Abstract
The primary cellular receptor for mouse hepatitis virus (MHV), a murine coronavirus, is MHVR (also referred to as Bgp1a or C-CAM), a transmembrane glycoprotein with four immunoglobulin-like domains in the murine biliary glycoprotein (Bgp) subfamily of the carcinoembryonic antigen (CEA) family. Other murine glycoproteins in the Bgp subfamily, including Bgp1b and Bgp2, also can serve as MHV receptors when transfected into MHV-resistant cells. Previous studies have shown that the 108-amino-acid N-terminal domain of MHVR is essential for virus receptor activity and is the binding site for monoclonal antibody (MAb) CC1, an antireceptor MAb that blocks MHV infection in vivo and in vitro. To further elucidate the regions of MHVR required for virus receptor activity and MAb CC1 binding, we constructed chimeras between MHVR and other members of the CEA family and tested them for MHV strain A59 (MHV-A59) receptor activity and MAb CC1 binding activity. In addition, we used site-directed mutagenesis to introduce selected amino acid changes into the N-terminal domains of MHVR and these chimeras and tested the abilities of these mutant glycoproteins to bind MAb CC1 and to function as MHV receptors. Several recombinant glycoproteins exhibited virus receptor activity but did not bind MAb CC1, indicating that the virus and MAb binding sites on the N-terminal domain of MHVR are not identical. Analysis of the recombinant glycoproteins showed that a short region of MHVR, between amino acids 34 and 52, is critical for MHV-A59 receptor activity. Additional regions of the N-terminal variable domain and the constant domains, however, greatly affected receptor activity. Thus, the molecular context in which the amino acids critical for MHV-A59 receptor activity are found profoundly influences the virus receptor activity of the glycoprotein.
Initial events in virus infection of a cell include attachment of the virus to the cell, entry, and disassembly of the virion. For most viruses, attachment is mediated through a specific interaction between the virus attachment protein and a cell surface receptor. Previous studies identified the murine biliary glycoprotein MHVR (also referred to as Bgp1a or C-CAM) as the primary cellular receptor for murine coronavirus mouse hepatitis virus strain A59 (MHV-A59) (20, 53). This glycoprotein, isolated from liver and intestinal brush border membranes of MHV-sensitive BALB/c mice, binds to MHV-A59 virions in a solid-phase viral overlay protein blot assay (9) and is recognized by an antireceptor monoclonal antibody (MAb CC1) that protects cells expressing MHVR from infection by MHV-A59 in vivo and in vitro (20, 52, 53). A cDNA encoding an allelic variant of MHVR, Bgp1b (also referred to as mmCGM2) (38), was isolated from cells of MHV-resistant SJL/J mice (18, 53), and a second murine biliary glycoprotein, Bgp2, which is expressed in the colons of both BALB/c and SJL/J mice, also has been characterized (38). MHVR and Bgp1b consist of an N-terminal immunoglobulin (Ig)-like variable domain, three Ig-like constant domains, a transmembrane domain, and a cytoplasmic tail. The Bgp2 glycoprotein exhibits a similar structure except that it contains only one constant domain. The Bgp1b and Bgp2 glycoproteins can serve as functional receptors for MHV-A59 when overexpressed in MHV-A59-resistant hamster cells in transient transfection assays, but these glycoproteins do not bind virus in solid-phase binding assays and are not recognized by MAb CC1 (18, 38). Natural splice variants of MHVR and Bgp1b yield glycoproteins containing the N-terminal and fourth Ig-like domains, the transmembrane domain, and the cytoplasmic tail (18, 21, 53).
A secreted three Ig domain murine glycoprotein called bCEA, a pregnancy-specific glycoprotein in the murine carcinoembryonic antigen (CEA) family, is expressed in C57BL/6 mouse brain and placenta and exhibits a low level of MHV-A59 receptor activity when expressed in COS-7 cells (11). To date, the only murine CEA-related glycoprotein shown to have no MHV receptor activity in transient transfection assays in MHV-A59-resistant hamster cells is Cea10 (formerly referred to as mmCGM3), a secreted glycoprotein consisting of two variable Ig-like domains that does not bind MHV-A59 or MAb CC1 (26, 32).
Deletion mutagenesis studies showed that MHV-A59 and MAb CC1 bind to the N-terminal Ig-like variable domain of MHVR (21). A recombinant chimeric glycoprotein containing the N-terminal domain of MHVR and the second, third, transmembrane, and cytoplasmic domains of the mouse poliovirus receptor (Pvr) homolog serves as a functional receptor for MHV-A59 when expressed in hamster cells (17). Furthermore, a soluble recombinant glycoprotein consisting of only the N-terminal domain of MHVR can inhibit MHV-A59 infectivity in a concentration-dependent manner (19). MAb CC1 recognizes both the MHVR/mph chimera and the soluble N-terminal domain of MHVR in immunoblot assays. A chimeric glycoprotein consisting of the N-terminal domain of Cea10, the three constant domains, transmembrane region, and cytoplasmic tail of MHVR, however, does not bind MHV-A59 or MAb CC1 (32).
Sequence analysis of the various receptor-like glycoproteins in the murine CEA family shows that the 108-amino-acid N-terminal domains of MHVR, Bgp1b, and Cea10 are significantly different, with 29 amino acid differences between MHVR and Bgp1b and 43 amino acid differences between MHVR and Cea10 (18, 26, 32). These glycoproteins also differ significantly in their receptor activities. A detailed analysis of the virus and MAb binding sites in the N-terminal domain of MHVR was done to elucidate the molecular basis for these observed differences in the receptor activities of the murine CEA-related glycoproteins. We have constructed a series of recombinant chimeric glycoproteins and tested their abilities to serve as functional receptors for MHV-A59 in transient transfection assays. The abilities of MAb CC1 to protect transfected cells from infection by MHV-A59 and to bind the recombinant glycoproteins in an immunoblot assay also were examined. Results of these assays indicate that amino acids 34 to 52 of the glycoprotein are critical for receptor activity and that binding of the MAb is very sensitive to any changes in the tertiary structure of MHVR. Site-directed mutagenesis studies confirmed the importance of these residues. Thus, this small region of the N-terminal domain of MHVR is a critical determinant of MHV receptor activity. These residues alone, however, are not sufficient for optimal receptor activity. Additional amino acids within the N-terminal domain of MHVR and the three Ig-like constant domains of MHVR also profoundly affect receptor activity. The data suggest that these domains either influence the conformation of the virus-binding site or affect events subsequent to virus binding that are required for infection.
MATERIALS AND METHODS
Viruses, cells, and antibodies.
MHV-A59 was propagated in 17Cl1 cells maintained in Dulbecco’s modified Eagle’s minimal essential medium supplemented with 10% fetal bovine serum and antibiotics and titered in L2 cells as previously described (22). Recombinant vaccinia virus strain vTF7-3 was provided by B. Moss (National Institutes of Health, Bethesda, Md.). The BHK-21 line of baby hamster kidney fibroblasts (American Type Culture Collection) was maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 10% tryptose phosphate broth, and antibiotics. MAb CC1 binds to the N-terminal domain of MHVR and protects cells expressing MHVR from MHV infection (18, 21, 52, 53). The polyclonal rabbit anti-MHVR antibody 655, which recognizes both MHVR and Bgp1b (20, 52, 53), was preabsorbed against paraformaldehyde-fixed or acetone-fixed BHK-21 cells prior to use for surface immunofluorescence or immunoblotting assays respectively as described below.
