles were hierarchically clustered. The distribution of H3K4me3 in the active genes is homogeneous.
(D) The H3K4me2 enrichment of the set of active genes was analyzed as in (C). This pattern reveals largely TSS-focused enrichment of H3K4me2 but somewhat less homogeneous than in H3k4me3.
(E) The H3K4me2 enrichment of the set of poised genes (me2+/me3−) was analyzed as in (C). Note the heterogeneous and apparently random pattern of H3K4me2 distribution in this set of genes.
The previous analysis revealed the mean enrichment of hundreds or thousands of genes in each gene subset. In order to gain insight into the contribution of individual genes within each subset, the enrichment profiles in the three gene sets (me2+/me3+, me2+/me3−, and me2−/me3−) were averaged (contiguous 700 bp windows) and organized using hierarchical clustering (Figures 3C–3E and Figures S10A–S10C). The H3K4me3 enrichment profiles of the H3K4me2+/me3+ genes were homogeneous, with most enrichment focused just downstream of the TSS, falling off rapidly away from the TSS (Figure 3C). The H3K4me2 enrichment of the same set was also focused on the TSS but displayed a larger subset of genes in which the enrichment extended substantially upstream and/or downstream (Figure 3D). Most interesting was the distribution of H3K4me2 on the set of me2+/me3− genes (Figure 3E). As the mean profiles had demonstrated, H3K4me2 enrichment showed no particular preference for the TSS in this group. Instead, the enrichment profiles at individual me2+/me3− genes were quite heterogeneous, with subsets of genes that were enriched upstream, downstream, or at the TSS, as well as others that were enriched throughout the entire region.
Considered together, our analyses reveal that (1) H3K4me3 is strictly and uniformly localized in close proximity to the TSS of active genes and(2)there is a topological difference between the targeting of H3K4me2 at poised (me2+/me3−) versus active (me2+/ me3+) genes. These data suggest that H3K4me2 and H3K4me3 are coordinately targeted to the TSSs of activated genes, whereas H3K4me2is targeted to poised (me2+/me3−) promoters in a temporally and mechanistically independent manner.
TSS-Independent H3K4me2 Distribution in me2+/me3− Genes Is Correlated with Hematopoietic Transcription Factor Binding Sites
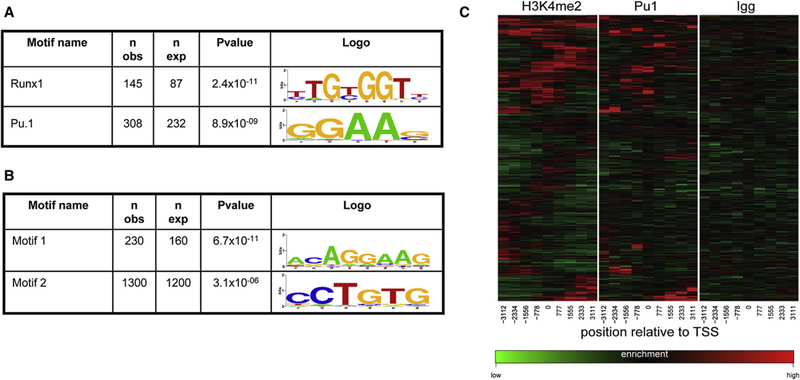
The heterogeneous pattern of H3K4me2 distribution in poised genes raised the question of how this mark was being targeted to the specific sequences. Given that some well-defined enhancers of hematopoietic genes colocalized with domains of H3K4me2 enrichment (data not shown), we hypothesized that DNA-binding transcription factors (TFs) may function to poise these genes by targeting H3K4me2 without fully activating transcription. Two approaches were taken to identify a relationship between the sites of H3K4me2 enrichment and the TF binding motifs. First, abundance of the consensus binding motifs was assessed for two critical hematopoietic TFs (Runx1/Aml1 and Pu.1) within the H3K4me2+ regions of the H3K4me2+/me3 genes. This analysis found statistically significant overabundance of both TF motifs within the H3K4me2+ domains relative to the H3K4me2 domains (Figure 4A). To verify this result, ChIP-chip assays were performed to identify true Pu.1 binding sites. A direct comparison of the H3K4me2 and Pu.1 enrichment patterns (Figure 4C) clearly demonstrated that Pu.1 targets strongly correlate with the H3K4me2+ domains of the H3K4me2+/me3− genes (partial correlation p value < 10−16).
