Abstract
To identify promoter regions that impart differential temporal regulation of channel catfish virus (CCV) genes, the transcriptional kinetics of an immediate-early gene and prospective early and late genes were characterized. A cDNA clone, designated IE3C, representing a third immediate-early transcript was identified. The 5′ end of the IE3C transcript was mapped to nucleotides 15,368 and 131,043 in the terminal repeat regions of the CCV genome. The full length of the transcript represented by the IE3C clone is 1,412 bp, and it most likely codes for the protein specified by open reading frame (ORF) 12. The putative product of ORF12 contains a consensus RING finger metal binding motif (C3HC4 structure). Temporal expression studies, in conjunction with protein synthesis and DNA replication inhibition, demonstrated that the IE3C transcript belongs to an immediate-early kinetic class, the ORF5 transcript is a member of the early kinetic class, and ORF39 and ORF46 are true late-kinetic-class genes. Additionally, we demonstrated that ORF38 transcription overlaps ORF39 and the products presumably share the same poly(A) signal. The 5′ ends of the transcripts encoding ORF38, ORF39, and ORF46 were mapped to nucleotides 44,862, 45,254, and 59,644, respectively, and potential transcriptional control elements were located.
Channel catfish virus (CCV), or ictalurid herpesvirus 1, is a cytopathic herpesvirus that causes a severe hemorrhagic disease in young channel catfish, Ictalurus punctatus (58). CCV is the most intensely studied herpesvirus of lower vertebrates. The entire genome has been sequenced, and it has been predicted to contain 79 genes, 14 of which are located in the terminal-repeat regions (10). The genomic structure of CCV is much different from those of identified herpesviruses of mammalian or avian species (10).
Herpesvirus gene expression is coordinately regulated and sequentially ordered such that the genes can be classified into three broad temporal groups, immediate-early (IE), early, and late genes. IE gene expression initiates the viral lytic cascade (28, 29). The induction of early and late viral genes depends on viral regulatory proteins encoded by IE genes that act in trans. The IE genes are expressed first and are defined as capable of being transcribed in the absence of de novo viral protein synthesis (28, 29).
The initiation of early and late-gene expression, unlike that of the IE genes, is not homogeneously defined. The early gene products function in nucleotide metabolism and viral DNA synthesis, and some downregulate the expression of IE genes (7, 37, 38, 42, 59). The late genes, expressed last and requiring prior early gene expression, generally encode the virion structural proteins. They either may be expressed in the absence of viral DNA synthesis (delayed early class) or may stringently require viral DNA synthesis (true late class) (31, 32, 36).
Kinetic analysis of the synthesis of CCV polypeptides revealed three distinct groups of proteins differing in their time of appearance and magnitude of synthesis. These results suggested that CCV protein synthesis is coordinately regulated (13). Recently, two IE transcripts were characterized and reported as representing open reading frame 8a (ORF8a) and ORF9 of the terminal-repeat regions of the CCV genome (53). However, the structure, function, and temporal expression class of most CCV-encoded proteins have yet to be determined. Because CCV is evolutionarily distant from mammalian herpesviruses, its conserved regulatory mechanism might provide insight into the factors influencing herpesvirus evolution. Exploring the differential temporal gene regulation of CCV is the first step in dissecting the virus-associated regulatory mechanisms controlling CCV infection. In this study, representative transcripts of the IE, early, and late genes of CCV were identified, their temporal regulation was characterized, and promoter regions were predicted from the CCV genome sequence. The results confirm that CCV gene expression is sequentially ordered and regulated in a manner similar to that of mammalian herpesviruses. We also identified a third IE gene.
MATERIALS AND METHODS
Cells and viruses.
The CCV strain used in this experiment was Auburn clone A (American Type Culture Collection). Channel catfish ovary (CCO) cells and their thymidine kinase (TK)-negative counterpart (CCOBr cells) were cultured as described previously (23). When a specific chemical inhibitor of transcription or DNA replication was used, the chemical was added to the cell culture medium and the cells were incubated with the chemical for 1 h before infection, as well as during infection and for a specified time postinfection.
The replication of CCV DNA in CCOBr cells over time or in the presence of acyclovir (ACV) (Sigma Chemical Co., St. Louis, Mo.) or phosphonoacetic acid (PAA) (Sigma Chemical Co.) was analyzed by the trichloroacetic acid (TCA) precipitation assay as described previously (49) and by slot blot DNA-DNA hybridization. For the TCA precipitation assay, the cells were cultured in medium containing 10 μCi of [methyl-3H]thymidine per ml (48 Ci/mmol) and infected with 10 PFU of CCV per cell. Three replicate cell samples were harvested and lysed at serial time points. Viral DNA was precipitated with 10% TCA on G6 glass fiber filters (Fisher Scientific, Pittsburgh, Pa.). The slot blot assays were performed by infecting monolayered CCO cells in 24-well plates with 3 PFU of CCV per cell in 150 μl of medium per well for 30 min, aspirating off the inoculum, and overlayering with 1 ml of fresh medium per well. At the appropriate time, the medium was aspirated off, the cells were lysed, and DNA was purified by using a Puregene kit (Gentra Systems Inc., Minneapolis, Minn.). One-half of the DNA purified from each cell culture well was transferred to a positively charged nylon membrane (Zeta Probe GT; Bio-Rad Laboratories, Hercules, Calif.) slot in a slot blot (1). CCV DNA was detected by using a nonradioisotopic DNA-DNA hybridization kit (ECL Direct Nucleic Acid Labelling Systems; Amersham International, Buckinghamshire, England) with EcoRI-digested cosmid pHCCV 386 (22) as a probe and Hyperfilm-ECL (Amersham International).
Isolation of RNA.
The CCO cells were infected with 10 PFU of CCV per cell in the presence or absence of 100 μg of cycloheximide per ml for 8 h. The total RNA was isolated by the guanidinium thiocyanate method (8). The mRNA was purified from total RNA by using a PolyATtract mRNA isolation system (Promega, Madison, Wis.).
