Abstract
The pseudorabies virus (PrV) gene homologous to herpes simplex virus type 1 (HSV-1) UL53, which encodes HSV-1 glycoprotein K (gK), has recently been sequenced (J. Baumeister, B. G. Klupp, and T. C. Mettenleiter, J. Virol. 69:5560–5567, 1995). To identify the corresponding protein, a rabbit antiserum was raised against a 40-kDa glutathione S-transferase–gK fusion protein expressed in Escherichia coli. In Western blot analysis, this serum detected a 32-kDa polypeptide in PrV-infected cell lysates as well as a 36-kDa protein in purified virion preparations, demonstrating that PrV gK is a structural component of virions. After treatment of purified virions with endoglycosidase H, a 34-kDa protein was detected, while after incubation with N-glycosidase F, a 32-kDa protein was specifically recognized. This finding indicates that virion gK is modified by N-linked glycans of complex as well as high-mannose type. For functional analysis, the UL53 open reading frame was interrupted after codon 164 by insertion of a gG-lacZ expression cassette into the wild-type PrV genome (PrV-gKβ) or by insertion of the bovine herpesvirus 1 gB gene into a PrV gB− genome (PrV-gKgB). Infectious mutant virus progeny was obtained only on complementing gK-expressing cells, suggesting that gK has an important function in the replication cycle. After infection of Vero cells with either gK mutant, only single infected cells or small foci of infected cells were visible. In addition, virus yield was reduced approximately 30-fold, and penetration kinetics showed a delay in entry which could be compensated for by phenotypic gK complementation. Interestingly, the plating efficiency of PrV-gKβ was similar to that of wild-type PrV on complementing and noncomplementing cells, pointing to an essential function of gK in virus egress but not entry. Ultrastructurally, virus assembly and morphogenesis of PrV gK mutants in noncomplementing cells were similar to wild-type virus. However, late in infection, numerous nucleocapsids were found directly underneath the plasma membrane in stages typical for the entry process, a phenomenon not observed after wild-type virus infection and also not visible after infection of gK-complementing cells. Thus, we postulate that presence of gK is important to inhibit immediate reinfection.
Herpesvirions are complex structures consisting of a nucleoprotein core, capsid, tegument, and envelope. They comprise at least 30 structural proteins (35). Pseudorabies virus (PrV), a member of the Alphaherpesvirinae, is an economically important animal pathogen, causing Aujeszky’s disease in swine. It is also highly pathogenic for most other mammals except higher primates, including humans (28, 45), and a wide range of cultured cells from different species support productive virus replication, reflecting the wide in vivo host range. Envelope glycoproteins play major roles in the early and late interactions between virion and host cell. They are required for virus entry and participate in release of free virions and viral spread by direct cell-to-cell transmission (27, 37). For PrV, 10 glycoproteins, designated gB, gC, gD, gE, gG, gH, gI, gL, gM, and gN, have been characterized (20, 27); these glycoproteins are involved in the attachment of virion to host cell (gC and gD), fusion of viral envelope and cellular cytoplasmic membrane (gB, gD, gH, and gL), spread from infected to noninfected cells (gB, gE, gH, gI, gL, and gM), and egress (gC, gE, and gI) (27, 37). Homologs of these glycoproteins are also present in other alphaherpesviruses (37). The gene coding for a potential 11th PrV glycoprotein, gK, has been described recently (3), but the protein and its function have not been identified.
The product of the homologous UL53 open reading frame (ORF) of herpes simplex virus type 1 (HSV-1) is gK (13, 32). gK was detected in nuclear membranes and in membranes of the endoplasmic reticulum but was not observed in the plasma membrane (14). Also, it did not appear to be present in purified virion preparations (15). The latter result was surprising since earlier studies identified several mutations in HSV-1 gK resulting in syncytium-inducing phenotypes (7, 14), which indicates participation of gK in membrane fusion events during HSV-1 infection. Moreover, HSV-1 mutants in gK exhibited a delayed entry into noncomplementing cells, which is difficult to reconcile with absence of gK from virions (31). Mutants deficient for gK expression have been isolated and investigated by different groups (16, 17). Mutant F-gKβ carries a lacZ gene insertion in the HSV-1 strain F gK gene, which interrupts the ORF after codon 112 (16). In mutant ΔgK, derived from HSV-1 KOS, almost all of the UL53 gene was deleted (17). Both mutants formed small plaques on Vero cells, and virus yield was reduced to an extent which varied with the different confluencies of the infected cells, cell types, and mutants used for infection. However, both HSV-1 gK mutants showed a defect in efficient translocation of virions from the cytoplasm to the extracellular space, and only a few enveloped virions were present in the extracellular space after infection of Vero cells (16, 17). The authors therefore suggested that HSV-1 gK plays a role in virion transport during egress.
Different routes of final envelopment and egress of alphaherpesvirions are discussed. It has been suggested that HSV-1 nucleocapsids acquire their envelope at the inner nuclear membrane and are transported as enveloped particles through the endoplasmic reticulum to the Golgi stacks, where glycoproteins are modified in situ during transport (5, 6, 19, 39), although other potential egress pathways cannot be excluded (4). In contrast, maturation of varicella-zoster virus and PrV involves primary envelopment at the nuclear membrane, followed by release of nucleocapsids into the cytoplasm and secondary envelopment in the trans-Golgi area (10, 12, 43). Final egress of virions appears to occur via transport vesicles containing one or more virus particles by fusion of vesicle and cell membrane. The possibility of different routes of virion egress is supported by studies of other proteins involved in egress, e.g., the UL20 proteins of HSV-1 and PrV and the PrV UL3.5 protein, which lacks a homolog in the HSV-1 genome (1, 8, 9). In UL20-negative HSV-1, virions accumulated in the perinuclear cisterna of Vero cells (1), while PrV UL20− virions accumulated and were retained in cytoplasmic vesicles (9). PrV UL3.5 is important for budding of nucleocapsids into Golgi-derived vesicles during secondary envelopment (8). Thus, there appear to be profound differences in the egress pathways. Since HSV-1 gK was also implicated in egress, we were interested in identifying the PrV homolog and analyzing its function.
MATERIALS AND METHODS
Cells and viruses.
Mutants are based on wild-type PrV strain Kaplan (PrV-Ka [22]). Virus was propagated on porcine kidney (PSEK) or African green monkey kidney (Vero) cells grown in Eagle’s minimum essential medium supplemented with 5% fetal calf serum. Construction of PrV-9112C2, which carries the bovine herpesvirus 1 (BHV-1) gB gene in the partially deleted PrV gB gene locus, is described elsewhere (24). PrV-1112, which carries the lacZ gene in the gG gene locus and exhibits growth properties similar to those of wild-type PrV (29), was used in experiments in which 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining was applied. Transfections were performed as described by Graham and van der Eb (11).
