Abstract
Key message
The transcriptomic, phenotypic and metabolomic analysis of transgenic plants overexpressing GhMPK31 in upland cotton revealed the regulation of H2O2 burst and the synthesis of defensive metabolites by GhMPK31.
Abstract
Mitogen-activated protein kinases (MAPKs) are a crucial class of protein kinases, which play an essential role in various biological processes in plants. Upland cotton (G. hirsutum) is the most widely cultivated cotton species with high economic value. To gain a better understanding of the role of the MAPK gene family, we conducted a comprehensive analysis of the MAPK gene family in cotton. In this study, a total of 55 GhMPK genes were identified from the whole genome of G. hirsutum. Through an investigation of the expression patterns under diverse stress conditions, we discovered that the majority of GhMPK family members demonstrated robust responses to abiotic stress, pathogen stress and pest stress. Furthermore, the overexpression of GhMPK31 in cotton leaves led to a hypersensitive response (HR)-like cell death phenotype and impaired the defense capability of cotton against herbivorous insects. Transcriptome and metabolomics data analysis showed that overexpression of GhMPK31 enhanced the expression of H2O2-related genes and reduced the accumulation of defensive related metabolites. The direct evidence of GhMPK31 interacting with GhRBOHB (H2O2-generating protein) were found by Y2H, BiFC, and LCI. Therefore, we propose that the increase of H2O2 content caused by overexpression of GhMPK31 resulted in HR-like cell death in cotton leaves while reducing the accumulation of defensive metabolites, ultimately leading to a decrease in the defense ability of cotton against herbivorous insects. This study provides valuable insights into the function of MAPK genes in plant resistance to herbivorous insects.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00299-024-03167-1.
Keywords: Cotton, MAPK gene family, HR-like cell death, H2O2, Insect resistance
Introduction
MAPK is an important serine/threonine protein kinase which forms a cascade of MAPK by sequential phosphorylation with MAP kinase kinase (MAPKK/MEK) and MAP kinase kinase kinase (MAPKKK/MEKK) (Hamel et al. 2006; Jagodzik et al. 2018). MAPK cascades are found extensively across eukaryotes, exhibiting high levels of conservation throughout evolution, and serving a crucial role in signal transduction pathways in plants (Group 2002). When plants are stimulated by the external environment, MAPKKK undergoes initial phosphorylation by upstream signals, and the activated MAPKKK proceeds to phosphorylates downstream MAPKK by targeting serine/threonine residues within the conserved S/T-X3-5-S/T motifs (Pitzschke 2015). Then, Activated MAPKKs phosphorylate downstream MAPKs through tyrosine and threonine residues in the conserved activation loop (T-loop). Finally, activated MAPKs can complete the signal transduction process by phosphorylating various downstream substrates (Zhang et al. 2018). As the terminal element of MAPK cascade, MAPK is responsible for relaying the signal to downstream components. In the face of various substrates such as cytoplasmic protein kinases, cytoplasmic cytoskeleton and nuclear transcription factors (TFs), MAPK shows more complexity and functional diversity (Xu and Zhang 2015).
Previous studies have demonstrated MAPKs involvement in diverse biological processes (Zhang and Zhang 2022). In terms of plant growth and development, AtMPK3/6 is known to be involved in stomatal development, floral organ abscission, meristem maintenance, early anther development and embryogenesis in Arabidopsis thaliana (Cho et al. 2008; Hord et al. 2008; Shao et al. 2020; Wang et al. 2007; Zhang et al. 2017). In the response of plants to abiotic stress, SlMPK1 negatively regulates the heat tolerance of tomatoes through directly phosphorylation of the serine-proline-rich protein homolog (SlSPRH1) (Ding et al. 2018). In the response of plants to pathogen stress, GhMPK20 negatively regulated the resistance of cotton to Fusarium oxysporum (Wang et al. 2018). MAPK can also regulate hypersensitive response (HR), a type of programmed cell death (PCD), which is commonly associated with plant disease resistance (Ren et al. 2002). Endogenous activation of MAPK represented by SIPK, Ntf4 and WIPK in tobacco can trigger HR-like cell death in the absence of pathogens infection (Jin et al. 2003; Ren et al. 2006). The absence of AtMPK4 results in the dwarfing of A. thaliana during the second to third leaf stage, but no necrotic lesions were observed. Silencing of GmMPK4s causes stunting in soybean plants, accompanied by spontaneous cell death on leaves and stems (Liu et al. 2011; Petersen et al. 2000). Silenced GmMPK6 in soybean resulted in leaf growth retardation and spontaneous cell death, while overexpression of GmMPK6 also resulted in HR-like cell death in tobacco and A. thaliana (Liu et al. 2014). Although the precise mechanism underlying plant dwarfing and HR-like cell death remains uncertain, it is strongly associated with the blast of reactive oxygen species (ROS).
However, an increasing amount of biochemical and genetic evidence suggests that MAPK plays a critical role in plant resistance against herbivores. The activation of MAPK is one of the earliest signaling events in plants when they are attacked by herbivores. Upon herbivore attack, the plant MAPK signaling pathway is activated, leading to alterations in plant hormone levels as well as reshaping of the transcriptome and proteome, which ultimately affect the plant's ability to defend against herbivores (Hettenhausen et al. 2015). Applying Manduca sexta oral secretions (OS) to leaf wounds in Nicotiana attenuata simulates M. sexta feeding and significantly induces the activity of MAPK family members SIPK and WIPK. When SIPK or WIPK is silenced, the levels of jasmonic acid (JA) induced by M. sexta feeding are significantly reduced. Transcriptome analysis reveals that SIPK and WIPK mediate the accumulation of many defense-related genes induced by M. sexta feeding (Wu et al. 2007). JA and ethylene (ET) play positive roles in regulating plant defenses against herbivores, with 1-aminocyclopropane-1-carboxylic acid synthase (ACS) being the rate-limiting ET biosynthesis (Chae et al. 2003). In A. thaliana, AtMPK6 increases the production of ET by directly phosphorylating ACS6 and ACS2 (Liu and Zhang 2004). In tomatoes, silencing MPK1/2 simultaneously results in a decrease in the levels of JA by lowering the expression of JA biosynthetic genes, thereby promoting better growth of M. sexta larva on MPK1/2 silenced plants (Kandoth et al. 2007). In rice, the silenced OsMPK3 reduced the herbivore-induced trypsin protease inhibitors (TrypPIs) and JA levels, and improved the performance of striped stem borer (SSB) larvae (Wang et al. 2013).
As the various members of MAPK exhibit distinct biological functions, there is increasingly necessary to identify MAPK family members in different species. Currently, 20, 17, 21, 16 and 17 MAPK gene members have been identified in A. thaliana, Oryza sativa, Camellia sinensis, Fagopyrum tataricum and Lactuca sativa, respectively (Colcombet and Hirt 2008; Liu et al. 2022; Reyna and Yang 2006; Wang et al. 2022; Yao et al. 2022). Being one of the most significant cash crops globally, cotton yields a substantial quantity of natural fibers and oils. In this study, we present a comprehensive analysis of MAPK gene family in cotton and provide functional analysis of GhMPK31. Overexpression of GhMPK31 in cotton results in a dwarfed plant architecture and HR-like cell death in the leaves, this phenotype was found to be associated with a greater accumulation of hydrogen peroxide (H2O2). To elucidate the regulatory mechanism of GhMPK31 on ROS burst in cotton, we observed that GhMPK31 can induce the expression of Respiratory Burst Oxidase Homolog B (GhRBOHB), also known as NADPH oxidase, which is one of the main enzymes responsible for extracellular ROS generation in plants (Jalmi and Sinha 2015). Therefore, we speculated that GhMPK31 plays a role in the regulation of H2O2 synthesis in cotton by influencing the expression of GhRBOHB. In addition, selective and non-selective feeding experiments were conducted on wilt type (WT) and OM31 plants using Helicoverpa armigera (cotton bollworm) and Spodoptera litura, revealing that overexpression of GhMPK31 enhances leaf feeding by these insects. This effect could be attributed to the regulatory role of GhMPK31 in the synthesis of defensive compounds.
In conclusion, the comprehensive analysis of GhMPKs gene family and the functional characterization of GhMPK31 enriched the functional study of MAPK in upland cotton and led the foundation for the improvement of upland cotton varieties.
