Abstract
Salmonid herpesvirus 1 (SalHV-1) is a pathogen of the rainbow trout (Oncorhynchus mykiss). Restriction endonuclease mapping, cosmid cloning, DNA hybridization, and targeted DNA sequencing experiments showed that the genome is 174.4 kbp in size, consisting of a long unique region (UL; 133.4 kbp) linked to a short unique region (US; 25.6 kbp) which is flanked by an inverted repeat (RS; 7.7 kbp). US is present in virion DNA in either orientation, but UL is present in a single orientation. This structure is characteristic of the Varicellovirus genus of the subfamily Alphaherpesvirinae but has evidently evolved independently, since an analysis of randomly sampled DNA sequence data showed that SalHV-1 shares at least 18 genes with channel catfish virus (CCV), a fish herpesvirus whose complete sequence is known and which is unrelated to mammalian herpesviruses. The use of oligonucleotide probes demonstrated that in comparison with CCV, the conserved SalHV-1 genes are located in UL in at least five rearranged blocks. Large-scale gene rearrangements of this type are also characteristic of the three mammalian herpesvirus subfamilies. The junction between two SalHV-1 gene blocks was confirmed by sequencing a 4,245-bp region which contains the dUTPase gene, part of a putative spliced DNA polymerase gene, and one other complete gene. The implications of these findings in herpesvirus taxonomy are discussed.
Herpesviruses are a large group of complex, double-stranded DNA viruses which infect vertebrates from teleost (bony) fish to humans. They exhibit narrow host specificites, most infecting only a single species in nature, and are thus considered likely to have evolved with their hosts. Comparisons of primary amino acid sequences predicted from complete genome sequences have shown that mammalian herpesviruses are genetically very divergent but nonetheless share a set of about 40 homologous genes, thus providing compelling evidence that these viruses evolved from a single ancestral herpesvirus (reviewed in reference 7). Moreover, genetic comparisons support the division of the family into three subfamilies, Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae, as proposed previously from biological criteria (15). The order of genes is largely conserved within each subfamily, whereas members of different subfamilies are more distantly related and exhibit several large-scale genomic rearrangements (4, 9). Viral phylogenies derived from rigorous sequence comparisons generally fit well with host phylogenies deduced from the fossil record, thus supporting the view that mammalian herpesviruses have cospeciated with their hosts, and this has allowed a time frame to be assigned (13, 14). Moreover, limited sequence data also indicate that avian herpesviruses fit readily into the subfamily Alphaherpesvirinae.
Nearly all research on herpesviruses has involved mammalian (and, to a lesser extent, avian) herpesviruses, and little is known about the many herpesviruses which infect cold-blooded vertebrates. The most extensively studied member of the latter group, channel catfish virus (CCV; ictalurid herpesvirus 1), was initially classified as a herpesvirus on the basis of its virion morphology and as a member of the Alphaherpesvirinae on the basis of its biological properties (15). Analysis of the complete genome sequence (6) indicated, however, that CCV has no specific relationship with mammalian herpesviruses at the level of primary amino acid sequence, in that no counterpart of a protein which is encoded only by mammalian herpesviruses, such as a structural protein, was detected in CCV. Thus CCV cannot be accommodated by the current taxonomy. The virus does encode several enzymes which are also specified by mammalian herpesviruses, such as DNA polymerase, dUTPase, and thymidine kinase. The genes encoding these proteins, however, are ubiquitous and could quite possibly have been acquired independently by the mammalian and fish herpesvirus lineages. Moreover, the CCV enzymes are no more closely related to their counterparts in other herpesviruses than to those in other organisms.
These findings may be interpreted in two ways. First, CCV and mammalian herpesviruses arose independently and have convergently acquired similar virion morphologies. Second, they evolved from an ancestral herpesvirus but have diverged so extensively over the 400 million years since their hosts separated that little sequence evidence remains. Several lines of evidence support the latter view, but it is fair to say that the case is not yet overwhelming. The best genetic indication for divergence rests in a single highly conserved protein which is encoded by two exons in the mammalian herpesviruses and three in CCV (open reading frames [ORFs] 62, 69, and 71). This protein apparently has a distant relative in bacteriophage T4 which functions as a subunit of the terminase involved in DNA packaging, but the fact that no cellular counterpart has yet been discovered highlights it as the best candidate for a gene which may have been inherited from a common ancestor rather than acquired via independent capture events. Moreover, despite the lack of conservation of the amino acid sequences of structural proteins, structural and functional congruences have been detected. Thus, the detailed three-dimensional structure of the CCV capsid is strikingly similar to that of herpes simplex virus type 1 (3). Also, local sequence features of the putative scaffold protein involved in CCV capsid formation suggest that it may be autoproteolytically processed via a pathway that is otherwise found only in mammalian herpesviruses (8).
Evidence for a herpesvirus lineage that lies outside the current taxonomic scheme has prompted investigations of its extent. Comparisons of CCV with salmonid herpesviruses appear useful in this respect, since the fossil record indicates that the three main subgroups of euteleosts (salmoniforms, neoteleosts, and ostariophysans, the latter including catfish) diverged around 130 million years ago (1). Salmonid fish are host to several herpesviruses, the principal of which are salmonid herpesviruses 1 and 2 (SalHV-1 and SalHV-2) (reviewed in reference 19). SalHV-1 was isolated on several occasions from a rainbow trout (Oncorhynchus mykiss) hatchery in the state of Washington in association with excessive mortality in young fish (20). The virus causes disease when injected into young rainbow trout maintained at 6 to 9°C but not in other salmonid species. SalHV-2 was isolated from Oncorhynchus masou, a landlocked Japanese form of Pacific salmon (11). It is serologically distinct from and has a wider host range than SalHV-1, causing virulent disease in the young of several Oncorhynchus species, including the rainbow trout. It also exhibits a higher temperature optimum for growth in cell culture than SalHV-1.
