Summary
Sperm motility is crucial to reproductive success in sexually-reproducing organisms. Impaired sperm movement causes male infertility, which is increasing globally. Sperm are powered by a microtubule-based molecular machine – the axoneme – but it is unclear how axonemal microtubules are ornamented to support motility in diverse fertilization environments. Here, we present high-resolution structures of native axonemal doublet microtubules (DMTs) from sea urchin and bovine sperm, representing external and internal fertilizers. We identify >60 proteins decorating sperm DMTs; at least 15 are sperm-specific and 16 are associated with infertility. By comparing DMTs across species and cell types, we define core microtubule inner proteins and analyze evolution of the tektin bundle. We identify conserved axonemal microtubule-associated proteins (MAPs) with unique tubulin-binding modes. Additionally, we identify a testis-specific serine/threonine-kinase that links DMTs to outer dense fibers in mammalian sperm. Our study provides structural foundations for understanding sperm evolution, motility, and dysfunction at a molecular level.
Graphical Abstract

Introduction
Sperm are among the most specialized metazoan cell types, adapted for a single function – to find and fuse with the egg. The odyssey of the sperm cell is remarkable; in mammals, sperm must swim distances over ~1000-fold their own length while contending with fluid viscosity, shear flows, and physical barriers 1. Sperm motility is therefore crucial to a successful life cycle in sexually reproducing organisms. Defects in sperm movement can lead to male infertility, which is on the rise globally, increasing demand for assisted reproductive technologies 2,3. However, a significant portion of male infertility cases remain unexplained 4. Sperm motility is also an important target for male contraception, for which there is clearly an unmet need as almost half of all pregnancies worldwide are unplanned 5. Progress towards these reproductive health pursuits hinges on improving our understanding of how the sperm tail is organized at a molecular level.
Sperm motility is powered by a microtubule-based molecular machine called the axoneme, a veritable molecular behemoth comprised of hundreds of different proteins, including dynein motors that drive movement and an extensive array of regulatory proteins that fine-tune motility. These proteins are anchored on axonemal microtubules, which consist of nine doublet microtubules (DMTs) arranged around a central pair of singlet microtubules. Axonemes are also found in the cilia of motile single-celled organisms, such as the green alga Chlamydomonas reinhardtii, and in the fluid-propelling cilia that line the brain ventricles and respiratory tracts of mammals 6–8.
Although the axoneme is highly conserved across evolution, structural variations exist across species and cell types 9–14. One particularly divergent feature is the vast repertoire of microtubule inner proteins (MIPs) that decorate the lumen of axonemal DMTs 15–18 and that stabilize the structure during beating 19. MIPs bind with varying repeat lengths that are multiples of the 8-nm tubulin repeat, but with an overall periodicity of 48-nm that is in register with the 96-nm periodicity of external axonemal complexes 13. Notably, mammalian sperm DMTs have significantly more MIP densities 12,20,21 than DMTs from Chlamydomonas flagella 15 or mammalian respiratory cilia 16,17, but the proteins that make up these densities are unknown.
The natural diversity of sperm presents a unique opportunity to study the evolution of the axoneme. This diversity reflects the variety of reproductive modes across the tree of life 22–24. For example, sperm cells of broadcast-spawning marine organisms voyage across open seawater, whereas sperm cells of internally fertilizing species navigate through the viscoelastic fluids of the female reproductive tract. These different environmental pressures have led to the evolution of structures that support sperm motility (Fig. 1A–B). Sperm tails from internal fertilizers like mammals and other amniotes have large accessory structures like outer dense fibers (ODFs) and the fibrous sheath 25,26, which are proposed to suppress buckling of the axoneme in high-viscosity fluids 27. In contrast, sperm tails from external fertilizers like sea urchins and zebrafish consist essentially of the axoneme surrounded by a plasma membrane 24. However, very little is known about how the core microtubule machinery of the axoneme varies across sperm from different species.
Figure 1. Cryo-electron microscopy of native sperm axonemal doublet microtubules (DMTs).

(A-B) Schematic diagrams of (A) sea urchin and (B) bovine sperm.
(C-D) Electron micrographs of (C) sea urchin and (D) bovine sperm. Scale bars: left panels – 10 μm, right panels – 50 nm.
(E-F) Cross sections (viewed from the plus end) of cryo-EM maps of the 48-nm DMT repeat from (E) sea urchin sperm, with a total of 55 microtubule inner proteins (MIPs) and microtubule-associated proteins (MAPs) colored individually; and from (F) bovine sperm, with 65 MIPs/MAPs colored individually. Unassigned proteins are colored in grey.
In this study, we use cryo-electron microscopy (cryo-EM) to determine high-resolution structures of native axonemal DMTs from sea urchin and bovine sperm, representing external and internal fertilizers respectively. Our reconstructions allow us to identify over 60 MIPs and microtubule-associated proteins (MAPs) decorating sperm DMTs. By comparing structures across species and cell types, we analyze the evolution of the axonemal DMT and define fundamental architectural principles of this core molecular machine. We also discover conserved axonemal MAPs and identify the link between DMTs and ODFs in mammalian sperm. Our analyses provide a structural foundation for understanding the molecular bases of male infertility and identifying novel targets for male contraception.
Results
Cryo-EM reveals elaborate sperm-specific ornamentation of axonemal DMTs
To gain insight into the structural specializations of the sperm axoneme, we imaged native sperm DMTs with cryo-EM. For sea urchin sperm, we simply demembranated sperm cells and blotted them onto a cryo-EM grid; surface tension in the thin aqueous film caused the axoneme to splay apart in the grid holes (Fig. 1C, Fig. S1A). For bovine sperm, we demembranated sperm cells, removed the mitochondrial sheath, and induced microtubule sliding with ATP 28. Without the mitochondrial sheath to restrain them, the DMTs buckled out of the midpiece while still attached to their respective ODFs, disintegrating the axoneme and exposing individual DMTs suitable for cryo-EM (Fig. 1D, Fig. S1B).
To reconstruct the 48-nm DMT repeats of sea urchin and bovine sperm, we applied the same strategy that was used for Chlamydomonas flagella and mammalian respiratory cilia 15,16. Firstly, we extracted particles with an 8-nm periodicity along each microtubule, then used 3D classification on the MIPs at the inner junction to retrieve 16-nm particles. Subsequently, we performed a second round of 3D classification on the MIPs bound to the A-tubule seam to retrieve 48-nm particles. To improve map quality, we carried out 36 focused refinements using masks covering 2-3 protofilaments and extending longitudinally along ~1/3 of the 48-nm repeat (Fig. S1C–D). This strategy resulted in overall resolutions of ~3.3 Å and ~3.6 Å for the sea urchin and bovine sperm DMTs, respectively (Fig. S1, Table S1).
Well-resolved side chain densities allowed us to unambiguously identify almost all the proteins that bind the DMTs. Proteins were identified from a list of candidates obtained by mass spectrometry of sea urchin and bovine sperm (Table S6) using one of three general strategies (see Methods for full details). The first strategy involved construction of polyalanine backbone models either manually using Coot 29 or automatically using ModelAngelo 30, followed by the use of findMySequence 31 to identify the most probable candidates from our sperm proteomes. Briefly, findMySequence objectively estimates side chain probabilities based on the density, from which it constructs a multiple sequence alignment that it uses as an input to a HMMER search 32 against sequence databases created from the sperm proteomes. The second approach used ModelAngelo 30 or DeepTracer 33 to automatically build models into our maps, followed by querying the predicted sequence against our sequence databases using protein-BLAST 34. The third strategy was applied to proteins with globular domains and relied on a structure-based approach in which backbone traces were used as inputs to either DeepTracer-ID 35 or the DALI server 36,37 to search against PDB or AlphaFold2 libraries. Top hits were then mutated to polyalanine, re-fit into the density map, and used as queries for findMySequence 31. To ensure the accuracy of the identified proteins, we manually evaluated all assignments to confirm their consistency with the experimental density. Examples of map densities and their models are provided in Data S1.
Using these strategies, we identified and built atomic models for over 60 proteins decorating sperm DMTs, including both MIPs and MAPs (Fig. 1E–F, Tables S2 and S3). By comparing these structures with DMTs from Chlamydomonas flagella 15 and mammalian respiratory cilia 16,17, we were able to define 25 “core-MIPs” present in all structures (Fig. 2A–B, Table S4). We also identified 14 MIPs that are specific to sea urchin sperm DMTs and 17 MIPs that are specific to bovine sperm, 9 of which are conserved between the two species (henceforth “sperm-MIPs”). Because many of the proteins we identify are poorly characterized and have uninformative placeholder names, we have consulted with the HUGO Gene Nomenclature Committee to rename them (Tables S2 and S3). We introduce the names “sperm-associated microtubule inner protein (SPMIP)” or “sperm microtubule-associated protein (SPMAP)” for MIPs/MAPs so far only seen in sperm DMTs; and the names “ciliary microtubule inner protein (CIMIP)” or “ciliary microtubule-associated protein (CIMAP)” for MIPs/MAPs seen in both sperm and tracheal DMTs.
Figure 2. Structural comparisons of DMTs from different species and cell types.

(A) Venn diagram of MIPs and MAPs identified in cryo-EM structures of DMTs from sea urchin and bovine sperm (this study), bovine respiratory cilia (PDB 7RRO) 16, and Chlamydomonas flagella (PDB 6U42) 15. Full lists of proteins corresponding to all regions of the Venn diagram are provided in Table S4.
(B) Lists of core-MIPs, sperm-MIPs, and MAPs identified in this study. Notes: (1) SAXO proteins are present in all DMT structures to date, although individual proteins may be species- or cell type-specific. (2) CCDC81 and SPATA45 are also present in our bovine sperm proteome, but definitive density is currently only resolved in the cryo-EM map of sea urchin sperm. (3) CIMAP3 (Pitchfork) density is weaker in cryo-EM structures of bovine sperm, but it is present in our proteomics data.
(C) Comparison of mammalian DMTs from different cell types, with MIPs specific to trachea colored in green and MIPs specific to sperm colored in pink.
(D) Comparison of sperm DMTs from different species, with MIPs resolved only in sea urchin colored in blue and MIPs resolved only in bovine colored in pink.
Overall, bovine sperm have more MIPs than sea urchin sperm (Fig. 2D). The B-tubule decorations are noticeably different, with mammalian sperm having striations of SPACA9 and sea urchin sperm having as-yet-unidentified MIPs projecting into the lumen (Fig. 2D, Fig. S2D). In addition to sperm-specific MIPs, mammalian sperm DMTs retain nearly all MIPs present in mammalian respiratory DMTs, with the exception of CIMIP2B (FAM166B), which is expressed at low levels in the testis 38 and is replaced by CIMIP2A (FAM166A) in sperm (Fig. 2C). Bovine sperm also have two additional copies of EFCAB6 per 48-nm repeat than in bovine trachea.
The structurally diverse Tektin-5 expands the tektin bundle in mammalian sperm
The mammalian sperm DMT features an elaborate tektin bundle that fills nearly the entire lumen of the A-tubule (Fig. 3A). Tektins are abundant ciliary proteins that form hyperstable filaments within axonemal DMTs 39,40. Of the five different tektins found in mammals, four form a pentagonal eight-tektin bundle within respiratory DMTs (Fig. 3B) 16, while the fifth, known as Tektin-5, is expressed only in the testes. Tektins are important for sperm motility and male fertility (Table S5). In mice, deficiencies in Tektin-2 or Tektin-4 cause infertility and subfertility respectively 41,42, whereas a lack of Tektin-3 leads to reduced sperm motility and forward progression 43. Genetic variants of Tektin-2 and the sperm-specific Tektin-5 have also been linked to human male infertility 44–46.
Figure 3. Evolution and structural expansion of the tektin bundle.

A simplified phylogenetic tree with nodes indicating the number of tektin genes found in each species.
(A-D) Top panels show cryo-EM maps of DMTs from (A) bovine sperm (this study), (B) bovine respiratory cilia (EMD-24664) 16, (C) sea urchin sperm (this study), and (D) Chlamydomonas (this study). Tektins and TEKTL1, a tektin-like protein, are colored. Middle panels show ribbon representations of the tektin bundle and TEKTL1 filament. Bottom panels show schematic diagrams of the tektin bundle.
(E) Longitudinal slice showing cryo-EM density colored by protein for the expanded tektin bundle in bovine sperm DMTs. TEKT5-F runs diagonally perpendicular to other tektins.
(F) Zoomed-in ribbon diagrams showing the intermolecular β-sheet formed by the N-termini of TEKT5-F, TEKT3-1, and CIMIP2A.
Our structure demonstrates that the additional tektin supercomplex in mammalian sperm – termed the “sickle” 21 – is formed by multiple copies of the testis-specific Tektin-5, which can be divided into seven groups (TEKT5-A to -G) based on their location and organization in the DMT (Fig. S3). TEKT5-A, -B, and -C form continuous filaments with 16-nm periodicity through head-to-tail interactions between neighboring molecules (Fig. 3E), similar to Tektins 1-4 in respiratory cilia 16. In contrast, TEKT5-D to -G adopt unexpected conformations and organizations (Fig. S3A). For example, TEKT5-D forms an interrupted filament in which two copies per 48-nm repeat adopt a “bent” conformation to accommodate NME-7 (Fig. S3B) with their L12 loops inserting into the putative nucleotide-binding pocket of NME-7 (Fig. S3C), which may render the kinase inactive. TEKT5-G is even more unusual as it has 48- rather than 16-nm periodicity and does not form filaments (Fig. 3A,E; Fig. S3A). Its density is also weaker than adjacent TEKT-5 molecules, suggesting that it may be loosely bound to its neighbors or present only in a subset of DMTs. However, the most atypical group is TEKT5-F, which binds diagonally perpendicular to the tektin bundle, with one copy every 16 nm, forming the handle of the “sickle” (Fig. 3A,E). By binding diagonally, TEKT5-F molecules may act as crossbeams to stabilize the expanded tektin bundle.
The structural diversity of Tektin-5 appears to be consequence of other MIPs preventing the correct ordering of the N-terminal 1A helix and the L12 and L2 loops that are needed for canonical filament formation 16. These elements are either disordered, as in TEKT5-E, TEKT5-F, and TEKT5-G (Fig. S3A, green arrowheads), or held by other MIPs in alternative conformations incapable of filament formation. For example, the L12 loop of the TEKT5-D group is ordered through its interactions with NME-7 (Fig. S3A, green arrowheads) whereas its L2 loop can be ordered in one copy every 48-nm by interactions with a neighboring TEKT5-D (Fig. S3B). In TEKT5-F, the 1A helix has folded back onto the central helical bundle and is held in place by an N-terminus that forms an intermolecular β-sheet with TEKT3-1 and CIMIP2A (FAM166A) (Fig. 3F).
