Abstract
We have analyzed the binding of recombinant human immunodeficiency virus type 1 nucleocapsid protein (NC) to very short oligonucleotides by using surface plasmon resonance (SPR) technology. Our experiments, which were conducted at a moderate salt concentration (0.15 M NaCl), showed that NC binds more stably to runs of d(G) than to other DNA homopolymers. However, it exhibits far more stable binding with the alternating base sequence d(TG)n than with any homopolymeric oligodeoxyribonucleotide; thus, it shows a strong sequence preference under our experimental conditions. We found that the minimum length of an alternating d(TG) sequence required for stable binding was five nucleotides. Stable binding to the tetranucleotide d(TG)2 was observed only under conditions where two tetranucleotide molecules were held in close spatial proximity. The stable, sequence-specific binding to d(TG)n required that both zinc fingers be present, each in its proper position in the NC protein, and was quite salt resistant, indicating a large hydrophobic contribution to the binding. Limited tests with RNA oligonucleotides indicated that the preferential sequence-specific binding observed with DNA also occurs with RNA. Evidence was also obtained that NC can bind to nucleic acid molecules in at least two distinct modes. The biological significance of the specific binding we have detected is not known; it may reflect the specificity with which the parent Gag polyprotein packages genomic RNA or may relate to the functions of NC after cleavage of the polyprotein, including its role as a nucleic acid chaperone.
A single protein species, the Gag polyprotein, is sufficient for assembly of retrovirus particles. Since this process includes the selective encapsidation of viral RNA, this protein is evidently capable of specific interactions with nucleic acids. The nature of these interactions is not well understood as yet. After the virion is released from the cell, the polyprotein is cleaved by the virus-encoded protease; one of the cleavage products, termed the nucleocapsid protein (NC), then binds to the genomic RNA, forming the ribonucleoprotein core of the mature particle (21, 35, 41).
The interaction between Gag and the genomic RNA is known to involve the NC domain of the polyprotein, since mutants within this domain of Gag are defective in RNA packaging (e.g., references 2, 16, 17, 24–27, 31, 36, 37, and 39) and since the specificity of encapsidation tends to be determined by the NC domain in chimeric Gag molecules (9, 18, 49). However, NC is a basic protein and has frequently been described as binding to single-stranded DNA or RNA in a sequence-independent manner. Indeed, it is probably capable of binding to any single-stranded nucleic acid under appropriate conditions. This binding activity appears to be crucial at several stages of virus replication (13, 19, 28, 46).
In the experiments described here, we have analyzed the binding of recombinant human immunodeficiency virus type 1 (HIV-1) NC to short oligonucleotides. These studies were performed at moderate ionic strengths, at which the nonspecific electrostatic interaction between NC and nucleic acids is minimized. We find that under these conditions, the protein exhibits profound sequence preferences. This sequence-specific binding is dependent upon the zinc fingers of the protein and has a strong hydrophobic component. The biological significance of this sequence specificity is not clear at present, but the results suggest that studies with very short oligonucleotides may provide important insights into NC function and perhaps functions of Gag as well.
MATERIALS AND METHODS
Reagents.
Oligonucleotides were synthesized by Marilyn Powers, NCI-Frederick Cancer Research and Development Center. For attachment to the surface plasmon resonance (SPR) surface, the oligonucleotides were synthesized with biotin at their 3′ ends by using biotin TEG CPG (Glen Research, Sterling, Va.).
HIV-1 NC was produced in bacteria and prepared as described previously (10, 46). The protein contained 55 residues, and its sequence was from the MN isolate of HIV-1 (29). “Finger switch” mutant NCs (25) were based on the NL4-3 NC sequence (1) and were a kind gift from Robert Gorelick. Wild-type NL4-3 NC was also prepared, and its properties in the current experiments were the same as those of the MN NC.
SPR experimental methods.
SPR was performed with the Biacore instrument manufactured by Pharmacia Biosensor AB (Uppsala, Sweden). SPR experiments were performed essentially as described previously (20). CM5 sensor chips were first modified by coupling the primary amino groups of streptavidin (Pierce Chemical Co., Rockford, Ill.) to surface carboxyl groups by using NHS/EDC coupling chemistry as indicated by Pharmacia. The biotinylated oligonucleotide was then injected onto the SPR sensor chip; the amount of immobilized nucleic acid bound to the chip was then determined by SPR analysis.
The buffer used in the SPR experiments was 0.15 M NaCl–10 mM HEPES (pH 7.5)–5 mM dithiothreitol (DTT)–0.05% Tween 20 (in the experiment shown in Fig. 8, 5 mM DTT was replaced with 100 mM β-mercaptoethanol). Unless otherwise noted, all experiments were initiated by passing buffer across an SPR chip containing a known amount of oligonucleotide for approximately 100 s at 8 μl/min, followed by injection, at a similar rate, of 20 μl of buffer containing NC solution. Injection of the NC sample was followed by injection of buffer (“washout”) for an additional 400 s. Finally, the surface was regenerated with two successive 5-μl pulses of 0.1% sodium dodecyl sulfate (SDS)–3 mM EDTA; the SPR response in this phase is not shown in the figures. The cycle could then be repeated with a more concentrated NC solution. Each SPR chip with its immobilized oligonucleotide was stable for hundreds of cycles of this type.
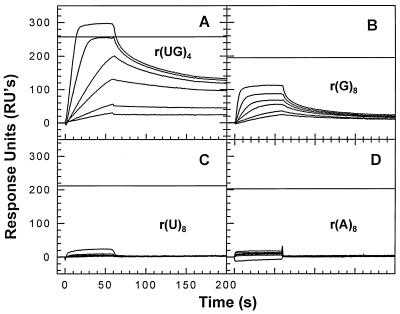
FIG. 8.