Materials and methods
Genome-wide comprehensive analysis of GhMPKs gene family
G. hirsutum and G. raimondii genomic data were obtained from Cottongen (https://www.cottongen.org/), the genomic data of A. thaliana and O. sativa were obtained from Ensembl Plants (http://plants.ensembl.org/index.html) and Rice Genome Annotation Project (http://rice.uga.edu/downloads_gad.shtml), respectively. MAPKs coding sequences of A. thaliana, O. sativa and G. raimondii were used as query sequences to search the genome of G. hirsutum (NBI (Zhang et al. 2015) and HAU (Wang et al. 2019)) by BLASTP program, and the local Hidden Markov Model-based searches (HMMER) were constructed according to their MAPKs coding sequences to identify GhMPKs. Then, the results of BLASTP and HMMER were compared, redundant sequences were removed, the last upload candidate genes to SMART (http://smart.embl.de/) and CDD (https://www.ncbi.nlm.nih.gov/cdd/) confirmed the existence of conservative domains. The amino acid numbers, molecular weight (MW) and isoelectric point (pI) of the GhMPKs were predicted by the ProtParam tool (https://web.expasy.org/protparam/). The subcellular localization, transmembrane domains and signal peptides of GhMPKs were predicted by DeepLoc-2.0 (https://services.healthtech.dtu.dk/services/DeepLoc-2.0/), DeepTMHMM (https://dtu.biolib.com/DeepTMHMM/) and SignalP-6 (https://biolib.com/DTU/SignalP-6/), respectively.
The phylogenetic tree of the four species (A. thaliana, O. sativa, G. hirsutum and G. raimondii) was constructed based on their full-length protein sequence with MEGA 7.0 software by the neighbor-joining (NJ) method, and the phylogenetic tree was visualized through EvolView (https://evolgenius.info//evolview-v2). The conserved motifs of GhMPKs were analyzed by the program of MEME and visualized by Tbtools (Chen et al. 2020). The gene structures of GhMPKs were analyzed with GSDS software (http://gsds.gao-lab.org/) by comparing coding sequences (CDSs) with the genome sequence corresponding to the gene.
Analysis of phosphorylation sites was completed by using NetPhos-3.1 (https://services.healthtech.dtu.dk/services/NetPhos-3.1/), and to analyze cis-acting in the promoter region of the GhMPK genes the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used, collinearity analysis between G. hirsutum with other three species was determined by MCScanX software, the visualization of all the above analyses was completed by TBtools. The localization images of GhMPK genes on chromosomes and their genome-wide gene duplication events were plotted by Circos software.
Expression of GhMPK genes in different stresses
The expression data of GhMPK genes in various tissues and abiotic stresses were downloaded from the transcriptome data of TM-1(Zhang et al. 2015), and the transcriptome data of GhMPK gene in response to H. armigera OS at different time periods were derived from NCBI Sequence Read Archive (accession number: PRJNA522889) (Si et al. 2020). Transcriptome data for stress of Verticillium dahliae was obtained from Ping Qiu’s study (Qiu et al. 2023).
Plant materials and growing conditions
The cultivated variety Jin668 of G. hirsutum was used as the stable conversion material. RNA interference (RNAi) and overexpression transgenic lines of GhMPK31 (IM31-19, IM31-28) and (OM31-11, OM31-24) were screened by expression levels for subsequent studies, and Jin668 was used as control. All materials were cultivated in the field during the regular growing season following the standard tillage practices of Wuhan. During the winter season, plants were grown in a controlled environment in a greenhouse maintained at 26 °C, under 16 h light/8 h dark photoperiod and 60% relative humidity.
Vector construction and agrobacterium-mediated genetic transformation
The full-length CDS and RNAi fragments of GhMPK31 were separately amplified and inserted into the pK2GW7 and pHellsgate 4 vectors using the Gateway cloning technology (Karimi et al. 2002; Liang et al. 2021; Luo et al. 2017; Tian et al. 2015; Wesley et al. 2001). The recombinant vectors for overexpression and RNAi were transformed into the cotton (cv. Jin668) hypocotyls by Agrobacterium tumefaciens (strain GV3101)-mediated transformation (Jin et al. 2006).
Analysis of RNA-sequencing data
The seeds of T0 generation OM31-11, OM31-24, IM31-19, IM31-28 and WT plants were germinated simultaneously and cultivated in a greenhouse maintained at 28 °C, under 16 h light/8 h dark photoperiod and 60% relative humidity. After 4 weeks, we measured the expression levels of GhMPK31 in the leaves of all T1 generation plants. We selected the T1 generation OM31-11 and OM31-24 plants with high GhMPK31 expression levels and the IM31-19 and IM31-28 plants with low GhMPK31 expression levels for subsequent experiments. The leaves were removed and divided into two portions, one for transcriptome sequencing and the other for Target-like metabolite assays, with 2 bio-replicates per sample. For RNA sequencing (RNA-seq) reads from different samples, the low-quality reads were filtered using Trimmomatic (v.0.39), and the clean reads were then mapped to the TM-1 reference genome (Zhang et al. 2015) by HISAT2 (v.2.2.1) (Kim et al. 2019) with default parameters. The expression level (transcripts per million; TPM) of genes was calculated by StringTie (v.2.1.4). A gene was considered to be expressed if its TPM was > 0. Subsequently, differentially expressed genes (DEGs) were identified by using the DESeq2 package with False Discovery Rate (FDR) < 0.05 and | log2 (fold change) |≥ 1 (Varet et al. 2016). All DEGs were tested for statistical enrichment in Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using the KOBAS 3.0 software (http://kobas.cbi.pku.edu.cn/index.php) with corrected pvalue < 0.05 (Li et al. 2016; Si et al. 2020). PCA plot of genes identified by RNA-Seq of OM31, IM31 and WT (Fig. S1).
Subcellular localization
The full length CDS sequence of GhMPK31 was cloned into the green fluorescent protein (GFP) vector pGWB405 via C-terminal fusion. The recombinant vectors pGWB405-GhMPK31 were transiently expressed in tobacco epidermal cells following Agrobacterium-mediated transfection. The localization of the protein was observed using Olympus FV1200 confocal microscope after 2 days of Agrobacterium transfection. CBL1: RFP was used as a plasma membrane marker, HY5: RFP was used as a nucleus marker.
Yeast two-hybrid, LCI and BiFC assays
To characterize the interaction between GhMPK31 and GhRBOHB proteins, the CDS of GhMPK31 was cloned into pGBKT7 using as bait vector and transformed into yeast strain Y2H. Then the CDS of GhRBOHB were constructed in pGADT7 as prey vectors and transformed into yeast strain Y187. The interactions between bait and prey were detected by the growth on SD -Leu-Trp (SD-2) medium and SD-Leu-Trp-His-Ade (SD-4) medium, respectively.
For the Luciferase Complementation Imaging (LCI) assays, the CDSs of GhMPK31 and GhRBOHB were cloned into the JW771 and JW772 vectors, respectively. The recombinant vectors were transformed into Agrobacterium tumefaciens GV3101 and transiently expressed in Nicotiana benthamiana leaves by needleless syringes. Fluorescence signals of LUC luminescence in LCI was observed by cryogenically cooled CCD camera (NightSHADE LB985). For Bimolecular Fluorescence Complementation (BiFC) assays, the CDSs of GhMPK31 and GhRBOHB were respectively constructed to the vector pxy104-cYFP and pxy106-nYFP. The vectors were transformed into A. tumefaciens strain GV3101 and transiently expressed in N. benthamiana leaves via needleless syringes. The fluorescence in the N. benthamiana epidermal cells was observed 60 h later using a confocal microscope (Olympus FV1200).
qRT-PCR
For each sample, 3 μg of RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega). The Real-time quantitative PCR (RT-qPCR) reactions were performed using the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems), and GhUB7 was used as an internal control for other genes.
DAB staining and H2O2 measurement
H2O2 was detected by an endogenous peroxidase-dependent in situ histochemical staining procedure using 3, 3ʹ-diaminobenzidine tetrahydrochloride (DAB). After the detached leaves were thoroughly immersed in DAB staining solution, a vacuum was applied for 15 min to facilitate entry of the staining solution into the intercellular space. After an 8 h soaked in darkness, it was exposed to light for 1 h. The remaining DAB staining solution on the leaves was rinsed with distilled water. Subsequently, the leaves were immersed multiple times in a decolorization solution (75% ethanol + 5% glycerin) under dark conditions at 37 °C until chlorophyll was completely removed. The presence of H2O2 production was evidenced by the formation of a reddish-brown precipitate in the transparent leaves. The concentration of H2O2 from leaves was determined using a H2O2 quantification kit (#C500069, Sangon Biotech, Shanghai, China).
