Abstract
We have previously demonstrated that open reading frame (ORF) 50 and ORF 57 encode transcriptional regulating genes in herpesvirus saimiri. ORF 50, a homolog of Epstein-Barr virus R protein, is a sequence-specific transactivator, whereas ORF 57 acts posttranscriptionally. In this report, we demonstrate that the ORF 57 gene is regulated by the ORF 50a gene product. We show that the ORF 57 gene is expressed at basal levels early in the virus replication cycle and that thereafter it is transactivated by the ORF 50a gene product, due to an increase in RNA levels. As it has been shown that the ORF 57 gene product downregulates ORF 50a due to the presence of its intron, these combined observations identify a feedback mechanism modulating gene expression in herpesvirus saimiri, whereby ORF 50a transcription is downregulated by the ORF 57 gene product, a gene which it specifically transactivates. Furthermore, we propose that the intron-containing ORF 57 gene downregulates itself by the same mechanism as that for ORF 50a, as both genes are downregulated at similar times during the replication cycle.
Herpesvirus saimiri (HVS) is a lymphotrophic Rhadinovirus (gamma-2 herpesvirus) of squirrel monkeys (Saimiri sciureus), which persistently infects its natural host without causing any obvious disease. However, HVS infection of other species of New World primates results in fulminant polyclonal T-cell lymphomas and lymphoproliferative diseases (8). HVS is also capable of transforming simian and human T lymphocytes to continuous growth in vitro (4). The genome of HVS (strain A11) consists of a unique internal low-G+C-content DNA segment (L-DNA) of approximately 110 kbp which is flanked by a variable number of 1,444-bp high-G+C-content tandem repetitions (H-DNA) (2). Analysis indicates that it has significant homology with the following herpesviruses: Epstein-Barr virus (EBV), bovine herpesvirus 4, Kaposi’s sarcoma-associated herpesvirus (also called human herpesvirus 8), and murine gammaherpesvirus 68 (1, 5, 11, 12, 29, 41, 49). The genomes of EBV, Kaposi’s sarcoma-associated herpesvirus, murine gammaherpesvirus 68, and HVS have been shown to be generally colinear, in that homologous sequences are found in approximately equivalent locations and in the same relative orientation. However, conserved gene blocks are separated by unique genes respective to each virus (1, 29, 30, 41, 49).
Gene expression during lytic replication is sequentially regulated and occurs in three main temporal phases: immediate-early (IE), delayed-early, and late (19). Two major IE transcripts encoded by the HindIII-G-IE gene (open reading frame [ORF] 14) and the IE 52-kDa gene (ORF 57) have been identified in HVS (2, 32, 33). Analysis of ORF 14 shows that it does not exhibit homology with any EBV-encoded protein but that it does contain homology with a putative superantigen (48). It has recently been shown to bind major histocompatibility complex class II molecules and to stimulate cell proliferation (54). The IE 52-kDa protein has been mapped to the EcoRI-IE fragments of HVS and is homologous to genes identified in all classes of herpesviruses. These include the EBV transactivator encoded by BMLFI, ICP27 of herpes simplex virus (HSV), BICP27 in bovine herpesvirus 1, ORF 4 encoded by varicella-zoster virus, UL69 in human cytomegalovirus, and ICP27 in equine herpesvirus 1 (7, 20, 32, 35, 47, 53, 55).
The ORF 57 gene product has transregulatory functions which are independent of the target gene promoter sequences and appear to be mediated at the posttranscriptional level, whereas repression of gene expression appears to correlate with the presence of introns, suggesting that ORF 57 is functionally homologous to ICP27 (52). In addition, the more widely studied homolog, ICP27, has been shown to be involved in the switch from early to late gene expression (24–26, 38, 42, 44) and in the downregulation of viral IE and early genes and is required for the expression of late genes (26, 38, 42, 45). Furthermore, ICP27 contributes to the shutoff of host cell protein synthesis and contributes to a decrease in cellular mRNA levels during infection, as deletion mutant infections result in higher levels of cellular protein synthesis and mRNA levels than do wild-type infections (15, 16, 18, 46).
The second transcriptional activator encoded by HVS is homologous to the EBV BRLF1 gene product, R (2, 31), a sequence-specific transactivator (14). The HVS R gene or ORF 50 produces two transcripts: the first is spliced, containing a single intron, and is detected at early times during the productive cycle, whereas the second is expressed later and is produced from a promoter within the second exon. The spliced transcript is fivefold more potent in activating the delayed-early ORF 6 promoter; the function of the nonspliced transcript is unclear (31, 50). Further analysis of ORF 50 indicates that it responds to DNA-specific sequences; gel retardation analysis has identified a consensus R-recognition sequence, CCN9GG, required for transactivation by both ORF 50 transcripts (51). Furthermore, the transactivating capability of the ORF 50a gene product (which is produced from a spliced transcript) is repressed by the IE ORF 57 gene product, whereas the transactivating capability of ORF 50b (an unspliced transcript) is slightly enhanced by ORF 57 (52).
In this report, we demonstrate that the ORF 50a gene product regulates ORF 57 gene expression in HVS. We show that the ORF 57 gene is expressed at basal levels up to 18 h postinfection (hpi); thereafter it is transactivated by the ORF 50a gene product, due to an increase in RNA levels. We have previously demonstrated that the ORF 57 gene product downregulates ORF 50a due to the presence of its intron (52). These combined observations lead to a feedback mechanism modulating gene expression in HVS, whereby ORF 50a is downregulated by a gene which it specifically transactivates. In addition, we propose that the intron-containing ORF 57 gene downregulates itself by the same mechanism by which it represses ORF 50a.