Binding of NC to RNA. SPR chips were prepared with 125.1 RU of r(UG)4 (A), 101.8 RU of r(G)8 (B), 104.0 RU of r(U)8 (C), or 101.6 RU of r(A)8 (D). They were then tested with 1, 5, 10, 25, 50, 100, and 200 nM NC solutions. In this experiment, the NC solutions contained 100 mM β-mercaptoethanol and the SPR buffer contained 5 mM β-mercaptoethanol instead of DTT. Also, the flow rate in this experiment was 64 rather than 8 μl/min.
Fluorescence methods.
Equilibrium binding isotherms for the association of NC with nucleic acids were obtained by monitoring changes in the fluorescence emission intensity of the single tryptophan residue in NC upon complex formation. Measurements were carried out in a Shimadzu 500U (Columbia, Md.) or a SPEX Fluorolog (Edison, N.J.) spectrofluorimeter, with excitation at 288 nm (3-nm bandwidth) and emission at 355 nm (10-nm bandwidth). Titrations were performed at 25°C in 10 mM sodium phosphate, pH 7.0, by stepwise addition of oligonucleotide to a solution of 0.8 or 3.0 μM NC in a dual-pathlength Suprasil quartz cuvette (0.2 by 1.0 cm; Uvonic Instruments, Plainview, N.Y.). Binding to nucleic acid results in quenching of the tryptophan fluorescence. The NC-nucleic acid complex was then disrupted by the progressive addition of NaCl to the cuvette, and the disruption was monitored by recovery of fluorescence. Corrections were made for dilution and inner filter effects.
RESULTS
The binding of NC to nucleic acids was measured in these experiments by SPR. In this procedure, a nucleic acid is immobilized on a chip. A solution of NC is then passed across the chip, and its binding is detected by the Biacore instrument as an increase in the mass (“response units” [RUs]) associated with the chip. (The amplitude of the RU signal is directly proportional to the amount of mass bound to the chip.) After ∼150 s of injection of NC solution, the solution is replaced with SPR buffer, and the removal of NC from the chip during this washout phase is observed. Each SPR figure contains a family of curves showing the kinetics of binding and washout obtained with a series of different NC concentrations; the curves are superimposed in the figures.
Binding of NC to single- and double-stranded DNA: initial characterization.
NC has traditionally been described as a single-stranded nucleic acid-binding protein (e.g., references 14 and 43). To determine whether this conclusion is accurate under SPR conditions, we initially measured the binding of recombinant HIV-1 NC to a single-stranded 28-base oligodeoxynucleotide. The sequence of this oligonucleotide (5′GACTTGTGGAAAATCTCTAGCAGTGCAT) contains 19 bases from the 3′ end of U5 (38) and is the same as that originally used in competitive hybridization experiments by Tsuchihashi and Brown (44). The results of SPR analysis using this oligonucleotide are shown in Fig. 1.
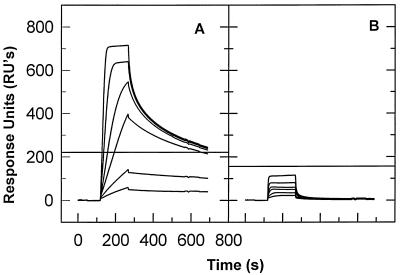
FIG. 1.
Binding of NC to single- and double-stranded DNA. (A) A chip containing 312 RU of biotinylated, immobilized 28-base single-stranded DNA (i.e., GACTTGTGGAAAATCTCTAGCAGTGCAT) was exposed successively to 5, 10, 25, 50, 100, and 200 nM NC solutions. In each cycle, buffer was applied to the chip for the first 160 s. This was followed by the NC solution, which was applied for the next 160 s (the washon phase). The NC solution was followed by SPR buffer, which was allowed to flow past the chip for 700 s (the washout phase). Finally, all NC was removed with SDS (not shown). The successive binding curves are all superimposed in the figure. The horizontal line shows the RU expected if one NC molecule bound to each oligonucleotide molecule. (B) A chip containing 441 RU of biotinylated 28-base-pair double-stranded DNA was tested with the same NC concentration series as that used for panel A.
Figure 1A shows that several NC molecules bound to the oligonucleotide (the horizontal line in the figure represents the response which would be observed if a single NC molecule bound to each oligonucleotide molecule). A portion of the bound NC was rapidly released during the washout phase. However, at later times, the washout curves became less steep, showing that a fraction of the NC is released quite slowly; further, this amount is practically the same in the curves obtained at NC concentrations between 25 and 200 nM. This observation suggests that there are a limited number of stable binding sites on the 28-base oligonucleotide; these sites are evidently almost fully occupied at NC concentrations as low as 25 nM. Additional binding can occur at higher NC concentrations, but this attachment is apparently at considerably lesser affinity, since this portion of the NC is easily removed during the washout. Extrapolation of the shallow portions of the washout curves (i.e., the portions representing removal of the tightly bound NC molecules, beginning at ∼600 s) to the beginning of the washout step and application of a simple exponential function (not shown) suggest that the stable binding involves two NC molecules per oligonucleotide.