Partial sequence data for two genes have previously indicated that SalHV-2 is related to CCV (2). In this report, I describe the genome structure and gene arrangement of SalHV-1 and show that this virus is evolutionarily related to SalHV-2 and CCV. The data indicate that the processes which have resulted in the generation of certain genome structures and large-scale gene rearrangements during mammalian herpesvirus evolution have parallels in fish herpesvirus evolution. They also imply that fish herpesviruses occupy a distinct evolutionary space of an size equivalent to that occupied by mammalian herpesviruses and urge an accommodation in the herpesvirus taxonomy.
MATERIALS AND METHODS
Growth of virus.
A rainbow trout gonad cell line (RTG-2; ATCC CCL 55) was grown in minimal essential medium containing Earle’s salts and Glutamax I (Life Technologies) supplemented with 10% fetal calf serum, nonessential amino acids, penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were subcultured at a ratio of 1:5 in trays, flasks, or roller bottles, becoming confluent in 2 to 3 weeks at 22 to 24°C.
Dilutions (100-μl aliquots) of a stock of SalHV-1 (ATCC-VR-868) with a stated titer of 105 PFU/ml (actual titer, about 103 PFU/ml) were absorbed to confluent monolayers of RTG-2 cells in 24-well trays for 3 h at 10°C, overlaid with 1 ml of medium, and incubated at 10°C for 1 week. Monolayers containing single plaques were harvested by scraping the cells into the medium, freeze-thawing, and sonicating. Dilutions (200-μl aliquots) of one of the single plaque harvests were adsorbed to monolayers in six-well trays, overlaid with 2 ml of medium, and incubated at 10°C for 1 week. The monolayers were washed and overlaid with fresh medium, and a single plaque was picked from a well containing a few well-separated plaques, placed into 400 μl of medium in a vial, freeze-thawed, and sonicated. This stock was grown progressively in a six-well tray, in a single 175-cm2 flask, and then in 24 175-cm2 flasks. Infected cells were pelleted by centrifugation, resuspended in 8 ml of medium, freeze-thawed, and sonicated to give a cell-associated virus stock (6.5 × 107 PFU/ml). The infected cell medium was centrifuged at a relative centrifugal force (RCF) (at an average radius [ravg]) of 14,750 (i.e., 12,000 rpm in a Sorvall GSA rotor) for 2 h at 4°C, and the pellet was resuspended in 1.5 ml of medium to give a cell-released virus stock (1.5 × 108 PFU/ml). Additional stocks of virus were prepared using inocula from the cell-released stock.
Preparation of virion DNA.
RTG-2 cells in batches of eight roller bottles were infected with SalHV-1 at a multiplicity of infection of approximately 0.01 PFU/cell. Cytopathic effect was complete after incubation at 10 to 12°C for 2 to 3 weeks. The infected cells were gently shaken into the medium and pelleted by centrifugation for 10 min at an RCF (at ravg) of 2,560 (i.e., 5,000 rpm in a Sorvall GSA rotor). Cell-released virus was pelleted from the supernatant by centrifugation at an RCF (at ravg) of 14,750 (i.e., 12,000 rpm in a Sorvall GSA rotor) for 2 h at 4°C and resuspended in a small volume of medium. Virions were purified by centrifugation on 5 to 15% (wt/vol) Ficoll gradients (18) and lysed by incubation for 3 h at 37°C in TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) containing 0.5% (wt/vol) sodium dodecyl sulfate and 0.5 mg of protease per ml. The DNA was extracted twice with TE-equilibrated phenol and once with 24:1 (vol/vol) chloroform-isoamyl alcohol, ethanol precipitated, resuspended in TE, and stored at −20°C. A roller bottle typically yielded 1 to 2 μg of DNA.
Construction of cosmids and plasmids.
A SalHV-1 cosmid library was constructed by using a modified form of Stratagene’s Supercos 1 vector (5), employing methods recommended by Stratagene. SalHV-1 DNA was digested partially with BamHI and ligated into the vector which had been digested with XbaI, dephosphorylated, and digested with BamHI. The DNA was packaged into bacteriophage λ particles and introduced into an appropriate Escherichia coli host, using Stratagene’s Gigapack II system. Recombinant cosmid DNA was prepared from ampicillin-resistant colonies by alkaline lysis of 1-ml cultures, and aliquots of the DNA (usually about 5% of the yield) were analyzed by restriction endonuclease digestion and agarose gel electrophoresis.
SalHV-1 BamHI P was isolated from an appropriate cosmid (cos2) and inserted into the BamHI site of pTZ19U (U.S. Biochemical), using standard procedures.
Isotopic labeling of DNA.
Oligonucleotides were labeled at their 5′ ends by treatment with bacteriophage T4 polynucleotide kinase in the presence of [γ-32P]ATP by using standard procedures and were purified in TE on small Sephadex G-50 columns. 32P-labeled probes were obtained from double-stranded DNA by using the Nonaprimer system (Appligene) and denatured prior to hybridization by heating at 100°C for 2 min.
DNA hybridization.
Restriction endonuclease fragments separated by agarose gel electrophoresis were denatured and passively transferred to a Hybond-N membrane (Amersham), using standard procedures. DNA was fixed to the membranes by irradiation in a UV Stratalinker 1800 (Stratagene). DNA hybridization was carried out for 3 to 16 h, using Rapid-hyb buffer (Amersham) in bottles rotating in a Hybaid Mark II hybridization oven. Standard hybridization temperatures were 65°C for double-stranded DNA probes and 42°C for oligonucleotide probes, but in some experiments temperatures were increased by up to 10°C for the former and 3°C for the latter. The membranes were then rinsed in washing buffer (20 mM Tris-HCl [pH 7.5], 0.3 M NaCl, 0.03 M trisodium citrate, 0.1% sodium dodecyl sulfate) heated to the temperature of hybridization, washed twice for 15 min in washing buffer at the temperature of hybridization, and finally rinsed in 10% washing buffer at room temperature. The membranes were air dried, reassembled, and autoradiographed.