The prominence of TEKT5 in mammalian sperm DMTs explains its importance for male fertility. A heterozygous deletion of TEKT5 was linked to azoospermia 46, and mice with reduced levels of TEKT5 have impaired sperm production 47. These observations suggest that TEKT5 plays a role in both the assembly and stability of the sperm tail.
Evolution and structural expansion of the tektin bundle
Comparison of the sperm and respiratory DMT structures demonstrates that the organization of the tektin bundle can vary substantially between cell types within an organism. Next, we asked how tektin organization varies between ciliated organisms, which can have considerable diversity in the number of tektin genes 40. To address this, we analyzed the tektin bundle in our cryo-EM structure of DMTs from sea urchin sperm, which feature three out of four sea urchin tektin genes (Fig. 3C).
The three sea urchin tektins (historically called TEKT-A, -B, and -C, corresponding to mammalian TEKT4, TEKT2, and TEKT1 respectively) form a relatively simple bundle of a tightly associated heterodimeric filament of TEKT4 and TEKT2 along with a more loosely associated filament of TEKT1 bound to the ribbon (protofilaments A12-A01) (Fig. 3C). This organization is consistent with previous biochemical studies 48. The smaller bundle means that tektin bundle-interacting MIPs TEKTIP1, DUSP21, SPMIP6 (SMRP1), and SPMIP9 (TEX37) are also absent from sea urchin sperm (Fig. S2B–C). These proteins may be adaptations to stabilize the larger tektin bundle in mammalian sperm.
The position and arrangement of the TEKT1, TEKT2, and TEKT4 filaments are nearly identical in sea urchin sperm, mammalian sperm, and mammalian respiratory cilia (Fig. 3A–C). Interestingly, of this core trimeric bundle only TEKT4 directly contacts the tubulin lattice through interactions with α-tubulin K40 loops on protofilament A12 (Fig. S3D), despite the 16-nm periodicity of tektins suggesting co-evolution with tubulin. Instead, the tektin filaments appear to be positioned by interactions with other MIPs. TEKT1 interacts with CIMIP2C (FAM166C) and SAXO4 (PPP1R32), which bridge the tektin bundle with tubulin protofilaments close to the inner junction (Fig. S2B). Likewise, the heterodimeric TEKT2/TEKT4 filament interacts with an uncharacterized protein bound to protofilaments A12/A13 that we name SAXO3 (Fig. S2C). These observations support a model in which core filaments of the tektin bundle are recruited to the ribbon through interactions with other MIPs, followed by outward expansion into the A-tubule lumen.
To expand our analysis of tektin evolution, we also determined a structure of a tektin-bound DMT from C. reinhardtii, which retains the ancestral state of having a single tektin gene (Fig. 3D). Our structures revealed that Chlamydomonas tektin binds to the B-tubule, where it assembles up to nine filaments 49, forming a beak-like structure 50. This observation explains previous biochemical studies showing a correlation between the appearance of the beak and the presence of tektins 51,52 and demonstrates that tektins can bind to either tubule. The characteristic tektin fold, head-to-tail polymerization, and assembly of filaments into larger complexes are all conserved in Chlamydomonas, indicating that these features were likely present in the ancestral tektin. However, Chlamydomonas tektin interacts mainly with β-tubulin as opposed to α-tubulin in the metazoan tektins (Fig. S3D).
TEKTL1 (CCDC105) is a tektin-like sperm-MIP
Our structures also identified CCDC105 as a conserved, sperm-specific, tektin-like molecule that forms a filament in the cleft between protofilaments A11 and A12 (Fig. 3A,C, Fig. 4) and binds several core- and sperm-MIPs (Fig. 4A–B). The tertiary structure of CCDC105 is similar to that of tektins (Fig. 4C, left panels) but with the equivalent of the tektin 1A helix split into two helices (which we call 1A′ and 1A″) separated by a loop (L1′) (Fig. 4C, upper left panel). Based on this structural similarity, we rename CCDC105 as “tektin-like 1 (TEKTL1)”. The mechanism by which TEKTL1 and tektins assemble into filaments differs slightly (Fig. 4C, right panels). Like tektin, the main contact points between neighboring TEKTL1 molecules are helical overhangs that extend beyond the central bundle. However, TEKTL1’s equivalent of the L12 loop does not clamp around the L2 loop of the neighboring molecule, instead looping around the 1A″ helix of the same protomer (Fig. 4C, arrowheads). Interactions between TEKTL1 protomers may be further stabilized by SPMIP10 (TEX43), which cradles the inter-TEKTL1 interface while interacting with protofilaments A11 and A12 (Fig. 4A–B). Functionally, little is known about TEKTL1 other than that it is highly expressed in testis 38,53 and that it is down-regulated in mice with structural defects in their sperm tails 54.
Figure 4. TEKTL1 is a conserved tektin-like sperm-MIP.

(A-B) Longitudinal slices from cryo-EM density maps of (A) sea urchin and (B) bovine sperm DMTs showing the molecular environment of the TEKTL1 filament. TEKTL1 interacts with both the outer junction core-MIP CFAP77 and the sperm-MIP SPMIP10 (TEX43).
(C) Comparison of the tertiary (left) and quaternary (right) structures of TEKTL1 (upper) and tektin filaments (lower, tektin 5-A as an example) from bovine sperm DMTs. In the left panels, proteins are colored in a rainbow palette from N- (blue) to C-terminus (red). TEKTL1 secondary structures are annotated by analogy to tektin. In the right panel, one copy of each protein is colored in a rainbow palette from N to C. The arrowheads indicate differences in the L12 loop at the inter-protomer interface. For the TEKTL1 filament, SPMIP10 molecules that bind at the inter-protomer interfaces are shown as transparent surfaces.
The Mn-motif is a universal microtubule-binding and stabilization motif
Like tektins, the SAXO (stabilizer of axonemal microtubules) family is ubiquitous in axonemal DMTs 15,17, as well in stable microtubules of neurons and parasites 55–57. The defining feature of SAXO proteins is an Mn-motif, which forms a 5- to 6-residue helix flanked by a Tyr/Phe residue on one end and by a Ser/Thr on the other that binds the shoulder of the α-tubulin S9/S10 loop 55. We used these signature features to systematically search for SAXO proteins at every possible tubulin binding site within the 48-nm DMT repeat (Fig. 5). Our analysis revealed that SAXO proteins are incredibly widespread. In bovine and sea urchin sperm, SAXO proteins are bound to 19 and 18 out of 23 protofilaments respectively (Fig. 5A–B), whereas Chlamydomonas DMTs have SAXO proteins bound to 14 protofilaments (Fig. S4A). Intriguingly, in Chlamydomonas, the SAXO proteins bound to protofilaments B02-B04 are only visible in classes with the tektin bundle (Fig. S4B). We also found SAXO proteins bound to all 13 protofilaments of singlet microtubules in the bovine sperm endpiece (Fig. 5C, Fig. S5). We could unambiguously identify many of the SAXO proteins based on sidechain density, but several remain unassigned due to incoherent averaging of Mn motifs. For bovine sperm, we tentatively assign SAXO proteins on protofilaments B02-B06 and in endpiece singlet microtubules as SAXO1 based on in-cell cross-linking with SPACA9 (Fig. S5, Table S6) and prior immunofluorescence and immunogold labelling studies 58.
Figure 5. The Mn-motif is a universal microtubule-binding motif.

(A-C) Location and identities of SAXO proteins in (A) sea urchin sperm DMTs, (B) bovine sperm DMTs, and (C) bovine sperm endpiece singlet microtubules (SMTs). SAXO proteins are classified based on how they bind the microtubule lattice: through a single Mn-motif (blue circles), through multiple Mn-motifs along a single protofilament (violet circles), or through multiple Mn-motifs across neighboring protofilaments (orange circles). Asterisks indicate that SAXO1 is tentatively assigned based on in-cell cross-links to SPACA9 (see also Fig. S5 and Table S6).
(D-F) Representative SAXO proteins from each of the aforementioned classes. Density and models are shown for selected sperm-MIPs from sea urchin sperm DMTs. Other examples are listed below and colored based on whether they are conserved (black), sperm-specific (pink), or Chlamydomonas-specific (green). Notes: (1) SAXO3 and SAXO4 (PPP1R32) are present in sea urchin sperm, bovine sperm, and bovine respiratory cilia, but not in Chlamydomonas. (2) CFAP68 and CFAP107 have only one Mn motif in Chlamydomonas.
(G) The sperm-MIP SAXO5 (TEX45) has 11 Mn-motifs in sea urchin (top) and Bos taurus (bottom) and 96-nm periodicity (as shown in Fig. S6).
We divide the SAXO proteins into three classes based on how they bind the tubulin lattice. In the first class, proteins bind using a single Mn motif (Fig. 5D). Although experiments had shown that a single Mn motif is sufficient for microtubule binding in vitro 59, our structures definitively show that proteins do use solitary Mn motifs to interact with microtubules. The second class contains proteins with multiple Mn motifs that bind longitudinally along a single protofilament (Fig. 5E). In such proteins, Mn motifs are spaced ~40 residues apart, corresponding to an apparent internal repeat of 8 nm. Based on the presence of several SAXO motifs, we have renamed two uncharacterized members of this class as SAXO4 (formerly PPP1R32) and SAXO5 (formerly TEX45). SAXO5 is notable within this class because it has 11 Mn motifs (Fig. 5G) and therefore has 96-nm periodicity, matching the external periodicity of the axoneme (Fig. S6A). The third class contains proteins with multiple Mn motifs that can bind across neighboring protofilaments (Fig. 5F). SAXO proteins that traverse protofilaments can have as few as two Mn motifs, like CFAP68 in which one Mn motif binds on each side of the seam (Fig. S4C). Mn motifs are conserved across species, even in proteins with only one or two motifs (Fig. S4C–D).
Thus, our structural analysis greatly expands the SAXO protein family and establishes the Mn motif as a ubiquitous microtubule-binding mode among MIPs. Given that minimal constructs containing only Mn motifs are sufficient to stabilize microtubules in vitro and in cell culture 58,59, the extensive array of SAXO proteins that decorates sperm DMTs likely stabilizes the tubulin lattice during sperm movement.
Identification of axonemal MAPs reveals unique tubulin-binding modes
Our structures also reveal conserved axonemal MAPs associated with nearly all protofilaments of sperm DMTs (Fig. 6). We identified outer dense fiber 3 (ODF3) or ODF3-like (ODF3L) proteins binding between protofilaments A05-A07, A08/A09, and B01-B07 (Fig. 6A–B, Fig. S7). The ODF3 family is so named because immunogold labelling localized them to the outer dense fibers of mammalian sperm 60. However, our observation of ODF3 orthologs in sea urchin sperm (Fig. 6A), which lack accessory structures, definitively show that the ODF3 family are bona fide MAPs of DMTs, although a second role within ODFs cannot be excluded. Because these proteins are also present in sperm lacking outer dense fibers, we instead rename them ciliary microtubule-associated protein 1 (CIMAP1).
Figure 6. Axonemal microtubule-associated proteins (MAPs) interact with DMTs through unique tubulin binding modes.

(A-B) Axonemal MAPs identified in cryo-EM structures of (A) sea urchin and (B) bovine sperm DMTs. MAPs are shown in licorice cartoon representation.
(C) Cross section views comparing microtubule-binding modes of neuronal MAPs Tau (PDB 6CVN) 63 and MAP7 (PDB 7SGS) 62 to the axonemal wedge-MAPs (CIMAP1, CIMAP2, CIMAP3, CFAP96) and arc-MAP (SPMAP2) identified in sperm DMTs.
(D) The axonemal MAP CFAP97D1 specifically recognizes the outer junction, a site of atypical inter-tubulin contacts.
(E) CFAP97D1 has an overall 24-nm repeat and consists of four helices joined by linkers that zig-zag between the surface helices of protofilament A10 and the neighboring protofilament B01.
(F) Binding of CFAP97D1 to protofilament B01 appears to be mediated by positively charged patches on CFAP97D1 interacting with negatively charged pockets at the minus end of β-tubulin molecules that are exposed by the unique inter-tubulin geometry at the outer junction.
The CIMAP1 family is characterized by six Pro-Gly-Pro motifs separated by ~40 residues 61 (Fig. S7D), similar to the spacing between Mn motifs, which gives the proteins an 8-nm repeat and an overall 48-nm periodicity (Fig. S7A). CIMAP1 proteins maintain their own periodicities through head-to-tail interactions (Fig. S7B), similar to filamentous MIPs 15. The C-termini of CIMAP1 proteins extend into the microtubule lumen, where they can interact directly with MIPs (Fig. S7C).
Another pair of axonemal MAPs, CIMAP2 (LEXM) and CIMAP3 (Pitchfork), occupy the inter-protofilament cleft between protofilaments A03/A04 in a manner similar to CIMAP1. However, these proteins repeat alternately along the DMT, resulting in an overall periodicity of 96 nm (Fig. S6B). Additionally, we found that the MAP binding between protofilaments B08/B09 is C4ORF47, which we refer to as CFAP96 since it is orthologous to FAP96 in Chlamydomonas.
CIMAP1, CIMAP2, CIMAP3, and CFAP96 have distinct tubulin-binding modes compared to other structurally characterized MAPs such as MAP7 62 and Tau 63, which bind to cytoplasmic microtubules in neurons (Fig. 6C). The axonemal MAPs bind deeper into the inter-protofilament cleft and make extensive contacts with flanking protofilaments (Fig. 6C). Expression of these “wedge-MAPs” is not restricted to the testis 38, suggesting that they may have general functions in cilia.
The axonemal MAP SPMAP2 (formerly testicular haploid enriched gene or THEG) also shows a unique tubulin-binding mode, stretching laterally across several protofilaments of the B tubule (Fig. 6A–B). SPMAP2 binds along the α/β tubulin interface and periodically dips into the inter-protofilament clefts (Fig. 6C). Given that this binding mode is analogous to how arc-MIPs bind laterally on the luminal surface of the microtubule 64, we classify SPMAP2 as an “arc-MAP”.