To test the preference of NC for single- rather than double-stranded nucleic acids under our experimental conditions, we also measured its ability to bind a double-stranded form of the 28-base oligonucleotide. An oligonucleotide complementary to the original 28-base sequence was added to an SPR chip like that used in the experiment shown in Fig. 1A. Hybridization to the single-stranded DNA already on the chip was confirmed by an increase in mass on the chip detected by SPR (data not shown). As shown in Fig. 1B, NC exhibited only negligible binding to this double-stranded DNA; it thus shows a strong preference for single-stranded DNA. (It might be suggested that NC displaces the hybridized DNA strand, with no net change in mass and consequently no effect on the SPR signal. However, this is not the case, since Fig. 1 shows a series of successive binding curves. If NC had displaced one strand in the first cycle, then after the removal of NC with SDS between the runs that generated the binding curves, a curve like that in Fig. 1A would have been obtained in the next cycle.)
Demonstration of sequence-specific binding of NC to single-stranded oligodeoxynucleotides; partial analysis of sequence preference.
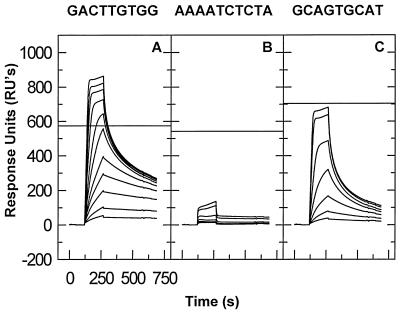
NC has frequently been described as binding to single-stranded nucleic acids at a ratio of one NC molecule per ∼7 or 8 nucleotide residues, with little or no sequence specificity (e.g., references 32, 33, and 48). Thus, it was surprising that the results presented above indicated the presence of two relatively stable binding sites in the 28-base oligonucleotide, rather than four. One possible explanation of these findings is that the occluded site size under our experimental conditions is ≥10 bases, rather than ∼7 bases; alternatively, NC might bind to the ∼7 base sites with significantly different stabilities and the oligonucleotide under study might contain only two sites which are bound tightly enough to be detected by the present methods. To test this possibility, we subdivided the 28-base sequence into three smaller sequences and tested binding to each of them. Three oligonucleotides, containing sequences from the 5′, middle, and 3′ regions of the 28-base sequence (designated sites I, II, and III, respectively), were analyzed, with the SPR results shown in Fig. 2A. Site I, i.e., GACTTGTGG, showed the most stable binding (Fig. 2A); site II, i.e., AAAATCTCTA, showed negligible binding (Fig. 2B); and site III, i.e., GCAGTGCAT, showed significant binding, but at a lower level and with a more rapid loss during the washout than was observed with site I (Fig. 2C). (One indication of the relative stabilities of the binding to the different oligonucleotides is the RU response near the end of the washout period. Thus, the response is approximately one-half of the stoichiometric level at 650 s in Fig. 2A but only about one-sixth of the stoichiometric level in Fig. 2C.) The results show clearly that there are profound differences in the stability with which NC binds to different sequences and that sequences somewhat shorter than 10 bases are sufficient for stable binding.
FIG. 2.
Binding of NC to oligonucleotides representing different regions of the 28-base oligonucleotide tested in Fig. 1. (A) A chip containing 286 RU of GACTTGTGG (site I) was tested with 5, 10, 20, 30, 40, 60, 80, 120, 160, 200, and 250 nM NC solutions. (B) A chip containing 358 RU of AAAATCTCTA (site II) was tested with 5, 10, 20, 40, 80, 160, and 200 nM NC solutions. (C) A chip containing 297 RU of GCAGTGCAT (site III) was tested as described for panel B.
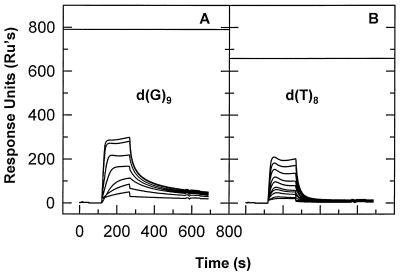
An inspection of the three tested sequences shows that they have quite different base compositions, with sites I and III being relatively G and T rich and site II being quite A rich. Thus, it seemed possible that the differences in binding to the three sequences were simply reflections of preferences of NC for different bases. To test this possibility, we also measured the binding of NC to the four homopolymeric oligodeoxynucleotides by SPR. Figure 3 shows the results obtained with d(G)9 and d(T)8. As can be seen, NC exhibited some binding to both of these oligonucleotides. The binding to d(G)9 was at a somewhat higher level and was somewhat more stable than the binding to d(T)8. However, both the extent of the binding to d(G)9 (relative to the horizontal line in the figure, which would be the SPR signal obtained upon the binding of one NC molecule to each oligonucleotide molecule on the chip) and the retention of NC during the washout were far lower than those seen with site I (Fig. 2A). The tests of d(A)9 and d(C)9 showed no detectable binding (data not shown), as in the tests of the corresponding oligoribonucleotides (see Fig. 8 below). The fact that the binding to site I is so much more stable than the binding to any homopolymer demonstrates that NC possesses a true sequence-specific component in its interaction with single-stranded DNA.
FIG. 3.
Binding of NC to homopolymeric oligodeoxynucleotides. (A) A chip containing 422 RU of d(G)9 was tested with 5, 10, 20, 40, 80, 160, and 200 nM NC solutions. (B) A chip containing 348 RU of d(T)8 was tested as described for panel A.