MATERIALS AND METHODS
Viruses, cell culture, and transfections.
HVS (strain A11) was propagated in owl monkey kidney (OMK) cells which were maintained in Dulbecco modified Eagle medium (Life Technologies) supplemented with 10% fetal calf serum. Plasmids used in the transfections were prepared by using Qiagen plasmid kits according to the manufacturer’s directions. OMK cells were seeded at 5 × 105 cells per 35-mm-diameter petri dish 24 h before transfection. Transfections were performed with DOTAP (Boehringer Mannheim) as described by the manufacturer, with 2 μg of the appropriate DNAs.
Immunofluorescence analysis.
OMK cells, seeded at 5 × 105 cells per 35-mm-diameter petri dish, were infected at a multiplicity of infection of 1. These cells were fixed with 4% formaldehyde in phosphate-buffered saline (PBS), washed in PBS three times, and permeabilized in 0.5% Triton X-100 for 5 min. The cells were rinsed in PBS and blocked by preincubation with 1% (wt/vol) nonfat milk powder for 1 h at 37°C. A 1:100 dilution of anti-ORF 57 SB antibody (a gift from Rick Randall) was layered over the cells and incubated for 1 h at 37°C. Fluorescein-conjugated anti-mouse immunoglobulin (Dako) at a 1:50 dilution was added for 1 h at 37°C. After each incubation step, cells were washed extensively with PBS. The immunofluorescence slides were observed with a Zeiss Axiovert 135TV inverted microscope with a Neofluar 40× oil immersion lens.
Plasmid constructs.
In order to generate an ORF 57 promoter reporter gene construct, the ORF 57 gene promoter was PCR amplified with primers 5′-CCC AAG CTT CTT TGC CTG CAG GTG TTG and 5′-ACG TCT AGA TTG GGC AGT TAG TCA CCA TAG. These oligonucleotides incorporated HindIII and XbaI restriction sites, respectively, for convenient cloning of the PCR product. The reaction (30 cycles of 1 min at 92°C, 1 min at 58°C, and 2 min at 72°C) was performed with 4 U of Pfu (Stratagene). This fragment was inserted upstream of the chloramphenicol acetyltransferase (CAT) coding region in pCATBasic (Promega) to derive pORF57CAT1. To generate a transfer vector containing the ORF 57 gene promoter and the ORF 57 coding region, viral DNA was digested with PstI and the appropriate fragment was cloned into pUC18 to derive pUCORF57. In order to generate an ORF 57 gene promoter reporter construct with deleted ORF 50 response elements, a smaller ORF 57 promoter construct was generated by PCR with the primer 5′-CCC AAG CTT GAT GGT CCA TTC TAT TAG and the 3′ primer previously used. This fragment was then ligated upstream of the CAT coding region to derive pORF57CAT2.
Plasmid and bacteriophage vectors containing restriction fragments representing the entire L-DNA of HVS have been previously described (21). The transactivator plasmids pORF50a and pAWHincII, which encode ORF 50a and 50b, respectively, have been previously described (50).
CAT assay.
Cell extracts were prepared 48 h after transfection and incubated with [14C]chloramphenicol in the presence of acetyl coenzyme A as described previously (13). The percentage of acetylation of chloramphenicol was quantified by scintillation counting (Packard) of appropriate regions of the thin-layer chromatography plate.
Total RNA extraction.
RNA was extracted from OMK cells infected with HVS (strain A11) at 5 PFU/cell at various times postinfection. Cells were lysed with Trizol reagent (Life Technologies). Chloroform (0.2 ml) was then added, and the solution was vortex mixed for 20 s and stored at 20°C for 15 min. Samples were centrifuged for 15 min at 4°C, and the aqueous phase containing nucleic acids was precipitated with 0.5 ml of isopropanol. Afterwards, the pellet was washed with 70% ethanol, resuspended in 50 μl of water, and stored at −70°C.
Primer extension analysis.
Primer extension was performed essentially as described by Sambrook et al. (43). First, a 20-bp oligonucleotide primer, 5′-GGT ACA TTG ACG AAC TGA CT, homologous to the CAT coding region 70 bp downstream of the initiation codon, was radiolabelled with [α-32P]ATP. A total of 5 μg of RNA was mixed with 300 ng of the radiolabelled primer; the samples were then boiled for 5 min and snap-chilled on ice. To the reaction mixtures were added 1 mM (each) dATP, dTTP, dCTP, and dGTP; buffer (final concentrations, 50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2), and 1 μl of reverse transcriptase (Life Technologies) in a final volume of 20 μl. The reaction was performed at 42°C for 60 min, and primer extension products were electrophoresed on a 6% acrylamide–7 M urea gel. After electrophoresis, the gel was dried and bands were visualized by exposure to X-ray film.
Gel retardation analysis.
The following sets of oligonucleotides were annealed and labelled with T4 polynucleotide kinase in the presence of [γ-32P]dATP: set 1, 5′-GCA ATG TAT CCA CTA ATT ATG GAA ACA GAT and 5′-ACT TGT TTC CAT AAT TAG TGG ATA CAT TGC; and set 2, GGT CCA TTC GAT GTA GGC ATC TAC CAT AAA and 5′-TTT ATG GTA GAT GCC TAC ATG CTT ACC TGG ACC. The radiolabelled oligonucleotides were incubated with nuclear extracts of untransfected cells or cells transfected with pORF50a and prepared by the method of Andrews and Faller (3). The binding reactions were performed in 20 μl of 100 mM KCl–20 mM HEPES (pH 7.3)–1% glycerol–0.2 mM EDTA–5 mM MgCl2–4 mM dithiothreitol–0.5 mM phenylmethylsulfonyl fluoride with 1 μg of poly(dI-dC) as an nonspecific competitor. The protein-nucleic acid complexes were separated on a 5% polyacrylamide gel, run in 1% Tris-borate-EDTA buffer, and detected by autoradiography.