Construction of an IE-enriched cDNA library.
A monolayer of CCO cells was treated with 50 μg of cycloheximide (Sigma Chemical Co.) per ml, infected with 10 PFU of CCV per cell, and incubated at 30°C for 6 h. The cells were then lysed, and mRNA was obtained by guanidinium thiocyanate lysis and oligo(dT) hybridization-mediated magnetic separation (PolyATtract System 1000; Promega). The cDNA was produced from poly(A) RNA by using Moloney murine leukemia virus reverse transcriptase (Promega) and oligo(dT) primer. The cDNA library was cloned into the specialized bacteriophage λ vector λ Zap II, using a kit (λ Zap Gold) as described by the manufacturer (Stratagene, La Jolla, Calif.). CCV-specific cDNA clones were identified by using nonradioisotopically labeled purified CCV DNA in plaque hybridization analyses with the ECL kit (Amersham Corporation, Arlington Heights, Ill.). Positive plaques were excised and bacteriophage was eluted. Then pBluescript SK portions of these clones were rescued by using ExAssist helper phage in the SOLR strain of Escherichia coli (Stratagene).
Nested DNA deletion and sequencing.
Nested deletion subclones of the IE3C cDNA were generated by using an exonuclease III-mung bean nuclease deletion kit (Stratagene) according to the manufacturer’s instructions after digestion with SacI and EcoRI, generating a unique 3′-overhang restriction site and a unique 5′ restriction site between the insert and the 3′ site chosen. These fragments were sequenced by using a Sequenase version 2.0 DNA sequencing kit (United States Biochemical, Cleveland, Ohio) with 1,000 Ci of [α-35S]dATP (Amersham Co.) per mmol and T3 or T7 primers according to manufacturer’s instructions. The sequence data were connected using Contig Manager of the DNASIS version 3.0 program (Hitachi Software Engineering America, Ltd., San Bruno, Calif.).
Identification of 5′ ends of transcripts.
The 5′ ends of IE and late transcripts were obtained by the 5′ rapid amplification of cDNA ends (RACE) method, using the 5′-AmpliFINDER RACE kit (Clontech Laboratories, Inc., Palo Alto, Calif.). For evaluation of the full-length IE3C transcript, first-strand cDNA was synthesized from 6 μg of total RNA isolated from cycloheximide-restricted CCV-infected CCO cells, using Moloney murine leukemia virus reverse transcriptase (Promega) and the P1 primer (for the primer sequence, see Table 1). The amplification step used the anchor primer (Clontech) and the P2 primer (Fig. 1B).
TABLE 1.
Sequences of primers used for cloning procedures
| Primer | Sequence |
|---|---|
| Anchor | 5′-CTGGTTCGGCCCACCTCTGAAGGTTCCAGAATCGATAG-3′ |
| P1 | 5′-CGGCGAGGATGGACACCA-3′ |
| P2 | 5′-CGGTGAGTGCGGGGAAGG-3′ |
| P3 | 5′-GTCATACTCCATCGGAAT-3′ |
| P4 | 5′-TCGTAGGTGGTGGTGGTC-3′ |
| P5 | 5′-GGTGATGGAGAGGGTGTT-3′ |
| P6 | 5′-TGAATCGATCCCACCCACCGTCTGTAG-3′ |
| CCV45342(+) | 5′-TAACGCACTCGCCAACAT-3′ |
| CCV45723(−) | 5′-TGAATCGATCCCACCCACCGTCTGTAG-3′ |
FIG. 1.
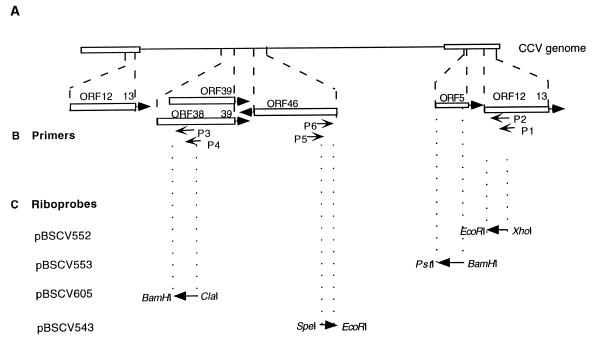
Diagram of the CCV genome demonstrating the locations and orientations of ORF5, ORF12, ORF39, ORF38-ORF39, and ORF46 and the primers and riboprobes used in this study. (A) The CCV genome with the unique region (solid line) and direct-repeat regions (open boxes), and expanded depictions of the regions covered by ORF5, ORF12, ORF38-ORF39, and ORF46, with arrows indicating the direction of transcription. (B) The primers used for cloning and 5′ RACE. (C) Riboprobes used for RNase protection assays and Northern blot analysis. The restriction sites used in the cloning procedures are indicated.
The 5′ transcription start sites of ORF39 and ORF46 were mapped similarly, using an avian myeloblastosis virus reverse transcriptase (Clontech)-generated cDNA template produced from 1 μg of mRNA from CCV-infected CCO cells. P4 and P5 were used for ORF39 and ORF46 cDNA synthesis, respectively. The custom primers P4 and P5 were used in the amplification step for ORF39 and ORF46, respectively (Fig. 1B).
Construction of plasmids.
Plasmid pBSCV552, containing the EcoRI-to-XhoI fragment of the IE3C cDNA, plasmid pBSCV553, containing the BamHI-to-PstI fragment of the CCV TK gene, and plasmid pBSCV543, containing the EcoRI-to-SpeI fragment of ORF46, were produced by cloning the respective fragments into the multiple cloning site of plasmid pBluescript SK− (Stratagene) (Fig. 1C). Plasmid pBSCV605 was constructed by insertion of the 363 bp of PCR-amplified ORF39 fragment, using primers CCV45342(+) and CCV45723(−) (Table 1), into SpeI and EcoRI sites of pBluescript SK−.
RNase protection assays and Northern blot analysis.