Construction of N-terminal domain chimeras.
All recombinant chimeric glycoproteins were generated from the full-length MHVR or Bgp1b cDNAs cloned into the HindIII-NotI site of pBlueScript SK+ (Stratagene, La Jolla, Calif.) (18, 20) or a construct which contained the cDNA encoding the N-terminal domain of Cea10 linked to the second, third, fourth, transmembrane, and cytoplasmic domains of MHVR. The amino acid sequences of the N-terminal domains of MHVR, Bgp1b, and Cea10 are shown in Fig. 1. Briefly, recombinant cDNAs were constructed by digesting the plasmids with the appropriate restriction endonucleases and separating the resulting DNA fragments by electrophoresis on agarose gels. DNA fragments of interest were eluted from the agarose either by passage through GenElute agarose spin columns (Supelco, Bellefonte, Pa.) or by elution with the Elu-Quick system (Schleicher & Schuell, Keene, N.H.) according to the manufacturers’ recommendations. Aliquots of the isolated DNA were run on agarose gels to estimate the amount of DNA recovered, and the fragments were then ligated into the pcDNA3 expression vector (InVitrogen, Carlsbad, Calif.), using Ready-to-Go T4 DNA ligase (Pharmacia Biotech, Piscataway, N.J.). The ligated material was transformed into Escherichia coli DH5α cells by using standard procedures, and recombinant plasmids were confirmed by restriction enzyme digestion and sequence analysis.
FIG. 1.
Comparison of the amino acid sequences of the N-terminal domains of the murine biliary glycoproteins MHVR (also referred to as Bgp1a or C-CAM), Bgp1b, and Cea10. For Bgp1b and Cea10, only amino acids that differ from MHVR are shown.
Chimeras were constructed by using the BamHI restriction site common to the cDNAs of MHVR, Bgp1b, and Cea10 which cleaves the cDNAs at nucleotides corresponding to amino acid 70. To further divide the N-terminal domains of MHVR and Cea10, KpnI and ClaI restriction sites were engineered into the cDNAs of MHVR and Cea10. These restriction enzyme sites allowed for the construction of recombinants at amino acids 34 (KpnI) and 52 (ClaI). Introduction of the ClaI site resulted in the amino acid changes S52N53M54F56 to IDRK in MHVR and N53R54K56 to DMF in Cea10. Introduction of the KpnI site resulted in the amino acid change K35 to Q in MHVR and Cea10. These amino acid changes had no effect on the virus receptor activities of the parental constructs as determined by immunofluorescence assays.
PCR-directed mutagenesis was used to introduce the KpnI and ClaI restriction sites into the MHVR and Cea10 cDNAs and to alter individual amino acids within the various chimeric cDNAs (48). Oligonucleotide primers for the PCR mutagenesis containing the nucleotide changes at their 5′ ends were synthesized with an Applied Biosystems DNA synthesizer (Applied Biosystems Inc., Foster City, Calif.). All mutations were confirmed by sequence analysis using a Taq DyeDeoxy Terminator Cycle Sequencing kit (ABI).
Detection of CEA-related glycoproteins in transfected BHK-21 cells by immunoblot analysis and flow cytometry.
To determine whether the recombinant chimeric cDNAs were producing glycoproteins with the appropriate molecular weights and immunoreactivities, protein extracts of transfected cells were analyzed. BHK-21 cells were infected with vaccinia virus encoding the T7 RNA polymerase (vTF7-3) at a multiplicity of infection of 10 (23). At 3 h postinfection, the cells were transfected with the cDNAs encoding MHVR, the chimeric recombinant or mutated glycoproteins, or empty plasmid in serum-free medium. At 24 h posttransfection, the cells were lysed with 0.3 ml of radioimmunoprecipitation assay buffer (0.1 M NaCl, 0.001 M EDTA [pH 7.4], 0.1% Nonidet P-40, 0.1% deoxycholate, 1% phenylmethylsulfonyl fluoride, 1% aprotinin). Thirty to forty microliters of the extract was mixed with sample treatment mix (53), boiled for 3 to 5 min, and separated by electrophoresis on sodium dodecyl sulfate–8 or 10% polyacrylamide gels. The proteins were transferred to nitrocellulose, which was blocked with 5% bovine serum albumin in B3 buffer (0.15 M NaCl, 0.001 M EDTA, 0.05 M Tris base, 0.05 M Tris-HCl, 0.05% Tween 20, 0.1% bovine serum albumin) and then probed with a 1:200 dilution of the polyclonal anti-MHVR antibody 655 or a 1:50 dilution of MAb CC1 followed by rabbit anti-mouse MAb. Proteins were detected with 125I-labeled staphylococcal protein A (New England Nuclear, New Bedford, Mass.) as previously described (38).
Flow cytometry was also used to confirm the expression of recombinant glycoproteins on the surface of transfected cells and to determine whether the glycoproteins could be recognized by MAb CC1. BHK-21 cells were transfected with recombinant cDNAs cloned into pcDNA3 or empty plasmid as described above. At 48 h posttransfection, the cells were trypsinized, and the abilities of receptor glycoproteins on the plasma membrane to bind MAb CC1 were determined by incubation of the cells with the MAb or an IgG1 MAb to an irrelevant antigen followed by R-phycoerythrin-conjugated affinity-purified anti-mouse IgG. Bound antibodies were detected in a fluorescence-activated cell sorter (FACS). Controls included cells without primary or secondary antibodies and cells incubated only with R-phycoerythrin.
Assays for virus receptor activity and MAb CC1 receptor blockade.
To determine if the recombinant chimeric and mutant glycoproteins could serve as functional receptors for MHV-A59, the glycoproteins were transiently expressed in BHK-21 cells as previously described (17, 18, 20, 21). Briefly, cells grown on glass coverslips were transfected with plasmids containing the cDNAs subcloned into pcDNA3 (InVitrogen) or the empty plasmid, using LipofectAMINE (Life Technologies, Gaithersburg, Md.) according to the manufacturer’s recommendations. Thirty-five hours posttransfection, the cells were incubated for 1 h with MHV-A59 at a multiplicity of infection of 1. At 16 h after virus inoculation, the cells were fixed with cold acetone. Viral antigens in the cytoplasm were detected with a 1:50 dilution of polyclonal convalescent mouse anti-MHV serum followed by a 1:50 dilution of rhodamine-labeled goat anti-mouse IgG and visualized in a Zeiss microscope. For recombinant molecules that showed virus receptor activity in this assay, the ability of MAb CC1 to block MHV-A59 infection was examined. Transfected cells were treated for 1 h with a 1:5 dilution of MAb CC1 hybridoma supernatant or a control antibody of the same isotype directed against an unrelated antigen and then inoculated with virus in the presence of MAb CC1. Virus receptor activity was then assayed as indicated above. To confirm expression of the recombinant glycoproteins at the plasma membrane, transiently transfected BHK-21 cells were fixed with 2% paraformaldehyde 48 to 72 h posttransfection, incubated with BHK-21 cell preadsorbed polyclonal antireceptor antibody 655 or normal rabbit serum followed by rhodamine-labeled goat anti-rabbit serum, and visualized in a Zeiss microscope (20).