Northern blot analysis.
Northern blot analysis was performed essentially as described by Sambrook et al. (43). Total RNA was isolated from HVS-infected OMK cells, harvested at intervals of 3 h up to 24 hpi and then at intervals of 6 h until 48 hpi, and separated by electrophoresis on a 1% denaturing formaldehyde agarose gel. The RNA was transferred to Hybond-N membranes and hybridized with 32P-labelled randomly primed probes specific for ORF 57 and ORF 50 coding sequences.
RESULTS
The ORF 57 gene product appears at 12 hpi.
In order to determine the temporal appearance of the ORF 57 gene product in HVS-infected cells, indirect immunofluorescence analysis of HVS-infected cells was performed. Infected cells were harvested at intervals of 3 h up to 24 hpi and then at intervals of 6 h until 48 hpi, incubated with an ORF 57-specific monoclonal antibody, and then stained with fluorescein-conjugated anti-mouse immunoglobulin (data not shown). This revealed strong fluorescence in the nuclei of infected cells, similar to previous observations with HVS-infected cells (36, 37). Results showed that the ORF 57 gene product appeared at between 9 and 12 hpi at very low levels; at 18 hpi, the amount of protein was greatly increased.
The ORF 57 gene is transactivated by ORF 50a.
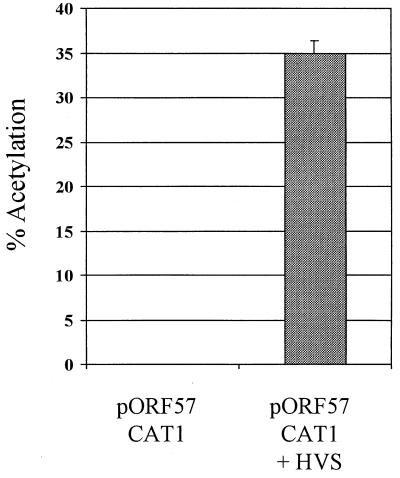
Because of this increase in the amount of ORF 57 gene product by 18 hpi, we were particularly interested in determining whether the ORF 57 gene promoter was transactivated by a virus gene product. Therefore, the ORF 57 gene promoter was generated by PCR amplification and ligated upstream of the CAT coding region to derive pORF57CAT1. OMK cells were transfected with pORF57CAT1; cells were then subsequently mock infected or superinfected with 50 PFU of HVS per cell at 24 h posttransfection. Cells were harvested at 48 h posttransfection, and the cell extracts were assayed for CAT activity (Fig. 1). The results show very low expression detected from the ORF 57 gene promoter in the absence of the superinfection; however, in the presence of HVS CAT activity was greatly enhanced, suggesting that the ORF 57 gene promoter was transactivated by a virus gene product.
FIG. 1.
Analysis of the ORF 57 promoter. OMK cell monolayers were transfected with 2 μg of pORF57CAT1 by using DOTAP (Boehringer Mannheim), as directed by the manufacturer; cells containing pORF57CAT1 subsequently either remained uninfected or were superinfected with 50 PFU of HVS (strain A11) per cell at 24 h posttransfection. Cells were harvested at 48 h posttransfection, and extracts were assayed for CAT activity. Products from these assays were separated by thin-layer chromotography and detected by autoradiography. Percentages of acetylation were calculated by the scintillation counting of the appropriate regions of the chromatography plate and are shown in graphic format.
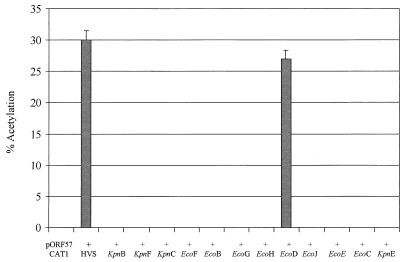
In order to identify the gene product responsible for ORF 57 transactivation, a series of cotransfection experiments were performed with pORF57CAT1 in the presence of a series of plasmids which contained the entire L-DNA region of HVS (strain A11) (21). Cells were harvested at 48 h posttransfection, and the cell extracts were assayed for CAT activity (Fig. 2). Results show that the ORF 57 gene promoter was transactivated only by a virus gene product contained within the EcoD fragment of the genome.
FIG. 2.
Mapping the gene responsible for ORF 57 transactivation. OMK cell monolayers were transfected with 2 μg of pORF57CAT1 and a series of plasmids which contained the entire L-DNA region of HVS (strain A11). Cells were harvested at 48 h posttransfection, and extracts were assayed for CAT activity. Products from these assays were separated by thin-layer chromotography and detected by autoradiography. Percentages of acetylation were calculated by the scintillation counting of the appropriate regions of the chromatography plate; error bars indicate the variations among three replicate assays.