Quasi-targeted metabolomics data acquisition
Tissues (100 mg) were individually grounded with liquid nitrogen and the homogenate was resuspended with prechilled 80% methanol by well vortex. Subsequently, the samples were incubated on ice for 5 min and centrifuged at 15,000 g, 4 °C for 20 min. A portion of the supernatant was diluted to a final concentration containing 53% methanol using LC–MS grade water. Next, the samples were transferred to a fresh Eppendorf tube and centrifuged again at 15,000 g, 4 °C for 20 min. Lastly, the supernatant was injected into the LC–MS/MS system for analysis (Want et al. 2013).
The detection of the experimental samples utilized MRM (Multiple Reaction Monitoring) and relied on the Novogene in-house database. Q3 was used for metabolite quantification. Q1, Q3, RT (retention time), DP (declustering potential) and CE (collision energy) were used for metabolite identification. The data files generated by HPLC–MS/MS were processed using SCIEX OS Version 1.4 for peak integration and correction. The main parameters were set as follows: minimum peak height (500), signal/noise ratio (5), and Gaussian smooth width (1). The area of each peak represents the relative content of the respective substance.
Quasi-targeted metabolomics data analysis
These metabolites were annotated using the KEGG database (http://www.genome.jp/kegg/), HMDB database (http://www.hmdb.ca/) and Lipidmaps database (http://www.lipid maps.org/). Principal components analysis (PCA) and Partial least squares discriminant analysis (PLS-DA) were performed using the metaX software (Luo et al. 2015) (Fig. S2). We applied univariate analysis (t-test) to calculate the statistical significance (p-value). Metabolites with VIP > 1 and p-value < 0.05, and fold change (FC) ≥ 2 or FC ≤ 0.5 were considered as differential metabolites. Volcano plots were used to identify metabolites of interest based on the Log2 (FC) and − log10 (p-value) of metabolites using the ggplot2 library in R language. Heat maps were generated using z-scores to normalize the intensity areas of differential metabolites and plotted using the Pheatmap package in R. The correlation between differential metabolites was analyzed using cor () in R language with the "Pearson" method. The statistical significance of the correlation between differential metabolites was then calculated using the cor.mtest () in R language. p-value < 0.05 was considered statistically significant. Correlation plots were generated using the corrplot package in R language. The functions of these metabolites and metabolic pathways were investigated by utilizing the KEGG database. Enrichment analysis of metabolic pathways for differential metabolites was conducted. Metabolic pathways were considered enriched when the ratio were satisfied by x/n > y/N, metabolic pathway was considered statistically significantly enriched when its p-value < 0.05.
Selective and non-selective feeding assays of H. armigera and S. litura
Both the T1 generation transgenic materials and the WT materials were cultivated under identical field conditions. Two types of leaves were selected from the OM31 lines: leaves without necrotic blotches and leaves showing necrotic blotches. Similarly, leaves from the corresponding parts of WT plants were selected as contrast. Choice feeding assays were performed in plastic boxes measuring 29 × 44 cm. First, we starved the third instar larvae for 6 h, then we symmetrically and evenly arranged the removed leaves. 12 starved larvae were placed in the middle of each plastic box and allowed to feed freely for 24 h. The consumption of cotton leaves was calculated by ImageJ software. For the non-selective feeding experiment with the moth, 9 cm round glass petri dishes were used to hold leaves from WT plants and OM31 separately. Second-instar S. litura larvae with comparable body weight were chosen and placed individually on the leaves, with one larva per dish. The body weight of the larvae was recorded at day 0 and day 5 (Hu et al. 2018).
Results
Comprehensive analysis of MAPK gene family in cotton
To comprehend the significance of the MAPK gene family in upland cotton, both BLASTP and Hidden Markov Model (HMM) searches against G. hirsutum (NBI and HAU) protein databases were conducted using A. thaliana, O. sativa and G. raimondii MAPK proteins as query sequences. After validation of the conserved domains, we successfully identified and characterized 55 putative GhMPK genes (Table S1). These putative GhMPK genes encoded proteins that varied in length from 275 to 716 amino acids, the proteins Mw ranging from 31.62 to 82.45 kDa, and the PI of the proteins fell within the range of 4.81 to 9.72. The subcellular localization prediction indicated that the majority of GhMPK proteins were located in the cytoplasm and nucleus. However, with the exception of GhMPK19, most MAPK proteins lack a signaling transmembrane domain, while GhMPK19 is the only one possessing two transmembrane domains. To investigate the evolutionary relationships of GhMPKs, we constructed a phylogenetic tree based on amino acid sequences of 55 GhMPKs from G. hirsutum, 20 AtMPKs from A. thaliana, 17 OsMPKs from O. sativa and 28 GrMPKs from G. raimondii (Fig. 1a). The findings indicated that all MAPKs were clustered into four groups: A, B, C and D. Notably, Group D constituted the largest cluster, featuring the highest count of MAPKs. In GhMPKs family, the TEY type of T-loop belongs to Groups A, B and C, whereas TDY type of T-loop is classified into Group D (Table S1).
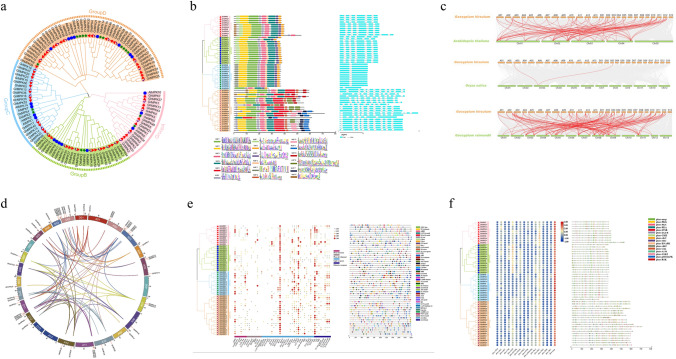
Fig. 1.
Comprehensive analysis of MAPK gene family in Gossypium hirsutum. a Phylogenetic analysis of MAPK gene families in G. hirsutum, A. thaliana, O. sativa and G. raimondii. A, B, C and D indicate different gene clusters (groups). The different shapes and colors of the symbols indicate different species. b Phylogenetic relationships, conserved protein motifs and gene structures of GhMPKs. On the left is the phylogenetic tree of the MAPK gene family in G. hirsutum. In the middle is the motif distribution of the MAPK protein. There are 18 motifs in total, displayed in different colored boxes, below is the sequence logo for MAPK proteins motifs. On the right is the exon–intron structure of the GhMPKs gene. The blue box represents the exon and the black line represents the intron. c Collinearity analysis of MAPK genes between GhMPKs and three other plants. Grey lines indicate collinear blocks within the G. hirsutum genome and other plant genomes, and the red curve indicates MAPK genes with collinearity. d Collinearity analysis and chromosomal location of the GhMPK gene family. The chromosome of G. hirsutum was distinguished by different color. Each GhMPK gene is marked with a short red line on the chromosome and collinear gene pairs are represented by a color curve. e Cis-acting elements in the promoter regions of GhMPKs. On the left is the phylogenetic tree of the MAPK gene family in G. hirsutum. In the middle is a heat map of the number of all cis-acting elements. All cis-elements are categorized by function into four categories, cis-elements with similar functions are displayed in the same color. On the right is the promoter acting element of the GhMPK genes. The black line indicates the promoter length of the GhMPK genes. The different colored boxes on the right represent cis-acting elements with different functions. f Prediction of phosphorylation sites of GhMPKs. On the left is the phylogenetic tree of the MAPK gene family in G. hirsutum. In the middle is a heat map of the number of all phosphorylation sites. On the right is the distribution of protein kinase phosphorylation sites in GhMPKs (colour figure online)
Moreover, we conducted an analysis of the distribution of motifs and the exon–intron structure of GhMPK proteins (Fig. 1b). A total of 18 conserved motifs were detected, which motifs 1, 2, 3, 4 and 6 appearing in nearly all GhMPK proteins, suggesting that GhMPKs were highly conserved. However, different groups contained its certain unique motifs. Motif 7 was found exclusively in groups A and B, whereas motif 11 was specific to group C. Additionally, group D contained unique motifs 5, 13, 14, 15, 16 and 17. These findings indicate that GhMPKs in different subgroups may possess distinct functions. The results of exon intron structure analysis showed that the gene structures of GhMPK within the same group exhibited a higher degree of conservation. For example, each member of group C contained 2 exons and 1 intron, and all GhMPKs of group B had the same intron–exon distribution pattern. However, group D exhibited a significantly higher number of exons and introns compared to other groups, with varied distribution patterns. These findings suggest that group D GhMPKs potentially demonstrates more complex functions. Chromosome mapping analysis revealed an uneven distribution of 47 GhMPKs across 22 out of 26 G. hirsutum chromosomes, with the remaining eight members distributed on the scaffold. Noticeably, GhMPKs were absent on chromosomes A06, D06, A13 and D13 (Fig. 1d). Additionally, our findings revealed that both subgroup A and subgroup D contained 23 and 24 members respectively, indicating an equal distribution of members within the GhMPKs family across these subgroups. By collinear analysis of the GhMPK genes family, 75 collinear pairs were identified on 22 chromosomes (Fig. 1d). The majority of the genes exhibited 1–2 collinear pairs, while a few had 4. In addition, the collinearity of G. hirsutum with A. thaliana, O. sativa and G. raimondii for MAPK was investigated and identified 77, 2 and 114 gene pairs, respectively (Fig. 1c). Homologous gene pairs demonstrate significant differences across species, suggesting a stronger evolutionary bias of GhMPK towards dicots (A. thaliana and G. raimondii) compared to monocots (O. sativa), thereby elucidating the underlying evolutionary mechanism of the GhMPK family in various species.