RESULTS
Identification of residues in the N-terminal domain of MHVR important for MHV-A59 receptor activity.
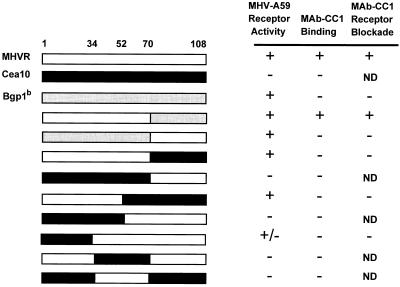
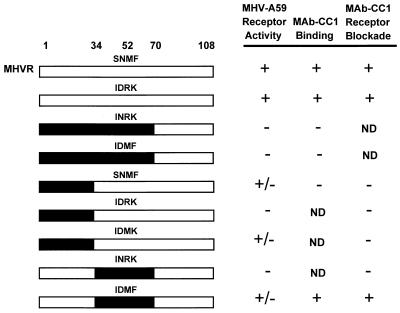
We previously demonstrated that the N-terminal Ig-like domain of MHVR binds MHV-A59 and is essential for virus infection (17, 19). When expressed at high levels in a transient transfection assay, Bgp1b also can serve as a functional receptor for MHV-A59, but this glycoprotein is not recognized by MHV-A59 in a solid-phase binding assay (9) and is not recognized by anti-MHVR MAb CC1 (18, 52). The N-terminal domain of Cea10 fused to the second, third, fourth, transmembrane, and cytoplasmic domains of MHVR cannot serve as a functional receptor for the virus when expressed in MHV-resistant BHK cells (32). To identify the regions of the MHVR N-terminal domain that are necessary for virus receptor activity, the abilities of MHVR/Bgp1b and MHVR/Cea10 N-terminal domain chimeras to serve as functional receptors for MHV-A59 were examined. Each of the N-terminal chimeras, shown schematically in Fig. 2, was followed by the second, third, fourth, transmembrane, and cytoplasmic domains of MHVR. cDNAs encoding the parental or chimeric glycoproteins in the expression vector pcDNA3 were transfected into MHV-resistant BHK-21 cells. Immunolabeling of transfected cells with anti-MHVR antibody 655 confirmed that each of the chimeric glycoproteins was expressed on the plasma membrane. Transfected cells were inoculated with MHV-A59 as described above. Constructs were scored as follows: +, positive (several cells expressing viral antigens were detected in every 60× field); +/−, weakly positive (5 to 50 cells expressing viral antigens were detected on the entire coverslip); or −, negative (fewer than 5 labeled cells were detected on the coverslip). Representative immunofluorescence data are shown in Fig. 3.
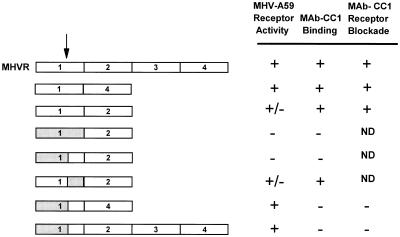
FIG. 2.
Functions associated with the N-terminal domain recombinant chimeric anchored glycoproteins. All chimeras also contained the second, third, fourth, transmembrane, and cytoplasmic domains of MHVR (not shown). Unshaded regions represent MHVR sequences, black regions represent Cea10 sequences, and gray regions represent Bgp1b sequences. Selected amino acid positions are indicated above. Receptor activity and MAb CC1 receptor blockade activity were determined by immunofluorescence as described in Materials and Methods. MAb CC1 binding was determined by immunoblot assay of cell lysates following infection with vaccinia virus vTF7-3 and transfection of recombinant plasmid. ND, not done.
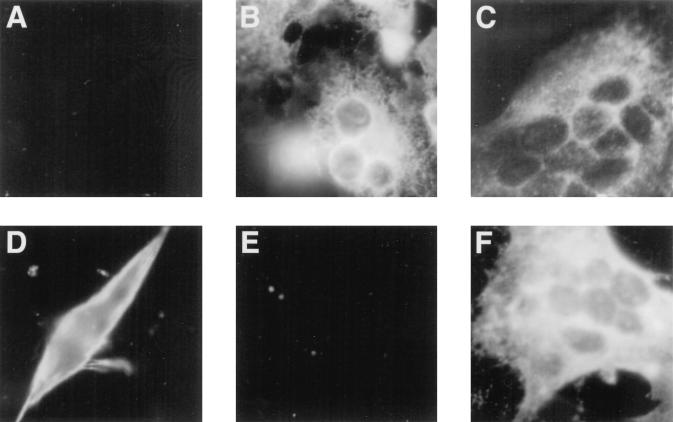
FIG. 3.
Detection of MHV antigens or chimeric glycoproteins in hamster cells transfected with plasmid containing the cDNA of MHVR, Cea10, or representative MHVR/Cea10 chimeras. BHK-21 cells grown on glass coverslips were transfected with recombinant MHVR cDNA in which the N-terminal domain consisted of Cea10 (A and D), MHVR (B and E), or MHVR1–52/Cea1053–108 (C and F). Following transfection, cells in panels A, B, C, E, and F were inoculated with MHV-A59, incubated for 16 h, and fixed in cold acetone. Cells in panel E and F were pretreated with MAb CC1. Viral antigens in the cytoplasm were detected with anti-MHV serum and rhodamine-labeled goat anti-mouse IgG. Cells in panel D were transfected, and surface expression of the transfected molecule was confirmed by immunofluorescence with the anti-MHVR polyclonal antibody 655.
Analysis of the receptor activities of the MHVR/Cea10 chimeric glycoproteins showed that every construct that served as a receptor for MHV-A59 contained amino acids 34 to 52 of MHVR (Fig. 2). For instance, one construct contained amino acids 1 to 52 of MHVR (MHVR1–52/Cea1053–108), while a second construct contained amino acids 34 to 108 of MHVR (Cea101–33/MHVR34–108). Both of these chimeras displayed receptor activity. In contrast, the construct containing amino acids 1 to 52 of Cea10 showed no receptor activity. Surprisingly, a chimeric glycoprotein in which the N-terminal domain contained only amino acids 34 to 70 of MHVR (Cea101–33,71–108/MHVR34–70) did not exhibit any MHV-A59 receptor activity. This finding demonstrates that while amino acids 34 to 52 of MHVR are essential determinants of MHV-A59 receptor activity, additional MHVR sequences, either from amino acids 1 to 33 or from amino acids 71 to 108, also affect MHV-A59 receptor activity.
To further determine which amino acids were important for MHV-A59 receptor activity, a series of point mutations was generated by site-directed mutagenesis. As shown in Table 1, amino acids in the N-terminal domain of MHVR were changed to alanine residues (I66I67/AA) or amino acids similar to those of Cea10 (L26A27L28A30A32/QTRVY, D42K43/HN, I66L74/TF), a rat biliary glycoprotein (E65/V, V85/A, T91E93/FQ, T98/I), or a human biliary glycoprotein (R96R97/VP) in an effort to abrogate receptor activity. None of these introduced mutations had any effect on receptor activity. A construct which contained an R64-to-S mutation and deletion of amino acids 55 to 58 and 65 to 71, however, exhibited no receptor activity. When the S64 mutation was reverted to R in this construct, weak but detectable receptor activity was restored. Modeling studies suggest that R64 is involved in formation of an intramolecular salt bridge (see Fig. 7) that probably is required for correct folding of the glycoprotein.