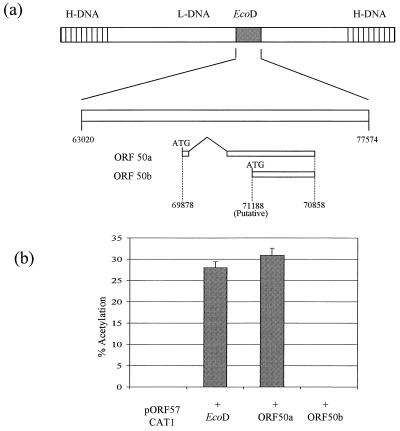
It has previously been shown that the EcoD fragment encodes the HVS R protein transactivator encoded by ORF 50 (31). This gene is homologous to the EBV R gene product (BRLF1) (2, 31). Therefore, we hypothesized that the ORF 57 gene promoter may be transactivated by one of the ORF 50 gene products. Cotransfection studies were performed to assess the effect of the ORF 50a and 50b gene products on the ORF 57 gene promoter. Results shown in Fig. 3 indicate that the ORF 57 gene promoter was transactivated only in the presence of the ORF 50a gene product, to the same extent as EcoD, suggesting that ORF 50a is the only gene responsible for ORF 57 transactivation; ORF 50b exerted no effect on the ORF 57 promoter.
FIG. 3.
ORF 57 is transactivated by the ORF 50a gene product. (a) Schematic representation of the location of ORF 50a and ORF 50b within the L-DNA of HVS. (b) OMK cell monolayers were transfected with 2 μg of pORF57CAT1 in the absence and presence of either pORF50a or pAWHincII, which encodes ORF 50a or ORF 50b, respectively. Cells were harvested at 48 h posttransfection, and extracts were assayed for CAT activity. Products from these assays were separated by thin-layer chromotography and detected by autoradiography. Percentages of acetylation were calculated by the scintillation counting of the appropriate regions of the chromatography plate; error bars indicate the variations among three replicate assays.
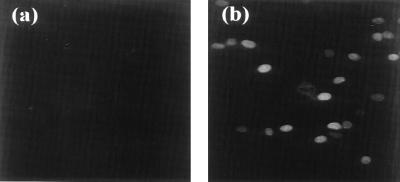
To further demonstrate that the ORF 57 gene promoter is transactivated by ORF 50a, pUCORF57, a plasmid containing the ORF 57 promoter and coding region, was cotransfected in the absence or presence of pORF50a. Cells were harvested at 48 h posttransfection, incubated with an ORF 57-specific antiserum, and then stained with fluorescein-conjugated anti-mouse immunoglobulin (Fig. 4). This revealed strong fluorescence of the nuclei of pUCORF57-cotransfected cells only in the presence of pORF50a, further implicating ORF 50a as the gene product responsible for ORF 57 transactivation.
FIG. 4.
Immunofluorescence analysis of ORF 57 transactivation. OMK cells were transfected with 2 μg of pUCORF57 in the absence (a) and presence (b) of pORF50a, incubated with an ORF 57-specific antiserum, and then stained with fluorescein-conjugated anti-mouse immunoglobulin.
ORF 50a transactivation of ORF 57 leads to an increase in mRNA levels.
It has been shown that the ORF 50a gene product increases the level of CAT activity produced from the ORF 57 promoter. To ascertain whether this rise is due to an increase in the levels of CAT mRNA in the presence of the transactivator, primer extension analysis was performed. Total RNA was isolated from untransfected OMK cells or from cells transfected with pORF57CAT1 or cotransfected with pORF57CAT1 and pORF50a, harvested at 24 hpi, and hybridized with a 32P-labelled primer homologous to the CAT coding region. Primer extension was then performed, and results are shown in Fig. 5. A primer extension product of 259 bp was observed with RNA harvested from cells transfected with pORF57CAT1, mapping the transcription start site of ORF 57 to bp 78272 of the published sequence. A similar-sized product was observed with RNA isolated from cells cotransfected with pORF57CAT1 and pORF50a, suggesting that the transactivation does not alter the mRNA initiation site. However, increased levels of primer extension product were produced upon transactivation with pORF50a, indicating that transactivation by ORF 50a on the ORF 57 gene promoter is due to an increase in RNA levels.
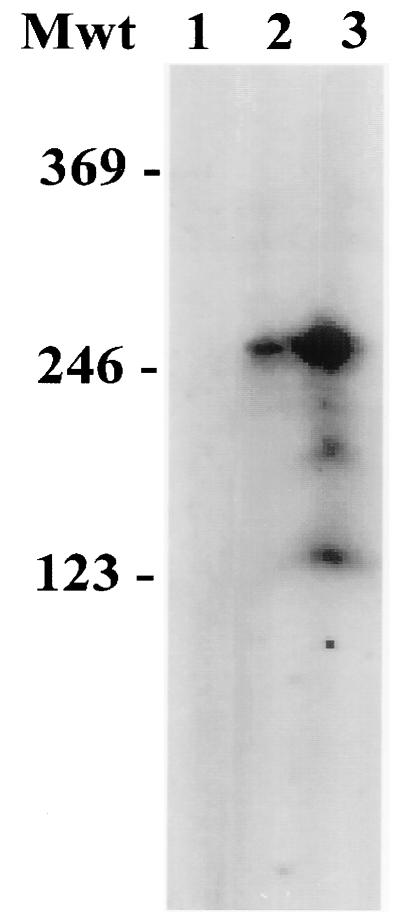
FIG. 5.
RNA analysis of ORF 57 transactivation. Total RNA was isolated from OMK cells (lane 1), OMK cells transfected with pORF57CAT1 (lane 2), and OMK cells cotransfected with pORF57CAT1 and pORF50a (lane 3) at 24 hpi and hybridized with a 32P-labelled primer homologous to the CAT coding region. Primer extension was performed, and the primer extension products were run on a 6% acrylamide–7 M urea gel and visualized by exposure to X-ray film. Mwt, molecular weight in thousands.