To further investigate the function of GhMPKs, the cis-acting elements in the promoter region of GhMPKs were identified and analyzed, which can be classified into four categories associated with plant hormones, plant development, photo response and defense/stress response (Fig. 1e). Among them, nearly all GhMPK genes contained elements related to light response, followed by stress response elements, with the least number of elements associated with the development of plants. These findings demonstrate that GhMPKs could potentially contribute to multiple pathways involved in plant growth and development. We also predicted a total of 2147 reliable phosphorylation sites across GhMPK gene family, with the number of phosphorylation sites for each member ranging from 22 to 67 (Fig. 1f). GhMPK49 exhibited the highest number of phosphorylated sites (67), whereas GhMPK8 contained the lowest number of phosphorylated sites (22). Nevertheless, we observed no positive correlation between the number of phosphorylation sites and the length of the amino acid sequence. As a crucial protein kinase class, the activation of MAPK typically occurs through phosphorylation. Therefore, analyzing phosphorylation sites plays a vital role in comprehending the mechanisms underlying GhMPK proteins.
Expression patterns of GhMPK genes under various abiotic and biotic stresses
Extensive research has demonstrated a close relationship between gene expression and functionality. By using the protein sequence of GhMPKs to construct a phylogenetic tree, which revealed that all GhMPKs cluster into four distinct subgroups (Fig. 2a). By analyzing the expression of GhMPKs between different subgroups under various stresses, a better exploration of the potential role of the MAPK gene family in upland cotton can be conducted. Initially, the expression of GhMPK genes were examined across various cotton tissues. Analysis of tissue expression patterns revealed widespread expression of all members of the GhMPKs in nearly all tested tissues. However, there were significant variations in their transcription levels, indicating functional distinctions within this gene family during growth and development (Fig. 2b). Afterwards, the expression patterns of GhMPKs were investigated in cotton leaves when subjected to four different abiotic stress conditions, namely heat, salt, cold and drought (Fig. 2c). Following high temperature (38 ℃) treatment, the expression of 96% GhMPKs in the experimental group was significantly down-regulated after 1 h. However, after 3 h, the expression levels of almost all GhMPKs were upregulated, with 31 genes showing higher expression levels than the control group, only GhMPK11 and GhMPK25 exhibited continued decline. After 1 h of salt (400 mM NaCl) and drought (20% PEG) treatment, 41 GhMPKs exhibited similar expression patterns, and compared with the control, GhMPK28 and GhMPK30 were significantly down-regulated under both treatments. After 1 h of low temperature (4 °C) treatment, the upregulated GhMPKs were mainly concentrated in subgroup D compared with the control, while most of the genes in subgroup A and C exhibited downregulation. Both GhMPK10 and GhMPK35 exhibited significant upregulation following exposure to cold and salt treatments, while displaying significant downregulation after heat treatment. These findings indicate that the GhMPK gene family experiences significant induction or inhibition in response to diverse abiotic stresses.
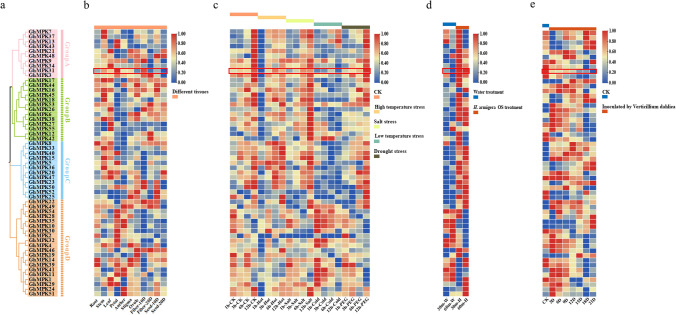
Fig. 2.
Expression patterns of the MAPK gene family under different tissues and stress conditions. a Phylogenetic tree of the MAPK gene family in G. hirsutum. Different colors represent different subgroups. b Analysis of expression profiling of GhMPKs in different tissues. From left to right are root, stem, leaf, petal, anther, stigma, ovule, fiber 10 days, fiber 20 days, seed 10 days and seed 20 days. Red indicates increased expression, blue indicates decreased expression. c Abiotic stress-induced expression profiles of GhMPKs under heat, cold, PEG and salt treatments. Red indicates increased expression, blue indicates decreased expression. d Expression patterns of GhMPK genes induced by H. armigera OS. Red indicates increased expression, blue indicates decreased expression. e Expression patterns of GhMPK genes induced by V. dahliae. Red indicates increased expression, blue indicates decreased expression (colour figure online)
Next, the transcriptome of this gene family were analyzed at different time points after treatment with H. armigera OS and V. dahliae infestation (Fig. 2d, e). The results revealed that 75.6% of GhMPKs exhibited up-regulation after 30 min of treatment with H. armigera OS, in comparison to the control group. Additionally, it was observed that 14 GhMPK genes displayed down-regulation, with 12 genes belonging to subgroup D, while all the genes in subgroup C exhibited up-regulation. GhMPK31 and GhMPK40 exhibited significant up-regulation in transcription upon treatment with H. armigera OS, showcasing high expression levels. Specifically, their transcription levels were up-regulated by more than 1.5-fold at both the 30-min and 60-min time points. After 3 days of V. dahliae inoculation, transcription levels of 53% GhMPKs genes increased compared with the control. GhMPK15 and GhMPK51 transcription levels were up-regulated by more than 1.5 times maintaining the high expression levels. The transcription of GhMPK14 and GhMPK39 was consistently inhibited for a period of 21 days following inoculation. These results indicate that the GhMPK gene family also plays an important role in response to biological stress.
Overexpression of GhMPK31 resulted in hypersensitive response (HR)-like cell death and dwarfing of cotton
GhMPK31 exhibits significant induction or inhibition in response to various biotic and abiotic stresses, particularly showing high expression levels under the treatment of H. armigera OS. Additionally, it shares homology with AtMPK6 (Yuasa et al. 2001), one of the extensively studied genes in plants. Consequently, we made the decision to conduct further research into the functionality of GhMPK31.
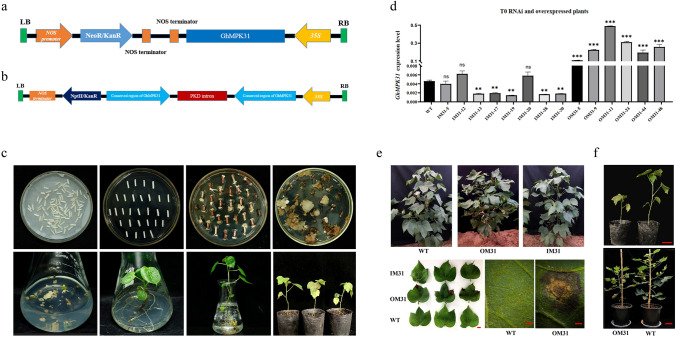
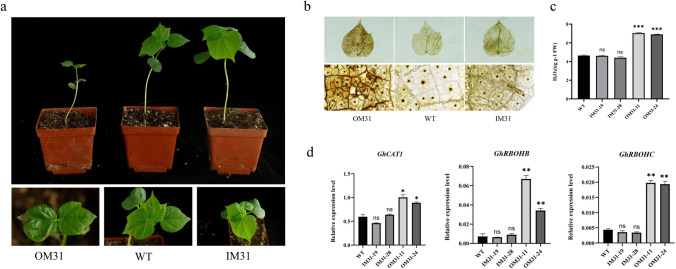
To determine the functional role of GhMPK31 in cotton, overexpression and RNAi vectors were constructed based on the CDS of GhMPK31, followed by the Agrobacterium-mediated genetic transformation, resulting in the generation of independent transgenic plants (Fig. 3a, b, c). Two independent RNAi lines (IM31-19 and IM31-28) and two independent overexpression lines (OM31-11 and OM31-24) were identified based on the expression level of GhMPK31 for subsequent studies (Fig. 3d). Phenotypic analysis of the T1 generation OM31 lines showed the presence of necrotic lesions on leaves and a dwarf plant architecture, whereas the T1 generation RNAi lines exhibited no discernible differences compared to the WT (Fig. 3e, f). We observed that necrotic blotches on the leaves of OM31 lines appeared during the seedling stag persisted throughout the entire growth period of cotton plants. Necrotic blotches were absent on the new leaves, but rather, they were typically found on the older leaves. Thus, these results suggest that the necrotizing spots are not caused by viruses but by a phenomenon known as HR-like cell death.