Membranes were routinely reprobed with labeled SalHV-1 DNA in order to locate all SalHV-1 DNA fragments. The previous probe was first removed by heating in 20 mM NaOH at 75°C for 10 min and rinsing extensively in water.
DNA sequencing.
The sequence of SalHV-1 BamHI P was determined by cloning random DNA fragments generated by sonication into M13mp19 and sequencing them by the dideoxynucleotide chain termination technology (10, 16). Autoradiographs were read with a Summagraphics Digitizer, and the database was compiled by using Staden’s sequence analysis program (17), having first eliminated M13 clones containing pTZ19U sequences by reading short sections of each clone. The sequence was edited by reference to autoradiographs and analyzed with programs from the Wisconsin package, version 9.0 (Genetics Computer Group, Madison, Wis.). The 4,245-bp sequence was determined an average of 8.2 times per nucleotide.
Random sequences from the SalHV-1 genome were generated by sonication and cloned into M13mp19 as described above. Sequences at the ends of SalHV-1 BamHI T and U were determined by cloning the fragments from appropriate cosmids (cos17 and cos35) into the BamHI site of M13mp19. Random SalHV-1 sequences and sequences at the ends of BamHI T and U were determined only on one strand and cannot be considered sufficiently accurate for deposition in the data library. They will eventually form part of the complete SalHV-1 genome sequence.
Nucleotide sequence accession number.
The sequence reported has been deposited with the GenBank data library under accession no. AF023673.
RESULTS
Size and structure of the SalHV-1 genome.
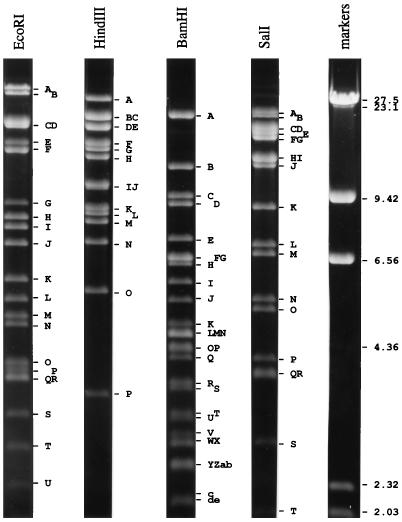
Figure 1 shows restriction endonuclease profiles of SalHV-1 DNA for four enzymes. The enzyme of primary interest in this study, BamHI, produced 48 fragments ranging in size from 0.33 to 19.5 kbp (Table 1), as determined from this gel and others of higher agarose concentration (not shown). The total size of the genome calculated from the sum of fragment sizes was 174.4 kbp. The BamHI map for the SalHV-1 genome shown in Fig. 2 was obtained by analyzing the restriction profiles of cosmid clones and from associated experiments as described below. The map was derived from a large amount of data, of which it is possible to present only a small proportion here.
FIG. 1.
Restriction fragments of SalHV-1 DNA visualized by short-wavelength UV irradiation on a 0.5% agarose gel stained with ethidium bromide. Sizes are indicated in kilobase pairs.
TABLE 1.
Sizes of SalHV-1 BamHI fragments
| Fragment | Size (kbp) | Fragment | Size (kbp) | Fragment | Size (kbp) | ||
|---|---|---|---|---|---|---|---|
| A | 19.5 | Q | 4.0 | g | 1.8 | ||
| B | 11.5 | R | 3.7 | h | 1.6 | ||
| C | 9.4 | S | 3.6 | i | 1.5 | ||
| D | 9.0 | T | 3.3 | j | 1.3 | ||
| E | 7.2 | U | 3.2 | k | 1.2 | ||
| F | 6.6 | V | 3.0 | l | 1.1 | ||
| G | 6.6 | W | 2.9 | m | 0.96 | ||
| H | 6.3 | X | 2.9 | n | 0.79 | ||
| I | 5.8 | Y | 2.6 | o | 0.76 | ||
| J | 5.3 | Z | 2.6 | p | 0.68 | ||
| K | 4.8 | a | 2.6 | q | 0.68 | ||
| L | 4.6 | b | 2.6 | r | 0.59 | ||
| M | 4.6 | c | 2.1 | s | 0.59 | ||
| N | 4.6 | d | 2.1 | t | 0.51 | ||
| O | 4.3 | e | 2.1 | u | 0.36 | ||
| P | 4.3 | f | 1.9 | v | 0.33 |
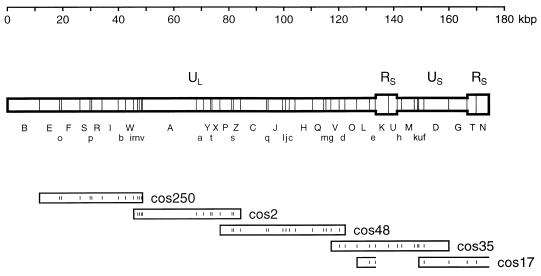
FIG. 2.
Genome structure of SalHV-1. One of the two genome isomers is shown and is defined as the prototype. BamHI sites are marked, and fragment nomenclature is shown below in two ranks (A to Z and a to v). The inverted repeat (RS) is shown in a wider format than nonreiterated regions (UL and US). The locations of five cosmids are shown as rectangles below the fragment nomenclature, and BamHI sites are indicated. cos17 arose from a genome molecule in which US was inverted and thus appears to proceed from the right end of UL, through RS and into the right end of US, as indicated by the open ends of the rectangles.
Digestion of each cosmid produced several BamHI fragments corresponding in electrophoretic mobility to fragments produced by BamHI digestion of SalHV-1 DNA, in addition to the 6.7-kbp vector fragment which was common to all cosmids. The 156 cosmids used in the analysis exhibited 72 different profile types. Since each cosmid insert consisted of a contiguous region of about 40 kbp of the SalHV-1 genome containing several BamHI fragments, it was possible to establish a linkage map along the genome by identifying fragments shared by different profile types. Local fragment orders within the regions containing fragments G and T, fragments E, F, and o, and fragments i, n, and r were ambiguous at this stage. Also, two fragments (B and N) did not occur in any cosmid. These fragments presumably originated from the genome termini.