Sequences within the ORF 57 gene promoter are essential for transactivation by the ORF 50a gene product.
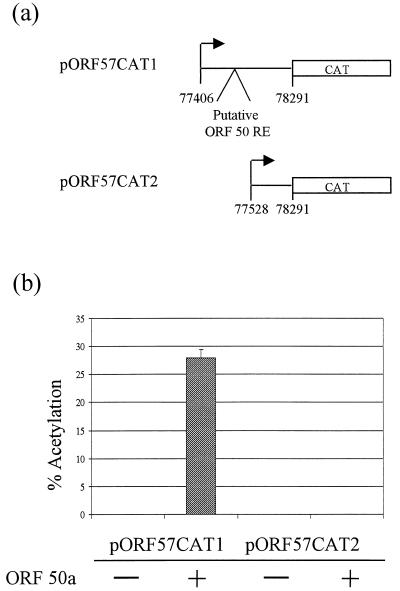
By deletion analysis and gel retardation assays, the ORF 50 responsive elements contained within the ORF 6 promoter were mapped to within 38 bp (51). Sequences homologous to the ORF 50 response element consensus sequence contained within the ORF 6 promoter are present within the ORF 57 promoter, corresponding to bp 77508 to 77521 of the published sequence. In order to determine if these sequences are essential for ORF 57 transactivation, an ORF 57 promoter in which the putative ORF 50 response elements were deleted was generated by PCR amplification. This fragment was ligated upstream of the CAT coding region to derive pORF57CAT2. Cotransfection studies were performed to assess the effect of the ORF 50a gene product on the altered ORF 57 gene promoter. The results of the cotransfection experiments with pORF57CAT2 in the absence or presence of ORF 50a are shown in Fig. 6 and demonstrate that ORF 50a transactivation of the ORF 57 gene promoter required the putative ORF 50 response elements.
FIG. 6.
ORF 57 transactivation requires the ORF 50 response elements contained within its promoter. (a) An ORF 57 promoter was generated by PCR amplification which deleted the putative ORF 50 response elements (RE); this fragment was ligated upstream of the CAT coding region to derive pORF57CAT2. (b) OMK cell monolayers were transfected with 2 μg of pORF57CAT2 in the absence and presence of pORF50a. Cells were harvested at 48 h posttransfection, and extracts were assayed for CAT activity. Products from these assays were separated by thin-layer chromotography and detected by autoradiography. Percentages of acetylation were calculated by the scintillation counting of the appropriate regions of the chromatography plate; error bars indicate the variations among three replicate assays.
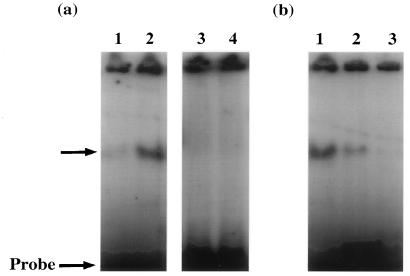
To determine which sequences within the ORF 57 gene promoter were necessary for ORF 50 responsiveness, gel retardation experiments were performed with a set of oligonucleotides which spanned the putative response elements. Radiolabelled probes were incubated with DNA-binding protein extracts from untransfected cells and from cells transfected with pORF50a, and the products were separated on a polyacrylamide gel (Fig. 7a). Results show the formation of a retarded complex with oligonucleotide set 1 and extracts of cells transfected with pORF50a. No other complex was identified with other oligonucleotides with untransfected or transfected cells, indicating that the sequences specific for ORF 50 responsiveness are contained within the 30 bp of oligonucleotide set 1, which contains bp 77500 to 77529 of the published sequence. Unlabelled oligonucleotide was shown to compete with this reaction (Fig. 7b), further suggesting that this 30-bp sequence was essential for ORF 57 transactivation and contained the ORF 50 response elements. However, at present we are unable to determine whether the ORF 50a gene product binds to the ORF 50 response elements directly or whether a protein(s) induced by ORF 50 promotes protein-DNA complex formation leading to transactivation, and this is currently being investigated.
FIG. 7.
Gel retardation analysis. To locate the ORF 50 response elements contained within the ORF 57 gene promoter, gel retardation assays were performed on oligonucleotides mapping to this region. (a) Set 1: untransfected cells (lane 1) and pORF50a-transfected cells (lane 2); set 2: untransfected cells (lane 3) and pORF50a-transfected cells (lane 4). (b) Gel retardation experiments were repeated in the presence of increased amounts of unlabelled set 1 oligonucleotides. The retarded complexes were separated on a 5% polyacrylamide gel, run in 1% Tris-borate-EDTA buffer, and detected by autoradiography.
Time of expression of ORF 57 and ORF 50a and 50b.
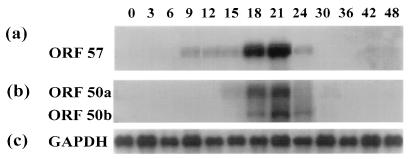
ORF 50a thus transactivates the ORF 57 gene due to an increase in RNA levels. To further examine this transactivation, we determined the time during the lytic replication cycle when ORF 57 and ORF 50a and 50b transcripts were produced. Northern blot analysis was performed with total RNA from infected OMK cells. Infected cells were harvested at intervals of 3 h up to 24 hpi and then at intervals of 6 h until 48 hpi, separated by electrophoresis on a 1% denaturing formaldehyde agarose gel, blotted onto a nylon membrane, and hybridized with radiolabelled probes specific for ORF 57 and ORF 50, respectively. Results indicate that low levels of ORF 57 are present from 9 hpi; however, levels dramatically increase from 18 hpi until 21 hpi, and thereafter ORF 57 levels are visibly reduced (Fig. 8a). ORF 50a became visible between 12 to 15 and 21 hpi, whereas ORF 50b appeared at between 18 and 24 hpi (Fig. 8b). These results further suggest that the ORF 50a gene product regulates ORF 57 gene expression.