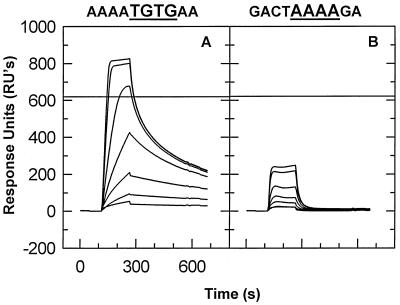
Comparison with binding to other oligonucleotides (data not shown) raised the possibility that the presence of the sequence TGTG within site I was responsible for the stable binding to this site. To test this possibility, we performed an SPR analysis of the binding to two additional oligonucleotides: one in which all of the bases in site I other than TGTG were replaced by A’s (runs of A were chosen because, as noted above, NC shows almost no binding to this sequence), and one in which the TGTG was replaced by AAAA. The results of these tests (Fig. 4) show that TGTG is necessary and sufficient for stable binding to a 10-base oligonucleotide, even if the other bases are all A’s, to which NC binds very poorly.
FIG. 4.
Significance of TGTG in site I. A chip containing 348 RU of AAAATGTGAA (A) or 296 RU of GACTAAAAGA (B) was tested as described for Fig. 3. These sequences are derived from site I by replacement of selected bases by A’s; an additional A is also present at the 3′ ends of these oligonucleotides.
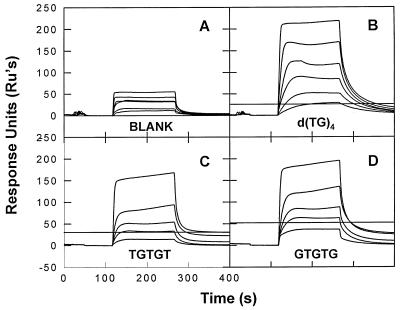
To determine the minimal length of an oligonucleotide composed of alternating T’s and G’s which was capable of stably retaining a molecule of NC, we tested different lengths of DNA containing only this sequence. As shown in Fig. 5C and D, we found that either of two pentanucleotides, i.e., TGTGT or GTGTG, constitute binding sites for NC, both binding a single NC molecule with roughly equivalent stabilities. Studies with longer oligonucleotides composed of alternating d(TG) sequences are summarized in Table 1; since two NC molecules bind stably to the decamer d(TG)5 and four bind to d(TG)10, the data show that with this alternating sequence five bases are sufficient for stable binding and that each NC molecule “occupies” a stretch of only five bases under our experimental conditions.
FIG. 5.
Stable binding of NC to pentanucleotides. SPR chips were prepared with streptavidin alone (A), or with 11.4 RU of (TG)4 (B), 9.8 RU of TGTGT (C), or 16.8 RU of GTGTG (D). These chips contained 0, 0.08, 0.2, and 0.11 mol of oligonucleotide per mol of streptavidin, respectively. The chips were then tested with 10, 25, 50, 100, 200, and 400 nM NC solutions (A and B) or with 10, 25, 50, 200, and 400 nM NC solutions (C and D). (Because of the extremely low levels of oligonucleotide on the chips in this experiment, a relatively large amount of NC bound in the nonsaturable mode. Thus, as noted in the Discussion section, the amplitude of binding during the washon does not reflect the affinity or stoichiometry of the high-affinity, saturable mode of binding of NC to an oligonucleotide.
TABLE 1.
Binding sites for NCa
| Oligonucleotide | No. of NC binding sites |
|---|---|
| d(TGTGT) | 1 |
| d(GTGTG) | 1 |
| d(TG)4 | 1 |
| d(TG)5 | 2 |
| d(TG)10 | 4 |
Analysis of the washout portion of SPR experiments shows that there are two independent systems for the binding of NC to short oligonucleotides. The more stable system achieves saturation but the less stable system does not. Stoichiometry for binding was determined by extrapolation of the shallow portion of the washout curve, where the more stable binding system predominates, by using a simple exponential function to find the number of apparent binding sites. This number was then rounded to the nearest integer.
Cross-linking of short oligonucleotides by NC.
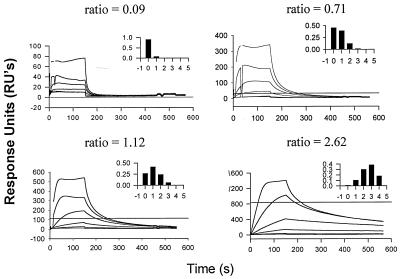
In the course of our experiments to define a minimal stable binding site for NC, we found that initial experiments with the tetranucleotide TGTG failed to give reproducible results. Further investigation suggested that the apparent affinity of NC for TGTG was a function of the concentration of the oligonucleotide during SPR analysis. This possibility was analyzed systematically in the following experiment. An SPR chip with four channels containing TGTG at different densities was made. For each channel, the ratio of TGTG molecules to streptavidin molecules was determined. Since a streptavidin molecule has four binding sites for biotinylated ligands, the ratio enabled us to calculate the expected number of streptavidin molecules occupied by zero, one, two, three, or four oligonucleotides. These frequency distributions are shown in the insets in Fig. 6. Finally, the ability of each channel to bind NC with a high degree of stability was determined by SPR. Inspection of the four curves in Fig. 6 shows clearly that stable binding is achieved at the highest density (Fig. 6D) and not at the lowest density (Fig. 6A). Quantitative analysis (data not shown) indicates that the level of stable binding seen in each channel can be completely accounted for by streptavidin molecules containing two or more oligonucleotides. In contrast, binding to longer oligonucleotides, including TGTGTGTG and site I, was essentially independent of the densities of these DNAs during SPR analysis (data not shown). We conclude that if two short oligonucleotides containing alternating T’s and G’s are close enough to each other (i.e., bound to the same streptavidin molecule), they can jointly participate in stable binding to NC.
FIG. 6.