The RNase protection assays were performed on lysates of CCOBr cell monolayers grown in 25-cm2 flasks, exposed to 10 PFU of CCV per cell, and harvested at serial time points, using a Lysate Ribonuclease Protection Assay Kit in accordance with the manufacturer’s directions (Ambion, Austin, Tex.) and 0.23 μCi of probe. After hybridization at 37°C overnight and RNase treatment, the protected fragments were electrophoresed on a 5% polyacrylamide gel containing 7% urea. The gel was autoradiographed with X-Omat film (Kodak, Rochester, N.Y.).
The riboprobes antisense to ORF12 (IE3C), ORF5 (TK), ORF39, and ORF46 were derived from vectors described above. Riboprobes were generated and labeled with [α-32P]UTP, using linearized plasmid DNA, and transcribed with either T3 or T7 RNA polymerase. Transcription of pBSCV552, pBSCV553, pBSCV605, and pBSCV543 generated riboprobes with lengths of 495, 624, 426, and 229 nucleotides (nt) for ORF12, ORF5, ORF39, and ORF46, respectively (Fig. 1C).
Northern blot analysis was performed as previously described (49). Following the electrophoresis, the mRNA was transferred to Zeta-probe GT blotting membranes (Bio-Rad Laboratories). The ORF39 and IE3C riboprobes described above were used. Hybridization was done at 70°C overnight in hybridization solution (0.25 M Na2HPO4 [pH 7.2], 7% sodium dodecyl sulfate [SDS]). The membrane was washed twice in 20 mM Na2HPO4 (pH 7.2) containing 5% SDS and then twice in 20 mM Na2HPO4 (pH 7.2) containing 1% SDS at 65°C. The sizes of Northern blot-detected transcripts were determined by comparing them to the RNA markers (Promega) that were run on the same gel.
RESULTS
Identification of a CCV IE gene.
To identify and clone CCV IE genes, a cDNA library was constructed from cycloheximide-restricted CCV-infected CCO cells. The cDNA library consisted of 4.2 × 105 PFU of recombinant bacteriophage λ, which was amplified to 3 × 108 PFU. Aliquots of 5.0 × 104 PFU were screened for clones containing CCV sequences by plaque hybridization with purified CCV DNA as a probe. The internal pBluescript plasmid of each of three isolated clones was rescued. Sequencing and comparison to the CCV genomic sequence localized the 5′-3′ coding strand sequence represented by one clone, IE3C, to regions 15,701 to 16,778 and 131,376 to 132,453 of the CCV genome. These regions span a portion of ORF12 through ORF13 of terminal-repeat portions of the CCV genome (10) (Fig. 1A).
Characterization of differential temporal transcripts.
To evaluate differential gene regulation, the IE3C clone was chosen as a representative IE transcript. The TK gene, ORF5 (10, 22), was chosen as a potential early gene. A putative glycoprotein gene, ORF46 (10), and ORF39, the major capsid protein gene (11), were chosen as potential late genes. The transcription of representative genes with predicted IE, early, and late regulation was characterized by RNase protection assays and Northern blot analysis. The assays were performed in conjunction with [3H]thymidine incorporation and slot blot DNA-DNA hybridization analysis of CCV DNA replication as a temporal reference to distinguish early and late expression.
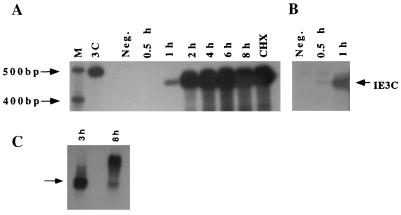
To characterize the production of the IE3C transcript, sequential extracts of CCV-infected cells obtained with or without cycloheximide were analyzed by RNase protection assays (Fig. 2). The IE3C riboprobe was protected by transcripts from lysates obtained as early as 0.5 h postinfection (p.i.) (Fig. 2B, lane 0.5 h). The concentration of transcripts from this region peaked at 2.0 h p.i. and remained persistently high through 8 h p.i. (Fig. 2A, lanes 1 h through 2 h). The IE3C transcripts were produced at high levels with cycloheximide inhibition (Fig. 2A, lane CHX).
FIG. 2.
RNase protection and Northern blot analyses indicating the kinetics of IE3C transcription. (A) Autoradiogram from an RNase protection assay of cell lysates collected from uninfected cells (Neg.), from cycloheximide treated infected cells (CHX), and from infected cells at 0.5, 1, 2, 3, 4, 6, and 8 h p.i. (lanes 0.5 h through 8 h). Bands represent the 453-nt antisense IE3C-protected RNA fragment (2-day film exposure) (see Fig. 1). Lane M, molecular size markers; lane 3C, undigested probe. (B) IE3C detection at 0.5 and 1 h p.i. (7-day film exposure). (C) Northern blotting of poly(A) RNA from CCV-infected cells harvested at 3 and 8 h p.i., using the IE3C antisense riboprobe. The 1,400-nt band corresponding to the IE3C transcript is indicated by an arrow. The predominant banding at 8 h p.i. ranges from 2,000 to 3,500 nt.
Because RNase protection assays cannot differentiate overlying transcripts, a Northern blot was performed on mRNA from CCV-infected cell lysates at 3 and 8 h p.i., using the IE3C riboprobe indicated in Fig. 1C. High-level expression of a 1,400-nt transcript predominated at 3 h p.i., and the expression of larger transcripts predominated by 8 h p.i. (Fig. 2C, lanes 3 h and 8 h, respectively). The results of the RNase protection assays indicate that the high level of transcription in the IE3C region at 8 h p.i. was apparently due to overlapping transcripts. These results demonstrate differential transcription of the CCV IE3C gene region as the infection progresses. Initial IE promoter expression is apparently reduced and production of early and/or late transcripts overlapping the IE3C ORF occurs during later phases of the infection.