TABLE 1.
Effects of point mutations in MHVR on MHV-A59 receptor activity, MAb CC1 receptor blockade activity, and MAb CC1 binding activitya
| Mutation(s) | MHV-A59 receptor activity | MAb CC1 receptor blockade | MAb CC1 binding |
|---|---|---|---|
| L26A27L28A30A32/QTRVY | + | − | − |
| D42K43/HN | + | − | − |
| E65/V | + | + | + |
| I66I67/AA | + | + | + |
| I66L74/TF | + | + | + |
| M77I78E93V106/AAGE | + | + | + |
| V85/A | + | + | + |
| M90/L | + | + | + |
| T91E93/FQ | + | + | + |
| R96R97/VP | + | + | + |
| T98/I | + | + | ND |
| R64/S, Δ55–58, Δ65–71 | − | − | − |
| Δ55–58, Δ65–71 | +/− | − | − |
All mutations were made in parental construct containing MHVR. Receptor activity and MAb CC1 protection activity were determined as described in Materials and Methods. MAb CC1 binding was determined in immunoblot assays. ND, not done.
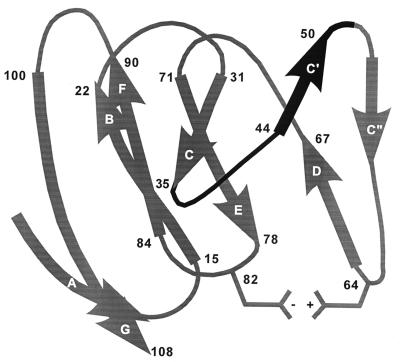
FIG. 7.
Structural model of the N-terminal domain of MHVR. Structure was determined based on sequence similarities between the N-terminal domains of MHVR and CD4. Beta sheets are indicated by wide ribbons and labeled. Numbers indicate amino acid positions. The amino acid region critical for MHV-A59 receptor activity is in black. A putative salt bridge between residues R64 and D82 is indicated.
In a second set of recombinants, mutations were introduced to change individual amino acids in Cea10 to the corresponding MHVR amino acids in an effort to generate a functional receptor molecule (Table 2). Because the chimeric glycoprotein studies described above indicated that amino acids 34 to 52 of MHVR were critical determinants of MHV-A59 receptor activity, we concentrated on these residues. The construct containing amino acids 1 to 70 of Cea10 and 71 to 108 of MHVR, which exhibited no receptor activity, was used as the parental construct for these mutagenesis studies (Fig. 2). When the amino acids S38G39G41 were mutated to TTI in Cea101–70/MHVR71–108, weak receptor activity was observed (Table 2). When the amino acid changes S38G39 to TT or G41 to I were introduced into the Cea101–70/MHVR71–108 chimera, however, no receptor activity was detected (Table 2). In addition, when the mutations S38G39G41 to TTI were introduced into a construct containing the entire N-terminal domain of Cea10, no receptor activity was observed (Table 2). These data show that amino acids 38 to 41 of MHVR are important determinants of MHV-A59 receptor activity when in the presence of amino acids 71 to 108 of MHVR but not amino acids 71 to 108 of Cea10.
TABLE 2.
Effects of point mutations in the N-terminal domain on MHV-A59 receptor activity and MAb CC1 binding activitya
| N-terminal domain | Mutation | MHV-A59 receptor activity | MAb CC1 binding |
|---|---|---|---|
| Cea101–70/MHVR71–108 | None | − | − |
| T50S51I52/PNS | − | − | |
| N43G46/KA | − | − | |
| S38G39G41/TTI | +/− | − | |
| S38G39/TT | − | − | |
| G41/I | − | − | |
| Cea101–108 | None | − | − |
| S38G39G41/TTI | − | − |
Mutations were designed to change Cea10 amino acids to their corresponding MHVR residues. MHV-A59 receptor activity and MAb CC1 binding activity were determined as described in Materials and Methods.
Introduction of a KpnI restriction enzyme site into the MHVR and Cea10 cDNAs resulted in a K35-to-Q amino acid change in both proteins. This change did not alter the receptor activity of MHVR, Cea10, or any of the chimeric recombinant glycoproteins (data not shown). Introduction of the ClaI restriction enzyme site into the cDNAs of MHVR and Cea10 resulted in S52N53M54F56 to IDRK changes in MHVR and N53R54K56 to DMF changes in Cea10. These amino acid changes also did not alter the receptor activities of either parental construct (Fig. 4). When these amino acid substitutions were introduced into certain chimeric glycoproteins, however, their receptor activities were affected. The chimeric glycoprotein containing amino acids 1 to 33 of Cea10 and 34 to 108 of MHVR exhibited weak MHV-A59 receptor activity (Fig. 2). When the amino acids S52N53M54F56 were mutated to IDRK in this chimera, however, MHV-A59 receptor activity was abrogated (Fig. 4). Conversely, although the chimeric glycoprotein containing Cea10 amino acids 34 to 70 in an MHVR background showed no receptor activity (Fig. 2), changing N53R54K56 to DMF in this construct restored weak MHV-A59 receptor activity and MAb CC1 binding activity (Fig. 4). These results strongly support the conclusions that amino acids 34 to 70 are critical determinants of MHV-A59 receptor activity and MAb CC1 binding and that additional sequences within the N-terminal domain outside this region also affect receptor activity.
FIG. 4.
Functions associated with the N-terminal recombinant chimeric anchored glycoproteins with and without ClaI restriction enzyme sites. All chimeras also contained the second, third, fourth, transmembrane, and cytoplasmic domains of MHVR (not shown). Unshaded regions represent MHVR sequences, and black regions represent Cea10 sequences. Mutated amino acids are indicated. Receptor activity and MAb CC1 receptor blockade activity were determined by immunofluorescence as described in Materials and Methods. MAb CC1 binding was determined by immunoblot assay of cell lysates following infection with vaccinia virus vTF7-3 and transfection of recombinant plasmid. ND, not done.
Role of MHVR constant domains in receptor activity.
Previous studies showed that a naturally occurring anchored two-domain splice variant of MHVR containing domains 1 and 4 (MHVR[1,4]) and a recombinant anchored two-domain variant of MHVR containing domains 1 and 2 (MHVR[1,2]) both can serve as functional receptors for MHV-A59 (18, 21). Because our analysis of the recombinant chimeric glycoproteins indicated that multiple regions within the N-terminal domain of MHVR can influence receptor activity, we also examined the effects of MHVR constant regions on receptor activity. As demonstrated previously (18, 21), MHVR[1,4] functioned as a viral receptor. MHVR[1,2], on the other hand, exhibited only weak MHV-A59 receptor activity in this assay (Fig. 5). In light of this result, we constructed several MHVR/Bgp1b N-terminal chimeras in which the recombinant N-terminal domains replaced the N-terminal domains of MHVR[1,4] or MHVR[1,2]. These chimeric glycoproteins were tested for MHV-A59 receptor activity and MAb CC1 binding and receptor blockade activities (Fig. 5). N-terminal domain chimeras that functioned as virus receptors when linked to the full-length MHVR also functioned as receptors when linked to MHVR[1,4]. In the context of MHVR[1,2], however, the N-terminal chimera Bgp1b1–70/MHVR71–108 did not serve as a functional receptor and the chimera MHVR1–70/Bgp1b71–108 functioned only weakly as a receptor for MHV-A59 (Fig. 5). These data show that the constant regions of MHVR also can influence receptor activity and suggest that optimal virus receptor activity requires the presence of domain 4.