Prokaryotic expression and preparation of antiserum.
For prokaryotic expression, plasmid pSal2 (Fig. 1B) was partially digested with BstXI, an 827-bp BstXI fragment was excised, and SalI linkers were added. After cleavage with SalI and XhoI, the 396-bp SalI/XhoI fragment was inserted into expression vector pGEX-4T-2 (Pharmacia, Freiburg, Germany) (Fig. 1E). The resulting plasmid contains codons 64 to 196 of the UL53 ORF fused to the glutathione S-transferase (GST) gene (Fig. 1E). Correct insertion was confirmed by restriction analysis and sequencing using pGEX-specific primers (Pharmacia). Bacterial expression was performed as described previously (23). The 40-kDa fusion protein was electroeluted (Amicon electroelutor; Amicon, Witten, Germany) after separation on a sodium dodecyl sulfate (SDS)–8% polyacrylamide gel and concentrated in Centricon microconcentrators (Amicon). One rabbit was immunized four times at 2-week intervals by intramuscular injection of 100 μg of the purified fusion protein emulsified in mineral oil. Sera collected before and after immunization were analyzed.
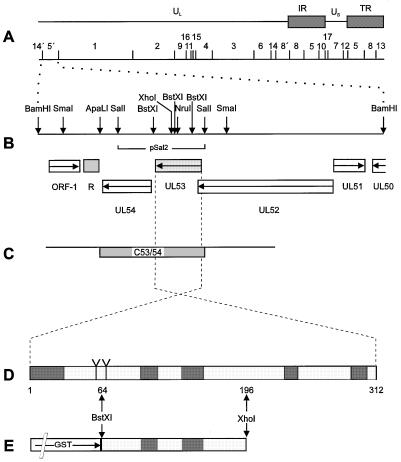
FIG. 1.
Construction of PrV gK mutants, complementing cell lines, and GST-gK fusion protein. (A) Schematic diagram of the PrV genome with the BamHI restriction fragment map. The PrV genome consists of a unique long region (UL) and a unique short region (US). The latter is flanked by inverted repeats (IR, internal repeat; TR, terminal repeat). (B) Enlargement of BamHI fragment 5′. Locations of the identified ORFs with transcriptional orientation, indicated by arrows, are given (R, region of reiterated sequences), and relevant restriction sites are marked, as is the location of fragment Sal2 used for cloning. (C) Part of viral BamHI fragment 5′ introduced into cell line C53/54. (D) Schematic diagram of gK. Indicated in dark grey are predicted signal sequence and transmembrane domains. (E) Prokaryotic expression of gK. In plasmid pGEX-Bst XI/Xho I, a fragment comprising codons 64 to 196 of the gK ORF was cloned downstream from and in frame with the GST gene (not drawn to scale) and expressed in E. coli for immunization of a rabbit.
Western blot analysis of virions and cell lysates.
Virions were purified as described previously (23). Infected cell lysates were harvested 24 h after infection by scraping into 1 ml of phosphate-buffered saline (PBS). Cells were collected by centrifugation at 4,000 rpm for 2 min in an Eppendorf centrifuge and resuspended in 100 μl of PBS, and the same volume of sample buffer was added. Five micrograms of purified virion protein or 10 μl of cell lysate was separated by SDS-polyacrylamide gel electrophoresis (PAGE) (25), electrotransferred onto polyvinylidene difluoride membranes (Immobilon-P; Millipore, Eschborn, Germany) (40), and reacted for 1 h at room temperature or overnight at 4°C with the gK-specific rabbit serum at a dilution of 1:1,000, with serum specific for PrV dUTPase (19) at a dilution of 1:1,000, with a gH-specific serum raised against bacterially expressed PrV gH (23) at a dilution of 1:70,000, or with a gC-specific monoclonal antibody (42) at a dilution of 1:100. After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was detected by enhanced chemiluminescence (ECL Western blot system; Amersham, Braunschweig, Germany) recorded on X-ray film.
Glycosidase digestions.
To remove N-linked carbohydrates, 5 μg of purified virion protein was incubated in 50 mM potassium phosphate buffer (pH 7.2)–50 mM EDTA–1.2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and either with 0.2 U of N-glycosidase F (PNGase F; Boehringer Mannheim, Mannheim, Germany), with 10 mU of endo-β-acetylglucosaminidase H (endo H; Boehringer Mannheim), or without enzyme for 16 h at 37°C before PAGE and Western blotting.
Construction of PrV gK mutants.
A 3.4-kb SmaI subfragment of BamHI fragment 5′ (BamHI 5′) (Fig. 1B), comprising the UL53 and UL54 genes and a region of reiterated sequences, was cloned into pUC18. After inactivation of the BamHI site in the vector, plasmid was cleaved with NruI, and BamHI linkers were added. Into this newly created BamHI site, a 3.5-kb gG-lacZ cassette (29) was inserted in parallel transcriptional orientation, thereby interrupting the UL53 ORF after codon 164 and resulting in plasmid pUL53Nru+Gal. Mutant virus was isolated after cotransfection of pUL53Nru+Gal with genomic DNA of PrV-Ka into gK-complementing cells (see below). Recombinant viruses were identified by their blue-plaque phenotype and were plaque purified by aspiration until all plaques appeared blue under an agarose overlay containing Bluo-Gal (Life Technologies, Eggenstein, Germany). One plaque isolate, designated as PrV-gKβ, was chosen for further analysis. Correct recombination was verified by Southern blot analysis of mutant virus DNA (data not shown).
To obtain a second, independent mutant, a 3.3-kb BamHI fragment comprising the gB gene of BHV-1 was inserted into the UL53 ORF (8) as described above, giving rise to plasmid pUL53gB(BHV). Following cotransfection with PrV gB− DNA (33), only recombinant viruses which had acquired the BHV-1 gB gene, inserted into the gene to be analyzed, were able to produce infectious progeny (heterologous cis complementation). After cotransfection of PrV gB− DNA and plasmid pUL53gB(BHV) into normal Vero cells, no infectious virus progeny was detectable, indicating that gK has an important function in the replicative cycle. Therefore, gK-expressing complementing cells which carry either the entire BamHI fragment 5′ (B5′-64) or the UL53 and UL54 transcription unit only (C53/54) were established (Fig. 1B and C). After cotransfection of pUL53gB(BHV) and PrV gB− DNA into either cell line, infectious progeny appeared. One randomly selected plaque isolate, designated PrV-gKgB, was further analyzed.
Construction of gK-expressing cell lines.