The aforementioned wedge- and arc-MAPs all bind between protofilament pairs that adopt canonical inter-tubulin contacts akin to those of cytoplasmic microtubules. However, the arrangement of protofilaments in the DMT also creates a site of unique inter-tubulin geometry at the outer junction (protofilaments A10/B01). Our structures allow us to identify CFAP97D1 as the axonemal MAP that binds specifically to the outer junction (Fig. 6D). Density for CFAP97D1 is also present in cryo-EM structures of DMTs from Chlamydomonas flagella (EMD-20631) 15 and bovine tracheal cilia (EMD-24664) 16, suggesting that it is widely conserved. Unlike CIMAP1, CFAP97D1 has a 24-nm repeat and consists of four helices joined by linkers (Fig. 6E). CFAP97D1 binds to the surface helices of protofilament A10 at a site similar to Tau (Fig. 6C). Its binding to the neighboring protofilament B01 appears to be stabilized by electrostatic interactions between positively charged patches on CFAP97D1 and negatively charged pockets at the minus end of β-tubulin molecules (Fig. 6F). Importantly, these electronegative pockets are only available to interact with CFAP97D1 because of the unique inter-tubulin geometry at the outer junction, explaining why CFAP97D1 binds specifically to this location in the DMT. Disrupting CFAP97D1 in mice leads to subfertility characterized by abnormal sperm motility and axonemal structural defects 65, suggesting that CFAP97D1 may support DMT stability by reinforcing the outer junction.
A testis-specific serine kinase links DMTs to ODFs in mammalian sperm
Mammalian sperm have large ODFs that surround and dwarf the axoneme. Recent cryo-ET studies and earlier classical EM work on mammalian sperm have shown that ODFs taper toward the endpiece and are directly coupled to axonemal DMTs from the principal piece onwards 12,66. Examination of our micrographs revealed DMT-ODF complexes that follow this trend, with ODFs associated with the DMT through a peripheral filamentous substructure (Fig. 7A).
Figure 7. Testis-specific serine kinase 6 (TSSK6) is a seam-binding axonemal MAP that links DMTs to outer dense fibers (ODFs) in mammalian sperm.
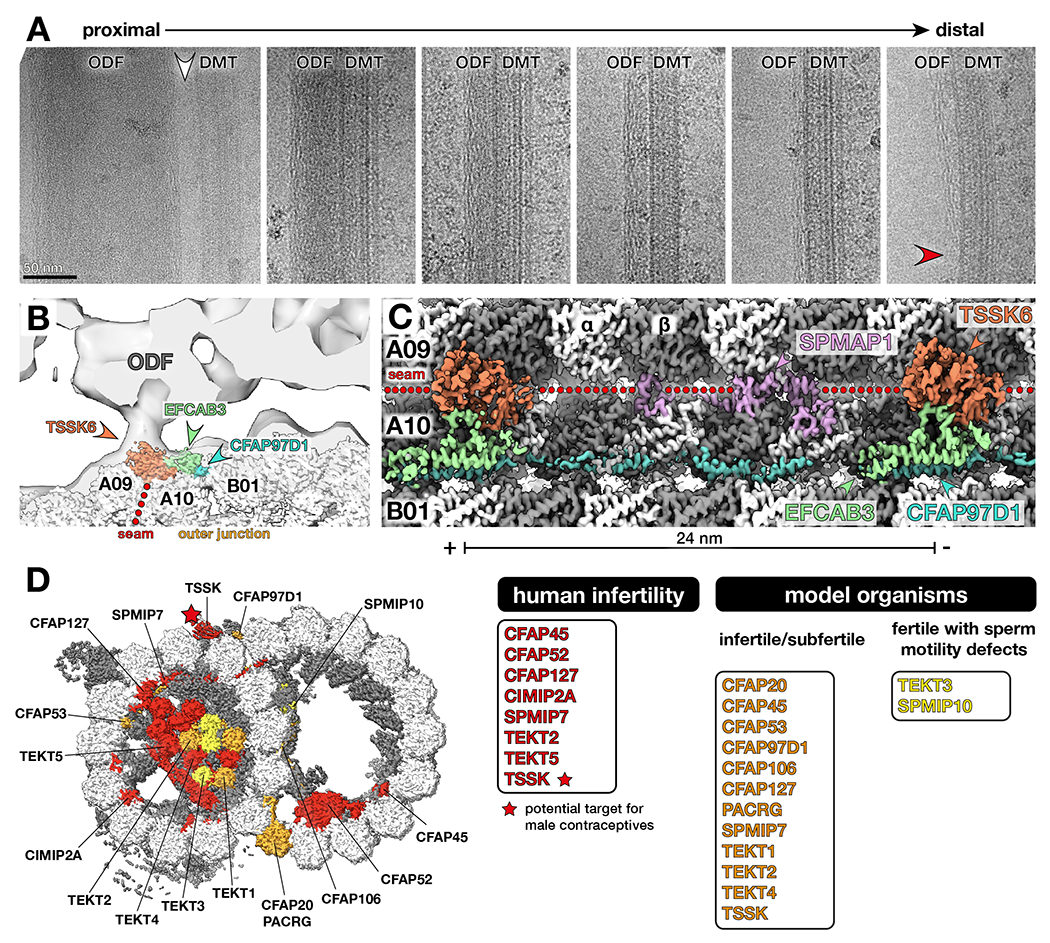
(A) Electron micrographs of DMT-ODF pairs from bovine sperm showing that ODFs possess a peripheral filamentous substructure that persists as the ODFs taper (red arrowhead). Note that the largest (and therefore most proximal) regions of the ODFs are not directly connected to DMTs (white arrowhead). Scale bar: 50 nm.
(B) Cryo-EM map of the bovine sperm DMT fit into an in-situ subtomogram average of porcine sperm DMTs (EMD-12071) 12. TSSK6 localizes to the base of the DMT-ODF linkage.
(C) Longitudinal view of cryo-EM density of bovine sperm DMTs showing MAPs at the seam and outer junction (TSSK6, SPMAP1, EFCAB3, and CFAP97D1).
(D) TSSK (starred) is linked to male infertility and a target for novel male contraceptives. Other proteins linked to human infertility are shown in red. Proteins that impair fertility when perturbed in model organisms are colored in orange. Proteins that do not reduce overall fertility but affect sperm motility are colored in yellow. See Table S5 for a full list of phenotypes.
Subtomogram averaging has shown that ODFs are connected to DMTs near the A-tubule seam (between protofilaments A09/A10) 12. To identify the molecular nature of this linkage, we used three-dimensional classification to enrich for DMTs with axonemal MAPs at the seam. This approach allowed us to identify three additional mammalian sperm-specific proteins: EFCAB3, SPMAP1 (C17ORF98), and testis-specific serine/threonine kinase 6 (TSSK6) (Fig. 7B–C). The 24-nm periodicity of these proteins precisely matches the periodicity observed for ODF-DMT linkages in in situ subtomogram averages from mammalian sperm12. TSSK6 and SPMAP1 interact directly with the tubulin lattice at the seam (Fig. 7B–C). EFCAB3, a potential calcium binding protein, acts as an adaptor between TSSK6 and the outer junction MAP, CFAP97D1.
A role for the TSSK family in linking ODFs to DMTs is supported by the phenotypes of TSSK knockout mice. Mice lacking TSSK6 67,68, TSSK3 69,70, or TSSK1/2 71 are infertile, whereas mice lacking TSSK4 are subfertile 72. Mouse knockouts for all TSSK family members show structural abnormalities of the sperm flagellum, including severe bends and disorganized ODFs. Several TSSK polymorphisms have also been linked to human infertility 73,74. These phenotypes are also consistent with the prevailing theory that ODFs stabilize the axoneme against the extreme forces required to bend the long, stiff flagella of mammalian sperm 75.
At least one member of the TSSK family (TSSK4) can interact with and phosphorylate ODF2 in in vitro cell culture assays 72,76. ODF2 is a putative protein component of the ODFs 77 and becomes phosphorylated during sperm capacitation 78, a series of biochemical changes that mammalian sperm must undergo in the female reproductive tract in order to become fertilization-competent 79–81. TSSK6 might therefore also phosphorylate ODF components, potentially in response to capacitation. Determining precisely how TSSK6 interacts with ODF components remains an important avenue for future structural work.
Discussion
Structures of sperm DMTs uncover general principles of axonemal architecture
Our cryo-EM structures have revealed shared features of DMTs across different organisms and cell types, including the identification of 25 core-MIPs present in all DMT structures studied to date (listed in Fig. 2B). Core-MIPs likely play fundamental roles in assembling or maintaining the specialized architecture of the DMT; indeed, notable core-MIPs include components of the inner and outer junctions (Fig. 2). Our analysis also reveals that SAXO proteins are a universal feature of axonemal DMTs, with SAXO motifs present in both core- and cell type-specific MIPs (Fig. 5). Interestingly, the identification of a SAXO MIP (SAXO5) with 96-nm periodicity (Fig. S6A) challenges the previous assumption that all MIPs within a DMT have a maximum of 48-nm overall periodicity. We hypothesize that SAXO proteins with 12 Mn motifs, such as SAXO1/2 in mammalian cilia and FAP236 in Chlamydomonas, also have 96-nm repeats as they are capable of binding 12 tandem copies of heterodimeric tubulin. The 32-nm periodicity of CFAP115 in sea urchin sperm (as depicted in Fig. S6C) and Tetrahymena DMTs 18 also only fits an overall periodicity of 96 nm, rather than 48 nm. The external and internal 96-nm periodicities may be directly connected by an external protein, CFAP91, that extends through the inner junction “hole”, which is present every 96 nm (Fig. S6D).
Our structures have also expanded our understanding of MAPs that decorate the external surfaces of axonemal DMTs. We have identified a broad class of proteins, known as axonemal wedge-MAPs, which bind deep into the outer inter-protofilament cleft and make extensive contacts with flanking protofilaments (Fig. 6). These proteins have orthologs in many ciliated organisms 82,83 and may account for the filamentous densities observed on the surface of the Tetrahymena DMT 18. Furthermore, they could be present on the microtubules of primary cilia 84. We propose that wedge-MAPs resist sliding between neighboring protofilaments, thereby allowing the microtubule to withstand and adapt to shear forces 85,86.
Insights into the evolution of the sperm tail
Our study reveals that bovine sperm DMTs are more extensively decorated than DMTs from respiratory cilia or sea urchin sperm. Specifically, the A-tubule lumen of bovine sperm is almost entirely filled with proteins, particularly tektin molecules (Fig. 1, Fig. 3). Although respiratory cilia also function in viscous environments, they beat with waveforms more similar to the asymmetric waveform of Chlamydomonas 87, and are neither as long nor as rigid as mammalian sperm flagella, so a less extensive MIP network may be sufficient to stabilize their DMTs. The appearance of extra sperm-MIPs may be linked with the evolution of accessory structures, such as the ODFs, in sperm cells of internally fertilizing species. This is supported by the fact that electron-dense A-tubules are found in the sperm of internally fertilizing fish species 88 but not in externally fertilizing zebrafish 89. The presence of electron-dense A-tubules in the sperm of internally fertilizing cephalopods, which independently evolved ODF-like accessory structures 90,91, provides further evolutionary support.
Accessory structures like the ODFs increase the stiffness and diameter of the sperm flagellum, enabling it to generate higher bending torque and entrain more dyneins with every bend 75 to facilitate swimming through the viscous fluids of the female reproductive tract. Expanded MIP networks could therefore act alongside the accessory structures to reinforce the microtubule lattice against the extreme mechanical stresses involved in bending the long, stiff flagella of mammalian sperm. This reinforcement may be particularly important as mammalian sperm DMTs need to withstand the increased forces generated during hyperactivation, where the sperm tail switches to a high-amplitude bending mode 26 in response to capacitation. Experimental testing will be required to investigate this hypothesis.
Insights into the evolution of the tektin bundle
Our cryo-EM structures also allow us to describe the expansion of the tektin bundle in relation to the evolution of the tektin gene family, from homomeric assemblies of the single tektin in Chlamydomonas to an elaborate heteromeric tektin network that fills nearly the entire A-tubule lumen in mammalian sperm (Fig. 3). The observation that tektins bind within the A-tubule lumen of mammalian and sea urchin sperm DMTs but in the B-tubule lumen of Chlamydomonas DMTs is surprising, given that all other core-MIPs have binding sites consistent between species. This difference may reflect the time of acquisition of the tektin gene during evolution. Chlamydomonas is proposed to have acquired the tektin gene by horizontal gene transfer 40. If this process occurred after the acquisition of other competing MIPs to the A-tubule, it may explain the relocation of tektin filaments to the B-tubule lumen. The asymmetric distribution of tektin bundles in Chlamydomonas, where they decorate only a subset of B tubules 50, may modulate the mechanical properties of the axoneme to enhance the swimming motility of Chlamydomonas.
In addition to different binding locations, tektins also form a range of higher-order structures, including a triangular prism in Chlamydomonas and a pentagonal prism in bovine DMTs. These higher-order arrangements cannot be predicted based on sequence alone, demonstrating the need for experimentally determined structures. The presence of an extensive tektin bundle does not seem to greatly affect the tubulin lattice, as inter-dimer distances are similar across DMTs from Chlamydomonas flagella, bovine respiratory cilia, and both sea urchin and bovine sperm (Fig. S4E–F).
Compounding the complexity of the tektin bundle is the extreme versatility of Tektin-5, which exists in both filamentous and monomeric forms within the bovine sperm DMT. The five tektins in extant mammals arose from duplications of the ancestral tektin present in the root opisthokont, with the most recent duplication occurring in early vertebrates giving rise to Tektin-3 and Tektin-5 40. Interestingly, the presence of a gene for Tektin-5 may correlate with whether a species has a DMT with a filled A-tubule lumen or not. For example, the sperm cells of birds and reptiles have electron-dense A-tubules 92–94, whereas the zebrafish Danio rerio and the frog Xenopus laevis, which have lost Tektin-5 40, do not 89,95. Tektins have independently duplicated in other non-vertebrate lineages that practice internal fertilization 40; future structural analyses of sperm DMTs from these taxa will help clarify possible links between expanded tektin families and elaborate MIP networks.
We suggest that any analysis of tektins should also include TEKTL1, a tektin-like sperm-MIP that appears to have already been present in early metazoans. CCDC105 shares many structural features with tektins, including the overall helix-turn-helix fold and head-to-tail association into filaments, but does not seem to assemble into the large oligomeric bundles associated with tektins. In both sea urchin and bovine sperm, TEKTL1 binds to the outer inter-protofilament cleft of A11/A12, where it may help stabilize the outer junction (Fig. 4). However, structures of sperm DMTs from diverse species are needed to determine if this is the sole binding location of TEKTL1, given the positional plasticity of tektins.