FIG. 5.
Functions associated with recombinant chimeric anchored glycoproteins containing various constant domains. All chimeras also contained the transmembrane and cytoplasmic domains of MHVR (not shown). Domains are numbered (1 = N-terminal domain). The arrow indicates approximate location of amino acid 70 in the N-terminal domain. Unshaded regions represent MHVR sequences, and gray regions represent Bgp1b sequences. Receptor activity and MAb CC1 receptor blockade activity were determined by immunofluorescence as described in Materials and Methods. MAb CC1 binding was determined by immunoblot assay of cell lysates following infection with vaccinia virus vTF7-3 and transfection of recombinant plasmid. ND, not done.
Analysis of antireceptor MAb CC1 binding domains.
The anti-MHVR MAb CC1 has been shown to protect cells expressing MHVR from infection by MHV-A59 (50, 51). To identify amino acids in the N-terminal domain of MHVR required for MAb CC1 binding activity, the ability of MAb CC1 to block infection by MHV-A59 of cells expressing receptor-positive recombinant chimeric glycoproteins was examined. In this receptor blockade assay, MAb CC1 did not block infection of cells expressing the chimeric glycoproteins MHVR1–70/Cea1071–108, MHVR1–52/Cea1053–108, or Cea101–33/MHVR34–108 (Fig. 2). FACS analysis of cells expressing these chimeric glycoproteins confirmed that MAb CC1 did not recognize the chimeric glycoproteins on the surface of these cells (data not shown), but all chimeric glycoproteins were expressed on the cell surface, as evidenced by surface labeling with the polyclonal antibody 655. While MAb CC1 did not block infection of cells expressing the chimeric glycoprotein MHVR1–70/Cea1071–108, it did block infection of cells expressing MHVR1–70/Bgp1b71–108. Cells expressing Bgp1b1–70/MHVR71–108, however, were not blocked by MAb-CC1 (Fig. 2). These data suggest that amino acids 1 to 70 of MHVR are critical for MAb CC1 binding activity. The context of these residues, however, also affects the antibody binding.
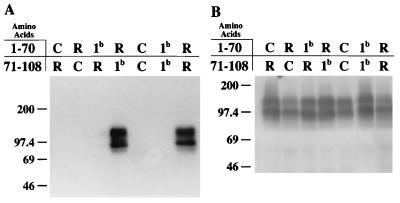
To determine if MAb CC1 could bind to the mutated recombinant glycoproteins in an immunoblot assay, cell lysates from vaccinia virus-infected, transfected cells were electrophoresed on polyacrylamide gels and transferred to nitrocellulose, and proteins were detected in a standard immunoblot assay. Representative results are shown in Fig. 6. None of the MHVR/Cea10 chimeric glycoproteins was recognized by MAb CC1. The antireceptor polyclonal antibody 655, on the other hand, detected all chimeric proteins in this assay (Fig. 6). Among MHVR/Bgp1b chimeras, MAb CC1 recognized the chimeric glycoprotein containing amino acids 1 to 70 of MHVR (MHVR1–70/Bgp1b71–108) but not the chimera containing the amino acids 1 to 70 of Bgp1b (Bgp1b1–70/MHVR71–108). These results confirm the importance of amino acids 1 to 70 of MHVR in MAb CC1 binding.
FIG. 6.
Recognition of MHVR, Bgp1b, Cea10, or representative chimeric glycoproteins by anti-MHVR antibodies. Cell lysates from BHK-21 cells infected with vaccinia virus vTF7-3 and transfected with cDNA encoding MHVR, Bgp1b, Cea10, or recombinant glycoproteins were separated by electrophoresis on sodium dodecyl sulfate–8 or 10% polyacrylamide gels, transferred to nitrocellulose, and incubated with either anti-MHVR MAb CC1 (A) or rabbit polyclonal anti-MHVR antibody 655 (B). Bound antibodies were detected by incubation of immunoblots with 125I-staphylococcal protein A. Sizes are indicated in kilodaltons. R, amino acids from MHVR; C, amino acids from Cea10; 1b, amino acids from Bgp1b.
To further analyze the binding characteristics of MAb CC1, the ability of this antibody to bind to the various point mutants in an immunoblot assay was determined. None of the recombinant glycoproteins in which Cea10 amino acids were changed to corresponding MHVR amino acids gained MAb CC1 binding activity (Table 2). We also examined the ability of MAb CC1 to bind to glycoproteins in which MHVR residues were changed to alanines or corresponding residues of Cea10, a rat biliary glycoprotein, or a human biliary glycoprotein. As shown in Table 1, most constructs in which one or a few MHVR amino acids were altered maintained MAb CC1 binding. In two constructs (D42K43 to HN and L26A27L28A30A32 to QTRVY), however, MAb CC1 binding was eliminated, thereby demonstrating the importance of amino acids 26 to 32, 42, and 43 in MAb CC1 binding.
DISCUSSION
Previous studies of MHV pathogenesis have shown that infection can lead to a variety of diseases (4, 49) and that the outcome is dependent both on the strain of virus and the genetic makeup of the host organism (1, 3, 5, 6, 15, 16, 25, 28, 29, 31, 43, 45, 50). Several reports have shown that, at least in cell culture assays where recombinant glycoproteins are expressed at high levels, multiple murine and human biliary glycoprotein-like molecules can serve as functional receptors for MHV-A59 (10, 11, 18, 38, 54, 55), and recent studies indicate that differences in binding affinity between the receptor and virus may determine the receptor effectiveness (39, 51). It is therefore possible that the various syndromes associated with MHV infection may, at least to some degree, be determined by molecular aspects of the receptor molecule.
To identify the amino acid residues of the MHVR glycoprotein necessary for receptor activity, we constructed a series of chimeric and mutated recombinant glycoproteins and analyzed the receptor activities and MAb binding properties of these molecules. All recombinant constructs were derived from MHVR, the principal MHV-A59 receptor molecule, which is also referred to as Bgp1a or C-CAM (20, 38, 52, 53), Bgp1b, an allelic variant derived from MHV-resistant SJL/J mice that exhibits receptor activity in cell culture assays (18), and Cea10, a closely related murine glycoprotein that exhibits no receptor activity (32). Analysis of the chimeric recombinant glycoproteins shows that all chimeric molecules possessing receptor activity contain amino acids 34 to 52 of MHVR, indicating the critical importance of these residues. Similar conclusions were reached by Rao et al., who showed that amino acids 38 to 43 of MHVR are critical for MHV binding (41). It is important to note, however, that these amino acids of MHVR are not sufficient for receptor activity. Our results show that a glycoprotein containing amino acids 34 to 70 of MHVR in a background of Cea10 does not serve as a functional receptor. Clearly, some amino acids between 1 and 33 or 71 and 108 of MHVR are required in conjunction with amino acids 34 to 52. We hypothesize that these additional amino acids are necessary to ensure the correct tertiary structure of the N-terminal domain and that structural differences between the N domains of MHVR and Cea10 explain the necessity for these additional MHVR residues.