FIG. 8.
Northern blot analysis of the expression of ORF 57 and ORF 50 genes. RNA was isolated from infected cells at intervals of 3 h up to 24 hpi and then at intervals of 6 h until 48 hpi, separated by electrophoresis on a 1% denaturing formaldehyde agarose gel, blotted onto a nylon membrane, and hybridized with radiolabelled probes specific for ORF 57 (a) and ORF 50 (b). Also shown are results of determination of RNA loading by hybridization with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (c).
DISCUSSION
In this report, we have further investigated the regulation of viral gene expression in HVS. Results presented in this report have identified a novel model of regulation of gene expression in HVS, summarized in Fig. 9. Central to this model are the virally encoded gene products ORF 57 and ORF 50a. The ORF 57 gene is produced at low levels from 9 hpi until it is transactivated by the early ORF 50a gene product at 18 hpi. Sequences within the ORF 57 promoter are essential for transactivation by the ORF 50a gene product, which results in an increase in RNA levels of the ORF 57 transcript. In addition, ORF 50a transactivates other genes which contain ORF 50 response elements within their promoters, for example, the major DNA-binding protein (31, 50, 51). Once transactivated by ORF 50a, the ORF 57 gene product has several functions. First, it has been shown to transactivate a range of HVS genes through posttranscriptional modification. Second, it downregulates ORF 50a, due to the presence of an intron within its coding region (52). Therefore, we believe that a feedback mechanism involving ORF 50a and ORF 57 is in operation, a mechanism which regulates gene expression in HVS, whereby a gene is downregulated by the product of the gene that it has previously transactivated. Third, we believe that the intron-containing ORF 57 gene is responsible for its own downregulation by the same mechanism by which it represses ORF 50a, as both genes are downregulated at similar times during the replication cycle.
FIG. 9.
Schematic representation of the role and interactions of the ORF 57 and ORF 50 genes which regulate gene expression in the HVS replication cycle. DE, delayed early; REs, response elements.
This series of events regulating gene expression in HVS differs from that in other herpesviruses. IE genes in all herpesviruses are defined as those which can be transcribed efficiently in the absence of de novo protein synthesis. Therefore, they mostly encode transcriptional regulators which are required for viral gene expression. However, despite their obvious role in virus replication the major IE genes are not conserved among herpesviruses. For example, during HSV replication five IE genes, ICP0, ICP4, ICP22, ICP27, and ICP47, are expressed in the absence of viral protein synthesis. Only one of these genes is conserved in HVS; ORF 57 is homologous to ICP27; this, however, may not be surprising, as HSV and HVS belong to different subfamilies of the herpesvirus genera. However, EBV, a member of the same subfamily as HVS, also differs from HVS in the IE genes which it encodes. Upon reactivation, two major IE genes which are the key transactivating genes in EBV are expressed. The first, the IE BZLF1 gene product, Z, is sufficient to trigger reactivation when overexpressed in latently infected cells (6, 10, 40). Z is able to transactivate several promoters containing Z-responsive elements, as well as to regulate its own promoter (10, 22, 34, 40). The second IE protein, the BRLF1 gene product, R, is also a sequence-specific transactivator. HVS does not encode a Z homolog; however, ORF 50 is homologous to the EBV R protein. Therefore, it suggests that the two genes encoded by HVS which are homologous to genes found in other herpesviruses play a critical role in the HVS replication cycle. In addition, we believe that a third, as yet unidentified gene may be responsible for the transactivation of ORF 50a.
Also of interest is the time at which the gene product of ORF 57 is produced in the HVS replication cycle. ORF 57 is believed to be an IE gene product due to the production of this transcript in cells infected with 25 PFU/cell and incubated in the continuous presence of cycloheximide, but it was undetectable in RNA samples infected with 5 PFU/cell (33). In addition, transfection experiments show very low or undetectable basal expression from the ORF 57 gene promoter. Furthermore, we have shown that ORF 57 is transactivated by the ORF 50a gene product. This contrasts with IE genes encoded by HSV, which appear much earlier in the replication cycle than the IE ORF 57 of HVS; maximum levels of these gene products occur at 2 to 4 hpi (17). It is also evident that the second transactivator, ORF 50, is expressed later in the replication cycle than is the homologous gene in EBV; ORF 50 is an early gene, whereas the EBV homolog is an IE gene. This suggests that the initiation of the replication cycle in HVS is significantly slower compared with that in other herpesviruses.
In addition, IE genes for many herpesviruses may have several methods for their own downregulation during the replication cycle. It is believed that HSV ICP4 turns off its own synthesis and that this autoregulation correlates with the binding of the protein to a cis-acting site across the transcription initiation site of the gene (28, 39). In addition, the IE EBV Z protein regulates a range of genes including its own promoter (by autoregulation). Its promoter contains several AP-1-like upstream elements that appear to mediate activation in the presence of low levels of Z protein and repression in the presence of high levels of Z (9, 23). We propose that the ORF 57 gene product may be responsible for its own downregulation due to the presence of an intron within its own coding region. We have shown that the ORF 57 gene product specifically downregulates ORF 50a due to the presence of its intron (52), at approximately 24 hpi, which is the same time at which ORF 57 is repressed to basal levels, suggesting that the same downregulation mechanism may be utilized in the presence of high levels of ORF 57.