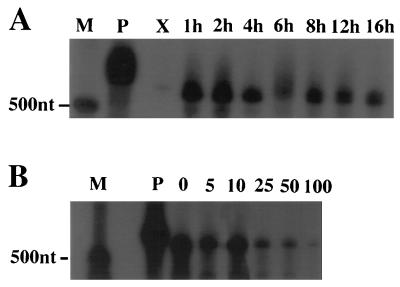
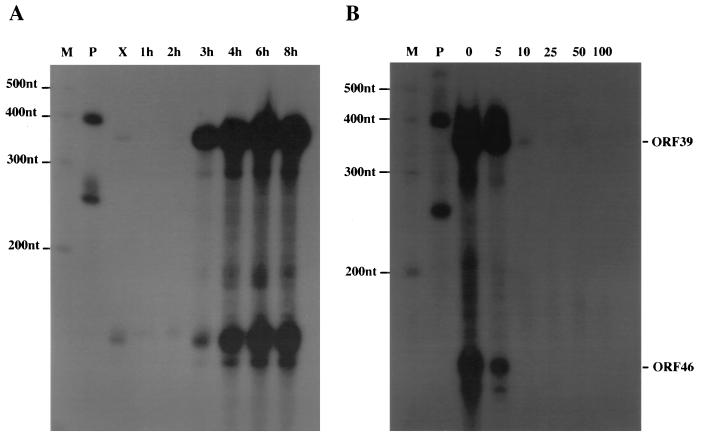
To determine the kinetic class of ORF5, ORF39, and ORF46 transcripts, RNase protection assays were performed on CCV-infected cell lysates harvested from 0.5 to 16 h p.i. The ORF5 (TK) transcript was first detected at 1 h p.i. The concentration of transcripts from this region was highest at 1 and 2 h p.i. and then decreased (Fig. 3A, lanes 1h, 2h, and 4h through 16h). Transcription of ORF5 was inhibited by cycloheximide (Fig. 3A, lane X). ORF39 and ORF46 transcripts were first detected at 3 h p.i., and the level of these transcripts continued to accumulate through 8 h p.i. (Fig. 4A, lanes 3h through 8h). The time of viral DNA replication was determined by analyzing [3H]dT incorporation in CCV-infected CCOBr cells, using lysates collected from 0.5 to 16 h p.i. Viral DNA replication was first detected at 2 h p.i. (230 ± 12.7 cpm) and peaked at 6 h p.i. (10,500 ± 120 cpm) compared to the background (89 ± 8.8 cpm). The TCA precipitation assay results were confirmed by DNA-DNA hybridization on slot blots of DNA purified from CCV-infected CCO cells harvested at 0, 1, 2, 3, 4, and 8 h p.i. The first detected signal above the 0-h-p.i. sample was at 3 h p.i.
FIG. 3.
RNase protection assays indicating the kinetics of TK gene transcription and the effect of cycloheximide and ACV on transcription. (A) RNase protection of ORF5 antisense riboprobe (see Fig. 1) when hybridized to infected-cell lysates harvested at various times from 1 to 16 h p.i. (lanes 1h to 16h) or when cultured in the presence of 100 μg of cycloheximide per ml for 8 h (X). (B) Cell lysate RNase protection of the same riboprobe in CCV-infected cell cultures incubated with incremental concentrations of ACV from 0 to 100 μM (lanes 0 through 100). Lane M, molecular size markers; lane P, undigested probe.
FIG. 4.
RNase protection assays indicating the kinetics of synthesis of the ORF39 and ORF46 transcripts. Cell lysates were collected from cycloheximide- or ACV-treated infected cells and from infected cells at various times p.i. (A) Riboprobes designed to detect ORF39 (363-nt protected fragment) and ORF46 (168-nt protected fragment) were hybridized with RNA in lysates of cells treated with 100 μM cycloheximide (X) and infected cells harvested at various times from 1 to 8 h p.i. (lanes 1h to 8h). (B) Riboprobes designed to detect ORF39 and ORF46 (see Fig. 1) were hybridized with infected-cell lysates, collected at 8 h p.i., from cultures incubated with ACV at concentrations from 0 to 100 μM (lanes 0 through 100). P, undigested probe; M, molecular size markers.
To distinguish true late gene expression from delayed early expression, viral DNA synthesis must be inhibited. Therefore, the effect of ACV and PAA on the synthesis of CCV DNA in CCOBr cells was examined by performing TCA precipitation assays. The amount of 3H-labeled DNA from 10 μM ACV-treated infected cells was reduced almost 90% at 12 h p.i. (mean ± standard deviation, 7,730 ± 746 cpm in the 10 μM ACV-treated infected cells versus 66,600 ± 6,210 cpm in untreated infected cells). The amount of 3H-labeled DNA was reduced only about 45% in the presence of 300 μg of PAA per ml (22,100 ± 6,100 cpm versus 48,900 ± 2,440 cpm in untreated infected cells at 8 h p.i.). The results indicate that 10 μM ACV inhibited CCV DNA synthesis. DNA-DNA hybridization on slot blots of DNA purified from CCV-infected CCO cells in 0, 5, and 10 μM ACV harvested at 8 h p.i. demonstrated low-level (in 5 μM ACV) or no (in 10 μM ACV) detectable CCV DNA replication compared to 0-h-p.i. controls.
When 10 μM ACV was used, production of the putative late transcripts in CCV-infected cells was inhibited (Fig. 4B, lane 10) while ORF5 transcripts continued to be produced (Fig. 3B, lane 10). Expression of the ORF5 transcript required de novo viral protein synthesis and was independent of CCV DNA replication. Therefore, the TK gene represented an early class of CCV gene. TK RNA synthesis was probably inhibited at 100 μM ACV because of detrimental effects on the host cell at these high concentrations. In comparison, the expression of ORF39 and ORF46 transcripts exhibited a stringent requirement for DNA replication, defining these genes as members of the true late class of CCV genes.
5′-End mapping of IE3C, ORF39, and ORF46 transcripts.