Mutational analysis of the chimeric recombinant glycoproteins confirmed the importance of amino acids 34 to 52 and showed that amino acids 52 to 56 also are important for receptor activity. The chimeric glycoprotein Cea101–70/MHVR71–108 exhibited no MHV-A59 receptor activity. When amino acids S38 G39 G41 were converted in this glycoprotein to the corresponding MHVR amino acids (TTI), the resulting chimeric glycoprotein exhibited weak but detectable receptor activity, thereby confirming the critical importance of this region of the molecule for MHV-A59 receptor activity. Furthermore, alteration of the amino acids 52, 53, 54, and 56 resulting from introduction of a restriction enzyme site into the cDNAs of MHVR and Cea10 also affected the receptor activities of certain chimeric glycoproteins. Our results show that simply converting these amino acids in Cea10 to the corresponding MHVR amino acid residues did not convert the N-terminal domain of Cea10 into that of a functional receptor. If these amino acids were changed in a chimeric glycoprotein that contained additional MHVR N-terminal regions, however, receptor activity was regained.
Molecular modeling studies suggest that residues R64 and D82 form a salt bridge in the N-terminal domain of the Bgp1 molecule. The two residues involved in the formation of this intramolecular salt bridge are conserved in mouse, rat, and human biliary glycoproteins. Mutational analysis of amino acid R64 confirms the importance of the tertiary structure of this molecule in receptor activity. An MHVR construct containing an R64-to-S mutation and deletion of residues 55 to 58 and 65 to 71 showed no virus receptor activity. When R64 was reintroduced into this deletion construct, weak but detectable receptor activity was restored.
Interestingly, these studies also showed that the constant domains of MHVR influence receptor activity. MHVR[1,2], which lacks the two C-terminal constant domains (domains 3 and 4), and N-terminal chimeras constructed with this molecule did not function as effective receptors for MHV-A59. In light of these findings, one could argue that the additional constant domains are required simply to project the virus-binding, N-terminal domain away from the cell surface, thereby permitting the virus unimpeded access to the receptor. As shown previously (18), though, and confirmed in this report, MHVR[1,4] functions quite well as a receptor and presumably extends from the cell surface a distance equivalent to MHVR[1,2]. Alternatively, one could argue that domain 4 but not domain 2 is necessary to ensure the correct conformation of the receptor-binding site. Previous analyses of the N-terminal domain (17, 19), however, make this scenario unlikely. Rather, we postulate that MHVR[1,2] is deficient in some, as yet undefined, postbinding event that is required for infection.
Two allelic variants of Bgp1, MHVR and Bgp1b, have been isolated from MHV-susceptible BALB/c and MHV-resistant SJL/J mice, respectively, and these isoforms differ noticeably in their abilities to serve as functional MHV receptors (18). A sequence comparison of the N-terminal domains of these Bgp1 allelic variants shows that the region identified as important for receptor activity also represents a highly variable region (18). While the amino acid sequences of the N-terminal domains of these two glycoproteins are identical from positions 1 to 37, 15 amino acid differences exist in the 22 residues from amino acid 38 to 60. This extensive difference in the presumptive virus-binding site provides a molecular explanation for the observed differences in receptor activity demonstrated by these two Bgp1 isoforms in vivo. The cellular function of these molecules is unknown, although related glycoproteins have been shown to function in vitro as intercellular adhesion molecules (12). Recent evidence suggests that this adhesion function in the rat C-CAM molecule is mediated by amino acids 63 to 67 of the mature protein (42). It is interesting that the domain associated with this biological function differs from the putative virus receptor site.
Our results show that binding of the antireceptor MAb CC1 to MHVR is very dependent on the tertiary structure of this glycoprotein. In contrast to the results obtained in the receptor activity assays, no CC1 antibody binding was detected to any of the MHVR/Cea10 recombinant glycoproteins in immunoblot, immunofluorescence, or FACS analysis, and MAb CC1 was unable to protect cells expressing MHVR/Cea10 chimeric recombinant glycoproteins from MHV-A59 infection. Analysis of MHVR/Bgp1b chimeric glycoproteins indicates that the MAb-CC1 binding site is within the first 70 amino acids of MHVR. Mutational analysis of MHVR confirms this finding. When the mutations L26A27L28A30A32 to QTRVY or D42K43 to HN were introduced into MHVR, antibody binding was eliminated, indicating that these residues or changes in the tertiary structure of the molecule resulting from changing these residues must be critical to MAb CC1 binding. These amino acids are in very close linear proximity to the amino acid residues identified as most important for receptor activity, suggesting that the binding sites of the virus and MAb may overlap. Such overlapping binding sites would explain the ability of MAb CC1 to protect cells expressing MHVR from infection. These findings also provide a molecular explanation for the previous observations that Bgp1b and Bgp2 can serve as functional receptors for MHV-A59 in in vitro assays but are not recognized by MAb CC1 (18, 38).
The murine glycoprotein Bgp1 has been identified as a member of the Ig superfamily (7, 20, 52, 53). Several other members of this family, including intercellular adhesion molecule 1, Pvr, and CD4, have been identified as receptors for viruses (receptors for rhinovirus, poliovirus, and human immunodeficiency virus, respectively) (14, 24, 27, 33, 35, 36, 44, 47). Initial mapping studies demonstrated for each of these pathogens that the virus recognizes the amino-terminal Ig-like domain of the receptor (17, 19, 21, 30, 34, 36, 44). For Pvr, CD4, and Bgp1, the amino-terminal domain represents a V-like Ig domain. More precise mapping studies of Pvr and CD4 suggest the importance of residues along the putative C′-C"-D edge of these two molecules in receptor activity (2, 8, 13, 37). Molecular modeling studies predict that this region represents a rather large, exposed face and also corresponds to the complementarity determining region 2. This finding has lead to speculation that the receptor-virus interaction may mimic at least some aspects of the antibody-antigen interaction (40). Our studies of Bgp1 show that residues in the putative C-C′ region of this glycoprotein are critical for receptor activity (Fig. 7). In fact, we constructed several point mutants in which single amino acids in the C"-D region of MHVR were altered. These changes resulted in no change in receptor activity. Based on our model of the structure of MHVR, it is interesting to speculate that the residues critical for receptor activity exist in a relatively sheltered location and that the MHV-Bgp1 interaction may not mirror the human immunodeficiency virus-CD4 or poliovirus-Pvr interactions.