Axonemal MIPs and MAPs are important for fertility in humans and model organisms
Of the MAPs and MIPs identified in our structures of sperm DMTs, mutations in 15 are linked to male infertility in either humans or model organisms (Fig. 7D, Table S5). These proteins include core-MIPs (CFAP20, PACRG, CFAP45, CFAP52, CFAP53, CFAP106, CFAP127), sperm-MIPs (CIMIP2A, SPMIP7), tektins, and axonemal MAPs (CFAP97D1, TSSK). Other sperm-MIPs like SPMIP6 and SPMIP8 do not appear to be strictly required for fertility, at least in mice 96. However, the absence of a mouse infertility phenotype does not exclude the possibility of more subtle effects on motility; for instance, mice deficient in TEKT3 and TEX43 were fertile but had sperm with reduced motility 43,97. Importantly, many of the proteins identified in our structures have yet to be systematically evaluated in the context of sperm function. Our structures will therefore serve as a blueprint to guide targeted molecular and genetic studies aimed at dissecting the roles of individual proteins on sperm motility. Given that TSSK proteins are already being actively explored as targets for male contraceptives 98, our structures will also help identify targets for the development of assisted reproductive technologies and novel contraceptives.
Limitations of the study
Our cryo-EM structures represent averages from all nine DMTs from along the length of the sperm tail. Cryo-ET has uncovered considerable asymmetry in radial spoke-associated complexes in mammalian sperm 99, but whether certain MIPs are distributed asymmetrically remains unclear. Although some weaker protein densities hint at partial decoration, relating these to genuine radial or longitudinal asymmetries of the axoneme will require high-resolution in situ cryo-ET data. While we identify nearly all MIPs, several SAXO densities remain unidentified. To assign these densities, it will be necessary to image and compare sperm from knockouts of different candidate SAXO genes. Knockout studies will also be necessary to evaluate the function of the identified proteins in fertility and sperm motility. For example, there are currently no knockout models for TEKT5 or TEKTL1, which are clearly important targets for further research. Knockout studies in different model organisms, along with additional cryo-EM studies with broader phylogenetic sampling, will help identify trends in the molecular evolution of the sperm tail.
STAR Methods
Resource Availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Dr. Tzviya Zeev-Ben-Mordehai (z.zeev@uu.nl).
Materials availability
Wild-type Chlamydomonas reinhardtii cells (CC-4402) are available from the Chlamydomonas Resource Center (University of Minnesota).
Data and code availability
Cryo-EM maps of the sea urchin sperm DMT, the bovine sperm DMT, the bovine sperm endpiece SMT, and Chlamydomonas DMTs with and without the tektin bundle have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-40619, EMD-17187, EMD-17188, EMD-40620, and EMD-40621 respectively. Atomic models of the 48-nm repeat of the sea urchin sperm DMT, the 48-nm repeat of the bovine sperm DMT, and one asymmetric unit of the 8-nm repeat of bovine sperm endpiece SMTs have been deposited in the Protein Data Bank (PDB) with accession codes 8SNB, 8OTZ, and 8OU0 respectively. Mass spectrometry data of bovine sperm is available from Proteomics Identifications Database (PRIDE) with accession code PXD035941.
Code used to refine and correct alignment parameters by FREALIGN is available at https://github.com/rui--zhang/Microtubule.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Models and Subject Details
Live sea urchins (Strongylocentrotus purpuratus) were purchased from Monterey Abalone Co., Monterey, California. Frozen bovine (Bos taurus, Groninger Blaarkop) semen straws were obtained from the Utrecht University Veterinary Faculty. Wild-type Chlamydomonas reinhardtii cells (CC-4402) were obtained from the Chlamydomonas Resource Center (University of Minnesota).
Method Details
Sperm preparation for cryo-EM
Sea urchin sperm
Sea urchin sperm were prepared following a previously established protocol 100. Spawning of adult male sea urchins was induced by injecting 200 μL of 0.5 M KCl into the peri-visceral cavity (one injection for each side). The sea urchins were gently shaken after the injection, and the sperm sample collected in a test tube. A small aliquot of sperm was diluted in artificial seawater (360 mM NaCl, 50 mM MgCl2, 10 mM CaCl2, 10 mM KCl, and 30 mM HEPES pH 8.0). The sample was pelleted by centrifugation at 1000×g and washed twice with HDMEKP buffer (30 mM HEPES pH 7.4, 50 mM KCl, 0.5 mM EDTA, 5 mM MgCl2, 1 mM DTT, Roche Protease Inhibitor), and then resuspended in HDMEKP buffer supplemented with 1% NP-40 to remove the flagellar membrane. After rotating the sample at 4°C for 1 h, demembranated sperm were pelleted by centrifugation at 1000×g, washed three times in HDMEKP buffer without NP-40, and resuspended to a final concentration of around 10 mg/ml for cryo-EM grid preparation. Samples were applied to glow-discharged Quantifoil R1.2/1.3 copper grids mounted in a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 16°C and 90% humidity. Following blotting for 4 s, the grids were plunge frozen in liquid ethane cooled to liquid nitrogen temperature.
Bovine sperm
Frozen bovine semen straws were thawed rapidly in a 37°C water bath. Sperm cells were washed twice in 1X Dulbecco’s phosphate-buffer saline (DPBS, Sigma) and counted. DMTs were exposed using a sliding disintegration protocol 28. To remove membranes and the mitochondrial sheath, sperm were diluted to 1-2 x 105 cells/mL in demembranation buffer (20 mM Tris-HCl pH 7.9, 132 mM sucrose, 24 mM potassium glutamate, 1 mM MgSO4, 1 mM DTT, and 0.1% Triton X-100), frozen at −20°C for 48-96 h, then thawed. To induce sliding disintegration, ATP (Sigma) was added to a final concentration of 1 mM and the solution incubated for 10-15 min at room temperature. Axoneme disintegration was verified under an inverted light microscope.
Approximately 4 μL of disintegrated sperm suspension was applied to glow-discharged Quantifoil R 2/1 200-mesh holey carbon grids, which were then blotted from the back for 5-6 s using a manual plunger (MPI Martinsried, Germany). Grids were plunged into a liquid ethane/propane mix (37% ethane) cooled to liquid nitrogen temperatures.
For imaging singlet microtubules (SMTs) in the bovine sperm endpiece, approximately 4 μL of whole untreated sperm cells (~1-3 x 106 cells/mL) was applied to glow-discharged Quantifoil R 2/1 200-mesh holey carbon grids. Grids were then blotted from the back for 4–6 s using a manual plunger (MPI Martinsried, Germany). Shear forces from blotting (or earlier pipetting and centrifugation steps) were sufficient to rupture the membrane around the endpiece. After blotting, grids were plunge-frozen as described above.
Chlamydomonas DMTs
Axonemes from wild type cells were isolated, dissociated into microtubules, and prepared for cryo-EM as previously described 15.
Cryo-electron microscopy
Sea urchin sperm and Chlamydomonas DMTs
Cryo-EM data of DMTs from sea urchin sperm flagella and Chlamydomonas flagella were collected using the Titan Krios microscopes (Thermo Fisher Scientific) at Case Western Reserve University (CWRU). Images were collected automatically using SerialEM 101 with parameters summarized in Table S1. Data used to calculate the cryo-EM map of the tektin-bound DMT from wild-type Chlamydomonas was reported in a previous study 15.
Bovine sperm DMTs
Cryo-EM data of DMTs from bovine sperm was collected using a Talos Arctica (Thermo Fisher Scientific) at the Utrecht University (UU) Electron Microscopy Centre and a Titan Krios at the Netherlands Centre for Electron Nanoscopy (NeCEN). A total of five datasets were collected from a total of 7 grids from 3 sperm straws. Imaging conditions are summarized in Table S1. Acquisition areas were identified manually and images were collected semi-automatically in SerialEM. Frames were motion-corrected on-the-fly using Warp 102 to monitor data quality during the session.
Bovine sperm SMTs
Cryo-EM data of SMTs from bovine sperm was collected using a Talos Arctica (Thermo Fisher Scientific) at the UU Electron Microscopy Centre. Data was collected over from a total of 8 grids from 3 sperm straws. Imaging conditions are summarized in Table S1. Acquisition areas were identified manually and images were collected semi-automatically in SerialEM. Frames were motion-corrected on-the-fly using Warp to monitor data quality during the session.
Cryo-EM data processing
Sea urchin sperm DMTs
A total of 2,900 movies were drift-corrected and dose weighted using patch motion correction in cryoSPARC 103. The parameters of contrast transfer function (CTF) for different patches of the drift-corrected micrographs were estimated using patch CTF estimation in cryoSPARC. DMTs were automatically picked using filament tracer in cryoSPARC. During the same tracing step, locations of individual DMT particles were marked using overlapping boxes with 8-nm step size. Next, DMT particles were extracted from the drift-corrected micrographs with 256-pixel box size (2x binning), and subject to two rounds of 2D classification in cryoSPARC to remove junk particles. Good DMT particles then entered structural refinement using Homogeneous Refinement (New) in cryoSPARC using a cryo-EM structure of the Chlamydomonas DMT (EMD-20631) 15 as the initial model.
We then exported particles and their associated alignment parameters to FREALIGN v9.11 104 and performed local refinement using customized scripts. During this step, most of the alignment errors could be identified and corrected, guided by the similarity score between the raw particle and the reference projection 105. The whole set of particles with improved alignment parameters was imported back into cryoSPARC and subjected to one round of local refinement, followed by tubulin signal subtraction.
To recover the 48-nm repeat from 8-nm particles, we performed 3D classification of the tubulin-subtracted DMT particles in Relion 3.1 106 using a soft-edged mask covering MIPs bound to protofilaments A08-A13. A similar strategy was used to further separate the 48-nm particles into 96-nm particles, using a soft-edged mask covering external axonemal complexes bound to protofilaments A01-A03. The coordinates of the 48-nm (and 96-nm) DMT particles were imported back into cryoSPARC, re-extracted with 512-pixel box size (no binning), and subjected to one round of local refinement followed by per-particle CTF refinement. This step produced a consensus 48-nm map.
To improve local resolution, we performed focused refinements in cryoSPARC using a set of masks modified from 15. These masks divide the DMT radially into 12 sub-regions (each encompassing ~2–3 protofilaments) and longitudinally into 3 segments for a total of 36 local refinements (Fig. S1C). To generate a composite map for model building and refinement, the 36 reconstructions were sharpened using deepEMhancer 107. Sharpened maps were multiplied by shorter versions of their corresponding masks (170 Å in length) and aligned to the consensus 48-nm map using the fit in map command in Chimera 108. Aligned maps were resampled onto the grid box of the consensus map and merged using the vop resample and vop maximum commands in Chimera. Close inspection of the resulting composite maps confirms that this process does not introduce any visible artefacts.
Bovine sperm DMTs
All data processing for bovine sperm DMTs was performed in Relion 3.1 106 based on strategies described in 15,16. Super-resolution frames were binned twice, motion-corrected, and dose-weighted using Relion’s implementation of MotionCor2. CTF parameters was estimated using CTFFIND4 109. DMTs were picked manually and particles were extracted every ~82 Å (the length of a tubulin dimer). For initial alignments, twice-binned particles were extracted with a box size of 336 (~700 Å, encompassing ~9 tubulin dimers).
Global alignment parameters were first determined for the 8-nm particles through a C1 helical auto-refinement in Relion 3.1. The cryo-EM map of the DMT from bovine respiratory cilia (EMD-24664) 16 filtered to ~30 Å was used as an initial reference. To enrich for fully decorated DMTs, tubulin signal was subtracted and 3D classification performed with a mask covering MIPs bound to protofilaments B02-B06. Classes with well-resolved density were selected for further processing. Particles offset by 4 nm were also identified at this stage and shifted back into register with the majority class. After checking for duplicate particles, the remaining particles were entered to a 3D auto-refinement, yielding a map of the 8-nm repeat. To retrieve the 16- and 48-nm repeats, tubulin signal was subtracted and 3D classification performed first with a mask covering MIPs at the inner junction, then with a mask covering MIPs at the seam of the A-tubule. The resulting 48-nm particles were re-extracted with a box size of 672 and refined. After CTF refinement (magnification anisotropy correction, followed by per-particle defocus estimation and aberration correction) and Bayesian polishing, the nominal overall resolution of the final map was ~4 Å.
To further improve resolution, we performed 36 separate local refinements, each with a cylindrical mask covering 2–3 protofilaments and extending along ~1/3 of the 48-nm repeat (Fig. S1D). To enrich for particles with MAPs bound to the seam (protofilaments A09/A10), we performed 3D classification with a soft-edged cylindrical mask around this region, then locally refined particles belonging to the MAP-bound class. We individually post-processed locally-refined maps for each subregion using deepEMhancer, then generated a composite map using the fit in map and volume maximum commands in ChimeraX 110.
Bovine sperm endpiece SMTs
All data processing was performed in Relion 3.1. Frames were motion-corrected and dose-weighted using Relion’s implementation of MotionCor2. CTF parameters were estimated using gctf 111. Microtubules were picked manually and particles were extracted every ~82 Å with a box size of 432 (~587 Å, encompassing ~7 dimers).
An initial C1 helical auto-refinement was performed using a ~20-Å subtomogram average of pig endpiece singlet microtubules (EMD-12068) 12 as a reference, resulting in a ~4.8-Å map. To improve particle alignments towards an improved C1 reconstruction, the microtubule Relion-based pipeline (MiRPv2) was used to perform rotation angle smoothing, XY shift smoothing, and seam correction 112,113. Aligned, seam-corrected particles were then subjected to a round of C1 helical auto-refinement with restricted searches and a mask encompassing the central ~40% of the microtubule. This resulted in a C1 reconstruction at ~4.6-Å resolution, which was improved to ~4.3 Å after two rounds each of CTF refinement and Bayesian polishing.
To improve MIP densities, symmetry-expansion was performed on the dataset based on helical parameters estimated from relion_helix_toolbox (rise = 9.57 Å, twist = −27.7°) 62. Particle subtraction was first run with a mask covering four tubulin dimers of the “good” protofilament opposite the seam (protofilament 7), followed by 3D classification of the resulting particles without image alignment. The class with the best MIP density was selected and entered into a local refinement with a mask on two central tubulin dimers. This resulted in a ~3.5-Å map with well-resolved side chain densities that we used for protein identification and model building.
Chlamydomonas DMTs
Cryo-EM structures of DMTs from wild-type C. reinhardtii were calculated as previously described 15. To isolate DMT particles containing tektin in the B-tubule, we subtracted tubulin signal from 48-nm DMT particles, then did 3D classification of tubulin-subtracted particles in Relion 3 using a soft-edged cylindrical mask covering the tektin densities. Two major classes were identified, corresponding to particles with and without tektin.