To finalize the map, five cosmids (cos250, cos2, cos48, cos35, and cos17) which contain the total genomic sequence except for BamHI B and N were used. The locations of the inserts in these cosmids are shown in Fig. 2. The order of BamHI i, n, and r (as i-r-n) was determined by sizing partial digestion products of cos250 and cos2 that had been labeled at AscI sites flanking the inserts and partially digested with BamHI (data not shown). Fragments at the left end of the genome were ordered by DNA hybridization experiments in which BamHI B and HindIII F (from virion DNA) and BamHI E, F, and o (from cos250) were radiolabeled and hybridized to digests of SalHV-1 DNA. The results supported the order B-E-o-F shown in Fig. 2. The order of BamHI G and T was determined by mapping a single EcoRI site in the cos17 insert (data not shown). The location of this site at the center of the insert and within BamHI G established the order as G-T. The location of T adjacent to K was also supported by the hybridization data in Fig. 3 which are described below.
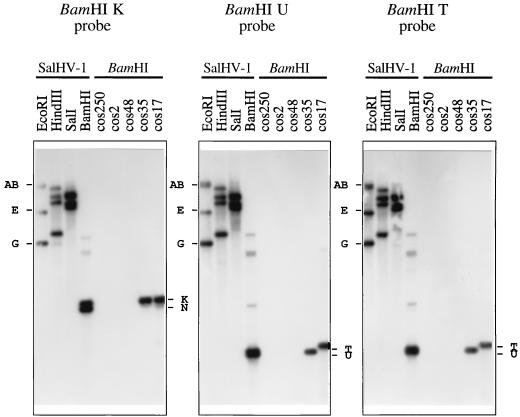
FIG. 3.
Examples of hybridization data for an inverting region (US) flanked by an inverted repeat (RS) in the SalHV-1 genome. Restriction fragments of SalHV-1 DNA or cosmids were transferred from a 0.6% agarose gel and probed with radiolabeled BamHI K, U, or T at 75°C. Hybridizing BamHI fragments are indicated to the right of each panel, and EcoRI fragments are indicated to the left.
Several lines of evidence indicated that the SalHV-1 genome comprises a long unique region (UL) in a single orientation and a short unique region (US) which is present in either orientation and is flanked by an inverted repeat (RS), as shown in Fig. 2. First, of the 26 cosmid profile types which extended to the right of BamHI K, about half (i.e., 14) proceeded through BamHI U (e.g., cos35) while the remainder proceeded through BamHI T (e.g., cos17). The latter group could be explained as originating from a proportion of virion DNA molecules containing the BamHI U-h-M-k-u-f-D-G-T region in the inverse orientation.
Second, when each of the five cosmids was radiolabeled and hybridized individually to BamHI digests of the five cosmids and SalHV-1 DNA, in addition to fragments known to be present in the cosmids, cos35 and cos17 hybridized to N, cos35 hybridized to T, and cos17 hybridized to U (data not shown). To identify the cross-hybridizing regions, purified BamHI fragments were hybridized to BamHI digests of the five cosmids and SalHV-1 DNA. The results (Fig. 3) show that K hybridized to N, U hybridized to T, and T hybridized to U, thus demonstrating that K shares sequences with N and that T shares sequences with U. To examine the sequences shared by T and U in more detail, sequences at the ends of each fragment were sequenced on one strand (data not shown). One end of T was identical to one end of U, and the other end of T was different from the corresponding end of U for the first 113 bp (93 bp in U) and was the same thereafter. These results are consistent with RS extending through the great majority of T (and U) into K (and N), as shown in Fig. 2. The strength of the hybridization signal between N and K indicates that the major part of the former fragment is repeated in the latter. The data do not prove, however, that RS extends to the genome terminus, although this assumption has been made in Fig. 2. The sequence data from the ends of T and U also showed that part of RS comprises short repetitive elements.
Third, certain restriction endonucleases produced submolar fragments from SalHV-1 DNA. In principle, enzymes that cleave in US but not in RS would produce four submolar fragments: for each orientation of US, one extending from within US to the right end of the genome and one extending from within US leftward through RS and into UL. If US is present in either orientation with equal probability, these fragments would be present at half the relative abundance of other fragments (i.e., half molar). This appears to be a feature of three of the digests of SalHV-1 DNA shown in Fig. 1. Indeed, EcoRI A, B, E, and G resolved from other fragments and were shown by densitometric scanning to be half molar. As expected, these fragments contain RS, since they hybridized to BamHI K, T, and U (Fig. 3). The profiles in Fig. 1 and the hybridization results in Fig. 3 indicated that A, C, E, and J and A, B, D, and G are corresponding submolar fragments for HindIII and SalI, respectively. Taking into account the presence of submolar fragments, the genome sizes estimated from the EcoRI, HindIII, and SalI profiles (172, 170, and 174 kbp, respectively) correspond well with that estimated from the BamHI profile, which lacks submolar fragments owing to the presence of a BamHI site in RS.
In summary, these studies indicate that the SalHV-1 genome is 174.4 kbp in size, consisting of an inverting region (US; 25.6 kbp) flanked by an inverted repeat (RS; 7.7 kbp, assuming that it is continuous and extends to the genome terminus) and a noninverting region (UL; 133.4 kbp) which is not flanked by a detectable repeat. This structure is shown in Fig. 2.
Relationship between SalHV-1 and other herpesviruses.
To assess the relationship between SalHV-1 and other herpesviruses, 298 M13 clones derived randomly from the genome were sequenced. In total, they represented 34% of the genome. Each DNA sequence was conceptually translated in all six reading frames by using Pepdata, producing a concatenated amino acid sequence. The amino acid sequences were then compared with protein sequence databases by using Fasta. Databases used included collections of protein sequences from mammalian herpesviruses, from CCV, and from the Swissprot database.