Our results have identified a small region of the MHVR glycoprotein critical for receptor activity. A more complete understanding of the MHV-Bgp1 interaction, however, will require an examination of the regions of the MHV spike glycoprotein involved in binding to the receptor. In initial experiments, Suzuki and Taguchi concluded that multiple, dissociated regions of the spike are involved in binding, suggesting that spike binding to the receptor is conformationally dependent (46). A three-dimensional structure of the spike alone or in conjunction with MHVR may provide insight into the regions of contact between these two moieties. Such detailed information could be useful in the development of antiviral agents that specifically interfere with this binding event. Finally, such detailed molecular information may provide insight into how slight variations in the receptor structure and viral attachment protein affect receptor activity, thereby influencing the pathogenesis of mammalian viruses.
ACKNOWLEDGMENTS
We are grateful to Alexis Basile and Jin Gao for excellent technical assistance and to David Wentworth, Bruce Zelus, Dianna Blau, and Dina Tresnan for critical reviews of the manuscript. The molecular model for the N terminal domain of MHVR was kindly provided by Stephen Harrison (Harvard University).
This research was supported by Uniformed Services University of the Health Sciences grant C074ET, NIH grants AI26075 and AI25231, and Medical Research Council of Canada grant 12036. D.R.W. was supported by NIAID fellowship AI08879.
REFERENCES
- 1.Arnheiter H, Baechi T, Haller O. Adult mouse hepatocytes in primary monolayer culture express genetic resistance to mouse hepatitis virus type 3. J Immunol. 1982;129:1275–1281. [PubMed] [Google Scholar]
- 2.Arthos J, Deen K C, Chaikin M A, Fornwald J A, Sathe G, Sattentau Q J, Clapham P R, Weiss R A, McDougal J S, Pietropaolo C, Axel R, Truneh A, Maddon P J, Sweet R W. Identification of the residues in human CD4 critical for the binding of HIV. Cell. 1989;57:469–481. doi: 10.1016/0092-8674(89)90922-7. [DOI] [PubMed] [Google Scholar]
- 3.Bang F B, Warwick A. Mouse macrophages as host cells for the mouse hepatitis virus and the genetic basis of their susceptibility. Proc Natl Acad Sci USA. 1960;46:1065–1075. doi: 10.1073/pnas.46.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthold S W. Mouse hepatitis virus: biology and epizootiology. In: Bhatt P N, Jacoby R O, Morse III H C, New A E, editors. Viral and mycoplasmal infections of laboratory rodents. Effects on biomedical research. Orlando, Fla: Academic Press; 1986. pp. 571–601. [Google Scholar]
- 5.Barthold S W, Beck D S, Smith A L. Mouse hepatitis virus nasoencephalopathy is dependent upon virus strain and host genotype. Arch Virol. 1986;91:247–256. doi: 10.1007/BF01314284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barthold S W, Smith A L. Mouse hepatitis virus strain-related patterns of tissue tropism in suckling mice. Arch Virol. 1984;81:103–112. doi: 10.1007/BF01309300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beauchemin N, Turbide C, Huang J Q, Benchimol S, Jothy S, Shirota K, Fuks A, Stanners C P. Studies on the function of carcinoembryonic antigen. In: Yachi A, Shively J E, editors. The carcinoembryonic antigen gene family. Amsterdam, The Netherlands: Elsevier; 1989. pp. 49–64. [Google Scholar]
- 8.Bernhardt G, Harber J, Zibert A, DeCrombrugghe M, Wimmer E. The poliovirus receptor: identification of domains and amino acid residues critical for virus binding. Virology. 1994;203:344–356. doi: 10.1006/viro.1994.1493. [DOI] [PubMed] [Google Scholar]
- 9.Boyle J F, Weismiller G G, Holmes K V. Genetic resistance to mouse hepatitis virus correlates with absence of virus-binding activity on target tissues. J Virol. 1987;61:185–189. doi: 10.1128/jvi.61.1.185-189.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D S, Asanaka M, Chen F S, Shively J E, Lai M M C. Human carcinoembryonic antigen and biliary glycoprotein can serve as mouse hepatitis virus receptors. J Virol. 1997;71:1688–1691. doi: 10.1128/jvi.71.2.1688-1691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D S, Asanaka M, Yokomori K, Wang F-I, Hwang S B, Li H-P, Lai M M C. A pregnancy-specific glycoprotein is expressed in the brain and serves as a receptor for mouse hepatitis virus. Proc Natl Acad Sci USA. 1995;92:12095–12099. doi: 10.1073/pnas.92.26.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung P H, Thompson N L, Earley K, Culic O, Hixson D, Lin S-H. Cell-CAM105 isoforms with different adhesion functions are coexpressed in adult rat tissues and during liver development. J Biol Chem. 1993;268:6139–6146. [PubMed] [Google Scholar]
- 13.Clayton L K, Hussey R E, Steinbrich R, Ramachandran H, Husain Y, Reinherz E L. Substitution of murine for human CD4 residues identifies amino acids critical for HIV-gp120 binding. Nature. 1988;335:363–366. doi: 10.1038/335363a0. [DOI] [PubMed] [Google Scholar]
- 14.Dalgleish A G, Beverley P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature (London) 1984;312:763. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 15.Dindzans V J, Skamene E, Levy G A. Susceptibility/resistance to mouse hepatitis virus strain 3 and macrophage procoagulant activity are genetically linked and controlled by two non-H2-linked genes. J Immunol. 1986;137:2355–2360. [PubMed] [Google Scholar]
- 16.Dupuy J M, Dupuy C, Decarie D. Genetically determined resistance to mouse hepatitis virus strain 3 is expressed in hematopoietic donor cells in radiation chimeras. J Immunol. 1984;133:1609–1613. [PubMed] [Google Scholar]
- 17.Dveksler G S, Basile A A, Cardellichio C B, Holmes K V. Mouse hepatitis virus receptor activities of an MHVR/mph chimera and MHVR mutants lacking N-linked glycosylation of the N-terminal domain. J Virol. 1995;69:543–546. doi: 10.1128/jvi.69.1.543-546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dveksler G S, Dieffenbach C W, Cardellichio C B, McCuaig K, Pensiero M N, Jiang G-S, Beauchemin N, Holmes K V. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J Virol. 1993;67:1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dveksler G S, Gagneten S E, Scanga C A, Cardellichio C B, Holmes K V. Expression of recombinant anchorless N-terminal domain of MHVR makes hamster or human cells susceptible to MHV infection. J Virol. 1996;70:4142–4145. doi: 10.1128/jvi.70.6.4142-4145.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dveksler G S, Pensiero M N, Cardellichio C B, Williams R K, Jiang G-S, Holmes K V, Dieffenbach C W. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dveksler G S, Pensiero M N, Dieffenbach C W, Cardellichio C B, Basile A A, Elia P E, Holmes K V. Mouse coronavirus MHV-A59 and blocking anti-receptor monoclonal antibody bind to the N-terminal domain of cellular receptor MHVR. Proc Natl Acad Sci USA. 1993;90:1716–1720. doi: 10.1073/pnas.90.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frana M F, Benke J N, Sturman L S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronaviruses: Host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuerst T R, Niles E G, Studier W F, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8128. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greve J M, Davis G, Meyer A M, Forte C P, Yost S C, Marlor C W, Kamarick M E, McClelland A. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 25.Hirano N, Murakami T, Taguchi F, Fujiwara K, Matumoto M. Comparison of mouse hepatitis virus strains for pathogenicity in weanling mice infected by various routes. Arch Virol. 1981;70:69–73. doi: 10.1007/BF01320795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keck U, Nédellec P, Beauchemin N, Thompson J, Zimmermann W. The CEA10 gene encodes a secreted member of the murine carcinoembryonic antigen family and is expressed in the placenta, gastrointestinal tract and bone marrow. Eur J Biochem. 1995;229:455–464. doi: 10.1111/j.1432-1033.1995.0455k.x. [DOI] [PubMed] [Google Scholar]