Map interpretation, model building, and refinement
Map interpretation was guided by atomic models of DMTs from Chlamydomonas flagella (PDB 6U42) 15 and bovine respiratory cilia (PDB 7RRO) 16. The positions of tubulin dimers and MIPs were manually adjusted by rigid-body fitting in ChimeraX 110, followed by real-space refinement in Coot v0.9.5 or v0.9.8.1 29 and in Phenix 114. Core-MIPs such as CFAP67 (NME7) and CFAP127 (MNS1) were similarly modelled this way, then subsequently adjusted based on the density. Likewise, tubulin C-terminal tails and the α-tubulin K40 loop were manually rebuilt based on the map density. For bovine sperm, α-tubulin sequences from PDB 7RRO were mutated to match the most abundant isotype identified in sperm (TUBA3); β-tubulin sequences already corresponded to the most abundant sperm isotype (TUBB4B). For sea urchin sperm, tubulin sequences were assigned to the most abundant sperm isotypes, tubulin alpha-1A (A0A7M7RGW6) and tubulin beta chain isoform X1 (A0A7M7NS69) and were confirmed by the side-chain densities (see “Mass spectrometry” section and Table S6).
Unknown MIPs were identified from databases of candidates obtained by mass spectrometry-based proteomics of sea urchin and bovine sperm (Table S6) using one of three approaches. In the first approach, polyalanine models were constructed either manually using Coot 29 or automatically using ModelAngelo 30. These backbone models were used as inputs to findMySequence 31, which estimates side chain probabilities based on map density and subsequently constructs a multiple sequence alignment that is used to perform a HMMER search 32 against sequence databases generated from our mass spectrometry data. In the second approach, models were built automatically into the cryo-EM maps using either ModelAngelo 30 or DeepTracer 33 and the predicted sequences queried against our sequence databases using protein-BLAST 34 in order to find the best match. In the third approach, which identified proteins with globular domains, backbone traces were used to query structure databases. For sea urchin sperm, we used DeepTracer-ID 35 to query a custom AlphaFold2 database 115 of the top 500 proteins in the sea urchin sperm proteome. For bovine sperm, we used the DALI server to query either the PDB or the human AlphaFold2 database 36,37. Top hits were then mutated to poly-Ala, re-fit into the density map, and used as queries for findMySequence. This approach identified DUSP21 and TSSK6 proteins in the bovine sperm DMT. All proteins identified by any of the three strategies were carefully evaluated based on sidechain densities to ensure accuracy.
Real-space refinements were performed in Phenix 1.20.1 114. For the first round of refinement, the 48-nm repeat was divided into several PDB files corresponding to each MIP or to each protofilament of the DMT. Each model was refined individually, then manually inspected in Coot. For subsequent rounds, PDB files were merged into a single model of the 48-nm repeat, which was refined as a whole against the composite map.
Mass spectrometry
Sea urchin sperm
Demembranated sea urchin sperm pellets were resuspended in HDMEKP buffer to a final concentration of 10 mg/mL. The frozen sample was sent for mass spectrometry (MS) analysis at the Proteomics and Metabolomics Facility at the University of Nebraska-Lincoln and digested with trypsin. All MS/MS samples were analyzed using Mascot version 2.7.0 (Matrix Science). Mascot was set up to search the common contaminants database (cRAP 20150130.fasta with 125 entries) and the sea urchin UniProt database (uniprot-refprot_UP000007110_Purple_sea_urchin_20211217 with 34,417 entries). Mascot was searched with a fragment ion mass tolerance of 0.6 Da and a parent ion tolerance of 10.0 PPM. A total of 641 proteins were identified in the sample. The annotated proteomics data are provided in Table S6.
Bovine sperm
Cross-linking, lysis, digestion, and peptide fractionation
All proteomics and cross-linking mass spectrometry (XL-MS) experiments were performed according to 116 on bovine sperm prepared as described above. The sperm cells were resuspended in 540 μL of PBS and supplemented with disuccinimidyl sulfoxide (Thermo Fisher Scientific) to a final concentration of 1 mM. The reaction was incubated for 30 min at 25°C with 700 rpm shaking in a ThemoMixer C (Eppendorf) and subsequently quenched for 20 min by adding Tris-HCl (final concentration 50 mM). Cells were centrifuged at 13800×g for 10 min at 4°C, and the supernatant was replaced with lysis buffer. Cells were resuspended in 1 mL of lysis buffer (100 mM Tris-HCl pH 8.5, 7 M Urea, 1% Triton X-100, 5 mM TCEP, 30 mM CAA, 10 U/mL DNase I, 1 mM MgCl2, 1% benzonase (Merck Millipore), 1 mM sodium orthovanadate, phosphoSTOP phosphatases inhibitors, and cOmplete Mini EDTA-free protease inhibitors) and lysed with the help of sonication (2 minutes with UP100H from Hielscher at 80% amplitude). The proteins were then precipitated and resuspended in digestion buffer (100 mM Tris pH 8.5, 1% sodium deoxycholate (Sigma-Aldrich), 5 mM TCEP, and 30 mM CAA). Trypsin and Lys-C proteases were added to a 1:25 and 1:100 ratio (weight/weight), respectively, and protein digestion performed overnight at 37°C shaking at 1300 rpm on ThemoMixer C. Peptides were then desalted with Oasis HLB plates (Waters) and fractionated with an Agilent 1200 HPLC pump system (Agilent) coupled to a strong cation exchange (SCX) separation column (Luna SCX 5 μm to 100 Å particles, 50×2 mm, Phenomenex), resulting in 24 fractions. Each fraction was then desalted with OASIS HLB plate.
Liquid chromatography with mass spectrometry
Before injecting each SCX fraction, 1,000 ng of peptides from each biological replicate were first injected onto an using an Ultimate3000 high-performance liquid chromatography system (Thermo Fisher Scientific) coupled online to an Orbitrap HF-X (Thermo Fisher Scientific). For this classical bottom-up analysis, we used the following parameters as in 117: Buffer A consisted of water acidified with 0.1% formic acid, while buffer B was 80% acetonitrile and 20% water with 0.1% formic acid. The peptides were first trapped for 1 min at 30 μL/min with 100% buffer A on a trap (0.3 mm by 5 mm with PepMap C18, 5 μm, 100 Å; Thermo Fisher Scientific); after trapping, the peptides were separated by a 50-cm analytical column packed with C18 beads (Poroshell 120 EC-C18, 2.7 μm; Agilent Technologies). The gradient was 9 to 45% B in 95 min at 400 nL/min. Buffer B was then raised to 55% in 10 min and increased to 99% for the cleaning step. Peptides were ionized using a spray voltage of 2 kV and a capillary heated at 275°C. The mass spectrometer was set to acquire full-scan MS spectra (350 to 1400 mass/charge ratio) for a maximum injection time of 120 ms at a mass resolution of 120,000 and an automated gain control (AGC) target value of 3 × 106. Up to 25 of the most intense precursor ions were selected for MS/MS. HCD fragmentation was performed in the HCD cell, with the readout in the Orbitrap mass analyzer at a resolution of 15,000 (isolation window of 1.4 Th) and an AGC target value of 1 × 105 with a maximum injection time of 25 ms and a normalized collision energy of 27%.
The SCX fractions were analyzed with same Ultimate HPLC and the same nano-column coupled on-line to an Orbitrap Lumos mass spectrometer (Thermo Fisher Scientific). For these runs, we used same gradient and LC setting of bottom up data with specific MS settings for DSSO cross-links: survey MS1 Orbitrap scan at 120,000 resolution from 350 to 1,400, AGC target of 250% and maximum inject time of 50 ms. For the MS2 Orbitrap scan we used 30,000 resolution, AGC target of 200%, and maximum inject time of 118 ms for detection of DSSO signature peaks (difference in mass of 37.972 Da). The four ions with this specific difference were analysed with a MS3 Ion Trap scans at AGC target of 200%, maximum inject time of 200 ms for sequencing selected signature peaks (representing the individual peptides).
Data processing for cross-linking mass spectrometry
Raw files were analyzed with MaxQuant version 1.6.17 118 with all the default settings adding Deamidation (N) as dynamic modification against the Bos taurus reference proteome (UniProt version of 02/2021 with 37,512 entries). With this search, we were able to calculate intensity-based absolute quantification values and created a smaller FASTA file to use for analysis of cross-linking experiments. Raw files for cross-linked cells were analyzed with Proteome Discoverer software version 2.5 (Thermo Fisher Scientific) with the incorporated XlinkX node for analysis of cross-linked peptides as described by 119. Data were searched against the smaller FASTA created in house with “MS2_MS3 acquisition strategy”. For the XlinkX search, we selected full tryptic digestion with three maximum missed cleavages, 10 ppm error for MS1, 20ppm for MS2, and 0.5 Da for MS3 in Ion Trap. For modifications, we used static Carbamidomethyl (C) and dynamic Oxidation (M), Deamidation (N), and Met-loss (protein N-term). The crosslinked peptides were accepted with a minimum score of 40, minimum score difference of 4, and maximum false discovery rate set to 5%; further standard settings were used. Annotated proteomics data are provided in Table S6 and XL-MS data are available from PRIDE.
Quantification and Statistical Analysis
Cryo-EM resolution estimates were performed in either cryoSPARC (for sea urchin sperm) or Relion (for bovine sperm) using the 0.143 FSC criterion proposed by 120. Validation metrics for atomic models (Table S1) were calculated with Phenix. Inter-dimer distances presented in Fig. S4 were calculated from the atomic models each cryo-EM structure using the mean of the Cα distances between α- and β-tubulin for each pair of corresponding residues based on the sequence alignment. Descriptive statistics (mean and standard deviation) were calculated with GraphPad Prism v9. Further details can be found in the figure legends.
Supplementary Material
Table S1. Cryo-EM data collection, refinement, and validation statistics, Related to STAR Methods
Table S2. Proteins identified in sea urchin sperm DMTs, Related to Figure 1.
Notes:
a CFAP91, CIMAP1A-like (A0A7M7G414), CIMAP2, CIMAP3, and SAXO5 have 96-nm periodicity and thus are not included in the atomic model of the 48-nm repeat.
b CFAP115 has 32-nm periodicity and are not included in the atomic model of the 48-nm repeat.
Table S3. Proteins identified in bovine sperm DMTs, Related to Figure 1.
Notes:
a CFAP91, CIMAP2, CIMAP3, and SAXO5 have 96-nm periodicity and thus are not included in the atomic model of the 48-nm repeat.
b SAXO1 is tentatively assigned based on in-cell cross-links to SPACA9, so it is only modelled as a poly-Ala trace.
Table S4. Comparing MIPs across different species and cell types, Related to Figure 2.
Table S5. Infertility and ciliopathy phenotypes, Related to Figure 7.
Table S6. Mass spectrometry data of sea urchin and bovine sperm, Related to STAR Methods.
Figure S1. Sample preparation and data processing workflows for cryo-electron microscopy of native sperm DMTs, Related to Figure 1.
(A-B) Schematic diagrams describing preparation of native sperm DMTs for cryo-EM. (A) Sea urchin sperm were demembranated with Nonidet P-40 and the mitochondria removed with dithiothreitol (DTT), after which cells were directly blotted onto EM grids. Surface tension caused the axonemes to splay apart in the holes. (B) Bovine sperm were demembranated with Triton X-100 and the mitochondrial sheath stripped with DTT and a freeze-thaw cycle. Microtubule sliding was initiated by adding ATP to thawed sperm, which after ~10-15 min disintegrated the axonemes sufficiently for cryo-EM.
(C-D) DMTs were divided into 36 sub-regions for local refinement (12 regions around the DMT covering 2-3 protofilaments and ~1/3 of the 48-nm repeat). Masks and resolution estimates for local refinements of the middle of the DMT are shown for (C) sea urchin and (D) bovine sperm.
(E-F) Fourier shell correlation (FSC) curves for each of the 36 local refinements performed for (C) sea urchin and (D) bovine sperm DMTs.
Figure S2. Longitudinal slices through cryo-EM density maps of sperm DMTs, Related to Figure 1.
(A-D) Longitudinal slices are shown for sea urchin sperm (left) and bovine sperm (right), with viewing directions indicated by red arrows in the insets. Notable sperm-MIPs and MIPs that are different between sea urchin and bovine are labelled.
Figure S3. Tektin-5 adopts diverse conformations in bovine sperm DMTs, Related to Figure 3.
(A) Positions and conformations of TEKT-5 groups in the bovine sperm DMT. For reference, the locations of tryptophan residues are indicated with black (Trp115, N-terminal region), blue (Trp155, 1A helix), and green (Trp336, 2A helix) arrows. A schematic of the tektin domain arrangement and corresponding color-code is provided at the bottom of the figure. TEKT5-A, B, and C all adopt the canonical tektin conformation. Note how the N-terminal region is not resolved in the other groups, except in TEKT5-F where part of it refolds (dark blue arrowhead). In the TEKT5-D group, the L12 loops are ordered by interaction with NME7 (compare green arrowheads, see also Fig. 2d). The L2 loop of the “straight” tektin is also resolved (compare orange arrowheads) because it is stabilized by interaction with a neighboring tektin.
(B) Interactions between NME7 and the TEKT5-D and TEKT5-E filaments. The monomers in the TEKT5-D filament adopt different conformations depending on whether their 2A/2B helices interact with NME7 (“bent” TEKT5-D) or a neighboring tektin (“straight” TEKT5-D).
(C) The L12 loop of the “straight” TEKT5-D monomer inserts into the putative nucleotide-binding pocket of NME7. The L12 of one “bent” TEKT5-D monomer interacts with the second copy of NME-7 in a similar fashion. Panel (C) is rotated 10° relative to the view in (B).
(D) Cryo-EM density of the interfaces between tektin-4 and protofilament A12 in sea urchin (left) and bovine sperm (middle), and between tektin and protofilament B05 in Chlamydomonas flagella (right). Tektin-4 interacts primarily with the K40 loop of α-tubulin, whereas Chlamydomonas tektin interacts mainly with β-tubulin.
Figure S4. SAXO proteins in the Chlamydomonas DMT and conservation of Mn-motifs across species, Related to Figure 5.
(A) Location and identities of SAXO proteins in the DMT from Chlamydomonas flagella. As in Fig. 5, SAXO proteins are classified and colored based on how they bind the microtubule lattice: via a single Mn-motif (blue), via multiple Mn-motifs along a single protofilament (violet), or via multiple Mn-motifs across neighboring protofilaments (orange).
(B) In wild-type Chlamydomonas, SAXO proteins bound to protofilaments B04-B06 are only visible in classes with the tektin bundle.
(C-D) Mn motifs are conserved across species in both (B) core-MIPs (CFAP68) and (C) sperm-MIPs (SPMIP4, formerly C7ORF31). Top panels show cartoon models with Mn motifs highlighted in red. Bottom panels show sequence alignments of Mn motifs in several animal species. NCBI accession numbers of CFAP68 sequences are as follows: NP_073598.1 (Hs, Homo sapiens), NP_001033266.1 (Bt, Bos taurus), NP_075972.2 (Mm, Mus musculus), XP_040546331.1 (Gg, Gallus gallus), XP_018081403.1 (Xl, Xenopus laevis), NP_001313533.1 (Dr, Danio rerio), XP_784695.1 (Sp, Strongylocentrotus purpuratus). Accession numbers for SPMIP4 (C7ORF31) sequences are as follows: NP_001358280.1 (Hs), XP_024846866.1 (Bt), NP_081840.1 (Mm), XP_025003204.2 (Gg), XP_018122881.1 (Xl).