Figure 4. Known and Putative Transcription Factor Binding Motifs Correlate with H3K4me2 Distribution in Poised Genes.
(A) The consensus motifs for Runx1 and Pu.1 are overrepresented within regions of H3K4me2 enrichment in poised genes.
(B) A de novo motif-finding algorithm identified primary sequence motifs that were overrepresented in the same regions of the poised genes as in the ChIP-chip analyses. Two of these motifs were found to be overrepresented within H3K4me2 domains within these genes. Motif 1 appears to be an Ets consensus binding site, while the identity of the factor(s) that binds to motif 2 is unknown.
(C) Experimentally obtained Pu.1-binding sites in U-EML cells correlate with H3K4me2 enrichment at poised genes (p < 10−16).
Second, a de novo motif search was performed in order to identify unique motifs. Several motifs were identified that were overrepresented among the 7 kb region of the poised genes; two of these motifs were found to be statistically correlated with the H3K4me2+ regions of the poised genes (Figure 4B). Of these two motifs, motif 1 is very similar to the consensus motif for Ets family TFs. Ets family members contain a winged helixloop-helix domain and include important regulators of hematopoiesis and leukemogenesis, including Pu.1, Elf-1, Mef, and Tel (Oikawa and Yamada, 2003; Oikawa, 2004). Motif 2 is novel, with no known binding partner. This intriguing finding may implicate additional regulatory TFs in the regulation of these H3K4me2+/me3 genes, though clearly further experimental testing of this hypothesis is required.
The Distribution of H3K4me2 and H3K4me3 at CGI and Non-CGI Genes Is Markedly Dissimilar
CGIs are short genomic regions that are highly enriched in CpG dinucleotides. Unlike those found throughout most of the genome, the cytosines of CpG dinucleotides located within CGIs tend to be unmethylated. Approximately 45% of genes in the mouse genome contain CGIs, most located at or near the TSS. Given the extraordinarily high coincidence of H3K4me3 enrichment and CGIs noted by others in undifferentiated ES cells (Bernstein et al., 2006), we determined the relationship between the methylation state of H3K4 and CGIs in the hematopoietic EML cell line.
The H3K4 methylation properties were starkly different for CGI and non-CGI genes (Figure 5). In both U-EML and E-EML cells, CGI genes were highly overrepresented among the me2+/ me3+ population of genes (78%, hypergeometric p value < 10−10), while the non-CGI genes were largely located in the me2−/me3− set of genes (80%, p value < 10−10).
Figure 5. H3K4 Methylation State Correlates with the Presence of CpG Islands.
(A) The distribution of CGI (left) and non-CGI (right) genes (orange) is superimposed on the H3K4me2 versus H3K4me3 scatterplot of all genes (gray) from U-EML cells. The patterns of H3K4 methylation are different in the two cases, with the majority of CGI genes in the me2+/me3+ quadrant and the majority of non-CGI genes in the me2−/me3− quadrant. CGI genes are depleted from the set of poised genes, while non-CGI genes are highly enriched in this set.
(B) The proportion of H3K4me2+ genes that are poised (H3K4me2+/me3−) is strongly correlated with CGI status.
The likelihood that an H3K4me2+ gene belongs to the poised set is dependent on its CGI status. While only 5% of H3K4me2+ CGI genes were poised, a full 52% of H3K4me2+ non-CGI genes were within the poised gene set (Figure 5B). Conversely, non-CGI genes were strongly overrepresented and CGI genes depleted among the poised genes (p value < 10−10). The substantially stronger tendency of H3K4me2+ CGI genes to be concomitantly marked by H3K4me3 suggests the intriguing possibility that CGIs may facilitate the activation of H3K4me2+/me3− genes by stimulating the trimethylation of H3K4me2 genes, thereby resulting in relatively few H3K4me2+/me3− CGI genes.