Binding of NC to SPR channels with different densities of TGTG. For each channel, the amount of streptavidin was measured by SPR analysis before the addition of TGTG. The amount of TGTG added to each channel was then quantitated by SPR. The ratio of TGTG to streptavidin was thus empirically determined for each channel: the four channels contained 0.09, 0.71, 1.12, or 2.62 mol of TGTG per mol of streptavidin. The chips were then tested with 10, 20, 40, 80, 160, and 200 nM NC solutions. The inset in each panel is a histogram showing the fraction of streptavidin molecules occupied by zero, one, two, three, or four TGTG molecules, as predicted from the ratio (shown above each panel) of moles of TGTG to moles of streptavidin by using the binomial expansion. The horizontal line in each panel is the RU expected if one NC molecule were to bind to each streptavidin molecule with two or three TGTG molecules and if two NC molecules were to bind to each streptavidin with four TGTG molecules.
Figure 6A also shows that NC shows a substantial degree of initial binding to TGTG, even when the latter is present at low density. Thus, the data demonstrate a difference between the level of binding and the stability of binding. This point will be considered further in the Discussion section.
Stable binding depends on zinc fingers in NC.
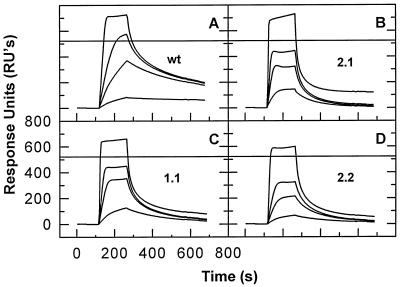
HIV-1 NC contains two zinc fingers. Mutational analysis shows that these structures are of crucial importance in vivo, participating both in the packaging of genomic RNA during virus assembly and in some additional step(s) during the infection process (2, 16, 26, 27). In contrast, there is little evidence for a significant role for the fingers in interactions of NC with nucleic acids in vitro. To test the importance of the fingers for the sequence-specific binding described in the present study, we analyzed the binding of three finger switch mutants of NC (25) to site I. These mutant proteins contain two zinc fingers, but in mutant 1.1, the C-terminal finger has been replaced with a second copy of the N-terminal finger. Mutant 2.2 has an analogous duplication of the C-terminal finger, while in mutant 2.1 both fingers are present but their positions in the protein have been reversed.
The results of this experiment are shown in Fig. 7. The data show that the mutant proteins all exhibit some binding to site I, but in every case the mutants (Fig. 7B to D) washed out considerably faster than the wild-type protein (Fig. 7A). Similar results have also been obtained with AAAATGTGAA (data not shown). (While the wild-type control used in this experiment was the NC of the MN isolate of HIV-1, similar results have been obtained in experiments using the NL4-3 wild-type NC, which is identical to the mutant proteins except for the exchanges of the fingers [25]) [data not shown]).
FIG. 7.
Binding of finger switch NC mutant proteins to site I. A chip with 286 RU of site I (GACTTGTGG) was prepared and tested with wild-type NC (A), mutant 2.1 (B), mutant 1.1 (C), and mutant 2.2 (D) (25) at 10, 50, 100, and 200 nM.
As an additional test of the role of the zinc fingers in the sequence-specific binding of NC to DNA, we performed SPR analysis in the presence of EDTA. No significant binding to the 28-base oligonucleotide was observed when 1 μM NC was tested in the presence of 3.3 mM EDTA (data not shown). This result supports the conclusion that the specific binding we observed is dependent upon intact zinc fingers within NC.
Analysis of binding by fluorescence.
As a completely independent way of studying the interaction between NC and oligonucleotides, we measured the change in tryptophan fluorescence upon the stepwise addition of oligonucleotide to NC solutions. The initial titration was performed in 0.01 M sodium phosphate, but after saturation of the protein with oligonucleotide, NaCl was added and the dissociation of the protein-nucleic acid complex was monitored by measuring fluorescence. This approach allowed us to determine the degree to which the binding was resistant to dissociation by high ionic strength. The midpoints of these titrations, i.e., the NaCl concentrations at which 50% recovery of the tryptophan fluorescence was achieved, are presented in Table 2.
TABLE 2.
Salt resistance of NC binding
| Oligonucleotide | Salt resistancea (mM) |
|---|---|
| d(A)8 | 15 |
| d(T)8 | 42 |
| d(I)8 | 115 |
| d(G)8 | 150 |
| d(GA)4 | 90 |
| d(IT)4 | 132 |
| d(TG)4 | 355 |
NaCl concentration at which 50% of NC is removed from the oligonucleotide, as determined by the recovery of tryptophan fluorescence.
The tests of homopolymers (Table 2) showed that the binding of NC to G’s is significantly more salt resistant than the binding to T’s or A’s. In addition, we measured the midpoints of the salt titrations for the binding of NC to oligodeoxynucleotides with alternating base sequences. We found that the midpoint for binding to d(GA)4, i.e., 90 mM Na+, falls approximately halfway between the midpoints for dA8 (15 mM) and dG8 (150 mM), as would be expected if the binding to the heteropolymer were not qualititavely different from the binding to simple polymers of its constituent bases. In contrast, the binding to d(TG)4, with a midpoint of 355 mM, is far more salt resistant than binding to either dT8 or dG8.
We further analyzed the specific binding properties of NC by measuring the salt resistance of its interaction with d(IT)4. Except for the fact that the alternating sequence begins with the purine rather than the pyrimidine, this oligonucleotide differs from d(TG)4 only by the absence of the exocyclic amino group from the purine residues. Table 2 shows that the midpoint for this interaction, i.e., 132 mM, is somewhat higher than the weighted average of the midpoints for dI8 and dT8 but almost threefold lower than that of the d(TG)4 titration.
High-affinity binding of NC to RNA.