Fig. 3.
Creation of GhMPK31 transgenic plants with overexpression and RNAi. a Construction of overexpression GhMPK31 vector. b Interference GhMPK31 vector construction. c Genetic transformation process of cotton. d Determination of GhMPK31 expression level in transgenic materials. The expression of GhUB7 was used as internal control. Means ± SE (n = 3). Significant difference analysis was done by Student’s t-tests (**, p < 0.01, ***, p < 0.001). e Phenotypes comparison between T1 transgenic materials and WT plants. Compared with WT plants, OM31 lines showed necrotic lesions in the leaves, a kind of HR-like cell death phenotype. The red line represents 1 cm. f Comparison of plant height between T1 generation OM31 and WT plants. The red line represents 10 cm (colour figure online)
RNA-seq analysis revealed dramatic gene expression difference among OM31, IM31 and WT cotton plants
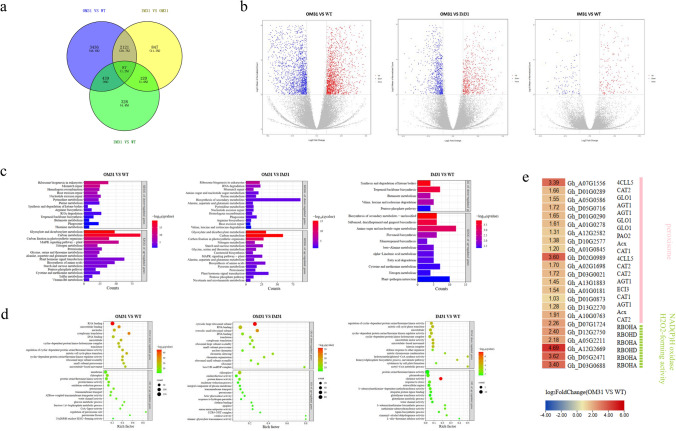
We investigated the expression differences among T1 generation OM31, IM31 and WT cotton plants using RNA-seq. A total of 6038 DEGs were identified in the OM31 lines compared with the WT cotton plants, of which 2844 DEGs were down-regulated and 3239 DEGs were up-regulated. A total of 3175 DEGs were identified in the comparison between OM31 and IM31 plants. In contrast, only 974 DEGs were identified in the comparison between IM31 and WT plants (p-adjust value < 0.05 and | log2 (fold change) |≥ 1) (Fig. 4a, b), suggesting that interference with GhMPK31 has no significant effect on the expression of other genes. GO term enrichment analysis and KEGG pathway enrichment analysis were separately conducted for the three groups: OM31 vs WT, OM31 vs IM31 and IM31 vs WT, based on the up-regulated and down-regulated DEGs (Fig. 4c, d). Interestingly, the top 15 enriched KEGG pathways in both the OM31 vs WT and OM31 vs IM31 groups exhibited up-regulation of the same pathways. These pathways encompassed Carbon fixation in photosynthetic organisms, Carbon metabolism, Nitrogen metabolism and the Peroxisome pathway. Moreover, the identical GO term was identified within the top 15 up-regulated GO terms in both groups, encompassing chloroplast, peroxisome, NADPH oxidase and H2O2-forming activity. In addition, we found that some key genes (RBOHA, RBOHD, CAT1 and CAT2) associated with H2O2 were significantly induced in OM31 plants (Fig. 4e). These results demonstrated that overexpression of GhMPK31 in plants may be related to processes such as carbon fixation, nitrogen metabolism and H2O2 generation and decomposition.
Fig. 4.
RNA-seq analysis revealed the effect of GhMPK31 on gene expression. a Venn diagram showing overlap of DEGs between three lines. b Differential gene volcano map of three groups. Red represents up-regulated DEGs, and blue represents down-regulated DEGs. (qvalue < 0.05 and | log2 FC|≥ 1). c Three groups of KEGG pathway enrichment up-regulated or down-regulated DEGs. (qvalue < 0.05). d Three groups of up-regulated DEGs or down-regulated DEGs enrichment top 15 GO terms (qvalue < 0.05). e The differentially expressed ROS-related genes were identified by RNA-seq method. The color in each cell represents the value of the log2 fold change (OM31 VS WT) (colour figure online)
H2O2 levels increase in GhMPK31-overexpression plants
Transcriptome results suggest that overexpression of GhMPK31 leads to the upregulation of genes related to H2O2 and the phenomenon of HR-like cell death in plants is often associated with the excessive accumulation of H2O2 (Liu et al.2007). Therefore, we hypothesize that the necrotic blotches phenotype observed in the leaves of OM31 plants is a result of the excessive accumulation of H2O2. To validate this hypothesis, the seeds of T1 generation OM31, IM31 and WT were simultaneously cultivated under identical environmental conditions (Fig. 5a). Six weeks later, DAB staining was conducted on the leaves from the corresponding parts of the three lines. The results revealed a significant accumulation of reddish-brown precipitate in the OM31 lines, however, neither IM31 lines nor WT showed any such accumulation (Fig. 5b). The H2O2 content test demonstrated significantly higher levels of H2O2 in the OM31 lines compared to the IM31 lines and WT (Fig. 5c). Additionally, using RT-qPCR, we identified several NADPH oxidase and catalase genes involved in H2O2 synthesis and decomposition. The results indicate significantly elevated expression levels of GhCAT1, GhRBOHB and GhRBOHC in the OM31 lines compared to both WT and IM31 lines (Fig. 5d). These results suggest that overexpression of GhMPK31 leads to an elevation in H2O2 content in cotton leaves, which might lead to HR-like cell death and dwarfing in OM31 plants.
Fig. 5.
Overexpression of GhMPK31 leads to accumulation of H2O2 in plants. a T2 generation OM31, IM31 and WT. b Presence of H2O2 in cotton leaves visualized by staining with DAB. Oxidized DAB formed a reddish-brown deposition. c H2O2 concentration in OM31, IM31 and WT leaves. Error bars represent ± standard errors of three biological replicates. Means ± SE (n = 3). d The relative expression of H2O2 related genes is in OM31, IM31 and WT backgrounds. The expression of GhUB7 was used as internal control. Means ± SE (n = 3). Statistical analyses were performed using Student’s t test. *, p < 0.05; **, p < 0.01. ***; p < 0.001. All of the experiments were repeated at least three times with similar results
GhMPK31 physically interacts with GhRBOHB revealed by Y2H, BiFC and LCI
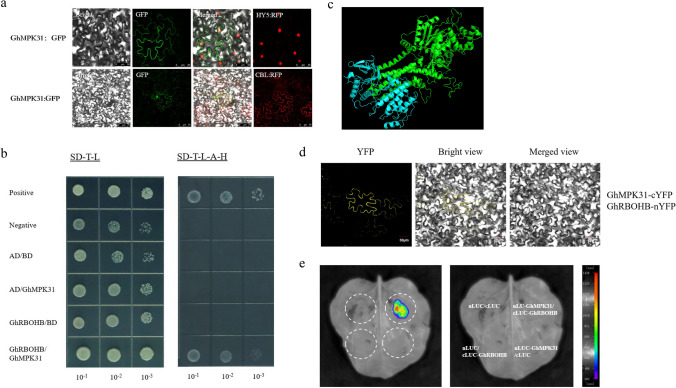
To further explore the mechanism of GhMPK31 regulating ROS, we firstly investigated the subcellular localization of GhMPK31. The full CDS length of GhMPK31 was fused to the N-terminus of GFP and transiently expressed in tobacco epidermis, and through confocal spectral microscope fluorescence signals were observed in both cell membrane and nucleus (Fig. 6a). Then, we carried out a yeast two-hybrid (Y2H) assay to screen potential interacting proteins and found the direct interaction of GhMPK31 protein with the NADPH oxidase (GhRBOHB) (Fig. 6b). We used Y2H-AOS protein interaction prediction analysis to construct the three-dimensional structure of the interaction model of these two proteins, and the protein interaction confidence score was 9.6, which proved that this interaction was highly possible (Fig. 6c). The interaction between GhMPK31 and GhRBOHB was further confirmed using the BiFC assay and LCI assay (Fig. 6d, e). These results revealed that GhMPK31 interacts with GhRBOHB in cotton.