(E) Comparisons of tubulin inter-dimer distances across DMTs from sea urchin sperm, bovine sperm, bovine trachea, and Chlamydomonas flagella. Bars represent means and standard deviations of measurements from six dimers, with each individual measurement represented as a circle.
(F) The bimodal distribution of tubulin inter-dimer distances at the ribbon is conserved. Inter-dimer distances are plotted for the six tubulin dimers of protofilaments A01-A03 and A11-A13.
Figure S5. Cryo-EM of native singlet microtubules in the bovine sperm endpiece, Related to Figure 5.
(A) Cryo-EM image of a frayed sperm endpiece on a holey carbon grid. Scale bar: 1 μm. Inset: High-magnification cryo-EM image of endpiece singlet microtubules (SMTs) captures a ladder-like luminal structure with an 8-nm periodicity. Scale bar: 100 nm.
(B) Cross section of the cryo-EM reconstruction of endpiece SMTs reveals discontinuous intraluminal spirals of SPACA9.
(C) Map obtained after symmetry expansion showing SPACA9 and a SAXO protein (likely SAXO1) bound to a tubulin heterodimer.
(D) Local resolution of the symmetry-expanded reconstruction, estimated by the “Local Resolution” job in Relion.
(E) Fourier shell correlation (FSC) plots of the full C1 reconstruction (black curve), the local refinement of protofilaments 7/8 (blue curve), and the protofilament reconstruction after symmetry expansion (pink curve).
(F) Examples of map quality in the symmetry-expanded map used for model building.
(G) Sequence alignment of the 12 Mn motifs within bovine SAXO1 shows the consensus Thr and Tyr residues (starred) demarcating the short helix that binds at the α/β-tubulin interface. Orange boxes mark lysine residues (K83 and K221) that cross-linked to SPACA9 and α-tubulin respectively.
(H) In-cell cross-linking mass spectrometry detects interactions between SAXO1 and α-tubulin (left); and between SAXO1 and SPACA9 (right). Red spheres indicate the cross-linked lysine residues. See also Table S6.
Figure S6. 96-nm features in sperm DMTs, Related to Figures 5 and 6.
96-nm maps were generated by classifying on the bases of external axonemal complexes bound to protofilaments A02-A03, followed by refining each half of the 96-nm repeat separately.
(A) SAXO5 (formerly TEX45) is a filamentous SAXO protein that binds along protofilament A04 with 96-nm periodicity.
(B) The wedge-MAPs CIMAP2 and CIMAP3 bind to the inter-protofilament cleft between A03/A04, where they alternate to yield an overall 96-nm repeat.
(C) CFAP115, found in sea urchin but not bovine sperm, has 32-nm periodicity.
(D) CFAP91, a component of the RS2 base with 96-nm periodicity, inserts into the inner junction hole formed by the absence of one copy of PACRG in every 96-nm repeat.
Figure S7. CIMAP1 (ODF3) proteins constitute a conserved family of axonemal MAPs, Related to Figure 6.
(A) Extracted density for CIMAP1A and neighboring protofilaments B06 and B07 from the bovine sperm cryo-EM map. The two CIMAP1A molecules are shown in blue-violet and purple, with Pro-Gly-Pro (PGP) motifs in red; α-tubulin is shown in white and β-tubulin in grey.
(B) Zoomed-in view showing head-to-tail interactions between neighboring CIMAP1A molecules. A PGP motif is indicated in red.
(C) The C-terminal tails of CIMAP1 proteins extend into the DMT lumen, where they can bind MIPs. For example, CIMAP1C (ODF3L1) between protofilaments A08/A09 interacts with the sperm-MIP SPMIP4 (C7ORF31).
(D) Sequence alignment of CIMAP1A and CIMAP1C proteins from several animal species, showing conservation of PGP motifs. NCBI accession numbers of CIMAP1A (ODF3) sequences are as follows: NP_444510.2 (Hs), XP_024854250.1 (Bt), NP_081295.2 (Mm), XP_024997911.1 (Gg), NP_001079370.1 (Xl), NP_956252.1 (Dr), NP_723368.1 (Dm), XP_003729878.1 (Sp). Accession numbers of CIMAP1C (ODF3L) sequences are as follows: NP_787077.1 (Hs), XP_005221973.2 (Bt), NP_001398329.1 (Mm), XP_024997911.1 (Gg), NP_001079644.1 (Xl), NP_001017836.1 (Dr), NP_651171.1 (Dm), XP_030833199.1 (Sp).
Acknowledgements
Electron microscopy of DMTs from Chlamydomonas flagella and sea urchin sperm was performed at either Washington University in St. Louis Center for Cellular Imaging (WUCCI) or Case Western Research University (CWRU). Imaging of DMTs from bovine sperm was performed at the Utrecht University (UU) Electron Microscopy Centre and at the Netherlands Centre for Electron Microscopy (NeCEN). We thank M. Rau (WUCCI), K. Li (CWRU), Ingr. C. Schneijdenberg, J. Meeldijk (UU), and Dr. W. Noteborn (NeCEN) for microscopy support. We acknowledge M. Naldrett and S. Alvarez at the Proteomics and Metabolomics Facility at the University of Nebraska-Lincoln for proteomics analysis of sea urchin sperm. We thank Dr. H. Henning and A. Rijneveld at the UU Veterinary Faculty for assistance with bovine sperm samples. We also thank Dr. M. Vanevic for computational support, Dr. S. Howes for valuable discussions on data collection and processing, Dr. H. Bloomfield-Gadelha for valuable discussions on microtubule biomechanics and Prof. A. Akhmanova for critical reading of early versions of the manuscript.
R.Z.C., J.F.H., and A.J.R.H. acknowledge support from the NWO X-omics Road Map program project 184.034.019. S.K.D. is supported by NIGMS grant R35-GM131909. A.B. is supported by NIGMS grant R01GM141109. This work was funded by NIGMS grant 1R01GM138854 (R.Z.) and NWO Start-Up Grant 740.018.007 (T.Z.).
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- 1.Suarez SS, and Pacey AA (2006). Sperm transport in the female reproductive tract. Hum. Reprod. Update 12, 23–37. 10.1093/humupd/dmi047. [DOI] [PubMed] [Google Scholar]
- 2.De Jonge C, and Barratt CLR (2019). The present crisis in male reproductive health: an urgent need for a political, social, and research roadmap. Andrology 7, 762–768. 10.1111/andr.12673. [DOI] [PubMed] [Google Scholar]
- 3.Ravitsky V, and Kimmins S (2019). The forgotten men: rising rates of male infertility urgently require new approaches for its prevention, diagnosis and treatment. Biol. Reprod 101, 872–874. 10.1093/biolre/ioz161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada A, Esteves SC, and Agarwal A (2011). Unexplained male infertility. Hum. Androl 1, 2–16. 10.1097/01.xha.0000397686.82729.09. [DOI] [Google Scholar]
- 5.Hemmerling A, Christopher E, and Young Holt B (2020). Towards a roadmap to advance non-hormonal contraceptive multipurpose prevention technologies: strategic insights from key stakeholders†. Biol. Reprod 103, 289–298. 10.1093/biolre/ioaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan KY (2018). Coordination of eukaryotic cilia and flagella. Essays Biochem 62, 829–838. 10.1042/EBC20180029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilpin W, Bull MS, and Prakash M (2020). The multiscale physics of cilia and flagella. Nat. Rev. Phys 2, 74–88. 10.1038/s42254-019-0129-0. [DOI] [Google Scholar]
- 8.Beeby M, Ferreira JL, Tripp P, Albers S-V, and Mitchell DR (2020). Propulsive nanomachines: the convergent evolution of archaella, flagella and cilia. FEMS Microbiol. Rev 44, 253–304. 10.1093/femsre/fuaa006. [DOI] [PubMed] [Google Scholar]
- 9.Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, and Ishikawa T (2012). Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J. Struct. Biol 178, 199–206. 10.1016/j.jsb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Yin W, Smith MC, Song K, Leigh MW, Zariwala MA, Knowles MR, Ostrowski LE, and Nicastro D (2014). Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat. Commun 5, 5727. 10.1038/ncomms6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imhof S, Zhang J, Wang H, Bui KH, Nguyen H, Atanasov I, Hui WH, Yang SK, Zhou ZH, and Hill KL (2019). Cryo electron tomography with Volta phase plate reveals novel structural foundations of the 96-nm axonemal repeat in the pathogen Trypanosoma brucei. Elife 8, 1–30. 10.7554/elife.52058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung MR, Roelofs MC, Ravi RT, Maitan P, Henning H, Zhang M, Bromfield EG, Howes SC, Gadella BM, Bloomfield-Gadêlha H, et al. (2021). The multi-scale architecture of mammalian sperm flagella and implications for ciliary motility. EMBO J. 40, 1–17. 10.15252/embj.2020107410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicastro D, Fu X, Heuser T, Tso A, Porter ME, and Linck RW (2011). Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci 108, E845–E853. 10.1073/pnas.1106178108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinskey JM, Lagisetty A, Gui L, Phan N, Reetz E, Tavakoli A, Fu G, and Nicastro D (2022). Three-dimensional flagella structures from animals’ closest unicellular relatives, the Choanoflagellates. Elife 11, 2022.02.24.481817. 10.7554/eLife.78133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, and Zhang R (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell 179, 909–922.e12. 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui M, Farley H, Anujan P, Anderson JR, Maxwell DW, Whitchurch JB, Botsch JJ, Qiu T, Meleppattu S, Singh SK, et al. (2021). De novo identification of mammalian ciliary motility proteins using cryo-EM. Cell 184, 5791–5806.e19. 10.1016/j.cell.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gui M, Croft JT, Zabeo D, Acharya V, Kollman JM, Burgoyne T, Höög JL, and Brown A (2022). SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules. Proc. Natl. Acad. Sci. U. S. A 119, e2207605119. 10.1073/pnas.2207605119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo S, Black C, Joachimiak E, Yang SK, Legal T, Peri K, Khalifa A, Ghanaeian A, Valente-Paterno M, Bellis C. De, et al. (2022). Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins. bioRxiv 2022.09.30. 10.1101/2022.09.30.510376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owa M, Uchihashi T, Yanagisawa H. aki, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi K. ichi, Ando T, and Kikkawa M (2019). Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat. Commun 10, 1143. 10.1038/s41467-019-09051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Touré A, et al. (2021). Tubulin glycylation controls axonemal dynein activity, flagellar beat, and male fertility. Science. 371, eabd4914. 10.1126/science.abd4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afzelius BA, Dallai R, Lanzavecchia S, and Bellon PL (1995). Flagellar structure in normal human spermatozoa and in spermatozoa that lack dynein arms. Tissue Cell 27, 241–247. 10.1016/S0040-8166(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 22.Kahrl AF, Snook RR, and Fitzpatrick JL (2021). Fertilization mode drives sperm length evolution across the animal tree of life. Nat. Ecol. Evol 5, 1153–1164. 10.1038/s41559-021-01488-y. [DOI] [PubMed] [Google Scholar]
- 23.Pitnick S, Hosken DJ, and Birkhead TR (2009). Sperm morphological diversity. In Sperm Biology (Elsevier Ltd; ), pp. 69–149. 10.1016/B978-0-12-372568-4.00003-3. [DOI] [Google Scholar]
- 24.Fawcett DW (1970). A comparative view of sperm ultrastructure. Biol. Reprod. Suppl 2, 90–127. 10.3138/9781442676824-011. [DOI] [PubMed] [Google Scholar]
- 25.Fawcett DW (1975). The mammalian spermatozoon. Dev. Biol 44, 394–436. 10.1016/0012-1606(75)90411-X. [DOI] [PubMed] [Google Scholar]
- 26.Lindemann CB, and Lesich KA (2016). Functional anatomy of the mammalian sperm flagellum. Cytoskeleton 73, 652–669. 10.1002/cm.21338. [DOI] [PubMed] [Google Scholar]
- 27.Gadêlha H, and Gaffney EA (2019). Flagellar ultrastructure suppresses buckling instabilities and enables mammalian sperm navigation in high-viscosity media. J. R. Soc. Interface 16, 20180668. 10.1098/rsif.2018.0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindemann CB, Fentie I, and Rikmenspoel R (1980). A selective effect of Ni2+ on wave initiation in bull sperm flagella. J. Cell Biol 87, 420–426. 10.1083/jcb.87.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamali K, Kimanius D, and Scheres S (2022). ModelAngelo: Automated Model Building in Cryo-EM Maps. 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chojnowski G, Simpkin AJ, Leonardo DA, Seifert-Davila W, Vivas-Ruiz DE, Keegan RM, and Rigden DJ (2022). FindMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM. IUCrJ 9, 86–97. 10.1107/S2052252521011088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eddy SR (2011). Accelerated profile HMM searches. PLoS Comput. Biol 7. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfab J, Phan NM, and Si D (2021). DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on cov-related complexes. Proc. Natl. Acad. Sci. U. S. A 118. 10.1073/pnas.2017525118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Chang L, Wang F, Connolly K, Meng H, Su Z, Cvirkaite-Krupovic V, Krupovic M, Egelman EH, and Si D (2022). DeepTracer-ID: De novo protein identification from cryo-EM maps. Biophys. J 121, 2840–2848. 10.1016/j.bpj.2022.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holm L (2022). Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215. 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, and Rosenström P (2010). Dali server: Conservation mapping in 3D. Nucleic Acids Res. 38, 545–549. 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science. 347. 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 39.Amos LA (2008). The tektin family of microtubule-stabilizing proteins. Genome Biol. 9. 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bastin BR, and Schneider SQ (2019). Taxon-specific expansion and loss of tektins inform metazoan ciliary diversity. BMC Evol. Biol 19, 1–25. 10.1186/s12862-019-1360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, Maekawa M, and Nishimune Y (2004). Mice Deficient in the Axonemal Protein Tektin-t Exhibit Male Infertility and Immotile-Cilium Syndrome Due to Impaired Inner Arm Dynein Function. Mol. Cell. Biol 24, 7958–7964. 10.1128/mcb.24.18.7958-7964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy A, Lin Y-N, Agno JE, DeMayo FJ, and Matzuk MM (2007). Absence of tektin 4 causes asthenozoospermia and subfertility in male mice. FASEB J. 21, 1013–1025. 10.1096/fj.06-7035com. [DOI] [PubMed] [Google Scholar]
- 43.Roy A, Lin YN, Agno JE, Demayo FJ, and Matzuk MM (2009). Tektin 3 is required for progressive sperm motility in mice. Mol. Reprod. Dev 76, 453–459. 10.1002/mrd.20957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S. hong, Zhang J. hui, Ding X. ping, Zhang S, Chen H. han, and Jing Y. ling (2016). Association of polymorphisms in tektin-t gene with idiopathic asthenozoospermia in Sichuan, China. J. Assist. Reprod. Genet 33, 181–187. 10.1007/s10815-015-0617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuccarello D, Ferlin A, Garolla A, Pati MA, Moretti A, Cazzadore C, Francavilla S, and Foresta C (2008). A possible association of a human tektin-t gene mutation (A229V) with isolated non-syndromic asthenozoospermia: Case Report. Hum. Reprod 23, 996–1001. 10.1093/humrep/dem400. [DOI] [PubMed] [Google Scholar]