For construction of complementing cell lines, the 7.4-kb BamHI fragment 5′ (Fig. 1B) was cloned into plasmid pSV2neo (36). In a second approach, the UL53/UL54 transcription unit was inserted into plasmid pRc/CMV (InVitroGen, Leek, The Netherlands). To this end, the leftmost 1.7-kb BamHI/SalI subfragment (pSal1) and the adjacent 1.9-kb SalI fragment (pSal2) were cloned in correct orientation into plasmid TN-77 (26), giving rise to plasmid pSal1/2. After BamHI/ApaLI digestion, the remaining 6.5-kb vector fragment comprising genes UL53 and UL54 was religated. The UL53 and UL54 transcription unit was then excised by HindIII/EcoRI digestion and cloned into the HindIII- and NotI-cleaved vector pRc/CMV. This insert comprises 107 bp upstream of the UL53 start codon, including putative transcriptional regulatory sequences, and ends 29 bp downstream of the polyadenylation signal following the UL54 ORF (3).
Both plasmids were transfected into Vero cells by calcium phosphate coprecipitation (11), and cell clones developing under selection medium (700 μg of Geneticin per ml; Life Technologies) were tested for the ability to allow replication of mutant PrV-gKgB. One cell clone each was selected for further studies and designated B5′-64 (BamHI fragment 5′) or C53/54 (UL53 and UL54), respectively.
Plaque assays, penetration analysis, and one-step growth curves.
For analysis of growth, complementing and noncomplementing cells were infected with serial dilutions of PrV-Ka, PrV-gKβ, and PrV-gKgB, overlaid with medium containing 0.8% methylcellulose, and incubated for 2 days at 37°C. Thereafter, cells were fixed for 10 min in 2% formaldehyde–0.2% glutaraldehyde–0.02% Nonidet P-40–0.01% sodium deoxycholate in PBS and stained with a substrate solution for β-galactosidase containing 300 μg of X-Gal (Roth, Karlsruhe, Germany) per ml, 16 mM potassium ferricyanide, 16 mM potassium ferrocyanide, 2 mM magnesium chloride, 0.02% Nonidet P-40, and 0.01% sodium deoxycholate in PBS.
For analysis of penetration kinetics each well of complementing cells in six-well culture dishes was infected with approximately 500 PFU of PrV-Ka or noncomplemented and complemented PrV-gKβ for 1 h at 4°C. The inoculum was then removed, and prewarmed medium was added to allow penetration. Immediately thereafter and after 5, 10, 20, and 30 min, remaining extracellular virus was inactivated by low-pH treatment. Cells were washed with PBS and overlaid with methylcellulose. After 2 days of incubation at 37°C, cells were fixed and stained. Plaques were counted, and the percentage of PFU surviving low-pH treatment compared to a PBS-treated control was calculated.
For analysis of one-step growth, normal and complementing Vero cells were infected with PrV-Ka and PrV-gKβ at a multiplicity of infection (MOI) of 10 or with PrV-9112C2 and PrV-gKgB at an MOI of 5 for 1 h at 4°C. Then prewarmed medium was added, and virus was allowed to penetrate during a 2-h incubation period at 37°C. Remaining extracellular virus was inactivated by low-pH treatment. Cells and supernatant were harvested immediately thereafter (0 h) or after 4, 8, 12, 24, or 36 h. Virus progeny was titrated on complementing cells.
Electron microscopy.
Normal and complementing Vero cells were infected at an MOI of 1. Infected cells were fixed 16 h postinfection (p.i.) with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Cells were gently scraped off the plate, collected by low-speed centrifugation, and enclosed in 2% low-melting-point agarose (Biozym, Oldendorf, Germany). Agarose pieces with cell pellets were postfixed with 1% osmium tetroxide, briefly washed, stained en bloc with uranyl acetate, dehydrated stepwise in ethanol, and embedded in glycid ether 100 (Serva, Heidelberg, Germany). Ultrathin sections were stained with uranyl acetate and lead citrate. Sections were examined with an electron microscope (EM 400T; Philips, Eindhoven, The Netherlands).
RESULTS
Identification of PrV UL53 protein.
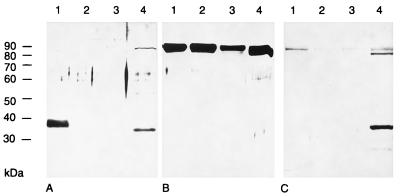
To identify PrV gK, a 396-bp BstXI/XhoI fragment comprising codons 64 to 196 of the UL53 ORF was expressed as GST fusion protein in Escherichia coli (Fig. 1E). The 40-kDa fusion protein was electroeluted after separation by SDS-PAGE (8% gel) and used for generation of a gK-specific antiserum in a rabbit. Using this serum, we investigated purified PrV-Ka, PrV-gKβ, and PrV-gKgB virions as well as PrV-Ka-infected Vero cell lysate by Western blot analysis (Fig. 2). This serum detected in PrV-Ka virion preparations (Fig. 2A, lane 1) a 36-kDa protein which was absent from preparations of PrV-gKβ (Fig. 2A, lane 2) and PrV-gKgB (Fig. 2A, lane 3). In PrV-Ka-infected cell lysate, a protein with an apparent molecular mass of 32 kDa was recognized (Fig. 2A, lane 4). This corresponds to the calculated molecular mass for the deduced protein after cleavage of a putative signal sequence (3). As a positive control, a parallel blot was incubated with a gH-specific serum (Fig. 2B). The 95-kDa gH was detectable in similar quantities in all virion preparations and in the infected cell lysate. Purity of the virion preparations was verified by electron microscopy (data not shown) as well as by Western blot analysis using serum specific for PrV dUTPase (Fig. 2C). PrV dUTPase is a nonstructural protein detectable in infected cells (Fig. 2C, lane 4) but not in the virion preparations (Fig. 2, lanes 1 to 3) (21). These results therefore indicate that PrV gK is a structural component of the virion.
FIG. 2.
Identification of PrV gK. Proteins of purified PrV-Ka (lanes 1), PrV-gKβ (lanes 2), or PrV-gKgB (lanes 3) virions or lysate of PrV-Ka-infected cells harvested 24 h p.i. (lanes 4) were separated by gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with the PrV gK-specific rabbit polyclonal serum (A), a gH-specific rabbit serum (B), or a dUTPase-specific serum (C). Bound antibody was visualized by chemiluminescence after incubation with a peroxidase-conjugated secondary antibody. Locations of molecular mass markers are indicated on the left.
PrV gK is N-glycosylated.