It was of interest to determine whether the sequences to which NC binds with high affinity in DNA would also constitute high-affinity sites in RNA. Therefore, we tested the binding of NC to the RNA analog of d(TG)4, i.e., r(UG)4. We found (Fig. 8A) that there was a substantial amount of stable binding to this oligonucleotide, as shown by the slow removal of NC during the washout. We also tested binding to homopolymeric oligoribonucleotides of G (panel B), C (panel C), and A (panel D); the results show, as in the case of oligodeoxyribonucleotides, a noticeable preference for G over the other bases, but far less stable binding than to r(UG)4. Thus, NC exhibits sequence-specific binding to RNA, as well as to DNA. It seems very likely that the binding characteristics of NC which we have described with short DNA molecules will be found to apply to RNA binding as well.
DISCUSSION
In the present study, we have used SPR to analyze the binding of recombinant HIV-1 NC to very short oligonucleotides. The results can be briefly summarized as follows. First, of the limited number of DNA sequences tested here, an alternating sequence of T’s and G’s gives the most stable binding detected. The stability with which NC binds to this sequence reflects a true sequence preference, rather than a preference for the bases T and/or G, since the binding is far more stable than that for T- or G-containing (or other) homopolymers (Fig. 3 and 4).
Second, this sequence-specific binding involves the zinc fingers in NC, since it is not observed with proteins with rearranged zinc fingers (Fig. 7) or when Zn2+ is removed from the wild-type protein with EDTA (data not shown). The failure of all three finger switch proteins to bind to site I with the same stability as that of wild-type NC demonstrates that both fingers must be present, each in its proper position in the protein, for the stable binding. The discovery of an in vitro activity of NC which is dependent on the zinc fingers seems quite significant, as these structures are of crucial importance in vivo (2, 16, 24–27, 36, 37) but are largely dispensable for most of the in vitro NC-nucleic acid interactions which have been analyzed previously (e.g., references 15, 19, and 46).
Third, the hydrophobic contribution to the binding seen with d(TG)4 (as measured by the resistance of the binding to dissociation by salt) is far greater than that seen for binding to other oligodeoxynucleotides, including d(GA)4 as well as a series of homopolymeric oligonucleotides (Table 2). Indeed, this contribution is significantly greater with d(TG)4 than with d(IT)4 (Table 2). This observation strongly suggests that the exocyclic amino group by which G differs from I is involved in the stable binding to d(TG)4. It seems possible that this amino group participates in a hydrogen bond with a residue in NC.
As part of our characterization of the binding of NC to oligodeoxynucleotides containing alternating T’s and G’s, we attempted to identify the minimum length required for stable binding. We found that pentanucleotides, i.e., either TGTGT or GTGTG, were capable of stable interaction with NC (Fig. 5), and the stoichiometry of binding showed that one NC molecule binds stably to every five nucleotides in a longer oligonucleotide (Table 1). This value of five as the minimum number of bases required for stable interaction is somewhat lower than 7 or 8, the value found in a number of studies for the “occluded site size,” or average number of bases per NC molecule when a nucleic acid is saturated with NC (e.g., references 32, 33, and 48). We submit that the value obtained in the present work is more precise than previous determinations, since we measured the binding to nucleic acid molecules of discrete, uniform lengths. Nevertheless, it is quite possible that the discrepancy between our results and those in the literature is real: perhaps NC is more compact or NC molecules can be crowded together more tightly when engaged in binding to a preferred sequence. It should also be noted that many of the previous studies were conducted with incompletely processed fragments of Gag containing NC, e.g., “p15” (11) and “NC71.” It has been shown (33) that the nucleic acid-binding properties, including the site size, of NC71 are quite different from those of the 55-amino-acid NC which we used here and which is the major species in mature HIV particles (29).
We also found that NC could bind stably to the tetranucleotide d(TG)2, but only if the TGTG molecules were in close proximity to each other (Fig. 6). This observation shows that a single NC molecule can simultaneously bind or cross-link two such oligonucleotides. The simultaneous binding of a single NC molecule to two tetranucleotides could mean that NC has a single binding site for nucleic acid, requiring at least five bases for stable interaction, and that two smaller oligonucleotides can cooperate to fill this site. Alternatively, it is possible that with the tetranucleotides, stability is attained by the simultaneous attachment of the two nucleic acid molecules to two distinct binding sites within the protein. To our knowledge, this is the first time SPR analysis has been used in this way.
Although nearly all of our experiments were performed with oligodeoxyribonucleotides, we also tested binding to a small sampling of RNA oligonucleotides. The results of these tests (Fig. 8) were completely congruent with the DNA results: NC shows a detectable preference for G over other bases but shows far more stable binding to an alternating (UG) sequence than to any homopolymer. It seems likely that all of the conclusions drawn from the DNA experiments will be applicable to the interaction with RNA as well.
The nature of the stable interaction between NC and nucleic acids appears remarkably complex. Thus, while NC binds with a high degree of stability to d(TG)4 (Table 1) and shows almost no binding to d(A)8 (Fig. 3), it binds with a low degree of stability to d(TG)2 when this oligonucleotide is present at low density on the SPR chip (Fig. 6A). This appears to represent a low-stability sequence-specific binding mode, different from the more stable type of binding seen with longer oligonucleotides.
We have subjected the SPR data obtained with d(TG)4 (see Fig. 5B) to a detailed quantitative analysis (unpublished data). Surprisingly, this analysis shows that NC can bind to this oligonucleotide in two distinct ways. These two binding systems are independent of each other, in essence competing with each other for the NC molecules. One of these systems exhibits a high level of affinity and is therefore primarily responsible for the slow washout from d(TG)4. The other represents the fraction of NC molecules which are released more rapidly during the washout phase; this system does not appear to be saturable under our experimental conditions. This binding system makes a major contribution to the amplitude of the SPR response during the “washon” part of the cycle, when NC is being applied to the chip. Therefore, the amplitude in this phase does not reflect the affinity or stoichiometry of the first, high-affinity system.