Fig. 6.
GhMPK31 physically interacts with GhRBOHB in cotton. a The subcellular localization of GhMPK31: GFP protein. GFP fluorescence was observed after transiently expressing GhMPK31: GFP protein in tobacco epidermal cells. CBL and HY5 were used as plasma membrane and nucleus markers, respectively. b Y2H assay for GhMPK31- GhRBOHB interaction. SD-2 (-Trp/-Leu), SD-4 (-Trp/-Leu/-His/-Ade). c 3D protein structural model of GhMPK31 and GhRBOHB interaction. d BiFC assay between GhMPK31-cYFP and GhRBOHB-nYFP in tobacco epidermal cells. Bars = 30 μm. e LCI analysis of GhMPK31-nLUC and GhRBOHB-cLUC in tobacco leaves
Overexpression of GhMPK31 reduces the accumulation of cotton defense metabolites and results in sensitive against herbivorous insects’ infestation
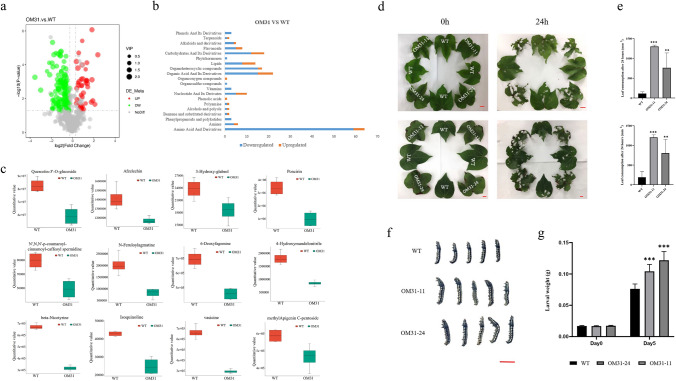
Secondary metabolites are widely recognized as key players in the plant's response to pest stress. Quasi-targeted metabolomics experiments were performed on T1 generation OM31 and WT plants to identify the variations in defense-related metabolites between them, and to investigate the impact of GhMPK31 on cotton's defense against herbivorous insects. A total of 698 metabolites were detected, with 185 of them exhibiting significant differences (p < 0.05). In comparison to WT plants, 76.7% of metabolites in OM31 lines were significantly decrease (Fig. 7a, S4). These differential metabolites were categorized into 21 distinct compound classes, most of which were classified as amino acids and their derivatives, carbohydrates and their derivatives, organic acids and their derivatives, organic heterocyclic compounds, etc. (Fig. S3). The known endogenous defense metabolites we focused on include flavonoids, alkaloids including their derivatives, phenols and its derivatives (Erb and Kliebenstein 2020), which significantly reduced in OM31 lines (Fig. 7b, c). We also assessed the expression levels of several genes associated with flavonoid synthesis using RT-qPCR. The results revealed a significant downregulation of GhCHS1, GhCHI, GhDFR and GhAN3 expression in OM31 lines compared to the WT (Fig. S5). To assess the impact of GhMPK31 on the defense capacity of cotton against insects, we conducted selective and non-selective feeding assays using H. armigera and S. litual. Two types of leaves were chosen for selective feeding according to the presence of necrotic blotches: leaves without necrotic blotche and leaves with visible necrotic blotches. Leaves were selected from corresponding parts of WT plants to serve as the control group. In the selection experiment, cotton leaves from both OM31 and WT plants were evenly distributed, and third-instar bollworm larvae were given unrestricted access to feed (Fig. 7d). After 24 h, we observed a significant increase in the consumption of both types of leaves in the OM31 plant lines compared to the WT plants. (Fig. 7e). In the non-selective experiment, second S. litual instar larvae were fed with leaves from both WT plants and OM31 lines for 5 consecutive days. It was observed that the larvae fed on OM31 leaves exhibited significantly greater weight compared to those fed on WT leaves (Fig. 7f, g). These results collectively demonstrate that the overexpression of GhMPK31 leads to a reduction in various metabolites, including defensive substances. Consequently, this reduction weakens the cotton plant's defense against herbivorous insects.
Fig. 7.
Overexpression of GhMPK31 reduced the accumulation of defensive metabolites and decreased the defense ability of cotton against herbivorous insects. a Volcanic map of differential metabolites between OM31 and WT. Red represents up-regulated metabolites and green represents down-regulated metabolites, (qvalue < 0.05). b Number of differential metabolites in each compound class in OM31 mutants. Orange represents up-regulated metabolites and green represents down-regulated metabolites (qvalue < 0.05). c Box diagram of defense-related metabolites between OM31 and WT. Green for WT, red for OM31. Error bars represent ± standard errors of four replicates. Means ± SE (n = 4). d Choice feeding assay of H. armigera. The third-instar H. armigera were pre-starved for 6 h and photographed at 0 h and 24 h. The red line represents 1 cm. e Leaf consumption in the choice feeding assay for H. armigera. Means ± SE (n = 6). f Comparison between the body-size of larvae fed on WT plants and OM31 lines leaves after 5 days of non-selective feeding. The red line represents 1 cm. g Average larval weight on days 0 and 5. Means ± SE (n = 12). Statistical analyses were performed using Student’s t test. *, p < 0.05; **, p < 0.01. ***; p < 0.001. All of the experiments were repeated at least three times with similar results
Discussion
MAPK gene family has been widely identified in different plant species, but detailed and comprehensive analysis in the G. hirsutum genome is relatively scarce. The analysis of phosphorylation sites and promoter cis-elements of GhMPKs provides valuable reference for functional analysis of MAPK gene family in upland cotton. In addition, GhMPK31, which is a homolog of AtMPK6, was screened out through the analysis of expression profiles under various stress conditions, warranting further investigation. AtMPK6 and its homologues in other plants have been extensively studied and are considered one of the earliest genes in the MAPK gene family (Doczi and Bogre 2018; Xu and Zhang 2015). This gene is characterized by its important functions in plants’ life cycle, such as growth and development, biotic and abiotic stresses and hormone signaling (Jia et al. 2016; Li et al. 2018; Majeed et al. 2023; Sun et al. 2018). Research on cotton has mainly focused on its response to ABA induction, participation in drought stress and regulation of fiber development (Chen et al. 2020b; Li et al. 2017; Luo et al. 2011). However, there have been no reported studies on the regulation of cotton leaf HR-like cell death and defense capabilities.
We overexpressed GhMPK31 in cotton, which can induce HR-like cell death in cotton leaves and dwarfing of cotton plants. This is typically associated with the outbreak of reactive ROS (Liu et al. 2007). ROS is an important second messenger in plants, which can be continuously produced in the cell through the respiration of mitochondria, chloroplasts and peroxisomes, including the superoxide anion (O2−), H2O2 and the hydroxyl radical (.OH). Meanwhile, it can also be eliminated by some scavenging enzymes, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), glutathione peroxidase (GPX) and CAT (Apel and Hirt 2004; Lamb and Dixon 1997). Maintaining a dynamic balance of reactive ROS is crucial for plant development. ROS serves as a crucial signaling molecule, mediating plant growth, development and responses to environmental stress. However, excessive levels of ROS can induce oxidative damage to cellular membranes, proteins, carbohydrates and DNA. Particularly, rapid bursts of H2O2 can trigger programmed cell death (Liu and He 2016; Moller et al. 2007). Based on our analysis of OM31 leaves, we observed an elevated H2O2 content compared to WT, so we hypothesized that GhMPK31 may promote HR-like cell death in cotton leaves by positively regulating H2O2 synthesis. Furthermore, the results of RNA-seq and RT-qPCR revealed a significant increase in the expression levels of genes associated with H2O2 synthesis and decomposition in OM31 leaves, providing further support for this hypothesis. The H2O2 content in IM31 plants was found to be comparable to that in WT plants, and RNA-seq analysis revealed a minimal number of DEGs between IM31 and WT. We believed that upland cotton, being a tetraploid crop, has functional redundancy of GhMPK31.