Significant similarities were found between SalHV-1 and CCV. Similarities were also found at a much lower level with a few mammalian herpesvirus enzymes which have counterparts in CCV (such as DNA polymerase and dUTPase), but in these cases the relationship to the cognate CCV protein was closer. It was clear from this initial analysis that of those herpesviruses for which complete sequence data are available, SalHV-1 is most closely related to CCV. The data were also analyzed by comparing individual CCV proteins with the collection of SalHV-1 sequences using Fasta. Scores above 80 were scrutinized as possibly significant. Particular cognizance was given to CCV proteins, such as DNA polymerase, which matched more that one SalHV-1 sequence in different regions. A lower score applied to one protein (dUTPase) but was considered significant since the conserved residues were limited to recognized functional motifs in this enzyme. As a result of this analysis, 20 ORFs representing 18 CCV genes were judged to have convincing counterparts in SalHV-1. Examples of alignments are shown in Fig. 4. The alignments for ORFs 37 (Fig. 4a) and 54 (Fig. 4b) were among the strongest and weakest obtained, respectively.
FIG. 4.
Amino acid sequence alignments of random SalHV-1 DNA sequences conceptually translated in appropriate reading frames with the CCV ORF 37 protein (residues 534 to 615) (a) and the CCV ORF 54 protein (residues 306 to 399) (b). Identical residues are indicated in the “con” line.
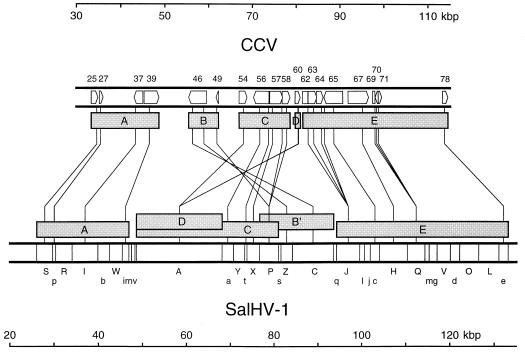
The conserved CCV ORFs are 25 (DNA helicase), 27 (capsid protein), 37, 39 (major capsid protein), 46 (putative membrane glycoprotein), 49 (dUTPase), 54, 56, 57 (DNA polymerase), 58, 60, 62 (first exon of the putative terminase), 63, 64, 65, 67 (tegument protein), 69 (second exon of the putative terminase), 70, 71 (third exon of the putative terminase), and 78 (putative zinc-binding protein). The analysis showed that each of the three CCV ORFs encoding the putative terminase has a counterpart in SalHV-1, and identification of potential splice sites in the SalHV-1 DNA sequences indicated that they are present as three exons.
The SalHV-1 DNA sequences responsible for the most convincing matches in this group of genes were then read again to remove any errors, and 30-mer oligonucleotides from sequences encoding the most conserved amino acid residues were synthesized. Of the 10 possible conserved amino acid residues in each case, 5 to 9 were identical between CCV and SalHV-1. A total of 25 oligonucleotides were used, since in some instances it was possible to derive two oligonucleotides from different regions of the same gene. The sequences of the oligonucleotides and the regions of the CCV genes corresponding to them are listed in Table 2. The oligonucleotides were 32P labeled at their 5′ termini and hybridized to BamHI digests of SalHV-1 and cosmid DNAs. The fragments to which the oligonucleotides hybridized are listed in Table 2, and examples of the data are shown in Fig. 5.
TABLE 2.
Summary of oligonucleotide hybridization data used to locate SalHV-1 genes
| Results of SalHV-1 hybridization
|
Corresponding CCV gene
|
|||
|---|---|---|---|---|
| Oligonucleotide probea | Hybridizing SalHV-1 BamHI fragment | Gene | Size of ORF (bp) | Location in ORFb (bp) |
| GCAATAAACGCCTACTGTGCCCAAGGAGCA | S | 25 | 1,494 | 1,240 |
| TATACTATGATCAACACTGGTGCAGAGAGC | p | 27 | 864 | 268 |
| ACACGACGCCGCCATGCGTTACAATACACG | I | 37 | 2,010 | 1,663 |
| AAGAGGTTGTTTATCACCGTGGGATCTGTA | i | 39 | 3,369 | 1159 |
| CTGACCAACCAACTGACTCAAGCGGCTCTG | C | 46 | 4,065 | 3,190 |
| ATTGTCAGGGGGACCACCTACAGTTGTATA | Z | 46 | 4,065 | 604 |
| CACTGGGGTGATTGATTCGGACTATAAAGG | P | 49 | 564 | 369 |
| GGCTTCAGGGGAGCCATCTTCCGGAGATTG | A | 54 | 1,821 | 1,003 |
| CGAGGGGACTTTAAGATACAAGATATGGCA | a | 56 | 3,537 | 2,131 |
| CCGAAAACTGGCGAATTGTCATTTAGAGGT | t | 56 | 3,537 | 106 |
| AAAAGAGAGGCTGCATTTGACATAGAGACG | X | 57 | 2,955 | 604 |
| TGTCATGTTGGTAAAAAACAGGTATGTTGA | P | 57 | 2,955 | 2,782 |
| AAATTTCTCGCTCACGTTTTGGTTGACATG | P | 58 | 1,797 | 988 |
| TGTGTGGTGATTGAACTCAAGACATACAAG | A | 60 | 1,179 | 655 |
| ATACACCACTCAGACTTGGCTCACTTGGCT | A | 60 | 1,179 | 717 |
| AAGGTCCACTTTCTGTCTTCTAGTCCACTG | J | 62 | 1,203 | 1,174 |
| GCTATGCCGGATAAGTTTTCAGCACGCTAC | J | 63 | 1,986 | 1,228 |
| GCCCACCGGATCATCTGGTTTAAACACCTG | J | 64 | 1,542 | 1,123 |
| TTGGACCGAAACCATTACGAGCGGCTAGTG | J | 65 | 4,302 | 4,192 |
| TGTGATCCTGTGTATGTGGGTCCGCTCACA | c | 65 | 4,302 | 2,068 |
| GTGATTGATACAAGATTACTCTTTGGTTTA | H | 67 | 4,668 | 3,235 |
| TCGTCCCTGGGCGTTTGCACCTTTGCGATA | Q | 69 | 558 | 508 |
| GTATATGCTGCAGTATGTGCCGTGGAGATG | Q | 70 | 681 | 337 |
| ATACTTGCCCTCGAGGAAATGCCGATCACC | Q | 71 | 795 | 1 |
| TGCTGTGACATAATCCTATGTGACATTTGC | e | 78 | 1,200 | 397 |
Hybridization data are given for 30-bp regions of randomly derived SalHV-1 sequences whose predicted translation products are related to CCV proteins. The list proceeds rightward along the CCV genome. Sequences correspond to the noncoding (upper) strand.