Previous studies have characterized the interaction of retroviral NC proteins with a wide variety of nucleic acids, including homo- and heteropolymeric oligonucleotides, ribozymes, and tRNAs (reviewed in reference 6). As noted above, NC has frequently been described as binding to single-stranded DNA or RNA in a sequence-independent manner. The results of the present report are fully consistent with this earlier work: while NC exhibits profound sequence preferences, it can undoubtedly bind with various degrees of affinity to virtually any single-stranded nucleic acid sequence.
As in the present work, some recent studies have also shown that NC binds to some sequences with greater affinities than others. In general, these experiments used much larger nucleic acids than the oligonucleotides used here, and the sequence preferences in some cases were demonstrated by competition rather than direct measurements of affinities (7, 8, 11, 12, 40, 45). It seems important that the experiments we performed used very short oligonucleotides, so that binding to specific sequences could be analyzed in the virtual absence of secondary or tertiary structure. It should be noted that some of these prior studies (7, 12, 40, 45) found a role for the zinc fingers in specific binding, in agreement with the present work.
Another important aspect of the experiments described here is that the analyses were performed in 0.15 M NaCl. The sequence-independent, electrostatic interaction between the positively charged NC protein and a nucleic acid molecule is obviously much weaker at this moderate ionic strength than in more dilute buffers, so that the differences in binding to different oligonucleotides are more apparent. Indeed, the analysis of the salt resistance of binding of NC to a series of oligonucleotides (Table 2) showed that the additional affinity of NC for a preferred sequence is largely hydrophobic.
Finally, two recent studies (4, 5) have used selection and amplification techniques to isolate RNA molecules for which NC has a very high affinity. Remarkably, the RNAs isolated in these two studies show no obvious resemblance to each other. However, the preferred sequence isolated by Berglund et al. contains runs of U and G and thus appears similar to the alternating sequence of T and G characterized as a preferred site here. It should be noted that we only tested a very small number of sequences, and other sequences may well bind NC even more stably than those found in this study. Selection experiments to detect such sequences among short oligodeoxynucleotides are now under way.
What is the biological significance of the sequence preferences exhibited by NC? There appear to be two types of possibilities. It is now clear that NC has at least one important function as a domain of the Gag polyprotein, viz., in RNA packaging during virus assembly (6). Clearly, this function involves a highly specific interaction with a nucleic acid molecule, i.e., genomic RNA. Thus, one possible explanation for the sequence specificity of NC is that it is simply a reflection, a “remnant,” of the specificity exhibited by the NC domain of Gag. (However, the specificity detected here for a simple, five-base alternating sequence seems insufficient to account for the exquisite selectivity of RNA encapsidation during virus assembly.)
Alternatively, the sequence preferences of NC may be important for the functions that the protein performs after it is cleaved from the Gag polyprotein. These functions are not yet understood. NC is complexed with the genomic RNA in the ribonucleoprotein core of the mature retrovirus particle, and in this structural role it may protect the RNA from nucleases or help to condense it into a small volume in the interior of the particle. It seems likely that, at the high RNA and protein concentrations in the viral core, NC is bound to the entire genomic RNA, regardless of sequence (21, 35, 41).
However, NC also has activity as a nucleic acid chaperone (reviewed in reference 30). That is, it lowers the energy barrier for breakage and reformation of base pairs in nucleic acids, enabling it to catalyze the rearrangement of a nucleic acid molecule into the structure with the maximum number of base pairs. This activity is known to be at work during virus maturation, when the dimeric genomic RNA undergoes a stabilization event (19, 22, 23, 42). In addition, recent in vitro studies strongly suggest that the chaperone activity is crucial during reverse transcription of the genomic RNA in the newly infected cell (3, 13, 28, 46). It seems possible that the binding of NC to preferred sequences is related to its functions as a nucleic acid chaperone. For example, the sequence preferences might serve to localize the protein at sites where its chaperone activity is required; conversely, the chaperone activity might result in the exposure of preferred binding sites, e.g., during reverse transcription.
It is interesting to note that alternating (UG)n sequences are found at several sites in HIV-1 genomic RNA; one of these is at the extreme 3′ end of U5 (most of the sequence of the 28-base oligodeoxynucleotide used in our initial experiments is from this portion of the genome). Another is in a more 5′ position in U5 (nucleotides 556 to 562 in the sequence of the NL4-3 isolate of HIV-1 [1]). The function of this highly conserved sequence is not known, but it must be crucial for the virus, since subtle changes lead to a profound diminution in replicative capacity (47). Similar sequences are also found in stem-loop 3, a region in the leader which appears to be important for encapsidation of the genome (34). The sequence selected by Berglund et al. (5) for highest-affinity NC binding resembles this sequence.
In summary, the binding of NC to nucleic acids exhibits profound sequence preferences. This sequence-specific interaction is remarkably complex at the biochemical level, and its biological significance is unknown at present. Ongoing work is directed at elucidating the questions remaining in both of these arenas. It seems possible that the stable, sequence-specific binding described here can be exploited in the development of new approaches to both detection and growth inhibition of HIV-1.
ACKNOWLEDGMENTS
We thank Sharon Bladen, Demetria Harvin, Donald Johnson, and Bradley Kane for technical assistance; Cheri Rhoderick for help with manuscript preparation; and Judith G. Levin for a thoughtful reading of the manuscript.