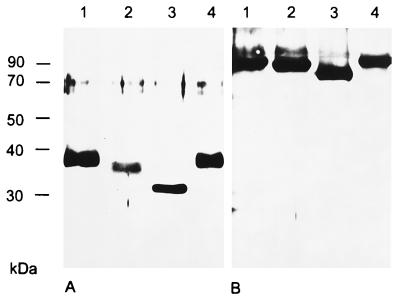
The deduced amino acid sequence of PrV gK revealed two consensus sequences for addition of N-linked glycans (3). To investigate whether the protein detected by the rabbit anti-gK serum is indeed N-glycosylated, purified virion preparations were incubated with endo H or PNGase F (Fig. 3). PNGase F, which cleaves nearly all N-linked glycans from the protein backbone (38), reduced the molecular mass of the detected protein to 32 kDa (Fig. 3A, lane 3). This deglycosylated form of gK exhibits the same electrophoretic mobility as the protein detected in infected cell lysates (Fig. 2A, lane 4). After incubation with endo H, which is specific for high-mannose forms of N-glycans (41), a 34-kDa protein was identified. These results indicate that mature virion gK contains high-mannose and complex N-glycans. As control, a parallel blot was incubated with the gH-specific serum (Fig. 3B).
FIG. 3.
Analysis of N-linked carbohydrates on PrV gK. Purified PrV-Ka virions were incubated either with endo H (lanes 2), with PNGase F (lanes 3), or without enzyme (lanes 4) or were separated without prior treatment (lanes 1). After gel electrophoresis and Western blotting, replica filters were incubated with serum specific for PrV gK (A) or gH (B). Locations of molecular mass markers are indicated on the left.
Plaque formation by PrV gK mutants.
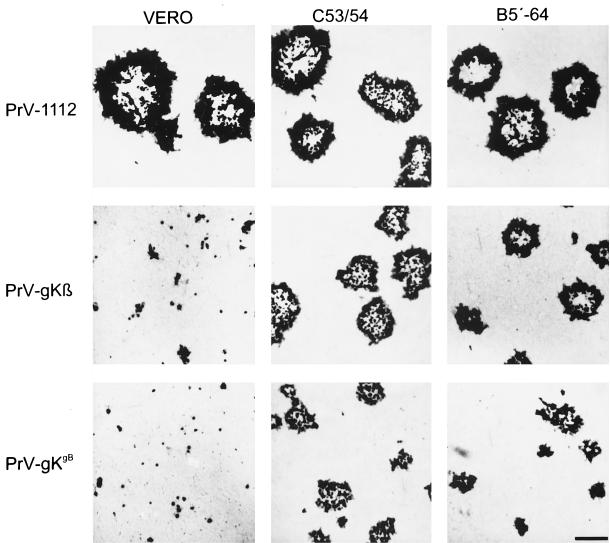
The fact that the mutants could be isolated only on complementing gK-expressing cells indicated that the marker gene insertion interfered with productive virus replication. To investigate the growth defect of the mutants, plaque assays were performed. Complementing and noncomplementing cells were infected with PrV-1112 (29), PrV-gKβ, and PrV-gKgB. Two days p.i. cells were fixed and stained with X-Gal (Fig. 4). PrV-1112, which carries the lacZ gene in the nonessential gG gene locus, forms plaques on all cells tested. In contrast, mutants PrV-gKβ and PrV-gKgB formed plaques only on complementing cells, while single infected cells or small foci of infected cells were observed after infection of noncomplementing Vero cells, indicating that PrV gK is necessary for efficient plaque formation. Plaques of mutant PrV-gKgB were smaller than those of PrV-gKβ, which is due to the exchange of PrV gB for BHV-1 gB as described for PrV-9112C2 (24).
FIG. 4.
Plaque morphology of PrV gK mutants. Noncomplementing Vero and complementing B5′-64 and C53/54 cells were infected with PrV-1112, PrV-gKβ, and PrV-gKgB under plaque assay conditions. Cells were fixed 2 days p.i., stained with X-Gal, and photographed. Bar = 500 μm.
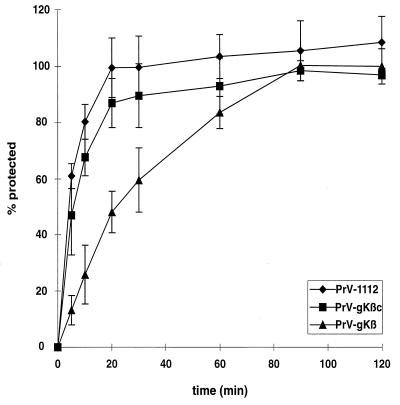
Penetration kinetics of PrV-gKβ.
PrV gK represents a structural component of virions, and HSV-1 gK has been implicated in penetration (31). Thus, we investigated whether the absence of gK interferes with efficient entry of PrV into host cells. For that purpose, we performed penetration assays on gK-complementing cells. Figure 5 shows the mean values from three independent experiments. While PrV-1112 entered cells with half-times of about 4 min, penetration of mutant PrV-gKβ was significantly delayed, with 50% PFU protected after only about 20 min. Phenotypic complementation of PrV-gKβ by propagation on complementing cells (PrV-gKβc) restored efficient penetration. These data indicate involvement of PrV gK in the entry process.
FIG. 5.
Penetration kinetics of PrV-1112 and PrV-gKβ. Complementing cells were infected with either PrV-1112 or PrV-gKβ grown on noncomplementing or complementing (PrV-gKβc) cells. After 1 h of incubation on ice, cells were overlaid with prewarmed medium to initiate penetration. At different times thereafter, remaining extracellular virus was inactivated by low-pH treatment. The percentage of PFU surviving low-pH treatment was calculated with reference to a PBS-treated control as percent penetration. Mean values and standard deviations of results of at least three independent experiments are shown.
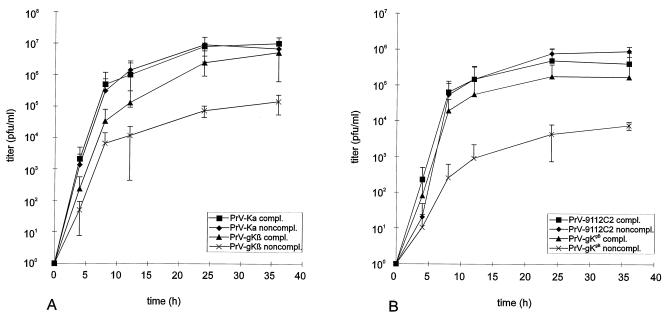
One-step growth and plating efficiency of PrV gK mutants.