In recent years, studies on MAPK6 regulating ROS in different species have been successively reported, although the specific mechanisms are not yet clear. In A. thaliana, AtMPK6 mediates ABA-induced CAT1 expression and H2O2 production (Xing et al. 2008). NTF6 regulates NADPH Oxidase-Dependent oxidative bursts in N. benthamiana (Asai et al. 2008), MPK6 mediates salt-induced iron superoxide dismutase gene expression and further induces ROS production (Xing et al.2 015). In soybean, GmMPK6 induces ROS generation through the transcriptional regulation of GmRbohI1 and increases salt tolerance in soybean (Son et al. 2023). Similarly, in this study, we detected an excessive amount of H2O2 in the leaves of OM31 plants and found direct evidence of interaction between GhMPK31 and GhRBOHB. Therefore, we speculate that GhMPK31 influences the generation of ROS through its interaction with GhRBOHB, which is also the direct cause of HR-like programmed cell death in cotton leaves. Furthermore, our wide comparative metabolomics analysis revealed a significant reduction in the levels of various defensive metabolites (such as flavonoids, alkaloids and their derivatives, phenols and their derivatives) in OM31 plants compared to the WT. This might be one of the factors to reduce OM31 defense capability shown in H. armigera selective feeding experiment, however, the regulatory mechanism of GhMPK31 in regulating these pathways was not clear which requires further investigation.
In conclusion, this study contributes to the existing knowledge on the mechanism by which MAPK regulates the outbreak of ROS, enhancing our understanding of how MAPK regulates plant defense against herbivores, we also proposed a model for GhMPK31 regulation of ROS outbreak and defense decline in cotton plants (Fig. 8). Additionally, it provides a foundation for further investigation into the functional characteristics of the MAPK gene family in upland cotton.
Fig. 8.
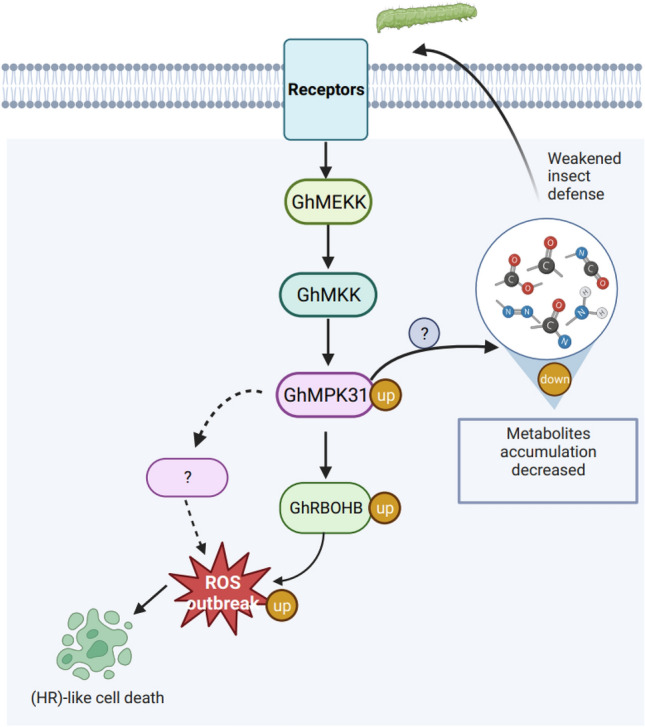
Model of GhMPK31 regulating ROS outbreak and defense decline in upland cotton. Enhanced expression of GhMPK31 triggers the expression of GhRBOHB and other genes, resulting in ROS outbreak and HR-like cell death in plant leaves. Simultaneously, it hampers the accumulation of defense metabolites, leading to a decline in cotton's defense ability against herbivorous insects
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Key R&D Program of China (2022YFF1001403) and Key Scientific and Technological Project of Henan Province (222102110151). We thank Sijia Liang at Academy of Industry innovation and Development, Huanghuai University, Zhumadian, Henan, China for providing genetically modified materials and for providing help in manuscript writing.
Author contributions
SXJ, XLZ and JLC provided experimental design and supervision. FQW performed the experiments and manuscript preparation. SJL provided genetically modified materials. QQW and ZPX Provided data analysis assistance. JL, GYW, BL, YBF, CYF, TYH, Muna Alariqi and Amjad Hussain provided suggestions and modified this manuscript.
Funding
National Key R& D Program of China, 2022YFF1001403, Zhongping Xu, Key Scientific and Technological Project of Henan Province, 222102110151, Sijia Liang.
Data availability
The RNA-seq data used in this study are available in the NCBI database under the accession PRJNA1087520.
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fuqiu Wang and Sijia Liang have contributed equally to this work and should be considered co-first authors.
Contributor Information
Jinglin Cao, Email: caojinglin670425@sohu.com.
Jian Li, Email: 17699436688@163.com.
Shuangxia Jin, Email: jsx@mail.hzau.edu.cn.
References
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Asai S, Ohta K, Yoshioka H. MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell. 2008;20:1390–1406. doi: 10.1105/tpc.107.055855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell. 2003;15:545–559. doi: 10.1105/tpc.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Wang NN, Zhang JB, Zheng Y, Li XB. Genome-wide identification of the mitogen-activated protein kinase (MAPK) family in cotton (Gossypium hirsutum) reveals GhMPK6 involved in fiber elongation. Plant Mol Biol. 2020;103:391–407. doi: 10.1007/s11103-020-00999-9. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. Regulation of floral organ abscission in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- Ding H, He J, Wu Y, Wu X, Ge C, Wang Y, Zhong S, Peiter E, Liang J, Xu W. The tomato mitogen-activated protein kinase SlMPK1 is as a negative regulator of the high-temperature stress response. Plant Physiol. 2018;177:633–651. doi: 10.1104/pp.18.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doczi R, Bogre L. The quest for MAP kinase substrates: gaining momentum. Trends Plant Sci. 2018;23:918–932. doi: 10.1016/j.tplants.2018.08.002. [DOI] [PubMed] [Google Scholar]
- Erb M, Kliebenstein DJ. Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 2020;184:39–52. doi: 10.1104/pp.20.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GroupM Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/S1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006;11:192–198. doi: 10.1016/j.tplants.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Hettenhausen C, Schuman MC, Wu JQ. MAPK signaling: a key element in plant defense response to insects. Insect Science. 2015;22:157–164. doi: 10.1111/1744-7917.12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CL, Sun YJ, Pillitteri LJ, Torii KU, Wang H, Zhang S, Ma H. Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol Plant. 2008;1:645–658. doi: 10.1093/mp/ssn029. [DOI] [PubMed] [Google Scholar]
- Hu Q, Min L, Yang X, Jin S, Zhang L, Li Y, Ma Y, Qi X, Li D, Liu H, Lindsey K, Zhu L, Zhang X. Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 2018;176:1808–1823. doi: 10.1104/pp.17.01628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagodzik P, Tajdel-Zielinska M, Ciesla A, Marczak M, Ludwikow A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front Plant Sci. 2018;9:1387. doi: 10.3389/fpls.2018.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalmi SK, Sinha AK. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front Plant Sci. 2015;6:769. doi: 10.3389/fpls.2015.00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Li B, Li S, Liang Y, Wu X, Ma M, Wang J, Gao J, Cai Y, Zhang Y, Wang Y, Li J, Wang Y. Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 2016;14:e1002550. doi: 10.1371/journal.pbio.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Liu Y, Yang KY, Kim CY, Baker B, Zhang S. Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J. 2003;33:719–731. doi: 10.1046/j.1365-313X.2003.01664.x. [DOI] [PubMed] [Google Scholar]
- Jin S, Zhang X, Nie Y, Guo X, Liang S, Zhu H. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol Plant. 2006;50:519–524. doi: 10.1007/s10535-006-0082-5. [DOI] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc Natl Acad Sci U S A. 2007;104:12205–12210. doi: 10.1073/pnas.0700344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Li J, Zhu L, Hull JJ, Liang S, Daniell H, Jin S, Zhang X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly) Plant Biotechnol J. 2016;14:1956–1975. doi: 10.1111/pbi.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Li M, Wang P, Cox KL, Jr, Duan L, Dever JK, Shan L, Li Z, He P. Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol. 2017;215:1462–1475. doi: 10.1111/nph.14680. [DOI] [PubMed] [Google Scholar]
- Li S, Han X, Yang L, Deng X, Wu H, Zhang M, Liu Y, Zhang S, Xu J. Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant Cell Environ. 2018;41:134–147. doi: 10.1111/pce.12984. [DOI] [PubMed] [Google Scholar]
- Liang S, Luo J, Alariqi M, Xu Z, Wang A, Zafar MN, Ren J, Wang F, Liu X, Xin Y, Xu H, Guo W, Wang Y, Ma W, Chen L, Lindsey K, Zhang X, Jin S. Silencing of a LIM gene in cotton exhibits enhanced resistance against Apolygus lucorum. J Cell Physiol. 2021;236:5921–5936. doi: 10.1002/jcp.30281. [DOI] [PubMed] [Google Scholar]
- Liu Y, He C. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 2016;35:995–1007. doi: 10.1007/s00299-016-1950-x. [DOI] [PubMed] [Google Scholar]