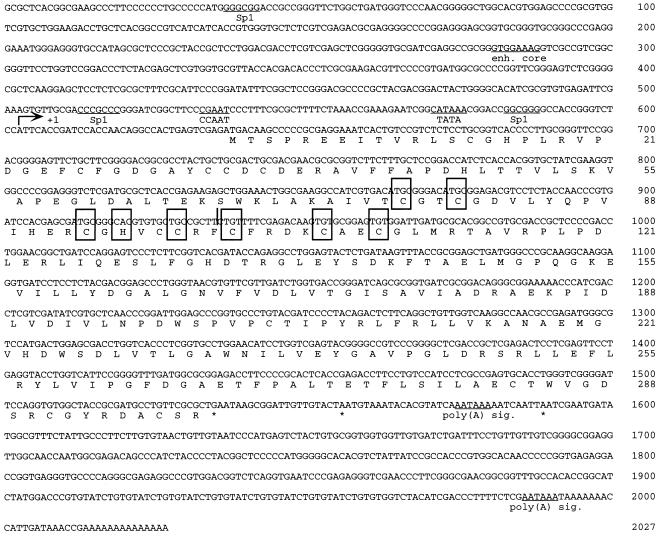
The 5′ terminus of the IE3C transcript was determined by the 5′ RACE method, using antisense nested primers P1 and P2 (Fig. 1B). The 3′ nucleotide of P2 corresponded to nt 529 of the IE3C cDNA. A single 854-bp PCR fragment which contained 327 bp upstream of the IE3C cDNA was amplified (Fig. 5). This placed the transcriptional start site of IE3C at positions 15,368 and 131,043 of the CCV genome within the terminal-repeat ends. The results of IE3C cDNA sequencing indicated that the full-length transcript is 1,412 nt long. It is unspliced and contains ORF12 and ORF13 (10). The putative start codon of ORF12 was located at nt +35 of the transcript. Sequence analysis of the region upstream of this transcriptional start site revealed one TATA-like sequence at nt −32, one core consensus sequence of enhancer at nt −321, and two Sp1 binding sites around the promoter (Fig. 6). The predicted protein encoded by the IE3C gene is 299 amino acids long and contains a RING finger (C3HC4) metal binding motif near the amino terminus (4, 20) (Fig. 6).
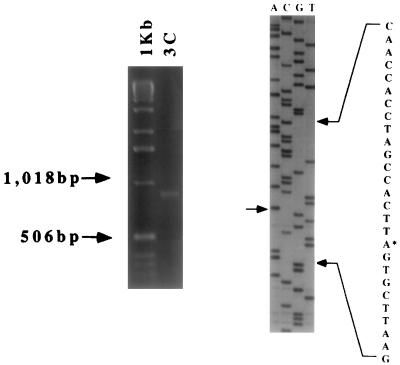
FIG. 5.
A 1.2% agarose electrophoretic gel of the product of 5′ RACE of IE3C transcripts, using anchor and P2 primers (left), and a sequencing gel of the cloned RACE product (right). The arrow and asterisk indicate the transcriptional start site. 1Kb, 1-kb DNA ladder (molecular size marker); 3C, 5′ RACE product of IE3C transcripts.
FIG. 6.
Sequence analysis of the IE3C transcript and its upstream promoter element. The TATA box, CCAT box, Sp1 element, putative HSV enhancer core (enh.), and poly(A) signals (sig.) are indicated. The RING finger (C3HC4) metal binding motif is indicated by the boxes (4, 20). The stop codon is indicated by an asterisk. The 5′ limit of the cDNA clone IE3C is indicated by a vertical line at nt 936. The arrow indicates the transcription start site.
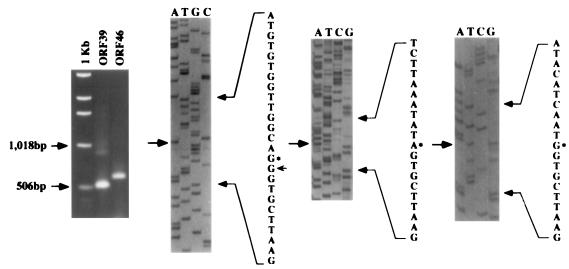
The 5′ end of the ORF46 transcript, mapped with nested primers P5 and P6 (Fig. 1B), generated a single 611-bp PCR fragment (Fig. 7). The 5′ end of the ORF46 transcript corresponded to nt 59,644 of the CCV genome, which is 139 nt upstream of the putative ORF46 start codon.
FIG. 7.
A 1.2% agarose electrophoretic gel of the product of 5′ RACE of ORF39 and ORF46 (left) and sequencing gels of the cloned RACE products, ORF38, ORF39, and ORF46 (from left to right). Arrows on gels and asterisks in sequences indicate the transcriptional start sites. The arrow on the ORF38 sequence indicates an extra guanosine. 1Kb, 1-kb DNA ladder (molecular size marker).
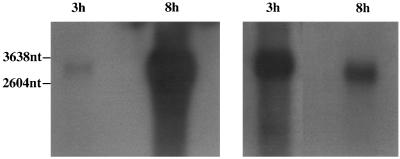
The 5′ end of the ORF39 transcript, mapped with nested primers P3 and P4 (Fig. 1B), generated 530- and 923-bp fragments (Fig. 7). Sequence analysis placed the 5′ end of the 530-bp fragment at nt 45,254 of the CCV genome, which is 63 bp upstream of the putative ORF39 start codon. The 5′ end of the 923-bp fragment was identified at nt 44,862 of the CCV genome, which is 40 nt upstream of the putative ORF38 start codon. The extra guanosine between the 5′ RACE anchor sequence and the CCV sequence (Fig. 7) is assumed to represent the methylated guanosine cap of the mRNA. The extra guanosine has been demonstrated in 5′ RACE on capped mRNA (2). Northern blot analysis of this region, using the ORF39 riboprobe on mRNA from CCV-infected cell lysates at 3 and 8 h p.i., demonstrated low-level expression of a slightly longer transcript at 3 h p.i. compared with that at 8 h p.i. (Fig. 8). This supported the results of the 5′ RACE indicating that the transcription of ORF38 overlaps ORF39.