H3K4me2+ Non-CGI Genes Are Hematopoietic Lineage-Specific Genes and H3K4me3− CGI Genes Are Developmental Regulators
We performed GO analysis on the CGI and non-CGI gene subsets based on their H3K4 methylation status. Among the me2+ non-CGI genes, the preponderance of hematopoietic categories was striking, as ~60% of the categories overrepresented in both the H3K4me2+/me3+ and H3K4me2+/me3− sets had a clear role in hematopoiesis or the function of hematopoietic lineages (see Tables S2 and S3 for selected category names and Tables S5 and S6 for complete lists). Many of the other categories were more generic and included hematopoietic genes within them. On the other hand, the GO categories enriched in the me2+ CGI genes were completely devoid of hematopoiesis-related categories. The me2+/me3+ CGI genes were predominantly housekeeping genes involved in the metabolic activities of the cell (Table S7). Interestingly, all me3− CGI genes, including the me2−/me3− genes and the small number of me2+/me3− genes, were dominated by developmental regulators and regulators of cell signaling and transcription (see Table S4 for selected categories in CGI me3− genes and Table S8 for a complete listing). Furthermore, 69% of H3K4me2−/me3− CGI genes in E-EML cells were among the set of “bivalent” (H3K4me3+/H3K27me3+) developmental regulatory genes in mES, as defined by Bernstein et al. (2006), whereas only 27% of all CGI genes are bivalent in mES cells.
This analysis indicates that multipotential hematopoietic cells have a limited number of euchromatic (H3K4me2+) non-CGI genes that are largely tissue-specific (hematopoietic) genes. The methylation state of H3K4 at these genes is highly dependent on the differentiation state of the cells because erythroid differentiation results in a dramatic alteration of the H3K4 methylation state of these genes, particularly among the poised (me2+/me3−) set of genes. Further, most me3− CGI genes are developmental regulators, while the active CGI genes are mainly involved in “housekeeping” functions.
The H3K4 Methylation State Is Strictly Correlated with CGI Status in Murine ES Cells
The data presented above illustrate that erythroid differentiation of multipotential hematopoietic cells results in the loss of H3K4me2 from nonerythroid lineage-specific hematopoietic genes. One possible explanation is that developmental potential is defined by, or at least reflected in, the distribution of H3K4me2 at lineage-specific genes throughout development. If this were true, ES cells with the potential to differentiate into all somatic cell types would maintain H3K4me2 at lineage-specific genes for all tissues of the body. To test this hypothesis, we assessed the enrichment of H3K4me2 at the panel of hematopoietic promoters in murine ES cells (Figure 6).
Figure 6. In ES Cells, the Distribution of H3K4me2 among Hematopoietic Genes Is Strongly Correlated with CGI Status and Not Developmental Potential.