- Liu YD, Zhang SQ. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ren D, Pike S, Pallardy S, Gassmann W, Zhang S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007;51:941–954. doi: 10.1111/j.1365-313X.2007.03191.x. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Horstman HD, Braun E, Graham MA, Zhang C, Navarre D, Qiu WL, Lee Y, Nettleton D, Hill JH, Whitham SA. Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 2011;157:1363–1378. doi: 10.1104/pp.111.185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Braun E, Qiu WL, Shi YF, Marcelino-Guimaraes FC, Navarre D, Hill JH, Whitham SA. Positive and negative roles for soybean MPK6 in regulating defense responses. Mol Plant Microbe Interact. 2014;27:824–834. doi: 10.1094/MPMI-11-13-0350-R. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhao M, Gu C, Jiang H, Sun J, Li J. Genome-wide identification of MAPK family genes and their response to abiotic stresses in tea plant (Camellia sinensis) Open Life Sci. 2022;17:1064–1074. doi: 10.1515/biol-2022-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Zhao LL, Gong SY, Sun X, Li P, Qin LX, Zhou Y, Xu WL, Li XB. A cotton mitogen-activated protein kinase (GhMPK6) is involved in ABA-induced CAT1 expression and H(2)O(2) production. J Genet Genomics. 2011;38:557–565. doi: 10.1016/j.jgg.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Luo P, Dai W, Yin P, Zeng Z, Kong H, Zhou L, Wang X, Chen S, Lu X, Xu G. Multiple reaction monitoring-ion pair finder: a systematic approach to transform nontargeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography-mass spectrometry. Anal Chem. 2015;87:5050–5055. doi: 10.1021/acs.analchem.5b00615. [DOI] [PubMed] [Google Scholar]
- Luo J, Liang S, Li J, Xu Z, Li L, Zhu B, Li Z, Lei C, Lindsey K, Chen L, Jin S, Zhang X. A transgenic strategy for controlling plant bugs (Adelphocoris suturalis) through expression of double-stranded RNA homologous to fatty acyl-coenzyme A reductase in cotton. New Phytol. 2017;215:1173–1185. doi: 10.1111/nph.14636. [DOI] [PubMed] [Google Scholar]
- Majeed Y, Zhu X, Zhang N, Ul-Ain N, Raza A, Haider FU, Si H. Harnessing the role of mitogen-activated protein kinases against abiotic stresses in plants. Front Plant Sci. 2023;14:932923. doi: 10.3389/fpls.2023.932923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE, Sharma SB, Klessig DF, Martienssen R, Mattsson O, Jensen AB, Mundy J. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/S0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- Pitzschke A. Modes of MAPK substrate recognition and control. Trends Plant Sci. 2015;20:49–55. doi: 10.1016/j.tplants.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Qiu P, Li J, Zhang L, Chen K, Shao J, Zheng B, Yuan H, Qi J, Yue L, Hu Q, Ming Y, Liu S, Long L, Gu J, Zhang X, Lindsey K, Gao W, Wu H, Zhu L. Polyethyleneimine-coated MXene quantum dots improve cotton tolerance to Verticillium dahliae by maintaining ROS homeostasis. Nat Commun. 2023;14:7392. doi: 10.1038/s41467-023-43192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Yang H, Zhang S. Cell death mediated by MAPK is associated with hydrogen peroxide production in Arabidopsis. J Biol Chem. 2002;277:559–565. doi: 10.1074/jbc.M109495200. [DOI] [PubMed] [Google Scholar]
- Ren D, Yang KY, Li GJ, Liu Y, Zhang S. Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defense response and its involvement in hypersensitive response-like cell death. Plant Physiol. 2006;141:1482–1493. doi: 10.1104/pp.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyna NS, Yang Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact. 2006;19:530–540. doi: 10.1094/MPMI-19-0530. [DOI] [PubMed] [Google Scholar]
- Shao Y, Yu X, Xu X, Li Y, Yuan W, Xu Y, Mao C, Zhang S, Xu J. The YDA-MKK4/MKK5-MPK3/MPK6 cascade functions downstream of the RGF1-RGI ligand-receptor pair in regulating mitotic activity in root apical meristem. Mol Plant. 2020;13:1608–1623. doi: 10.1016/j.molp.2020.09.004. [DOI] [PubMed] [Google Scholar]
- Si H, Liu HL, Sun YW, Xu ZP, Liang SJ, Li B, Ding X, Li JY, Wang QQ, Sun L, Zhang XL, Jin SX. Transcriptome and metabolome analysis reveal that oral secretions from Helicoverpa armigera and Spodoptera litura influence wound-induced host response in cotton. Crop J. 2020;8:929–942. doi: 10.1016/j.cj.2019.12.007. [DOI] [Google Scholar]
- Son S, Kim J, An CS, Kim SL, Lee H, Im JH. GmMPK6 positively regulates salt tolerance through induction of GmRbohI1 in soybean. Antioxidants (basel) 2023;12(3):601. doi: 10.3390/antiox12030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Nitta Y, Zhang Q, Wu D, Tian H, Lee JS, Zhang Y. Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 2018 doi: 10.15252/embr.201745324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Cheng L, Qi X, Ge Z, Niu C, Zhang X, Jin S. Transgenic cotton plants expressing double-stranded RNAs TARGET HMG-CoA Reductase (HMGR) gene inhibits the growth, development and survival of cotton bollworms. Int J Biol Sci. 2015;11:1296–1305. doi: 10.7150/ijbs.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet H, Brillet-Gueguen L, Coppee JY, Dillies MA. SARTools: a DESeq2- and EdgeR-Based R Pipeline for comprehensive differential analysis of RNA-Seq Data. PLoS ONE. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li J, Hu L, Zhang T, Zhang G, Lou Y. OsMPK3 positively regulates the JA signaling pathway and plant resistance to a chewing herbivore in rice. Plant Cell Rep. 2013;32:1075–1084. doi: 10.1007/s00299-013-1389-2. [DOI] [PubMed] [Google Scholar]
- Wang C, He X, Li Y, Wang L, Guo X, Guo X. The cotton MAPK kinase GhMPK20 negatively regulates resistance to Fusarium oxysporum by mediating the MKK4-MPK20-WRKY40 cascade. Mol Plant Pathol. 2018;19:1624–1638. doi: 10.1111/mpp.12635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tu L, Yuan D, Zhu D, Shen C, Li J, Liu F, Pei L, Wang P, Zhao G, Ye Z, Huang H, Yan F, Ma Y, Zhang L, Liu M, You J, Yang Y, Liu Z, Huang F, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019;51:224–229. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- Wang T, Liu M, Wu Y, Tian Y, Han Y, Liu C, Hao J, Fan S. Genome-wide identification and expression analysis of MAPK gene family in lettuce (Lactuca sativa L) and functional analysis of LsMAPK4 in high-temperature-induced bolting. Int J Mol Sci. 2022;23(19):11129. doi: 10.3390/ijms231911129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, Shockcor J, Loftus N, Holmes E, Nicholson JK. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat Protoc. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313X.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell. 2007;19:1096–1122. doi: 10.1105/tpc.106.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Jia W, Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- Xing Y, Chen WH, Jia W, Zhang J. Mitogen-activated protein kinase kinase 5 (MKK5)-mediated signalling cascade regulates expression of iron superoxide dismutase gene in Arabidopsis under salinity stress. J Exp Bot. 2015;66:5971–5981. doi: 10.1093/jxb/erv305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Zhang S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015;20:56–64. doi: 10.1016/j.tplants.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Yao Y, Zhao H, Sun L, Wu W, Li C, Wu Q. Genome-wide identification of MAPK gene family members in Fagopyrum tataricum and their expression during development and stress responses. BMC Genomics. 2022;23:96. doi: 10.1186/s12864-022-08293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa T, Ichimura K, Mizoguchi T, Shinozaki K. Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 2001;42:1012–1016. doi: 10.1093/pcp/pce123. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhang S. Mitogen-activated protein kinase cascades in plant signaling. J Integr Plant Biol. 2022;64:301–341. doi: 10.1111/jipb.13215. [DOI] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L acc TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33:531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- Zhang M, Wu H, Su J, Wang H, Zhu Q, Liu Y, Xu J, Lukowitz W, Zhang S. Maternal control of embryogenesis by MPK6 and its upstream MKK4/MKK5 in Arabidopsis. Plant J. 2017;92:1005–1019. doi: 10.1111/tpj.13737. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su J, Zhang Y, Xu J, Zhang S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr Opin Plant Biol. 2018;45:1–10. doi: 10.1016/j.pbi.2018.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data used in this study are available in the NCBI database under the accession PRJNA1087520.