FIG. 8.
Northern blotting of poly(A) RNA from CCV-infected cells harvested at 3 and 8 h p.i., using the ORF39 riboprobe. (Left panel) Autoradiograph demonstrating expression levels at 3 and 8 h p.i. X-ray film was exposed for 2 h. (Right panel) Use of different film exposure times to demonstrate the size shift of the riboprobe-specific mRNA. The 3-h-p.i. portion was autoradiographed for 14 h, and the 8-h-p.i. portion was autoradiographed for 20 min. The positions of molecular size markers are shown on the left.
DISCUSSION
In this study, we demonstrated the presence of a third CCV-encoded IE transcript. The total transcript sequence is 1,412 bp long. The 5′ end of the transcript is located at nt 15,368 and 131,043 in the CCV genome. Two ORFs were predicted to lie in this region: ORF12 and ORF13 (10). The identified transcript likely encodes the ORF12 polypeptide. It is interesting that ORF12 and ORF13 are in the same frame but read-through translation is not likely due to the presence of three stop codons between the two ORFs (Fig. 6). As reported previously (10), both putative polypeptides contain a potential zinc metal binding motif near the amino terminus. The zinc binding motifs were shown to be involved in protein-protein interactions and in binding DNA and RNA (5). Two herpes simplex virus type 1 (HSV-1) IE proteins, ICP0 and ICP27, contain zinc metal binding domains (17, 54, 55). The metal binding domain of the ORF12 product is in the C3HC4 RING finger form similar to ICP0 in HSV-1 and its homologs in other alphaherpesviruses (17, 20, 57). The presence of a putative RING finger metal binding motif in the protein product and the high level of transcription without de novo protein synthesis (cycloheximide inhibition) suggest that ORF12 belongs to the IE family of CCV genes that may be involved in regulating the expression of other virus products.
Many IE proteins of herpesviruses have been shown to be important for the transactivation of viral early genes and the progression of the lytic replication cycle (16, 38, 47). HSV-1 encodes five IE genes: α0, α4, α22, α27, and α47. The α4 gene encodes a major regulatory protein, ICP4, which binds to viral DNA and regulates viral genes both positively and negatively (12, 14, 18, 33, 35, 39–41). The α0 gene encodes a promiscuous transactivator (15, 43, 46). The α22 gene product is associated with viral replication and optimal expression of α0 and late genes (6). The α27 gene encodes a protein which regulates the processing of viral RNA (50). Only the α47 gene encodes a protein which does not have a known regulatory function. It blocks the presentation of viral peptides to major histocompatibility complex class I-restricted CD8+ T lymphocytes by associating with peptide transporters (TAP) in the endoplasmic reticulum (21, 25, 60). It is also well understood that ICP4 is required for the induction of TK expression (12, 44, 45). The HSV TK promoter is often used to evaluate the regulation of eukaryotic gene expression as well as its trans induction by viral regulatory proteins (9). We have attempted to evaluate the effect of the product of ORF12 on the expression of a lacZ reporter gene under the control of the CCV TK promoter, using the plasmid pBSCV464 (61) and a construct containing ORF12 in the plasmid pBK-CMV in cotransfection, transient expression assays; however, the assays were hampered by the poor transfection efficiency in CCO cells. Cotransfection, transient expression assays were also performed with the COS cell line. The results indicated a decrease in lacZ expression in the presence of the ORF12 product, but because of the unnatural mammalian cell system used, the results were equivocal. Considerable additional research on this gene and its potential regulatory protein product is needed to determine their roles in the progression of CCV gene expression.
Identification of the transcriptional start sites for ORF12, ORF38–39, ORF39, and ORF46 has allowed comparison of upstream promoter sequences of IE and late genes, as well as of well-characterized HSV IE- and late-promoter regulatory regions. The putative IE3C promoter includes a possible TATA box (CATAAA), a likely CCAAT box (CGAAT), and two Sp1 elements surrounding the TATA box. A core consensus transcriptional enhancer sequence, 5′-GTGGAAAG-3′, was found within the nt −321 to −315 region of the IE3C mRNA (Fig. 9). The core sequence is within the enhancer domains of a number of viruses, including HSV IE gene promoter regions (34, 56). The AT-rich homologs and GC-rich enhancer-like elements present in the IE3C promoter region may be critical for transcription of the IE3C (α) gene.
FIG. 9.
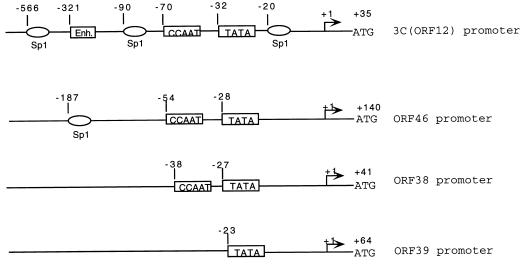
Comparison of CCV IE3C, ORF46, ORF38, and ORF39 promoter regions. Arrows indicate the positions of transcription start sites. The scale, in nt upstream of the mRNA start sites, is indicated at the top. The TATA box, CCAAT box, Sp1 element, and putative HSV enhancer core (Enh.) are indicated.
Inspection of the region upstream of the ORF46 transcriptional start site revealed that an Sp1 element and a CCAAT box were located upstream of a TATA-like sequence (TATTAA) (Fig. 9). It is unusual for a true late gene to contain two recognized upstream transcription-regulatory sequences. The well-characterized true late HSV-1 promoters contain only one TATA-like sequence with no other recognizable upstream cis-acting regulatory elements (3, 19, 26, 27, 31, 52). The structure of the ORF46 promoter is similar to that of the well-characterized HSV early promoters of UL23 (TK), UL9, UL8, and UL29 (ICP8) (3).