However, despite differences between the HVS transregulatory genes, their homologs do have similarities. For example, ORF 50 and its EBV homolog are both sequence-specific transactivators (51). We have also demonstrated that repression of gene expression mediated by ORF 57 is dependent on the presence of an intron within the target gene coding region (52). Similar results have been demonstrated with HSV-1 ICP27; repression of CAT constructs by ICP27 correlated with the presence of introns 5′ or 3′ to the target gene coding region (45). Homologs of ORF 57 are present in all classes of herpesviruses, and sequence analysis demonstrates that these genes are conserved only at the C-terminal region of the protein. This region has been shown to be essential for the inhibitory effect of ICP27 (44). We believe that the ORF 57 gene product contains a functional domain within the C terminus which is required for the repressor function of this protein. At present, further studies are being undertaken to determine if this domain is essential for the repressor activity of ORF 57. In addition, to further assess and confirm the results presented in this report about the roles of ORF 57 and the ORF 50 gene product in the viral replication cycle, viral mutants are required. Therefore, attempts are being made to produce recombinant viruses with deletions in these genes.
In summary, we have analyzed the interactions between the two known transcriptional regulatory genes encoded by HVS. We have demonstrated that the IE ORF 57 gene is expressed at basal levels early in the virus replication cycle; thereafter, it is transactivated by the ORF 50a gene product, which binds to specific sequences contained within the promoter region of ORF 57. Therefore, in conjunction with previous work demonstrating that the ORF 57 gene product downregulates ORF 50a due to the presence of its intron, we propose a feedback mechanism which modulates gene expression in HVS, whereby ORF 50a is downregulated by ORF 57, a gene which is specifically transactivates.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Yorkshire Cancer Research Campaign, the Medical Research Council, and the Wellcome Trust.
We thank Rick Randall and Ralph Grassman for providing the SB monoclonal antibody and the library of HVS-11 genomic clones, respectively.
REFERENCES
- 1.Albrecht J C, Fleckenstein B. Structural organization of the conserved gene block of herpesvirus saimiri coding for DNA polymerase, glycoprotein B, and major DNA binding protein. Virology. 1990;174:533–542. doi: 10.1016/0042-6822(90)90107-3. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittman S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beisinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bublot M, Manet E, Lequarre A S, Albrecht J C, Nicholas J, Fleckenstein B, Pastoret P P, Thiry E. Genetic relationships between bovine herpesvirus 4 and the gamma-herpesviruses Epstein-Barr and herpesvirus saimiri. Virology. 1992;190:654–665. doi: 10.1016/0042-6822(92)90903-3. [DOI] [PubMed] [Google Scholar]
- 6.Buisson M, Manet E, Trescol-Biemont M-C, Gruffat H, Durand B, Sergeant A. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J Virol. 1989;63:5276–5284. doi: 10.1128/jvi.63.12.5276-5284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison A J, Scott J E. The complete sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 8.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 253–332. [Google Scholar]
- 9.Flemington E, Speck S H. Epstein-Barr virus BZLF1 trans activator induces the promoter of a cellular cognate gene, c-fos. J Virol. 1990;64:4549–4552. doi: 10.1128/jvi.64.9.4549-4552.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furnari F B, Zacny V, Quinlivan E B, Kenney S, Pagano J S. RAZ, an Epstein-Barr virus transdominant repressor that modulates the viral reactivation mechanism. J Virol. 1994;68:1827–1836. doi: 10.1128/jvi.68.3.1827-1836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gompels U A, Craxton M A, Honess R W. Conservation of gene organization in the lymphotrophic herpesvirus saimiri. J Virol. 1988;62:757–767. doi: 10.1128/jvi.62.3.757-767.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gompels U A, Craxton M A, Honess R W. Conservation of glycoprotein H (gH) in herpesvirus: nucleotide sequence of the gH gene from herpesvirus saimiri. J Gen Virol. 1988;69:2819–2829. doi: 10.1099/0022-1317-69-11-2819. [DOI] [PubMed] [Google Scholar]
- 13.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence specific DNA binding protein. Nucleic Acids Res. 1990;18:6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and the regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay J, Ruyechan W T. Regulation of herpes simplex virus type 1 gene expression. Curr Top Microbiol Immunol. 1992;179:1–14. doi: 10.1007/978-3-642-77247-4_1. [DOI] [PubMed] [Google Scholar]
- 18.Hibbard M K, Sandri-Goldin R M. Arginine-rich regions succeeding the nuclear localization region of herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenney S, Holley-Guthrie E, Mar E-C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knust E, Schirm S, Dietrich W, Bodemer W, Kolb E, Fleckenstein B. Cloning of the herpesvirus saimiri DNA fragments representing the entire L-region of the genome. Gene. 1983;25:281–289. doi: 10.1016/0378-1119(83)90232-9. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The Zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) usage and the action of the immediate-early protein IE61 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLauchlan J, Simpson S, Clements J B. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell. 1989;59:1093–1105. doi: 10.1016/0092-8674(89)90765-4. [DOI] [PubMed] [Google Scholar]