This research was sponsored in part by the National Cancer Institute, DHHS, under contract with ABL.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldovini A, Young R A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen P, Collins B, Brown D, Hostomsky Z, Gold L. A specific RNA structural motif mediates high affinity binding by the HIV-1 nucleocapsid protein (NCp7) Virology. 1996;225:306–315. doi: 10.1006/viro.1996.0605. [DOI] [PubMed] [Google Scholar]
- 5.Berglund J A, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busch L K. Production of HIV-1 (MN) nucleocapsid protein (p7) by recombinant DNA technology. M.S. thesis. Frederick, Md: Hood College; 1994. [Google Scholar]
- 11.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darlix J L, Lapadat-Tapolsky M, de Rocquigny H, Roques B P. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 14.Davis J, Scherer M, Tsai W P, Long C. Low-molecular weight Rauscher leukemia virus protein with preferential binding for single-stranded RNA and DNA. J Virol. 1976;18:709–718. doi: 10.1128/jvi.18.2.709-718.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M, Roques B, Darlix J-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupraz P, Oertle S, Meric C, Damay P, Spahr P F. Point mutations in the proximal Cys-His box of Rous sarcoma virus nucleocapsid protein. J Virol. 1990;64:4978–4987. doi: 10.1128/jvi.64.10.4978-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupraz P, Spahr P F. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J Virol. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher R J, Fivash M, Casas-Finet J, Erickson J W, Kondoh A, Bladen S V, Fisher C, Watson D K, Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994;3:257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleissner E, Tress E. Isolation of a ribonucleoprotein structure from oncornaviruses. J Virol. 1973;12:1612–1615. doi: 10.1128/jvi.12.6.1612-1615.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Chabot D J, Rein A, Henderson L E, Arthur L O. The two zinc fingers in the human immunodeficiency virus type 1 nucleocapsid protein are not functionally equivalent. J Virol. 1993;67:3207–3211. doi: 10.1128/jvi.67.7.4027-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorelick R J, Henderson L E, Hanser J P, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorelick R J, Nigida S M, Jr, Bess J W, Jr, Arthur L O, Henderson L E, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Henderson L E, Bess J, Kane B, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein promotes efficient strand transfer and specific viral DNA synthesis by inhibiting TAR-dependent self-priming from minus-strand strong-stop DNA. J Virol. 1997;71:5178–5188. doi: 10.1128/jvi.71.7.5178-5188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 31.Housset V, de Rocquigny H, Roques B P, Darlix J L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karpel R L, Henderson L E, Oroszlan S. Interaction of retroviral structural proteins with single-stranded nucleic acids. J Biol Chem. 1987;262:4961–4967. [PubMed] [Google Scholar]
- 33.Khan R, Giedroc D P. Nucleic acid binding properties of recombinant Zn2 HIV-1 nucleocapsid protein are modulated by COOH-terminal processing. J Biol Chem. 1994;269:22538–22546. [PubMed] [Google Scholar]
- 34.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2080. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meric C, Darlix J L, Spahr P F. It is Rous sarcoma virus protein P12 and not P19 that binds tightly to Rous sarcoma virus RNA. J Mol Biol. 1984;173:531–538. doi: 10.1016/0022-2836(84)90396-6. [DOI] [PubMed] [Google Scholar]
- 36.Meric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meric C, Spahr P F. Rous sarcoma virus nucleic acid-binding protein p12 is necessary for viral 70S RNA dimer formation and packaging. J Virol. 1986;60:450–459. doi: 10.1128/jvi.60.2.450-459.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, Ivanoff L, Petteway S R, Pearson M L, Lautenberger J A, Papas T S, Ghrayeb J, Chang N, Gallo R C, Wong-Staal F. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 39.Rein A, Harvin D P, Mirro J, Ernst S M, Gorelick R J. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J Virol. 1994;68:6124–6129. doi: 10.1128/jvi.68.9.6124-6129.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakaguchi K, Zambrano N, Baldwin E T, Shapiro B A, Erickson J W, Omichinski J G, Clore G M, Gronenborn A M, Appella E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc Natl Acad Sci USA. 1993;90:5219–5223. doi: 10.1073/pnas.90.11.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart L, Schatz G, Vogt V M. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5092. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoltzfus C M, Snyder P N. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975;16:1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sykora K W, Moelling K. Properties of the avian viral protein p12. J Gen Virol. 1981;55:379–391. doi: 10.1099/0022-1317-55-2-379. [DOI] [PubMed] [Google Scholar]
- 44.Tsuchihashi Z, Brown P O. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tummino P J, Scholten J D, Harvey P J, Holler T P, Maloney L, Gogliotti R, Domagala J, Hupe D. The in vitro ejection of zinc from human immunodeficiency virus (HIV) type 1 nucleocapsid protein by disulfide benzamides with cellular anti-HIV activity. Proc Natl Acad Sci USA. 1996;93:969–973. doi: 10.1073/pnas.93.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W, Henderson L E, Copeland T D, Gorelick R J, Bosche W J, Rein A, Levin J G. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada O, Kraus G, Sargueil B, Yu Q, Burke J M, Wong-Staal F. Conservation of a hairpin ribozyme sequence in HIV-1 is required for efficient viral replication. Virology. 1996;220:361–366. doi: 10.1006/viro.1996.0324. [DOI] [PubMed] [Google Scholar]
- 48.You J C, McHenry C S. HIV nucleocapsid protein: expression in Escherichia coli, purification, and characterization. J Biol Chem. 1993;268:16519–16527. [PubMed] [Google Scholar]
- 49.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]