The ORF38 promoter region is similar to ORF46 in that it has a CCAAT-like sequence upstream of the respective TATA-like sequence, TATTAA (Fig. 9). The structure of the ORF38 promoter is closer to that of herpesvirus early promoters, which contain cis-acting regulatory elements. The early-promoter characteristic of ORF38 is supported by Northern blot analysis data, which showed low-level expression of ORF38 during early gene expression.
The putative ORF39 promoter includes a TATA-like sequence, TAATTT, with no other recognizable upstream cis-acting regulatory elements (Fig. 9), similar to HSV true late-promoter sequences (3, 19, 26, 27, 31, 52). ORF39 encodes the major capsid protein of CCV (11). In this study, we also demonstrated that the expression of the ORF39 transcript stringently requires viral DNA synthesis, suggesting that ORF39 belongs to the true late family of CCV genes.
CCV DNA sequence analysis by Davison (10) indicated that ORF38 and ORF39 share one poly(A) signal, AATAAA, located at nt 48,759 (10). Our results support this. Two transcripts were detected by 5′ RACE using ORF39 primers P3 and P4 (Fig. 1B), and both were detected by Northern blot analysis. Overlapping transcription of the ORF38-ORF39 gene region is likely, due to the absence of poly(A) signals between ORF38 and ORF39.
Temporal transcriptional evaluation and the associated effects of viral protein synthesis and DNA replication inhibition demonstrate that CCV transcriptional regulation is similar to that of mammalian herpesviruses. The identification of a third IE gene indicates that CCV may have a complex, interactive form of IE-mediated gene regulation similar to that of HSV-1 (47), bovine herpesvirus 1 (51), equine herpesvirus 1 (24), and varicella-zoster virus (48) but in contrast to that of pseudorabies virus, which has only one IE gene (30).
ACKNOWLEDGMENTS
This research was primarily supported by the National Research Initiative Competitive Grant Program/USDA (94-37204-0853). Support was also provided by Mississippi Agricultural and Forestry Experiment Station (MAFES) project MISV-0892 and the College of Veterinary Medicine, Mississippi State University.
We thank Mary Rudis and Suzana Marinovic for technical assistance.
Footnotes
Mississippi Agricultural and Forestry Experiment Station publication J-9180.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1996. [Google Scholar]
- 2.Bähring S, Sandig V, Lieber A, Strauss M. Mapping of transcriptional start and capping points by a modified 5′ RACE technique. BioTechniques. 1994;16:807–808. [PubMed] [Google Scholar]
- 3.Baradaran K, Dabrowski C E, Schaffer P A. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC. J Virol. 1994;68:4251–4261. doi: 10.1128/jvi.68.7.4251-4261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow P N, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 5.Berg J M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 6.Carter K L, Roizman B. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J Virol. 1996;70:172–178. doi: 10.1128/jvi.70.1.172-178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Coen D M, Weinheimer S L, McKnight S L. A genetic approach to promoter recognition during trans induction of viral gene expression. Science. 1986;234:53–59. doi: 10.1126/science.3018926. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 11.Davison A J, Davison M D. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology. 1995;206:1035–1043. doi: 10.1006/viro.1995.1026. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon R A F, Farber F E. Channel catfish virus: physicochemical properties of the viral genome and identification of viral polypeptides. Virology. 1980;103:267–278. doi: 10.1016/0042-6822(80)90186-5. [DOI] [PubMed] [Google Scholar]
- 14.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D. A detailed analysis of an HSV-1 early promoter: sequences involved in trans-activation by viral immediate-early gene products are not early-gene specific. Nucleic Acids Res. 1984;12:3037–3056. doi: 10.1093/nar/12.7.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D. The regulation of transcription of viral and cellular genes by herpesvirus immediate-early gene products. Anticancer Res. 1987;7:589–604. [PubMed] [Google Scholar]
- 17.Everett R D, Barlow P, Milner A, Luisi B, Orr A, Hope G, Lyon D. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038–1047. doi: 10.1006/jmbi.1993.1657. [DOI] [PubMed] [Google Scholar]
- 18.Faber S W, Wilcox K W. Association of herpes simplex virus regulatory protein ICP4 with sequences spanning the ICP4 gene transcription initiation site. Nucleic Acids Res. 1988;16:555–570. doi: 10.1093/nar/16.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan W M, Papavassiliou A G, Rice M, Hecht L B, Silverstein S, Wagner E K. Analysis of the herpes simplex virus type 1 promoter controlling the expression of UL38, a true late gene involved in capsid assembly. J Virol. 1991;65:769–786. doi: 10.1128/jvi.65.2.769-786.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freemont P S. The ring finger. A novel sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 21.Früh K, Ahn K, Djaballah H, Sempe P, Endert P M V, Tampe R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 22.Hanson L A, Kousoulas K G, Thune R L. Channel catfish herpesvirus (CCV) encodes a functional thymidine kinase gene: elucidation of a point mutation that confers resistance to Ara-T. Virology. 1994;202:659–664. doi: 10.1006/viro.1994.1387. [DOI] [PubMed] [Google Scholar]
- 23.Hanson L A, Thune R L. Characterization of thymidine kinase encoded by channel catfish virus. J Aquat Anim Health. 1993;5:199–204. [Google Scholar]
- 24.Harty R N, Colle C F, O’Callaghan D J. Equine herpesvirus type 1 gene regulation: characterization of transcription from the immediate-early gene region in a productive infection. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 319–337. [Google Scholar]
- 25.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 26.Homa F L, Glorioso J C, Levine M. A specific 15-bp TATA box promoter element is required for expression of a herpes simplex virus type 1 late gene. Genes Dev. 1988;2:40–53. doi: 10.1101/gad.2.1.40. [DOI] [PubMed] [Google Scholar]
- 27.Homa F L, Otal T M, Glorioso J C, Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (γ2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol Cell Biol. 1986;6:3652–3666. doi: 10.1128/mcb.6.11.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci USA. 1975;72:1276–1280. doi: 10.1073/pnas.72.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ihara S, Feldman R, Watanabe S, Ben-Porat T. Characterization of the immediate-early functions of pseudorabies virus. Virology. 1983;131:437–454. doi: 10.1016/0042-6822(83)90510-x. [DOI] [PubMed] [Google Scholar]