- 46.Wyrwoll MJ, Wabschke R, Röpke A, Wöste M, Ruckert C, Perrey S, Rotte N, Hardy J, Astica L, Lupiáñez DG, et al. (2022). Analysis of copy number variation in men with non-obstructive azoospermia. Andrology, 1–12. 10.1111/andr.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aoki N, and Matsui Y (2019). Comprehensive Analysis of Mouse Cancer/Testis Antigen Functions in Cancer Cells and Roles of TEKT5 in Cancer Cells and Testicular Germ Cells. Mol. Cell. Biol 39. 10.1128/MCB.00154-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, and Nicastro D (2014). Insights into the structure and function of ciliary and flagellar doublet microtubules: tektins, Ca2+-binding proteins, and stable protofilaments. J. Biol. Chem 289, 17427–17444. 10.1074/jbc.M114.568949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dymek EE, Lin J, Fu G, Porter ME, Nicastro D, and Smith EF (2019). PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. Mol. Biol. Cell 30, 1805–1816. 10.1091/mbc.e19-01-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoops HJ, and Witman GB (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol 97, 902–908. 10.1083/jcb.97.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagisawa H, and Kamiya R (2004). A Tektin Homologue Is Decreased in Chlamydomonas Mutants Lacking an Axonemal Inner-Arm Dynein. Mol. Biol. Cell 15, 2105–2115. 10.1091/mbc.e03-11-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanagisawa HA, Mathis G, Oda T, Hirono M, Richey EA, Ishikawa H, Marshall WF, Kikkawa M, and Qin H (2014). FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. Mol. Biol. Cell 25, 1472–1483. 10.1091/mbc.E13-08-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabach Y, Golan T, Hernández-Hernández A, Messer AR, Fukuda T, Kouznetsova A, Liu JG, Lilienthal I, Levy C, and Ruvkun G (2013). Human disease locus discovery and mapping to molecular pathways through phylogenetic profiling. Mol. Syst. Biol 9, 1–17. 10.1038/msb.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Zhang F, Li F, Jiang X, Yang Y, Li X, Li W, Wang X, Cheng J, Liu M, et al. (2019). Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun 10, 1–15. 10.1038/s41467-018-08182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Fu Y, Beatty WL, Ma M, Brown A, David Sibley L, and Zhang R (2021). Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins. Nat. Commun 12, 3065. 10.1038/s41467-021-23351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bosc C, Andrieux A, and Job D (2003). STOP Proteins. Biochemistry 42, 12125–12132. 10.1021/bi0352163. [DOI] [PubMed] [Google Scholar]
- 57.Cuveillier C, Delaroche J, Seggio M, Gory-Fauré S, Bosc C, Denarier E, Bacia M, Schoehn G, Mohrbach H, Kulić I, et al. (2020). MAP6 is an intraluminal protein that induces neuronal microtubules to coil. Sci. Adv 6. 10.1126/sciadv.aaz4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dacheux D, Roger B, Bosc C, Landrein N, Roche E, Chansel L, Trian T, Andrieux A, Papaxanthos-Roche A, Marthan R, et al. (2015). Human FAM154A (SAXO1) is a microtubule-stabilizing protein specific to cilia and related structures. J. Cell Sci 128, 1294–1307. 10.1242/jcs.155143. [DOI] [PubMed] [Google Scholar]
- 59.Lefèvre J, Savarin P, Gans P, Hamon L, Clément MJ, David MO, Bosc C, Andrieux A, and Curmi PA (2013). Structural basis for the association of MAP6 protein with microtubules and its regulation by calmodulin. J. Biol. Chem 288, 24910–24922. 10.1074/jbc.M113.457267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petersen C, Aumüller G, Bahrami M, and Hoyer-Fender S (2002). Molecular Cloning of Odf3 Encoding a Novel Coiled-Coil Protein of Sperm Tail Outer Dense Fibers. Mol. Reprod. Dev 61, 102–112. 10.1002/mrd.1136. [DOI] [PubMed] [Google Scholar]
- 61.De Carvalho CE, Tanaka H, Iguchi N, Ventelä S, Nojima H, and Nishimune Y (2002). Molecular cloning and characterization of a complementary DNA encoding sperm tail protein SHIPPO 1. Biol. Reprod 66, 785–795. 10.1095/biolreprod66.3.785. [DOI] [PubMed] [Google Scholar]
- 62.Ferro LS, Fang Q, Eshun-Wilson L, Fernandes J, Jack A, Farrell DP, Golcuk M, Huijben T, Costa K, Gur M, et al. (2022). Structural and functional insight into regulation o kinesin-1 by microtubule-associated protein MAP7. Science. 375, 326–331. 10.1126/science.abf6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, and Nogales E (2018). Near-atomic model of microtubule-tau interactions. Science. 360, 1242–1246. 10.1126/science.aat1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gui M, Wang X, Dutcher SK, Brown A, and Zhang R (2022). Ciliary central apparatus structure reveals mechanisms of microtubule patterning. Nat. Struct. Mol. Biol 29, 483–492. 10.1038/s41594-022-00770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oura S, Kazi S, Savolainen A, Nozawa K, Castañeda J, Yu Z, Miyata H, Matzuk RM, Hansen JN, Wachten D, et al. (2020). Cfap97d1 is important for flagellar axoneme maintenance and male mouse fertility. PLoS Genet. 16, 1–22. 10.1371/JOURNAL.PGEN.1008954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lesich KA, de Pinho TG, Dang L, and Lindemann CB (2014). Ultrastructural evidence that motility changes caused by variations in ATP, Mg2+, and ADP correlate to conformational changes in reactivated bull sperm axonemes. Cytoskeleton 71, 649–661. 10.1002/cm.21199. [DOI] [PubMed] [Google Scholar]
- 67.Sosnik J, Miranda PV, Spiridonov NA, Yoon S-Y, Fissore RA, Johnson GR, and Visconti PE (2009). Tssk6 is required for Izumo relocalization and gamete fusion in the mouse. J. Cell Sci 122, 2741–2749. 10.1242/jcs.047225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiridonov NA, Wong L, Zerfas PM, Starost MF, Pack SD, Paweletz CP, and Johnson GR (2005). Identification and Characterization of SSTK, a Serine/Threonine Protein Kinase Essential for Male Fertility. Mol. Cell. Biol 25, 4250–4261. 10.1128/mcb.25.10.4250-4261.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nayyab S, Gervasi MG, Tourzani DA, Caraballo DA, Jha KN, Teves ME, Cui W, Georg GI, Visconti PE, and Salicioni AM (2021). TSSK3, a novel target for male contraception, is required for spermiogenesis. Mol. Reprod. Dev 88, 718–730. 10.1002/mrd.23539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nozawa K, Garcia TX, Kent K, Leng M, Jain A, Malovannaya A, Yuan F, Yu Z, Ikawa M, and Matzuk MM (2022). Testis-specific serine kinase 3 is required for sperm morphogenesis and male fertility. Andrology, 1–14. 10.1111/andr.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu B, Hao Z, Jha KN, Zhang Z, Urekar C, Digilio L, Pulido S, Strauss JF, Flickinger CJ, and Herr JC (2008). Targeted deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency. Dev. Biol 319, 211–222. 10.1016/j.ydbio.2008.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Wei Y, Fu G, Li H, Saiyin H, Lin G, Wang Z, Chen S, and Yu L (2014). Tssk4 is essential for maintaining the structural integrity of sperm flagellum. Mol. Hum. Reprod 21, 136–145. 10.1093/molehr/gau097. [DOI] [PubMed] [Google Scholar]
- 73.Su D, Zhang W, Yang Y, Zhang H, Liu YQ, Bai G, Ma YX, Peng Y, and Zhang SZ (2010). C.822+126TG/C: A novel triallelic polymorphism of the TSSK6 gene associated with spermatogenic impairment in a Chinese population. Asian J. Androl 12, 234–239. 10.1038/aja.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su D, Zhang W, Yang Y, Deng Y, Ma Y, Song H, and Zhang S (2008). Mutation screening and association study of the TSSK4 gene in Chinese infertile men with impaired spermatogenesis. J. Androl 29, 374–378. 10.2164/jandrol.107.004598. [DOI] [PubMed] [Google Scholar]
- 75.Lindemann CB (1996). Functional significance of the outer dense fibers of mammalian sperm examined by computer simulations with the geometric clutch model. Cell Motil. Cytoskeleton 34, 258–270. . [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Li H, Fu G, Wang Y, Du S, Yu L, Wei Y, and Chen S (2016). Testis-specific serine/threonine protein kinase 4 (Tssk4) phosphorylates Odf2 at Ser-76. Sci. Rep 6, 1–8. 10.1038/srep22861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoyer-Fender S, Petersen C, Brohmann H, Rhee K, and Wolgemuth DJ (1998). Mouse odf2 cDNAs consist of evolutionary conserved as well as highly variable sequences and encode outer dense fiber proteins of the sperm tail. Mol. Reprod. Dev 51, 167–175. . [DOI] [PubMed] [Google Scholar]
- 78.Harrison RAP (2004). Rapid PKA-Catalysed Phosphorylation of Boar Sperm Proteins Induced by the Capacitating Agent Bicarbonate. Mol. Reprod. Dev 67, 337–352. 10.1002/mrd.20028. [DOI] [PubMed] [Google Scholar]
- 79.Austin CR (1952). The capacitation of the mammalian sperm. Nature 170, 326. 10.1038/170326a0. [DOI] [PubMed] [Google Scholar]
- 80.Chang MC (1959). Fertilization of rabbit ova in vitro. Nature 184, 466–467. 10.1038/184466a0. [DOI] [PubMed] [Google Scholar]
- 81.Yanagimachi R (1970). The movement of golden hamster spermatozoa before and after capacitation. Reproduction 23, 193–196. 10.1530/jrf.0.0230193. [DOI] [PubMed] [Google Scholar]
- 82.Konno A, Padma P, Ushimaru Y, and Inaba K (2010). Multidimensional Analysis of Uncharacterized Sperm Proteins in Ciona intestinalis : EST-Based Analysis and Functional Immunoscreening of Testis-Expressed Genes. Zoolog. Sci 27, 204–215. 10.2108/zsj.27.204. [DOI] [PubMed] [Google Scholar]
- 83.Sigg MA, Menchen T, Lee C, Johnson J, Jungnickel MK, Choksi SP, Garcia G, Busengdal H, Dougherty GW, Pennekamp P, et al. (2017). Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Dev. Cell 43, 744–762.e11. 10.1016/j.devcel.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kinzel D, Boldt K, Davis EE, Burtscher I, Trümbach D, Diplas B, Attié-Bitach T, Wurst W, Katsanis N, Ueffing M, et al. (2010). Pitchfork Regulates Primary Cilia Disassembly and Left-Right Asymmetry. Dev. Cell 19, 66–77. 10.1016/j.devcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pampaloni F, Lattanzi G, Jonáš A, Surrey T, Frey E, and Florin EL (2006). Thermal fluctuations of grafted microtubules provide evidence of a length-dependent persistence length. Proc. Natl. Acad. Sci. U. S. A 103, 10248–10253. 10.1073/pnas.0603931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taute KM, Pampaloni F, Frey E, and Florin EL (2008). Microtubule dynamics depart from the wormlike chain model. Phys. Rev. Lett 100, 1–4. 10.1103/PhysRevLett.100.028102. [DOI] [PubMed] [Google Scholar]
- 87.Dutcher SK (2020). Asymmetries in the cilia of Chlamydomonas. Philos. Trans. R. Soc. B Biol. Sci 375. 10.1098/rstb.2019.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Deurs B, and Lastein U (1973). Ultrastructure of the spermatozoa of the teleost Pantodon buchholzi Peters, with particular reference to the midpiece. J. Ultrasructure Res 42, 517–533. 10.1016/S0022-5320(73)80024-3. [DOI] [PubMed] [Google Scholar]
- 89.Yamaguchi H, Oda T, Kikkawa M, and Takeda H (2018). Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly. Elife 7, 1–25. 10.7554/elife.36979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Linck RW, Olson GE, and Langevin GL (1981). Arrangement of tubulin subunits and microtubule-associated proteins in the central-pair microtubule apparatus of squid (Loligo pealei) sperm flagella. J. Cell Biol 89, 309–322. 10.1083/jcb.89.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu JQ, Yang WX, You ZJ, and Jiao HF (2005). The ultrastructure of the spermatozoon of Octopus tankahkeei. J. Shellfish Res 24, 1203–1207. 10.2983/0730-8000(2005)24[1203:TUOTSO]2.0.CO;2. [Google Scholar]
- 92.Thurston RJ, and Hess RA (1987). Ultrastructure of spermatozoa from domesticated birds: comparative study of turkey, chicken and guinea fowl. Scanning Microsc. 1, 1829–1838. [PubMed] [Google Scholar]
- 93.Hess RA, Thurston RJ, and Gist DH (1991). Ultrastructure of the turtle spermatozoon. Anat. Rec 229, 473–481. 10.1002/ar.1092290406. [DOI] [PubMed] [Google Scholar]
- 94.Scheltinga DM, Jamieson BGM, Espinoza RE, and Orrell KS (2001). Descriptions of the mature spermatozoa of the lizardsCrotaphytus bicinctores,Gambelia wislizenii (Crotaphytidae), andAnolis carolinensis (Polychrotidae) (Reptilia, Squamata, Iguania). J. Morphol 247, 160–171. . [DOI] [PubMed] [Google Scholar]
- 95.Bernardini G, Stipani R, and Melone G (1986). The ultrastructure of Xenopus spermatozoon. J. Ultrastruct. Res. Mol. Struct. Res 94, 188–194. 10.1016/0889-1605(86)90065-0. [DOI] [Google Scholar]
- 96.Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A, Kiyozumi D, Kriseman ML, Mashiko D, Matsumura T, et al. (2016). Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc. Natl. Acad. Sci 113, 7704–7710. 10.1073/pnas.1608458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyata H, Oura S, Morohoshi A, Shimada K, Mashiko D, Oyama Y, Kaneda Y, Matsumura T, Abbasi F, and Ikawa M (2021). SPATA33 localizes calcineurin to the mitochondria and regulates sperm motility in mice. Proc. Natl. Acad. Sci. U. S. A 118, 1–9. 10.1073/pnas.2106673118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Salicioni AM, Gervasi MG, Sosnik J, Tourzani DA, Nayyab S, Caraballo DA, and Visconti PE (2020). Testis-specific serine kinase protein family in male fertility and as targets for non-hormonal male contraception. Biol. Reprod 103, 264–274. 10.1093/biolre/ioaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen Z, Greenan GA, Shiozaki M, Liu Y, Skinner WM, Zhao X, Zhao S, Yan R, Yu Z, Lishko PV, et al. (2023). In situ cryo-electron tomography reveals the asymmetric architecture of mammalian sperm axonemes. Nat. Struct. Mol. Biol, 2022.05.15.492011. 10.1038/s41594-022-00861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin J, Heuser T, Carbajal-González BI, Song K, and Nicastro D (2012). The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton 69, 88–100. 10.1002/cm.21000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 102.Tegunov D, and Cramer P (2019). Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152. 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). CryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 104.Grigorieff N (2007). FREALIGN: High-resolution refinement of single particle structures. J. Struct. Biol 157, 117–125. 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 105.Zhang R, and Nogales E (2015). A new protocol to accurately determine microtubule lattice seam location. J. Struct. Biol 192, 245–254. 10.1016/j.jsb.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, and Scheres SHW (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, 1–22. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanchez-Garcia R, Gomez-Blanco J, Cuervo A, Carazo JM, Sorzano COS, and Vargas J (2021). DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol 4, 1–8. 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 109.Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, and Ferrin TE (2018). UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci 27, 14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cook AD, Manka SW, Wang S, Moores CA, and Atherton J (2020). A microtubule RELION-based pipeline for cryo-EM image processing. J. Struct. Biol 209, 107402. 10.1016/j.jsb.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cook AD, Roberts AJ, Atherton J, Tewari R, Topf M, and Moores CA (2021). Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition. J. Biol. Chem 297, 101063. 10.1016/j.jbc.2021.101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr 74, 531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leung MR, Zenezini Chiozzi R, Roelofs MC, Hevler JF, Ravi RT, Maitan P, Zhang M, Henning H, Bromfield EG, Howes SC, et al. (2021). In-cell structures of conserved supramolecular protein arrays at the mitochondria–cytoskeleton interface in mammalian sperm. Proc. Natl. Acad. Sci 118, 1–10. 10.1073/pnas.2110996118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krstic J, Reinisch I, Schindlmaier K, Galhuber M, Riahi Z, Berger N, Kupper N, Moyschewitz E, Auer M, Michenthaler H, et al. (2022). Fasting improves therapeutic response in hepatocellular carcinoma through p53-dependent metabolic synergism. Sci. Adv 8. 10.1126/sciadv.abh2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 119.Klykov O, Steigenberger B, Pektaş S, Fasci D, Heck AJR, and Scheltema RA (2018). Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc 13, 2964–2990. 10.1038/s41596-018-0074-x. [DOI] [PubMed] [Google Scholar]
- 120.Rosenthal PB, and Henderson R (2003). Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-particle Electron Cryomicroscopy. J Mol Biol 333, 721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Cryo-EM data collection, refinement, and validation statistics, Related to STAR Methods
Table S2. Proteins identified in sea urchin sperm DMTs, Related to Figure 1.