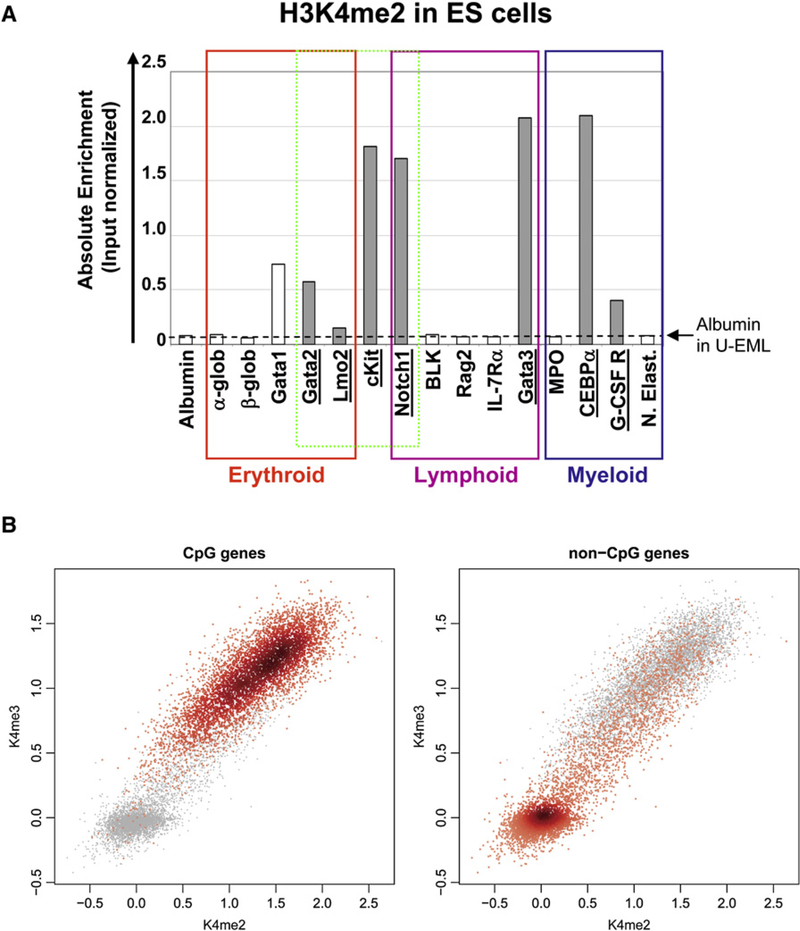
(A) The enrichment of H3K4me2 at the lineage-specific hematopoietic promoters was determined in murine ES cells. Although these cells clearly have hematopoietic potential, half of the hematopoietic genes lacked H3K4me2. Enrichment of H3K4me2 was more highly correlated with the presence of CpG islands (gray bars), with enrichment found at 7 of 7 CGI but only 1 of 9 non-CGI genes (white bars). The dashed line depicts the enrichment of albumin in H3K4me2 ChIP assays performed in parallel on U-EML cells.
(B) The genome-wide distribution of H3K4me2 and H3K4me3 is very highly correlated with CGI status in undifferentiated mES cells. Note the complete absence of the poised gene population (H3K4me2+/me3−) in these cells.
Contrary to the extrapolative model proposed above, approximately one-half of the hematopoietic genes lacked H3K4me2 in mES cells and, therefore, must acquire it during development. Interestingly, seven of eight H3K4me2+ genes were CGI genes, whereas all eight of eight H3K4me2− genes were non-CGI genes. Rather than developmental potential, the presence of CGI in the gene promoters appeared to be the major determinant of H3K4me2 enrichment in ES cells. Genome-wide analyses of the distribution of H3K4me2 and H3K4me3 in mES cells revealed that H3K4 methylation is, in fact, strictly correlated with CGI status. Approximately 93% of CGI genes were H3K4me2 and H3K4me3 enriched, while only 24% of non-CGI genes displayed H3K4 methylation. Strikingly, the population of H3K4me2+/me3− genes was completely absent in mES cells, further implicating this uniquely marked subset of genes in the regulation of lineage-specific genes.
DISCUSSION
The mechanisms that define cellular identity, both in the embryo and adult stem cells, are of great biological and clinical importance. These epigenetic mechanisms maintain a record of each cell’s unique developmental history. The data presented here provide insights into the genome-wide regulation of H3K4 methylation state in a multipotential hematopoietic cell line, demonstrating that H3K4me2 can occur independent of H3K4me3, resulting in a population of transcriptionally poised genes. The H3K4 methylation state of these poised genes is regulated throughout erythroid differentiation of these cells in a manner that reflects their developmental potential. The genes that lose H3K4me2 upon differentiation are almost all among the small minority of H3K4me2+ genes that lacked H3K4me3 and were transcriptionally inactive. The apparent low-level transcription of some of these genes may account for the phenomenon of “lineage priming” that results in the promiscuous expression of lineage-specific genes in primitive hematopoietic cells in a manner that reflects their developmental potential (Hu et al., 1997; Miyamoto et al., 2002).