Nucleotide in the CCV ORF which corresponds to the first residue of the oligonucleotide.
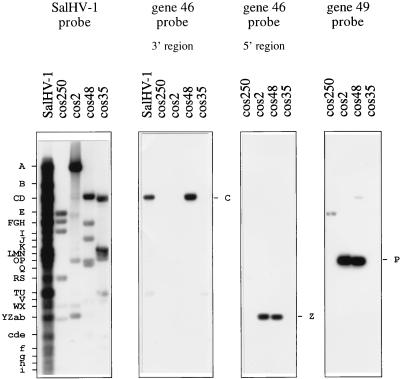
FIG. 5.
Examples of oligonucleotides hybridizing to SalHV-1 DNA. BamHI fragments of SalHV-1 or cosmids were transferred from a 0.8% agarose gel and probed with radiolabeled SalHV-1 DNA at 65°C or oligonucleotides at 42°C. Fragments to which oligonucleotides hybridized are shown to the right of each panel, and SalHV-1 BamHI fragments are shown on the left.
Knowledge of the SalHV-1 BamHI map enabled the genes under analysis to be located in the SalHV-1 genome. It was also possible to determine the orientation of most SalHV-1 genes for which two oligonucleotides were used. The organization of conserved genes in the two genomes is summarized in Fig. 6. It is clear that the conserved genes are not present in the same order in the two genomes. On the contrary, the gene order is related by rearrangement in UL of at least five sequence blocks, one of which (B) is inverted. The smallest block (D) was identified by a single gene (60) and is located at or near the left end of block C. The level of similarity detected for gene 60 in screening random sequences was convincing (data not shown), and the possibility of nonspecific hybridization of the relevant oligonucleotide was ruled out by using a different probe from the same gene (Table 2). Within each block, the gene spacing is approximately equivalent in the two genomes, except for ORFs 37 and 39, which are further apart in SalHV-1.
FIG. 6.
Scale representation of relative gene order in SalHV-1 and CCV. The locations and orientations of the 18 genes in the 30- to 115-kbp region of the 134-kbp CCV genome are shown at the top; ORFs 62, 69, and 71 are exons of a single gene. BamHI fragments in the 20- to 135-kbp region of the SalHV-1 genome are shown at the bottom. Regions in the CCV genome corresponding to SalHV-1 oligonucleotide probes are connected by lines to the centers of the SalHV-1 fragments to which they hybridized. Five gene blocks (A to E) are shown as shaded rectangles below the CCV genome, and corresponding blocks are shown above the SalHV-1 genome (B′ indicating an inversion).
Although SalHV-1 is clearly related to CCV, three random SalHV-1 sequences were also found to be related to SalHV-2 in two genes for which partial data are available (ORF 46 [2] and ORF 62 [EMBL data library entry OMHVORF62]). The levels of protein sequence identity between SalHV-1 and SalHV-2 are 79 and 77%, respectively, for the regions of ORF 46 shown in Fig. 7a and b. The levels of DNA sequence identity are 67 and 61%, respectively. The levels of similarity for the region of ORF 62 shown in Fig. 7c, which proceeds to the splice donor site at the 3′ end, were greater: 98% at the amino acid sequence level and 85% at the DNA sequence level. CCV is more distantly related to SalHV-1 and SalHV-2 in these regions of ORFs 46 and 62.
FIG. 7.
Amino acid sequence alignments of conceptual translation products of random SalHV-1 DNA sequences with their counterparts in the SalHV-2 genome and the CCV gene 46 protein at residues 180 to 246 (a) and residues 759 to 837 (b) and the CCV ORF 62 protein at residues 345 to 401 (c). Residues conserved between SalHV-1 and SalHV-2 are indicated in the “con” line; those conserved between both viruses and CCV are indicated in the “CON” line.
BamHI P sequence.