- 26.McLauchlan J, Phelan A, Loney C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael N, Roizman B. Repression of the herpes simplex virus 1 α4 gene by its gene product takes place within a context of viral genome and is associated with all three cognate sites. Proc Natl Acad Sci USA. 1993;90:2286–2290. doi: 10.1073/pnas.90.6.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholas J, Cameron K R, Coleman H, Newman C, Honess R W. Analysis of nucleotide sequence of the rightmost 43 kbp of herpesvirus saimiri (HVS) L-DNA: general conservation of genetic organization between HVS and Epstein-Barr virus. Virology. 1992;188:296–310. doi: 10.1016/0042-6822(92)90759-i. [DOI] [PubMed] [Google Scholar]
- 31.Nicholas J, Coles L S, Newman C, Honess R W. Regulation of the herpesvirus saimiri (HVS) delayed-early 110-kilodalton promoter by HVS immediate-early gene products and a homolog of the Epstein-Barr virus R trans activator. J Virol. 1991;65:2457–2466. doi: 10.1128/jvi.65.5.2457-2466.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas J, Gompels U A, Craxton M A, Honess R W. Conservation of sequence and function between the product of the 52-kilodalton immediate-early gene of the herpesvirus saimiri and the BMLF1-encoded transcriptional effector (EB2) of the Epstein-Barr virus. J Virol. 1988;62:3250–3257. doi: 10.1128/jvi.62.9.3250-3257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholas J, Smith E P, Coles L S, Honess R W. Gene expression in cells infected with gammaherpesvirus saimiri: properties of transcripts from two immediate-early genes. Virology. 1990;179:189–200. doi: 10.1016/0042-6822(90)90288-3. [DOI] [PubMed] [Google Scholar]
- 34.Packham G, Economou A, Rooney C M, Rowe D T, Farrell P J. Structure and function of the Epstein-Barr virus BZLF1 protein. J Virol. 1990;64:2110–2116. doi: 10.1128/jvi.64.5.2110-2116.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perera L P, Kaushal S, Kinchington P R, Mosca J D, Hayward G S, Straus S E. Varicella-zoster virus open reading frame 4 encodes a transcriptional activator that is functionally distinct from that of herpes simplex virus homolog ICP27. J Virol. 1994;68:2468–2477. doi: 10.1128/jvi.68.4.2468-2477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall R E, Honess R W, O’Hare P. Proteins specified by herpesvirus saimiri: identification and properties in virus-specific polypeptides in productively infected cells. J Gen Virol. 1983;64:19–35. doi: 10.1099/0022-1317-64-1-19. [DOI] [PubMed] [Google Scholar]
- 37.Randall R E, Newman C, Honess R W. A single major immediate-early virus gene product is synthesized in cells productively infected with herpesvirus saimiri. J Gen Virol. 1984;65:1215–1219. doi: 10.1099/0022-1317-65-7-1215. [DOI] [PubMed] [Google Scholar]
- 38.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of the herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts M S, Boundy A, O’Hare P, Pizzorno M C, Ciufo D M, Hayward G D. Direct correlation between a negative autoregulatory response element at the cap site of the herpes simplex virus type 1 IE175 (α4) promoter and a specific binding site for the IE175 (ICP4) protein. J Virol. 1988;62:4307–4320. doi: 10.1128/jvi.62.11.4307-4320.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney C M, Rowe D T, Ragot T, Farrell P J. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus production cycle. J Virol. 1989;63:3109–3116. doi: 10.1128/jvi.63.7.3109-3116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russo J J, Bohenzhy R A, Chein M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequences of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 46.Schoder H C, Falke D, Weise K, Bachman M, Carmo-Fonseca M, Zaubitzer T, Muller W E G. Change of processing and nucleoplasmic transport of mRNA in HSV-1 infected cells. Virus Res. 1989;13:61–78. doi: 10.1016/0168-1702(89)90087-7. [DOI] [PubMed] [Google Scholar]
- 47.Singh M, Fraefel C, Bello L J, Lawrence W C, Schwyzer M. Identification and characterisation of BICP27, an early protein of bovine herpesvirus 1 which may stimulate mRNA 3′ processing. J Gen Virol. 1996;77:615–625. doi: 10.1099/0022-1317-77-4-615. [DOI] [PubMed] [Google Scholar]
- 48.Thompson B J, Nicholas J. Superantigen function. Nature (London) 1991;351:530. doi: 10.1038/351530a0. [DOI] [PubMed] [Google Scholar]
- 49.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitehouse A, Carr I M, Griffiths J C, Meredith D M. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J Virol. 1997;71:2550–2554. doi: 10.1128/jvi.71.3.2550-2554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitehouse A, Stevenson A J, Cooper M, Meredith D M. Identification of a cis-acting element within the herpesvirus saimiri ORF6 promoter that is responsive to the HVS.R transactivator. J Gen Virol. 1997;78:1411–1415. doi: 10.1099/0022-1317-78-6-1411. [DOI] [PubMed] [Google Scholar]
- 52.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transcriptional activator. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao Z, Maraskovsky E, Spriggs M K, Cohen J I, Armitage R J, Alderson M R. Herpesvirus saimiri open reading frame 14, a protein encoded by a T lymphotrophic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J Immunol. 1996;56:3260–3266. [PubMed] [Google Scholar]
- 55.Zhao Y, Holden V R, Smith R H, O’Callaghan D J. Regulatory function of the equine herpesvirus 1 ICP27 gene product. J Virol. 1995;69:2786–2793. doi: 10.1128/jvi.69.5.2786-2793.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]