The distribution of H3K4me2 at me2+/me3− genes is independent of their TSSs (unlike at fully activated genes) and may be acquired by mechanisms different from those proposed for TSS-dependent accumulation of H3K4me2/me3 (Ruthenburg et al., 2006; Wysocka et al., 2005). Our results suggest that H3K4me2 enrichment is strongly correlated with binding of hematopoiesis-specific TFs. This was confirmed experimentally for Pu.1. We also expect additional TFs to be involved in the targeting of H3K4me2 to poised genes. The importance of these marks in the regulation of hematopoiesis was recently demonstrated by the targeted deletion of the H3K4me½-specific demethylase lsd1 in hematopoietic cell lines (Saleque et al., 2007).
Finally, these data reveal a striking difference in H3K4 methylation state of CGI and non-CGI genes in both U-EML and E-EML cells. The H3K4me2 enrichment of CGI genes is almost always accompanied by H3K4me3, with fully active (me2+/me3+) genes annotated predominantly with housekeeping functions and me3– CGI genes responsible for regulatory functions, such as development and cellular signaling pathways. Consistent with this dichotomy, the H3K4me2−/me3− CGI genes in E-EML cells are highly enriched in genes that were shown by others to be “bivalent” (H3K4me3+/H3K27me3+) in mES cells, a set of genes that has been implicated in the regulation of embryonic development (Bernstein et al., 2006). In contrast, non-CGI genes enriched for H3K4me2 are annotated with various lineage-specific hematopoietic functions. The paucity of me2+/me3− CGI genes may reflect efficient H3K4 trimethylation through association of an H3K4-specific histone methyltransferase, for example, with CGIs through its CXXC domain (Ayton et al., 2004). Conversely, poised non-CGI genes might require additional regulatory factors to facilitate their activation.
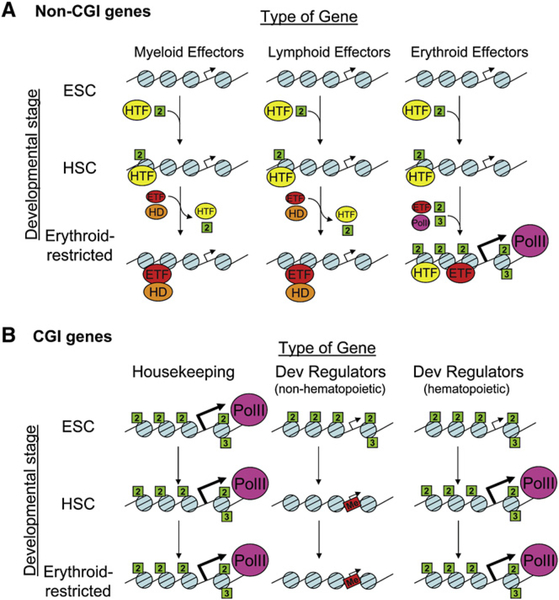
The data presented here, along with those published previously, suggest a model for the epigenetic regulation of CGI and non-CGI genes throughout hematopoietic development (Figure 7). As a group, non-CGI genes are highly enriched for lineage-specific genes and lack the euchromatic H3K4me2/me3 marks in ES cells (Figure 5 and Bernstein et al., 2006; Tanay et al., 2007). During development, hematopoietic non-CGI genes acquire TSS-independent H3K4me2, poising these genes for future expression. TSS-independent H3K4me2 appears to occur at many hematopoietic promoters by the time the cell acquires the hematopoietic stem cell fate and may be targeted by panhematopoietic TFs, such as Pu.1 and Runx1. Subsequent cell fate choices result in the activation of some genes and the loss of H3K4me2 from H3K4me2+/me3− genes affiliated with alternative cell lineages (i.e., those lineages down which the developing cell can no longer differentiate). This would explain the hematopoietic defects that result from the loss of the histone demethylase Lsd1 (Saleque et al., 2007).