To further investigate the growth defect of the mutant viruses, one-step growth analysis was performed. Complementing and noncomplementing cells were infected at an MOI of 10 with PrV-Ka and PrV-gKβ or at an MOI of 5 with PrV-9112C2 and PrV-gKgB. Mutant PrV-9112C2 carries the BHV-1 gB gene inserted into the partially deleted PrV gB gene (24) and was used as a control for phenotypic alterations solely due to the heterologous BHV-1 gB. Cells and supernatant were harvested at different times p.i. as indicated, and virus progeny was titrated on complementing cells. Mean values as well as standard deviations from two independent experiments were plotted. As shown in Fig. 6A, growth of PrV-gKβ on complementing cells was similar to growth of PrV-Ka either on complementing or noncomplementing cells, with a steep increase in virus titer between 4 and 12 h p.i. and comparable final titers. In contrast, PrV-gKβ exhibited a growth defect after infection of Vero cells at all times p.i., with about 30-fold-lower final titers. A similar reduction in virus yield was found for mutant PrV-gKgB on noncomplementing cells, which is compensated for on gK-complementing cells (Fig. 6B). Due to the gB substitution, maximum titers of PrV-9112C2 and PrV-gKgB were generally lower than those of PrV-Ka (24) and PrV-gKβ. Plating efficiencies of noncomplemented or complemented PrV-gKβ were similar on complementing and noncomplementing cells, as shown in Table 1, indicating that PrV gK is not required for virion infectivity. However, due to the impairment of plaque formation, only single infected cells or small foci of infection developed on the noncomplementing cell monolayer.
FIG. 6.
One-step growth analysis. Complementing and noncomplementing cells were infected at an MOI of 10 with PrV-Ka or PrV-gKβ (A) or at an MOI of 5 with PrV-9112C2 or PrV-gKgB (B) for 1 h at 4°C. After an additional 2 h at 37°C, remaining extracellular virus was inactivated by low-pH treatment. Immediately thereafter (0 h) and at the time points indicated, cells and supernatant were harvested, and progeny virus was titrated on complementing C53/54 cells. Mean values and standard deviations of results of two independent experiments are shown.
TABLE 1.
Plating efficiency of PrV gK mutantsa
| Virus | Cell type used for propagation of virus | Titer determined on:
|
|
|---|---|---|---|
| Complementing cells | Noncomplementing cells | ||
| PrV-Ka | Noncomplementing | 8.8 × 106 | 2.7 × 106 |
| Complementing | 6.2 × 106 | 3.6 × 106 | |
| PrV-gKβ | Noncomplementing | 8.2 × 104 | 1.2 × 105 |
| Complementing | 2.0 × 106 | 2.0 × 106 | |
| PrV-gKgB | Noncomplementing | 8.9 × 103 | 6.1 × 103 |
| Complementing | 3.0 × 105 | 2.6 × 105 | |
Viruses were grown on either gK-complementing or noncomplementing cells, and resulting virus stocks were titrated on complementing and noncomplementing cells. Both gK mutants produced only single infected cells or small foci of infectivity on noncomplementing cells, which were counted as infectious events.
Electron microscopy.
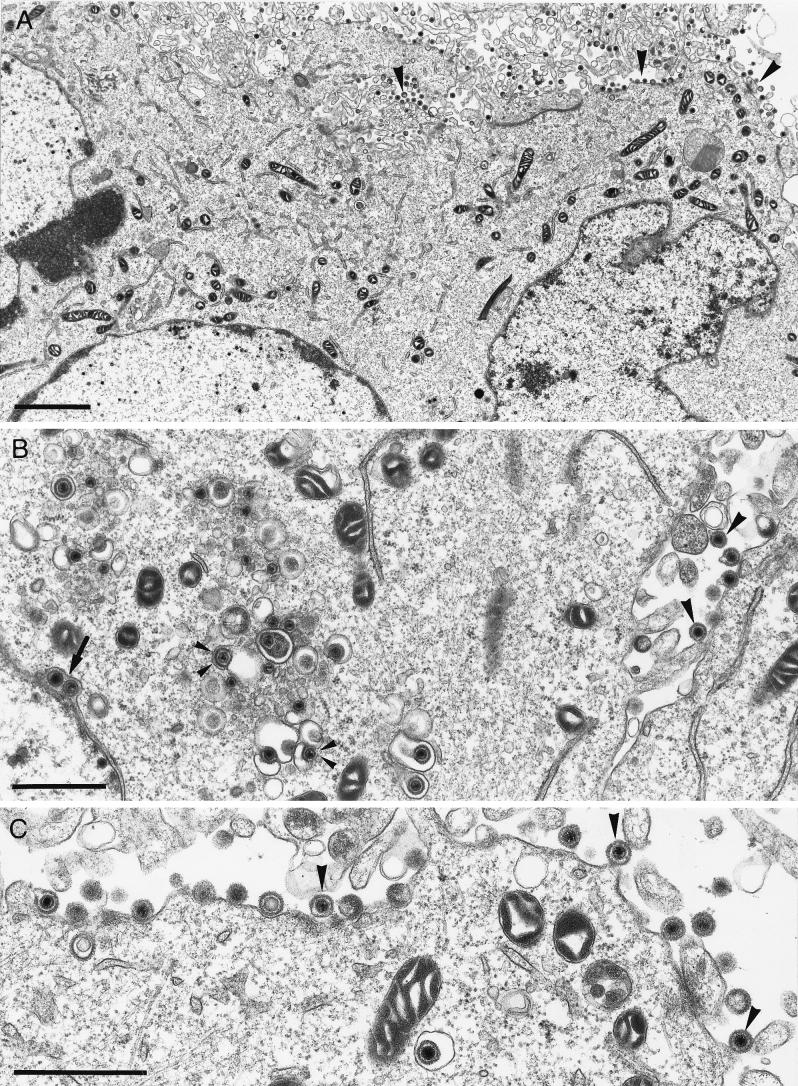
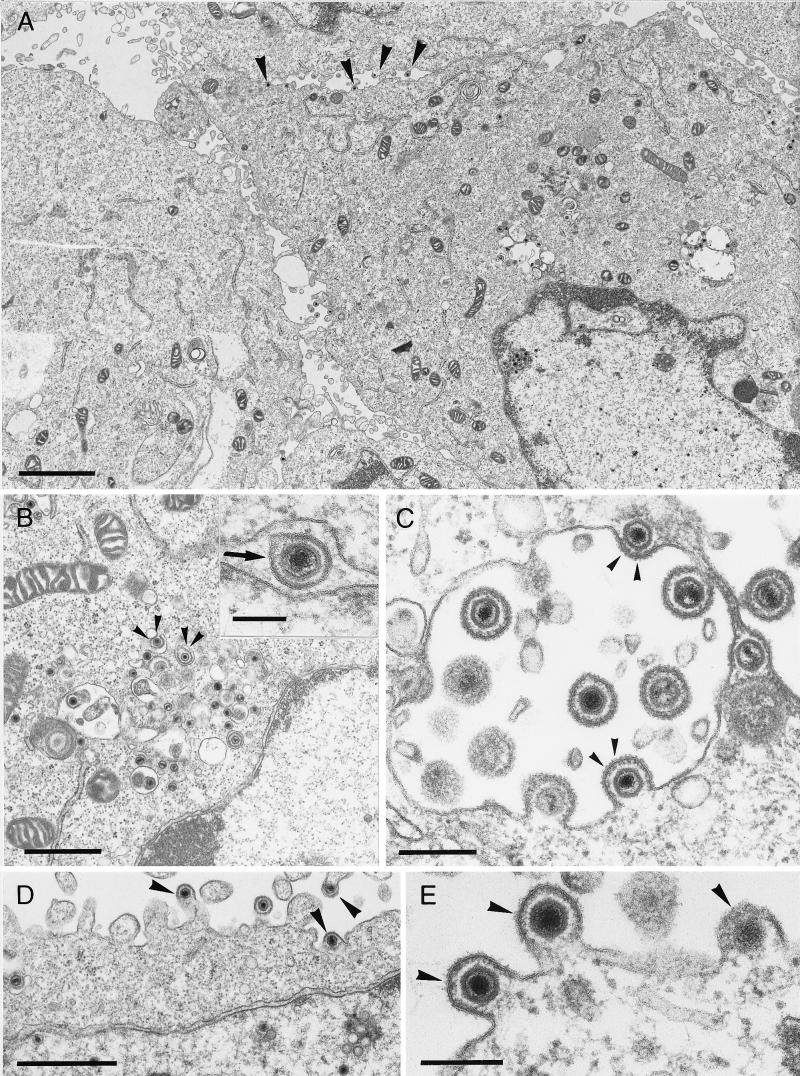
Since for HSV-1 gK mutants a defect in envelopment of nucleocapsids, as well as translocation of infectious virions to the extracellular space, has been described (16, 17), PrV-gKβ- and PrV-gKgB-infected complementing and noncomplementing cells were fixed 16 h p.i. and embedded, and ultrathin sections were investigated under the electron microscope. On complementing cells infected either with mutant PrV-gKβ (Fig. 7) or with mutant PrV-gKgB (data not shown), all stages of herpesvirus morphogenesis were found as described for PrV-Ka (12). Morphologically intact virus particles were visible in the extracellular space attached to or in proximity to the plasma membrane (Fig. 7C). In contrast, only few free virions were found outside the infected noncomplementing Vero cells (Fig. 8), although virion morphogenesis, including nucleocapsid formation in the nucleus (Fig. 8A), primary envelopment at the inner lamella of the nuclear membrane (Fig. 8B), and