Notes:
a CFAP91, CIMAP1A-like (A0A7M7G414), CIMAP2, CIMAP3, and SAXO5 have 96-nm periodicity and thus are not included in the atomic model of the 48-nm repeat.
b CFAP115 has 32-nm periodicity and are not included in the atomic model of the 48-nm repeat.
Table S3. Proteins identified in bovine sperm DMTs, Related to Figure 1.
Notes:
a CFAP91, CIMAP2, CIMAP3, and SAXO5 have 96-nm periodicity and thus are not included in the atomic model of the 48-nm repeat.
b SAXO1 is tentatively assigned based on in-cell cross-links to SPACA9, so it is only modelled as a poly-Ala trace.
Table S4. Comparing MIPs across different species and cell types, Related to Figure 2.
Table S5. Infertility and ciliopathy phenotypes, Related to Figure 7.
Table S6. Mass spectrometry data of sea urchin and bovine sperm, Related to STAR Methods.
Figure S1. Sample preparation and data processing workflows for cryo-electron microscopy of native sperm DMTs, Related to Figure 1.
(A-B) Schematic diagrams describing preparation of native sperm DMTs for cryo-EM. (A) Sea urchin sperm were demembranated with Nonidet P-40 and the mitochondria removed with dithiothreitol (DTT), after which cells were directly blotted onto EM grids. Surface tension caused the axonemes to splay apart in the holes. (B) Bovine sperm were demembranated with Triton X-100 and the mitochondrial sheath stripped with DTT and a freeze-thaw cycle. Microtubule sliding was initiated by adding ATP to thawed sperm, which after ~10-15 min disintegrated the axonemes sufficiently for cryo-EM.
(C-D) DMTs were divided into 36 sub-regions for local refinement (12 regions around the DMT covering 2-3 protofilaments and ~1/3 of the 48-nm repeat). Masks and resolution estimates for local refinements of the middle of the DMT are shown for (C) sea urchin and (D) bovine sperm.
(E-F) Fourier shell correlation (FSC) curves for each of the 36 local refinements performed for (C) sea urchin and (D) bovine sperm DMTs.
Figure S2. Longitudinal slices through cryo-EM density maps of sperm DMTs, Related to Figure 1.
(A-D) Longitudinal slices are shown for sea urchin sperm (left) and bovine sperm (right), with viewing directions indicated by red arrows in the insets. Notable sperm-MIPs and MIPs that are different between sea urchin and bovine are labelled.
Figure S3. Tektin-5 adopts diverse conformations in bovine sperm DMTs, Related to Figure 3.
(A) Positions and conformations of TEKT-5 groups in the bovine sperm DMT. For reference, the locations of tryptophan residues are indicated with black (Trp115, N-terminal region), blue (Trp155, 1A helix), and green (Trp336, 2A helix) arrows. A schematic of the tektin domain arrangement and corresponding color-code is provided at the bottom of the figure. TEKT5-A, B, and C all adopt the canonical tektin conformation. Note how the N-terminal region is not resolved in the other groups, except in TEKT5-F where part of it refolds (dark blue arrowhead). In the TEKT5-D group, the L12 loops are ordered by interaction with NME7 (compare green arrowheads, see also Fig. 2d). The L2 loop of the “straight” tektin is also resolved (compare orange arrowheads) because it is stabilized by interaction with a neighboring tektin.
(B) Interactions between NME7 and the TEKT5-D and TEKT5-E filaments. The monomers in the TEKT5-D filament adopt different conformations depending on whether their 2A/2B helices interact with NME7 (“bent” TEKT5-D) or a neighboring tektin (“straight” TEKT5-D).
(C) The L12 loop of the “straight” TEKT5-D monomer inserts into the putative nucleotide-binding pocket of NME7. The L12 of one “bent” TEKT5-D monomer interacts with the second copy of NME-7 in a similar fashion. Panel (C) is rotated 10° relative to the view in (B).
(D) Cryo-EM density of the interfaces between tektin-4 and protofilament A12 in sea urchin (left) and bovine sperm (middle), and between tektin and protofilament B05 in Chlamydomonas flagella (right). Tektin-4 interacts primarily with the K40 loop of α-tubulin, whereas Chlamydomonas tektin interacts mainly with β-tubulin.
Figure S4. SAXO proteins in the Chlamydomonas DMT and conservation of Mn-motifs across species, Related to Figure 5.
(A) Location and identities of SAXO proteins in the DMT from Chlamydomonas flagella. As in Fig. 5, SAXO proteins are classified and colored based on how they bind the microtubule lattice: via a single Mn-motif (blue), via multiple Mn-motifs along a single protofilament (violet), or via multiple Mn-motifs across neighboring protofilaments (orange).
(B) In wild-type Chlamydomonas, SAXO proteins bound to protofilaments B04-B06 are only visible in classes with the tektin bundle.
(C-D) Mn motifs are conserved across species in both (B) core-MIPs (CFAP68) and (C) sperm-MIPs (SPMIP4, formerly C7ORF31). Top panels show cartoon models with Mn motifs highlighted in red. Bottom panels show sequence alignments of Mn motifs in several animal species. NCBI accession numbers of CFAP68 sequences are as follows: NP_073598.1 (Hs, Homo sapiens), NP_001033266.1 (Bt, Bos taurus), NP_075972.2 (Mm, Mus musculus), XP_040546331.1 (Gg, Gallus gallus), XP_018081403.1 (Xl, Xenopus laevis), NP_001313533.1 (Dr, Danio rerio), XP_784695.1 (Sp, Strongylocentrotus purpuratus). Accession numbers for SPMIP4 (C7ORF31) sequences are as follows: NP_001358280.1 (Hs), XP_024846866.1 (Bt), NP_081840.1 (Mm), XP_025003204.2 (Gg), XP_018122881.1 (Xl).
(E) Comparisons of tubulin inter-dimer distances across DMTs from sea urchin sperm, bovine sperm, bovine trachea, and Chlamydomonas flagella. Bars represent means and standard deviations of measurements from six dimers, with each individual measurement represented as a circle.
(F) The bimodal distribution of tubulin inter-dimer distances at the ribbon is conserved. Inter-dimer distances are plotted for the six tubulin dimers of protofilaments A01-A03 and A11-A13.
Figure S5. Cryo-EM of native singlet microtubules in the bovine sperm endpiece, Related to Figure 5.
(A) Cryo-EM image of a frayed sperm endpiece on a holey carbon grid. Scale bar: 1 μm. Inset: High-magnification cryo-EM image of endpiece singlet microtubules (SMTs) captures a ladder-like luminal structure with an 8-nm periodicity. Scale bar: 100 nm.
(B) Cross section of the cryo-EM reconstruction of endpiece SMTs reveals discontinuous intraluminal spirals of SPACA9.
(C) Map obtained after symmetry expansion showing SPACA9 and a SAXO protein (likely SAXO1) bound to a tubulin heterodimer.
(D) Local resolution of the symmetry-expanded reconstruction, estimated by the “Local Resolution” job in Relion.
(E) Fourier shell correlation (FSC) plots of the full C1 reconstruction (black curve), the local refinement of protofilaments 7/8 (blue curve), and the protofilament reconstruction after symmetry expansion (pink curve).
(F) Examples of map quality in the symmetry-expanded map used for model building.
(G) Sequence alignment of the 12 Mn motifs within bovine SAXO1 shows the consensus Thr and Tyr residues (starred) demarcating the short helix that binds at the α/β-tubulin interface. Orange boxes mark lysine residues (K83 and K221) that cross-linked to SPACA9 and α-tubulin respectively.
(H) In-cell cross-linking mass spectrometry detects interactions between SAXO1 and α-tubulin (left); and between SAXO1 and SPACA9 (right). Red spheres indicate the cross-linked lysine residues. See also Table S6.
Figure S6. 96-nm features in sperm DMTs, Related to Figures 5 and 6.
96-nm maps were generated by classifying on the bases of external axonemal complexes bound to protofilaments A02-A03, followed by refining each half of the 96-nm repeat separately.
(A) SAXO5 (formerly TEX45) is a filamentous SAXO protein that binds along protofilament A04 with 96-nm periodicity.
(B) The wedge-MAPs CIMAP2 and CIMAP3 bind to the inter-protofilament cleft between A03/A04, where they alternate to yield an overall 96-nm repeat.
(C) CFAP115, found in sea urchin but not bovine sperm, has 32-nm periodicity.
(D) CFAP91, a component of the RS2 base with 96-nm periodicity, inserts into the inner junction hole formed by the absence of one copy of PACRG in every 96-nm repeat.
Figure S7. CIMAP1 (ODF3) proteins constitute a conserved family of axonemal MAPs, Related to Figure 6.
(A) Extracted density for CIMAP1A and neighboring protofilaments B06 and B07 from the bovine sperm cryo-EM map. The two CIMAP1A molecules are shown in blue-violet and purple, with Pro-Gly-Pro (PGP) motifs in red; α-tubulin is shown in white and β-tubulin in grey.
(B) Zoomed-in view showing head-to-tail interactions between neighboring CIMAP1A molecules. A PGP motif is indicated in red.
(C) The C-terminal tails of CIMAP1 proteins extend into the DMT lumen, where they can bind MIPs. For example, CIMAP1C (ODF3L1) between protofilaments A08/A09 interacts with the sperm-MIP SPMIP4 (C7ORF31).
(D) Sequence alignment of CIMAP1A and CIMAP1C proteins from several animal species, showing conservation of PGP motifs. NCBI accession numbers of CIMAP1A (ODF3) sequences are as follows: NP_444510.2 (Hs), XP_024854250.1 (Bt), NP_081295.2 (Mm), XP_024997911.1 (Gg), NP_001079370.1 (Xl), NP_956252.1 (Dr), NP_723368.1 (Dm), XP_003729878.1 (Sp). Accession numbers of CIMAP1C (ODF3L) sequences are as follows: NP_787077.1 (Hs), XP_005221973.2 (Bt), NP_001398329.1 (Mm), XP_024997911.1 (Gg), NP_001079644.1 (Xl), NP_001017836.1 (Dr), NP_651171.1 (Dm), XP_030833199.1 (Sp).
Data Availability Statement
Cryo-EM maps of the sea urchin sperm DMT, the bovine sperm DMT, the bovine sperm endpiece SMT, and Chlamydomonas DMTs with and without the tektin bundle have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-40619, EMD-17187, EMD-17188, EMD-40620, and EMD-40621 respectively. Atomic models of the 48-nm repeat of the sea urchin sperm DMT, the 48-nm repeat of the bovine sperm DMT, and one asymmetric unit of the 8-nm repeat of bovine sperm endpiece SMTs have been deposited in the Protein Data Bank (PDB) with accession codes 8SNB, 8OTZ, and 8OU0 respectively. Mass spectrometry data of bovine sperm is available from Proteomics Identifications Database (PRIDE) with accession code PXD035941.
Code used to refine and correct alignment parameters by FREALIGN is available at https://github.com/rui--zhang/Microtubule.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