Figure 7. A Proposed Model of the Developmental Regulation of H3K4 Methylation at CGI and Non-CGI Genes throughout Development.
(A) The distribution of H3K4me2 and H3K4me3 is tightly linked to CGIs in undifferentiated ES cells. Non-CGI genes, including hematopoietic lineage-restricted genes, lack these marks at the earliest stages of embryonic development. By the time cells have acquired hematopoietic specification, lineage-specific non-CGI genes become poised for expression by the association of critical regulators of hematopoietic development, such as Runx1, Pu.1, Gata2, or Scl (hematopoietic transcription factors [HTFs]). These factors induce localized, TSS-independent deposition of H3K4me2. Upon final lineage specification and differentiation, lineage-specific TFs (e.g., the erythroid-specific TF [ETF] Gata1) induce changes in both erythroid and nonerythroid genes. Nonerythroid genes lose H3K4me2, likely through the recruitment or activation of histone demethylases (HD), effectively committing the developing cells to the erythroid lineage, while erythroid genes become transcriptionally activated and acquire TSS-targeted H3K4me2/3.
(B) Conversely, CGI genes are all marked by H3K4me2/me3 in undifferentiated ES cells. However, many of these genes are not expressed, due, in part, to the concomitant association of H3K27me3 (data not shown). The activation state of a large proportion of CGI genes is not affected by development because these genes are involved in housekeeping functions required in all cells. However, the subset of developmental regulatory genes are gradually inactivated, potentially by DNA methylation of the CGIs (red boxes), in order to prevent the misexpression of these powerful developmental modulators.
Conversely, CGI genes, which are largely composed of either developmental regulators (encoding mainly TFs) or constitutively active housekeeping genes, are H3K4me2+/me3+ in embryonic stem cells (Figure 5; Bernstein et al., 2006). It is the regulated expression of the developmental regulatory subset of CGI genes that guides cells through hematopoietic development by making cell fate choices. Our data suggest that these developmental regulatory TFs (e.g., Pu.1) may poise and activate the lineage-specific non-CGI genes in more differentiated cell types. Given their apparent intrinsic tendency for activation, we believe that the expression of these CGI-containing developmental regulatory genes must be actively repressed in stem cells in order to prevent differentiation and allow for self-renewal. This is consistent with previously published reports of “bivalent” histone marks (i.e., polycomb-associated repressive H3K27me3 and activating H3K4me3 mark) on developmental regulatory genes in murine ES cells (Bernstein et al., 2006) and a recent report demonstrating that many H3K4me3+ genes are not expressed in ES cells due to the lack of transcriptional elongation (Guenther et al., 2007). Furthermore, our data demonstrate that the set of bivalent (H3K27me3+/H3K4me3+) genes in U-EML cells is composed almost entirely of CGI genes (data not shown).
In conclusion, this study provides a genome-scale view of epigenetic changes that occur during hematopoietic development that reveals a complex interdependence between DNA sequence, histone modifications, and developmental gene function. Incomplete methylation of H3K4 identifies a set of lineage-specific genes that are transcriptionally poised in multipotential hematopoietic stem/progenitor cells and have the potential to become activated during the process of terminal differentiation. If generalizable, these results provide insight into the reciprocal nature of the large-scale epigenetic transition that occurs during development.
EXPERIMENTAL PROCEDURES
Purification of Primary Hematopoietic Cells
For multipotential and erythroid primary cells, bone marrow was harvested from femurs, tibias, iliac crests, and vertebrae of 12 C57/B6 mice, and cell populations were isolated by FACS based on the following immunophenotypes: Sca+/cKit+/lineage− for multipotential cells and Ter119+ for erythroid cells. For mature naive B cells, spleens were harvested from two mice and the B220+/IgMlow/CD23+ isolated by FACS.