secondary envelopment in the cytoplasm (Fig. 8B and C), as well as virions found in transport vesicles (Fig. 8B and C), seemed to be undisturbed. Remarkably, numerous nucleocapsids were found directly underneath the plasma membrane (Fig. 8A, D, and E). These stages are found in PrV-Ka-infected cells at very early (1 to 2 min after onset of penetration) but never at late times after infection (12). We interpret these events as reentry and suggest that PrV gK is necessary for inhibition of reinfection. Identical effects were observed in mutant PrV-gKgB (data not shown), further indicating that the observed phenotype is due to the mutagenized gK.
FIG. 7.
Electron microscopy of PrV-gKβ on complementing cells. B5′-64 cells were infected with PrV-gKβ at an MOI of 1 and analyzed 16 h p.i. The arrow shows primary envelopment in the perinuclear cisterna (B), small arrowheads indicate secondary envelopment in the Golgi area (B), and large arrowheads point to free virions in the extracellular space (A to C). Bars: A, 2.5 μm; B, 1 μm; C, 1 μm.
FIG. 8.
Electron microscopy of PrV-gKβ on noncomplementing cells. Noncomplementing Vero cells were infected with PrV-gKβ at an MOI of 1 and analyzed 16 h p.i. The arrow in panel B (inset) indicates primary envelopment by budding into the perinuclear cisterna, small arrowheads in panels B and C point to secondary envelopment in the cytoplasm, and large arrowheads in panels A, D, and E show fusion stages at the cytoplasmic membrane which are typical for PrV gK mutants. Bars: A, 2.5 μm; B, 1 μm; inset, 150 nm; C, 300 nm; D, 1 μm; E, 200 nm.
DISCUSSION
While many of the alphaherpesvirus glycoproteins are now well characterized, information on gK, which has only recently been identified in HSV-1 (13), is limited. In this study, we describe characterization of PrV gK, the second gK protein in the alphaherpesviruses to be identified, and analyze the growth characteristics of two PrV gK mutants.
By using an antiserum directed against prokaryotically expressed amino acids 64 to 196 of PrV gK, a 36-kDa protein was detected in purified virion preparations. Thus, PrV gK clearly represents a virion structural component which has implications for its possible functional role in infection events which include interaction of whole virions with host cells. In HSV-1, gK has not been detected in virions (15), which appears somewhat puzzling since phenotypic alterations in penetration, i.e., fusion between virion envelope and target cell cytoplasmic membrane, have been attributed to mutations in the UL53 gene (7, 14, 31). Considering our observation, it appears possible that HSV-1 gK also represents a component of the virion, which due to lack of reactivity with the antiserum or the presence of only small amounts in the virus particle might have escaped detection. Although the possibility that PrV and HSV-1 virions differ in this respect cannot be excluded, the presence of gK in the HSV-1 virion would help to explain its influence on penetration kinetics (31). We demonstrate here that deletion of gK in PrV results in a significant delay in penetration, which can be rescued by phenotypic complementation after propagation of gK mutant viruses on gK-expressing cells. Thus, PrV gK represents the 11th PrV glycoprotein to be described and is the 10th glycoprotein identified as a structural component of the virion.
Like HSV-1 gK, PrV gK is predicted to comprise five hydrophobic regions long enough to span the lipid bilayer. The first hydrophobic domain could serve as signal sequence, which is indicated by results of in vitro transcription-translation reactions in the presence of canine microsomal membranes (2). This leaves four putative transmembrane regions in the mature protein. However, so far there is no direct proof that PrV gK is indeed localized in the virion envelope or in intracellular membranes, nor is intramembrane orientation known for any gK protein. HSV-1 gK has been detected in the nuclear membrane and in membranes of the endoplasmic reticulum but not in the plasma membrane or in virions (15, 16).