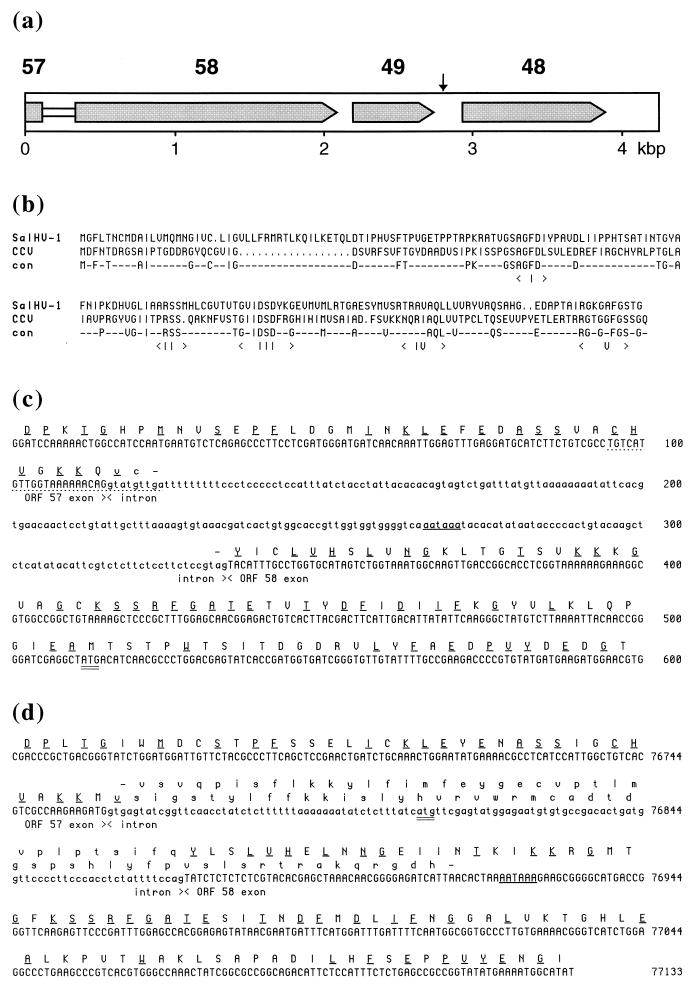
To confirm the presence of rearranged gene blocks, BamHI P was sequenced. Oligonucleotide hybridization experiments showed that this fragment contains portions of ORFs 49 (dUTPase), 57 (DNA polymerase), and 58. The sequence is 4,245 bp long, and the deduced gene arrangement is shown in Fig. 8a.
FIG. 8.
(a) Summary of gene order in SalHV-1 BamHI P displayed in the same orientation as the genome layout depicted in Fig. 2. Predicted protein coding regions are shaded, and the proposed intron linking ORFs 57 and 58 is shown as a white rectangle. An AATAAA element potentially involved in polyadenylation is shown as a vertical arrow. (b) Alignment of the putative amino acid sequences of SalHV-1 and CCV dUTPases. Conserved residues are shown in the “con” line. Five recognized dUTPase motifs (I to V) are indicated (12). The CCV protein is shown as commencing at the second ATG codon in the relevant reading frame. (c and d) Locations of potential splice sites linking ORFs 57 and 58 in SalHV-1 (c) and CCV (d). The first 600 bp of SalHV-1 BamHI P and the corresponding region in the CCV genome are shown, proceeding from the 3′ end of ORF 57 through the 5′ end of ORF 58. Potential exons and introns are marked. Translated sequences are bracketed by stop codons which define the 3′ and 5′ limits of ORFs 57 and 58, respectively; the two ORFs overlap in CCV. The first ATG codon in ORF 58 is doubly underlined in each sequence. Conserved amino acid residues are singly underlined. Nucleic acid and amino acid residues outside the putative exons are shown in lowercase. AATAAA elements that could signal polyadenylation of transcripts from ORF 57 are underlined. The sequence corresponding to the oligonucleotide used to locate the 3′ end of SalHV-1 ORF 57 is underlined by dots.
An alignment of the dUTPases of CCV and SalHV-1 is shown in Fig. 8b to convey the impression that greatest conservation is confined to recognized motifs of dUTPases and that, although the two proteins are related, the relationship is not close. The dUTPases of mammalian herpesviruses are unusual in that they appear to have arisen by gene duplication, divergence, and fusion to give a protein which is about twice the size of dUTPases from other organisms and contains the conserved motifs in a different order (12). The SalHV-1 dUTPase, like that of CCV, has not followed this evolutionary route. It is similar in size and motif order to dUTPases of nonherpesvirus genomes.
Examination of the region between the 3′ end of ORF 57 and the 5′ end of ORF 58 in the sequences of BamHI P and the CCV genome provided evidence for splicing (Fig. 8c and d). The 3′ extremity of CCV ORF 57 extends 49 codons beyond that of SalHV-1, and amino acid residues encoded upstream from the first ATG codon in SalHV-1 ORF 58 (Fig. 8c) show significant similarity to residues downstream from the first ATG codon in CCV ORF 58 (Fig. 8d). In addition, in both viruses potential splice donor and acceptor sites are present at identical locations near the 3′ end of ORF 57 (CAG:GTATGT in SalHV-1 and ATG:GTGAGT in CCV; splice donor consensus MAGGTAAGT, where bold residues are absolutely conserved and M is A or C) and the 5′ end of ORF 58 (TCTCCTTCTCCGTAG:T in SalHV-1 and ACCTCTATTTTCCAG:T in CCV; splice acceptor consensus Y11NYAGG, where Y is T or C), in positions that would facilitate fusion of the translation products of ORFs 57 and 58 with inclusion of residues that are conserved and exclusion of those that are not. The possibility that ORF 57 is expressed without splicing to ORF 58 is not ruled out, since each sequence has an AATAAA element that could be involved in polyadenylation of an ORF 57 transcript: in the intron in SalHV-1 and near the 5′ end of ORF 58 in CCV. The presence of an AATAAA element downstream from ORF 49 suggests that transcripts specified by ORFs 57/58 and 49 are 3′ coterminal (Fig. 8a).
The protein encoded by SalHV-1 ORF 48 was most similar to the CCV ORF 48 protein when screened by using Fasta, but amino acid sequence similarity is weak. The two proteins have similar hydrophobicity profiles (data not shown). The coding situation at the right end of BamHI P is unclear. A counterpart of CCV ORF 47, which encodes a putative subtilisin-like protease, was not detected in this region.