- 31.Johnson P A, Everett R D. DNA replication is required for abundant expression of a plasmid-borne late US11 gene of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:3609–3625. doi: 10.1093/nar/14.9.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kibler P K, Duncan J, Keith B D, Hupel T, Smiley J R. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J Virol. 1991;65:6749–6760. doi: 10.1128/jvi.65.12.6749-6760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristie T M, Roizman B. α4, the major regulatory protein of herpes simplex virus type 1, is stably and specifically associated with promoter-regulatory domains of α genes and of selected other viral genes. Proc Natl Acad Sci USA. 1986;83:3218–3222. doi: 10.1073/pnas.83.10.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lang J C, Spandidos D A, Wilkie N M. Transcriptional regulation of a herpes simplex virus immediate early gene is mediated through an enhancer-type sequence. EMBO J. 1984;3:389–395. doi: 10.1002/j.1460-2075.1984.tb01817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leopardi R, Michael N, Roizman B. Repression of the herpes simplex virus 1 α4 gene by its gene product (ICP4) within the context of the viral genome is conditioned by the distance and stereoaxial alignment of the ICP4 DNA binding site relative to the TATA box. J Virol. 1995;69:3042–3048. doi: 10.1128/jvi.69.5.3042-3048.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mavromara-Nazos P, Roizman B. Activation of herpes simplex virus 1 γ2 gene by viral DNA replication. Virology. 1987;161:593–598. doi: 10.1016/0042-6822(87)90156-5. [DOI] [PubMed] [Google Scholar]
- 37.McGeoch D J, Dalrymple M A, Dolan A, McNab D, Perry L J, Taylor P, Challberg M D. Structures of herpes simplex virus type 1 genes required for replication of virus DNA. J Virol. 1988;62:444–453. doi: 10.1128/jvi.62.2.444-453.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKnight J L C, Kristie T M, Silver S, Pellet P E, Mavromara-Nazos P, Campadelli-Fiume G, Arsenakis M, Roizman B. Regulation of herpes simplex virus 1 gene expression: the effect of genomic environments and its implications for model systems. In: Botchan M, Grodzicker T, Sharp P A, editors. Cancer cells. 4. DNA tumor viruses: control of gene expression and replication. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. pp. 163–173. [Google Scholar]
- 39.Michael N, Roizman B. Binding of the herpes simplex virus major regulatory protein to viral DNA. Proc Natl Acad Sci USA. 1989;86:9808–9812. doi: 10.1073/pnas.86.24.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michael N, Roizman B. Repression of the herpes simplex virus 1 α4 gene by its gene product occurs within the context of the viral genome and is associated with all three identified cognate sites. Proc Natl Acad Sci USA. 1993;90:2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller M T. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J Virol. 1987;61:858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olivo P D, Nelson N J, Challberg M D. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J Virol. 1989;63:196–204. doi: 10.1128/jvi.63.1.196-204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perry L J, Rixon F J, Everett R D, Frame M C, McGeoch D J. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986;67:2365–2380. doi: 10.1099/0022-1317-67-11-2365. [DOI] [PubMed] [Google Scholar]
- 44.Post L E, Mackem S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 45.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979;29:275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan M P, Knipe D M. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 48.Ruyechan W T, Ling P, Kinchington P R, Hay J. The correlation between varicella-zoster virus transcription and the sequence of the viral genome. In: Wagner E K, editor. Herpesvirus transcription and its regulation. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 319–337. [Google Scholar]
- 49.Sambrook T, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Sandri-Goldin R M, Mendosa G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 51.Seal B S, Whetstone C A, Zamb T J, Bello L J, Lawrence W C. Relationship of bovine herpesvirus 1 immediate-early, early, and late gene expression to host cellular gene transcription. Virology. 1992;188:152–159. doi: 10.1016/0042-6822(92)90744-a. [DOI] [PubMed] [Google Scholar]
- 52.Shapira M, Homa F L, Glorioso J C, Levine M. Regulation of the herpes simplex virus type 1 late (γ2) glycoprotein C gene: sequences between base pairs −34 and +29 control transient expression and responsiveness to transactivation by the products of the immediate early (α) 4 and 0 genes. Nucleic Acids Res. 1987;15:3097–3111. doi: 10.1093/nar/15.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverstein P S, Bird R C, van Santen V L, Nusbaum K E. Immediate-early transcription from the channel catfish virus genome: characterization of two immediate-early transcripts. J Virol. 1995;69:3161–3166. doi: 10.1128/jvi.69.5.3161-3166.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith I L, Sekulovich R E, Hardwicke M A, Sandri-Goldin R M. Mutations in the activation region of herpes simplex virus regulatory protein ICP27 can be trans dominant. J Virol. 1991;65:3656–3666. doi: 10.1128/jvi.65.7.3656-3666.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaughan P J, Thibault K J, Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus immediate-early protein ICP27 encodes a potential metal binding domain and binds zinc in vitro. Virology. 1992;189:377–384. doi: 10.1016/0042-6822(92)90720-a. [DOI] [PubMed] [Google Scholar]
- 56.Weiher H, König M, Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983;219:626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- 57.Wirth U V, Fraefel C, Vogt B, Vlc̆ek C, Pac̆es V, Schwyzer M. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J Virol. 1992;66:2763–2772. doi: 10.1128/jvi.66.5.2763-2772.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolf K, Darlington R W. Channel catfish virus: a new herpesvirus of ictalurid fish. J Virol. 1971;8:525–533. doi: 10.1128/jvi.8.4.525-533.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H G, Hanson L A. Recombinant channel catfish virus (Ictalurid herpesvirus 1) can express foreign genes and induce antibody production against the gene product. J Fish Dis. 1996;19:121–128. [Google Scholar]