Deduced amino acid sequence indicated presence of two N-glycosylation motifs in the PrV gK protein (3). Enzymatic deglycosylation showed that PrV gK is indeed modified by addition of N-glycans which contribute ca. 4 kDa to the apparent molecular mass of mature 36-kDa gK. Interestingly, virion gK contains both high-mannose and complex forms of N-glycans, as demonstrated by digestion with PNGase F or endo H. Digestion with endo H reduces the apparent molecular mass to ca. 34 kDa, whereas complete removal of all N-glycans by PNGase F leads to a reduction to 32 kDa, which equals the size of the intracellular gK protein detected by the antiserum in infected cell lysates and also correlates with the in vitro translation product obtained in the presence of microsomal membranes (2). High-mannose and complex forms of N-glycans have also been detected on mature PrV gH. For PrV gH, incomplete processing of glycans can, at least in part, be explained by complex formation with gL, which inhibits terminal modification of at least one of the three putative N-glycans (23). The reason for the incomplete modification on gK is unknown. So far, no protein complex comprising gK has been identified, although the overall hydrophobic character of gK might render it prone to aggregation. Remarkably, HSV-1 gK also contains endo H-sensitive glycans (15). It is, however, interesting that intracellularly we were able to detect only the 32-kDa precursor form, whereas in virion preparations only the 36-kDa protein was found.
To investigate the function of PrV gK, we constructed two mutant viruses carrying insertions within the UL53 gene. Mutant PrV-gKβ contains a gG-lacZ expression cassette, and mutant PrV-gKgB contains the BHV-1 gB gene inserted after codon 164 of the UL53 gene. BHV-1 gB has been shown to complement the lethal defect associated with lack of PrV gB (24). Insertion of the heterologous gB gene into the genome of a gB− PrV mutant therefore restores replication competence. This procedure, called heterologous cis complementation, has previously been applied successfully for easy construction and isolation of PrV mutant viruses (8). Exchange of PrV gB for BHV-1 gB reduces final titers and plaque size to a limited degree but has no overall negative effect on PrV replication (24). Although our approach resulted in virus mutants which, at least theoretically, are able to express the amino-terminal 164 amino acids of gK, we did not detect any truncated gK product in infected cells or virions with our antiserum. Since the prokaryotic expression product against which the antiserum was made contains amino acids 64 to 196 of gK, any C-terminally truncated product should be recognized. Thus, the amino terminus of gK is either not expressed in our mutants, present in undetectable amounts, or rapidly degraded. We consider the last explanation the most likely, although we cannot completely rule out a contribution by the putative truncated form of gK on the observed phenotypes.
For HSV-1, two different gK mutants have been isolated. F-gKβ contains a lacZ insertion after codon 112 in a strain F background (16); in mutant ΔgK, the UL53 gene has been deleted from strain KOS (17). Both mutants produced only small plaques on Vero cells, and virus titers were reduced. Differences between the two mutants in the ability to fuse 143TK− cells were found and were attributed to the putative fusogenic properties of the C-terminally truncated possible expression product of F-gKβ. Also, the plaquing efficiency of F-gKβ was drastically reduced in comparison to that of ΔgK on Vero cells. Although these differences could be due to the different mutations within the gK gene, it has to be emphasized that HSV-1 strains F and KOS differ in their biological properties, and it cannot be excluded that strain background has a profound effect on the phenotypes associated with gK mutations. Inactivation of PrV gK in PrV-gKβ resulted in a dramatic reduction in the capability of the virus to produce plaques. Only single or small foci of infected cells were observed on noncomplementing cells, but plaque formation was restored on complementing cells. These results clearly link the gK mutation to the defect in plaque formation. A similar phenotype was observed for PrV-gKgB. In one-step growth, the two PrV gK mutants showed similar characteristics; i.e., on noncomplementing cells, production of infectious virus was decreased at all time points compared to that of complementing cells or wild-type virus-infected cells, resulting in a ca. 30-fold decrease in final titer. Plating efficiencies of gK-complemented and noncomplemented viruses on Vero cells were identical, however, which demonstrates that gK is not required for infectious entry of PrV into target cells, although it is required for plaque formation by direct cell-to-cell spread. This again shows that penetration and direct cell-to-cell spread are distinct and separable events in herpesvirus infection. It is important to note that whereas gK is dispensable for entry and necessary for plaque formation, PrV gD exhibits the opposite properties; i.e., it is required for entry but dispensable for plaque formation (30, 34).
Most importantly, however, both HSV gK mutants exhibited a defect in nucleocapsid envelopment and in the translocation of virions from the cytoplasm to the extracellular space (16, 17). Electron microscopic examinations of complementing and noncomplementing cells infected with PrV-gKβ or PrV-gKgB revealed no difference in intracellular morphogenesis and transport of virions. Intranuclear and intracytoplasmic capsids as well as secondary envelopment stages in the trans-Golgi area and enveloped particles within vacuoles which were postulated to represent transport vesicles (12) were observed. However, a striking difference was also detected. Compared to numerous extracellular virions after infection of complementing cells, on noncomplementing cells very few extracellular virus particles were observed. Closer examination revealed the presence of nucleocapsids immediately beneath the cytoplasmic membrane in stages reminiscent of entry of virions during initial penetration or budding into vesicles during secondary envelopment (12). Although the electron micrographs do not prove directionality of the observed events, we consider it highly unlikely that these represent budding stages. Herpesviruses do not acquire their envelope from the cytoplasmic membrane, and we never observed budding stages at the cytoplasmic membrane in numerous electron microscopic analyses of PrV maturation (12). Since secondary envelopment in the trans-Golgi region appears to be unimpaired (Fig. 8), we hypothesize that these stages represent reentry of released virus particles and postulate that PrV gK is required to prevent fusion between virion envelope and cytoplasmic membranes of PrV-infected cells, thus leading to final release of infectious viruses. We are currently establishing cell lines constitutively expressing gK to more directly analyze the possible interference function of PrV gK. Interference with infection has so far been attributed solely to gD (18). HSV-1 gD binds cell surface receptor(s) (44), and intracellular sequestration of these receptors by gD has been postulated to denude the cell surface of necessary receptor proteins (18). However, it is conceivable the interference works at different levels during entry, i.e., attachment (receptor binding) and fusion. Since HSV-1 gK has been shown to regulate fusion during syncytium formation, i.e., deregulation by syncytium-inducing mutations leads to extensive fusion events (14), it appears reasonable to assume that gK-mediated interference is at the level of fusion.