Cell Lines
The EML cell line was acquired from the ATCC and grown (according to the instructions of Dr. Schickwann Tsai) in IMDM plus 20% heat-inactivated donor equine serum (Hyclone), 16% SCF-conditioned medium (from BHK-MKL cell line, a gift from Dr. Tsai), and L-glutamine (Tsai et al., 1994). All other cell lines were grown in the following culture conditions: MEL cells in DMEM plus 10% FBS; 32D cells in RPMI plus 10% FBS, recombinant mouse IL-3 (10 ng/ml, R&D Systems), and L-glutamine; A20 cells in RPMI plus 10% FBS and L-glutamine; V6.5 murine embryonic stem cells on irradiated murine embryonic fibroblasts in standard ES cell medium (DMEM [Invitrogen], 15% ES qualified serum (Hyclone), LIF (100 ng/ml, Peprotech), 100 μM β-mercaptoethanol, and 2 mM L-glutamine.
Chromatin Immunoprecipitations
ChIP assays were performed essentially as described by Upstate. A more detailed protocol can be found in the Supplemental Data. ChIP assays that were analyzed by qPCR were performed on 200 μl aliquots of cell lysate (2 × 106 cell equivalents) that were diluted with 1.8 ml ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl, 30 mM sodium butyrate, and HALT protease inhibitor) using 3.5 μl antiserum. Primary cell ChIP assays were performed on lysate from 3–5 × 105 cells. ChIP assays that were used for genome-wide location analysis were performed similarly except that each ChIP was performed on 1 ml of cell lysate (2 × 107 cell equivalents) with either 5 μl of antiserum or 5 mg of purified IgG.
qPCR Analysis
Quantitative PCR of genomic loci and cDNA was performed as described in Supplemental Data.
Erythroid Differentiation of EML Cells
EML cells were plated in 45 ml of normal growth medium plus 10 U/ml of human erythropoietin (Epo; Ortho) at 75,000 cells/ml in a T75 culture flask and cultured for 2 days. The cells were then plated in T150 flasks (25,000 cells/ml and 90 ml total) in SCF reduced-growth medium (this is the same as growth medium except with only 4% BHK-MKL conditioned medium) plus 10 U/ml Epo. After 3 days, an additional 10 U/ml of Epo was added, without changing the medium. After 2 days, the number of differentiated (c-Kit−) cells ranged from 50% to 75% of the culture.
FACS Sorting for ChIP Assays
Approximately 3 × 108 undifferentiated or differentiated EML cells were labeled with either anti-Sca-1-PE (undifferentiated) or anti-c-Kit-FITC (differentiated) for 25 min, washed in cold PBS/2% FBS, filtered through 0.30 μm mesh, and FACS sorted. The top 80% of Sca+ undifferentiated cells and the c-Kit- erythroid cells (based on an unstained control) were sorted. The cells were maintained at 4°C presort and postsort. Due to the loss of certain histone modifications during the sorting procedure, we placed these cells back in culture in the appropriate growth conditions for 2 hr prior to processing for ChIP assays, as described above.
Gene Expression Profiling
Total RNA was purified from 2 × 107 EML cells using the QIAGEN RNEasy kit (QIAGEN, Chatsworth). The RNA concentration, purity, and integrity were evaluated by UV spectrophotometry and an RNA-nano Bioanalyzer (Agilent, Palo Alto, CA). Probe synthesis and hybridization to mouse 430 GeneChip DNA microarrays (Affymetrix, Santa Clara, CA) were performed following the manufacturer’s instructions. Expression values were calculated using the Affymetrix MAS5 algorithm. For each cell type, expression values for each gene were calculated by averaging the expression values of each probe set across experiments. For genes that correspond to multiple probe sets, the probe set with the highest mean expression value was selected.
Genome-wide Location Analysis
ChIP-chip experiments were performed using both Nimblegen and Agilent microarray platforms. See Supplemental Data and Experimental Procedures for detailed methodology.