- 27.Klatzman D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 28.Knobler R L, Linthicum D S, Cohn M. Host genetic regulation of acute MHV-4 viral encephalomyelitis and acute experimental autoimmune encephalomyelitis in (BALB/cKe × SJL/J) recombinant-inbred mice. J Neuroimmunol. 1985;8:15–28. doi: 10.1016/S0165-5728(85)80044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamontagne L M, Dupuy J M. Natural resistance of mice to mouse hepatitis virus type 3 infection is expressed in embryonic fibroblast cells. J Gen Virol. 1984;65:1165–1171. doi: 10.1099/0022-1317-65-7-1165. [DOI] [PubMed] [Google Scholar]
- 30.Landau N R, Warton M, Littman D R. The envelope glycoprotein of the human immunodeficiency virus binds to the immunoglobulin-like domain of CD4. Nature (London) 1988;334:159–162. doi: 10.1038/334159a0. [DOI] [PubMed] [Google Scholar]
- 31.Le Prevost C, Levy-Leblond E, Virelizier J L, Dupuy J M. Immunopathology of mouse hepatitis virus type 3 infection. Role of humoral and cell-mediated immunity in resistance mechanisms. J Immunol. 1975;114:221–225. [PubMed] [Google Scholar]
- 32.Lu J-H. Ph.D. thesis. Bethesda, Md: Uniformed Services University of the Health Sciences; 1995. [Google Scholar]
- 33.Maddon P J, Dalgleish A G, McDougal J S, Clapham P R, Weiss R A, Axel R. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell. 1986;47:333–348. doi: 10.1016/0092-8674(86)90590-8. [DOI] [PubMed] [Google Scholar]
- 34.McClelland A, DeBear J, Connolly Yost S, Meyer A M, Marlor C W, Greve J M. Identification of monoclonal antibody epitopes and critical residues for rhinovirus binding in domain 1 of intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1991;88:7993–7997. doi: 10.1073/pnas.88.18.7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDougal J S, Mawle A, Cort S P, Nicholson J K A, Cross D, Schleppler-Campbell J A, Hicks D, Sligh J. Cellular tropism of the human retrovirus HTLV/LAV. I. Role of T cell activation and expression of the T4 antigen. J Immunol. 1985;135:3151–3162. [PubMed] [Google Scholar]
- 36.Mendelson C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 37.Moebius U, Clayton L K, Abraham S, Harrison S C, Reinherz E L. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic studies of mutants in conjunction with a high-resolution CD4 atomic structure. J Exp Med. 1992;176:507–517. doi: 10.1084/jem.176.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nédellec P, Dveksler G S, Daniels E, Turbide C, Chow B, Basile A A, Holmes K V, Beauchemin N. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J Virol. 1994;68:4525–4537. doi: 10.1128/jvi.68.7.4525-4537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohtsuka N, Yamada Y K, Taguchi F. Differences in virus-binding activity of two distinct receptor proteins for mouse-hepatitis virus. J Gen Virol. 1996;77:1683–1692. doi: 10.1099/0022-1317-77-8-1683. [DOI] [PubMed] [Google Scholar]
- 40.Peterson A, Seed B. Genetic analysis of monoclonal antibody and HIV binding sites on the human lymphocyte antigen CD4. Cell. 1988;54:65–72. doi: 10.1016/0092-8674(88)90180-8. [DOI] [PubMed] [Google Scholar]
- 41.Rao P V, Kumari S, Gallagher T M. Identification of a contiguous 6-residue determinant in the MHV receptor that controls the level of virion binding to cells. Virology. 1997;229:336–348. doi: 10.1006/viro.1997.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sippel C J, Shen T, Perlmutter D H. Site-directed mutagenesis within an ectoplasmic ATPase consensus sequence abrogates the cell aggregating properties of the rat liver canalicular bile acid transporter/ecto-ATPase/cell CAM 105 and carcinoembryonic antigen. J Biol Chem. 1996;271:33095–33104. doi: 10.1074/jbc.271.51.33095. [DOI] [PubMed] [Google Scholar]
- 43.Smith M S, Click R E, Plagemann P G. Control of mouse hepatitis virus replication in macrophages by a recessive gene on chromosome 7. J Immunol. 1984;133:428–432. [PubMed] [Google Scholar]
- 44.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;56:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 45.Stohlman S A, Frelinger J A. Resistance to fatal central nervous system disease by mouse hepatitis virus, strain JHM. I. Genetic analysis. Immunogenetics. 1978;6:277–281. [Google Scholar]
- 46.Suzuki H, Taguchi F. Analysis of the receptor-binding site of murine coronavirus spike protein. J Virol. 1996;70:2632–2636. doi: 10.1128/jvi.70.4.2632-2636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomassini J E, Graham D, DeWitt C M, Lineberger D W, Rodkey J A, Colonno R J. CDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc Natl Acad Sci USA. 1989;86:4907–4911. doi: 10.1073/pnas.86.13.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vallejo A N, Pogulis R J, Pease L R. Mutagenesis and synthesis of novel recombinant genes using PCR. In: Dieffenbach C W, Dveksler G S, editors. PCR primer. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 603–612. [Google Scholar]
- 49.Wege H, Siddell S, Ter Meulen V. The biology and pathogenesis of coronaviruses. Curr Top Microbiol Immunol. 1982;99:165–200. doi: 10.1007/978-3-642-68528-6_5. [DOI] [PubMed] [Google Scholar]
- 50.Weiser W, Vellisto I, Bang F B. Congenic strains of mice susceptible and resistant to mouse hepatitis virus. Proc Soc Exp Biol Med. 1976;152:499–502. doi: 10.3181/00379727-152-39426. [DOI] [PubMed] [Google Scholar]
- 51.Wessner, D. R., B. Zelus, and K. V. Holmes. Unpublished data.
- 52.Williams R K, Jiang G-S, Holmes K V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sci USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams R K, Jiang G-S, Snyder S W, Frana M F, Holmes K V. Purification of the 110-kilodalton glycoprotein receptor for mouse hepatitis virus (MHV)-A59 from mouse liver and identification of a nonfunctional homologous protein in MHV-resistant SJL/J mice. J Virol. 1990;64:3817–3823. doi: 10.1128/jvi.64.8.3817-3823.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokomori K, Lai M M C. Mouse hepatitis virus utilizes two carcinoembryonic antigens as alternative receptors. J Virol. 1992;66:6194–6199. doi: 10.1128/jvi.66.10.6194-6199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokomori K, Lai M M C. The receptor for mouse hepatitis virus in the resistant mouse strain SJL is functional: implications for the requirement of a second factor for viral infection. J Virol. 1992;66:6931–6938. doi: 10.1128/jvi.66.12.6931-6938.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]