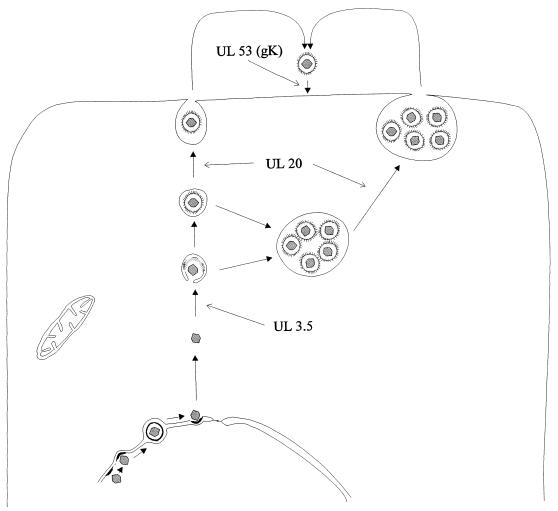
It is currently unclear whether a similar function is also executed by HSV-1 gK. HSV-1 gK has been implicated in the translocation of virions from the cytoplasm to the extracellular space, and electron micrographs show an accumulation of mutant virions in large vacuoles (17). For PrV and varicella-zoster virus, egress of virions from infected cells has been postulated to involve primary envelopment at the inner nuclear membrane, loss of primary envelope by fusion at the outer lamella of the nuclear membrane, and secondary envelopment of intracytoplasmic nucleocapsids by budding into intracytoplasmic vesicles which then fuse with the cytoplasmic membrane, leading to release of enveloped virions into the extracellular space. This hypothesis is based on electron microscopic observations (10, 12), inhibitor studies (43), and analysis of PrV mutants with lesions in the UL3.5 and UL20 genes (8, 9). In PrV-infected cells, gK would function last in a cascade of events which involve (i) the UL3.5 protein, important for secondary envelopment, (ii) the UL20 protein, whose absence leads to accumulation of virions in large vacuoles, and (iii) gK, which inhibits immediate reinfection (Fig. 9). HSV-1 lacks a UL3.5 homolog, and the UL20 protein appears to function in a different intracellular compartment (1) than PrV UL20 (9). Therefore, it would not be surprising to also see different roles for the gK homologs in virus egress.
FIG. 9.
Diagram of proposed events and involvement of viral proteins during egress of PrV.
In summary, we describe PrV gK as a 36-kDa virion component which is required for productive virus replication. PrV gK plays an important role in egress, probably by preventing immediate reentry of released infectious virions. However, it is dispensable for entry, which further emphasizes differences in the entry and egress pathways of herpesviruses.
ACKNOWLEDGMENTS
This study was supported by grant Me 854/3 from the Deutsche Forschungsgemeinschaft.
We thank A. Petersohn and E. Mundt for help with preparation of the antiserum, J. Schmidt for help with artwork, E. Weiland for monoclonal antibodies, and U. Hartwig and C. Möller for excellent technical assistance.
REFERENCES
- 1.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumeister, J., and B. G. Klupp. Unpublished data.
- 3.Baumeister J, Klupp B G, Mettenleiter T C. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J Virol. 1995;69:5560–5567. doi: 10.1128/jvi.69.9.5560-5567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne H, Bell S, Minson T, Wilson D W. An endoplasmatic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume G, Farabegoli F, DiGaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DiLazzaro C, Campadelli-Fiume G, Torrisi M R. Intermediate forms of glycoconjugates are present in the envelope of herpes simplex virions during their transport along the exocytic pathway. Virology. 1995;214:619–623. doi: 10.1006/viro.1995.0073. [DOI] [PubMed] [Google Scholar]
- 7.Dolter K E, Ramaswamy R, Holland T C. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs W, Klupp B G, Granzow H, Rziha H-J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs W, Klupp B G, Granzow H, Mettenleiter T C. The UL20 gene product of pseudorabies virus functions in virus egress. J Virol. 1997;71:5639–5646. doi: 10.1128/jvi.71.7.5639-5646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 12.Granzow H, Weiland F, Jöns A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchinson L, Goldsmith K, Snoddy D, Gosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutchinson L, Graham F L, Cai W, Debroy C, Person S, Johnson D C. Herpes simplex virus (HSV) glycoproteins B and K inhibit cell fusion induced by HSV syncytial mutants. Virology. 1993;196:514–531. doi: 10.1006/viro.1993.1507. [DOI] [PubMed] [Google Scholar]
- 15.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet it is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson D, Burke R, Gregory T. Soluble forms of herpes simplex virus glycoprotein D bind to a limited number of cell surface receptors and inhibit virus entry into cells. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jöns A, Granzow H, Kuchling R, Mettenleiter T C. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J Virol. 1996;70:1237–1241. doi: 10.1128/jvi.70.2.1237-1241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jöns A, Mettenleiter T C. Identification and characterization of pseudorabies virus dUTPase. J Virol. 1996;70:1242–1245. doi: 10.1128/jvi.70.2.1242-1245.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan A, Vatter A. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 23.Klupp B G, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J Virol. 1992;66:2754–2762. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1979;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter, T. C. Unpublished data.
- 27.Mettenleiter T C. Initiation and spread of α-herpesvirus infections. Trends Microbiol. 1994;2:1–33. doi: 10.1016/0966-842x(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 28.Mettenleiter T C. Pseudorabies (Aujeszkys disease) virus: state of the art. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 29.Mettenleiter T C, Rauh I. A glycoprotein gX-βgalactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 30.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertel P E, Spear P G. Modified entry and syncytium formation by herpes simplex virus type 1 mutants selected for resistance to heparin inhibition. Virology. 1996;226:222–233. doi: 10.1006/viro.1996.0624. [DOI] [PubMed] [Google Scholar]
- 32.Ramaswamy R, Holland T C. In vitro characterization of the HSV-1 UL53 gene product. Virology. 1992;186:579–587. doi: 10.1016/0042-6822(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 33.Rauh I, Weiland F, Fehler F, Keil G M, Mettenleiter T C. Pseudorabies virus mutants lacking the essential glycoprotein gII can be complemented by glycoprotein gI of bovine herpesvirus 1. J Virol. 1991;65:621–631. doi: 10.1128/jvi.65.2.621-631.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roizman B, Derosiers R, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 36.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 region promoter. J Mol Appl Gen. 1982;1:327–341. [PubMed] [Google Scholar]
- 37.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–189. [Google Scholar]
- 38.Tarentino A L, Gomez C M, Plummer T H. Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry. 1985;24:4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 39.Torrisi M R, DiLazzaro C, Pavan A, Pereira L, Campadelli-Fiume G. Herpes simplex virus envelopment and maturation studies by fracture label. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trimble R B, Maley F. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal Biochem. 1984;141:515–522. doi: 10.1016/0003-2697(84)90080-0. [DOI] [PubMed] [Google Scholar]
- 42.Weiland, E., and B. Klupp. Unpublished data.
- 43.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1992;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittmann G, Rziha H-J. Aujeszky’s disease (pseudorabies) in pigs. In: Wittmann G, editor. Herpes disease of cattle, horses and pigs. Boston, Mass: Kluwer; 1989. pp. 230–325. [Google Scholar]