GO Category Analysis
Statistical overrepresentation of GO Biological Process categories was determined using the BiNGO software package with Benjamini and Hochberg false discovery rate correction (Maere et al., 2005).
CpG Islands
The CpG islands were identified using a 500 bp sliding window with 5 bp step. The windows with GC fraction over 0.55 and CpG observed/expected ratio above 0.62 were called CpG islands. A gene was classified as CpG island-containing if it contained at least one CpG island.
Sequence Motifs
Detailed descriptions of bioinformatics analyses of genomic sequence motifs are provided in the Supplemental Data.
Pu.1 Genome-wide Location Analysis
Pu.1 ChIP-chip experiments were performed on 2 × 107 U-EML cells using 5 mg of normal rabbit IgG or 5 μg of anti-Pu.1 as described in the Supplemental Experimental Procedures. The significance of correlation with the experimentally determined Pu.1-binding pattern was assessed by calculating partial correlation between binned (700 bp) H3K4me2+ enrichment and Pu.1-binding patterns, controlling for the IgG pattern. The statistical significance was calculated based on Fisher’s z transform.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the thoughtful discussions and insights from Bob Kingston and members of his lab and for the help of Andre Catic and Heather Fleming in the preparation of the manuscript. We greatly appreciate access to the Agilent genome-wide promoter microarray designs granted by Rick Young’s laboratory and the technical expertise of Tom Volkert. The EML and BHK-MKL cell lines were graciously provided by Schickwann Tsai, and the MEL cell line was provided by Stuart Orkin. K.O. is supported by the MGH Fund for Medical Discovery fellowship and by a K08 Development Award from NIDDK; P.K. is supported by a fellowship in biomedical informatics from NLM; P.J.P. is supported by a K25 award from NIGMS. This work was supported in part by funds from the NHLBI and NIDDK to D.T.S.
Footnotes
ACCESSION NUMBERS
Gene expression and Agilent ChIP-chip data are available via the Gene Expression Omnibus with accession number GSE11044.
SUPPLEMENTAL DATA
Supplemental Data include ten figures, eight tables, and Supplemental Experimental Procedures and can be found with this article online at http://www.developmentalcell.com/cgi/content/full/14/5/798/DC1/.
REFERENCES
- Ayton PM, Chen EH, and Cleary ML (2004). Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol. Cell. Biol 24, 10470–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, et al. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120, 169–181. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, and Allis CD (2003). Histone and chromatin cross-talk. Curr. Opin. Cell Biol 15, 172–183. [DOI] [PubMed] [Google Scholar]
- Fisher AG (2002). Cellular identity and lineage choice. Nat. Rev. Immunol 2, 977–982. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, and Young RA (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, and Enver T. (1997). Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11, 774–785. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, and Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, and Reinberg D. (2005). The key to development: interpreting the histone code? Curr. Opin. Genet. Dev 15, 163–176. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, and Henikoff S. (2005). Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet 37, 1090–1097. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, and Akashi K. (2002). Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3, 137–147. [DOI] [PubMed] [Google Scholar]
- Nightingale KP, O’Neill LP, and Turner BM (2006). Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr. Opin. Genet. Dev 16, 125–136. [DOI] [PubMed] [Google Scholar]
- Oikawa T. (2004). ETS transcription factors: possible targets for cancer therapy. Cancer Sci. 95, 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, and Yamada T. (2003). Molecular biology of the Ets family of transcription factors. Gene 303, 11–34. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, and Verdine GL (2006). Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol 13, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, and Orkin SH (2007). Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell 27, 562–572. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, and Kouzarides T. (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Tanay A, O’Donnell AH, Damelin M, and Bestor TH (2007). Hyperconserved CpG domains underlie Polycomb-binding sites. Proc. Natl. Acad. Sci. USA 104, 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J, Glass C, and Rosenfeld MG (1998). Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol 10, 373–383. [DOI] [PubMed] [Google Scholar]
- Tsai S, Bartelmez S, Sitnicka E, and Collins S. (1994). Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 8, 2831–2841. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, and Allis CD (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.