DISCUSSION
The mapping data described in this report showed that the 174.4-kbp SalHV-1 genome consists of an inverting region (US; 25.6 kbp) flanked by an inverted repeat (RS; 7.7 kbp) and a noninverting region (UL; 133.4 kbp) which is not flanked by a detectable repeat. This arrangement is characteristic of mammalian alphaherpesviruses in the Varicellovirus genus, such as pseudorabies virus, equine herpesvirus 1, bovine herpesvirus 1, and varicella-zoster virus. Given the great divergence between SalHV-1 and mammalian herpesviruses implied by the absence of a specific genetic relationship, it appears that this genome structure has evolved independently in the mammalian and fish herpesvirus lineages. Similarly, the genome structure of CCV, which consists of a UL region flanked by a substantial direct repeat, is the same as that of betaherpesviruses in the Roseolovirus genus (human herpesviruses 6 and 7) and one of the gammaherpesviruses ostensibly in the Rhadinovirus genus (equine herpesvirus 2) and again has apparently evolved independently.
Analysis of randomly sampled DNA sequence data indicated that SalHV-2 is the closest relative of SalHV-1. This conclusion is based on limited regions of two genes, and information on the overall level of sequence similarity and on gene order is not available. SalHV-1 is also clearly related to CCV in at least 18 genes at the level of amino acid sequence. Nevertheless, the distance between SalHV-1 and CCV is rather substantial, qualitatively of the same order as that between members of different mammalian herpesvirus subfamilies. Moreover, the gene orders in the two viruses are different, being related by rearrangement of at least five sequence blocks in UL. Large-scale gene rearrangement in UL is characteristic of the mammalian herpesvirus subfamilies and again appears to have occurred independently in fish herpesviruses.
Detailed sequence analysis confirmed that SalHV-1 BamHI P contains the junction between two gene blocks. It also resulted in the speculation that the DNA polymerases of SalHV-1 and CCV may be encoded by spliced mRNAs. This interpretation is not pivotal to this report, but subsequent confirmation from transcript mapping data would lead to the prediction that CCV DNA polymerase is 173 kDa in size, somewhat larger than mammalian herpesvirus DNA polymerases (approximately 140 kDa) which are not expressed via splicing. The role of the putative ORF 58 domain is a matter for more extreme speculation, given that recognizable CCV DNA polymerase motifs are all located in the ORF 57 domain.
Fish herpesviruses form a distinct and largely unrecognized part of the herpesvirus family which we now see as possibly similar in evolutionary breadth to herpesviruses of mammals and birds. The striking parallels observed in genome structure and gene rearrangement indicate that similar evolutionary mechanisms have operated independently in the two lineages. Thus, it is likely that it will be feasible to classify fish herpesviruses at the level of subfamily (e.g., as probably represented by SalHV-1 and CCV) and genus (e.g., as perhaps represented by SalHV-1 and SalHV-2), as has been undertaken for mammalian herpesviruses. Set in this context, it is apparent that the present position of the family name (Herpesviridae) is inappropriate by the criterion of genetic relatedness, since it encompasses only herpesviruses of mammals and birds. This name should be raised by one taxonomic level (to superfamily or order), and novel names should be developed to represent the mammalian/avian and fish herpesvirus lineages at what is currently the family level. It is possible that further accommodations of a more or less radical nature will eventually be necessary to fit herpesviruses of amphibians, reptiles, and invertebrates into this scheme as data become available.
ACKNOWLEDGMENTS
I am grateful to Duncan McGeoch for criticizing the manuscript and Ross Reid for synthesizing oligonucleotides.
This work was supported in part by EC FAIR contract CT95-0850.
REFERENCES
- 1.Benton M J. Vertebrate palaeontology. London, England: Harper Collins Academic; 1990. pp. 123–144. [Google Scholar]
- 2.Bernard J, Mercier A. Sequence of two Eco RI fragments from salmonis herpesvirus 2 and comparison with ictalurid herpesvirus 1. Arch Virol. 1993;132:437–442. doi: 10.1007/BF01309552. [DOI] [PubMed] [Google Scholar]
- 3.Booy F P, Trus B L, Davison A J, Steven A C. The capsid architecture of channel catfish virus, an evolutionary distant herpesvirus, is largely conserved in the absence of discernible sequence homology with herpes simplex virus. Virology. 1996;215:134–141. doi: 10.1006/viro.1996.0016. [DOI] [PubMed] [Google Scholar]
- 4.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, III, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 6.Davison A J. Channel catfish virus: a new type of herpesvirus. Virology. 1992;186:9–14. doi: 10.1016/0042-6822(92)90056-u. [DOI] [PubMed] [Google Scholar]
- 7.Davison A J. Herpesvirus genes. Rev Med Virol. 1993;3:237–244. [Google Scholar]
- 8.Davison A J, Davisol M D. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology. 1995;206:1035–1043. doi: 10.1006/viro.1995.1026. [DOI] [PubMed] [Google Scholar]
- 9.Davison A J, Taylor P. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J Gen Virol. 1987;68:1067–1079. doi: 10.1099/0022-1317-68-4-1067. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J, Telford E A R. Large scale DNA sequencing by manual methods. In: Dale J W, Sanders P G, editors. Methods in gene technology. Vol. 2. London, England: JAI Press; 1994. pp. 151–175. [Google Scholar]
- 11.Kimura T, Yoshimizu M, Tanaka M, Sannohe H. Studies on a new virus (OMV) from Oncorhynchus masou. I. Characteristics and pathogenicity. Fish Pathol. 1981;15:143–147. [Google Scholar]
- 12.McGeoch D J. Protein sequence comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGeoch D J, Cook S. Molecular phylogeny of the Alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 14.McGeoch D J, Cook S, Dolan A, Jamieson F E, Telford E A R. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 15.Roizman B. The family Herpesviridae: general description, taxonomy, and classification. In: Roizman B, editor. The herpesviruses. Vol. 1. New York, N.Y: Plenum Press; 1982. pp. 1–23. [Google Scholar]
- 16.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staden R. Computer handling of DNA sequencing projects. In: Bishop M J, Rawlings C J, editors. Nucleic acid and protein sequence analysis: a practical approach. Oxford, England: IRL Press; 1987. pp. 173–217. [Google Scholar]
- 18.Szilágyi J F, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 19.Wolf K. Biology and properties of fish and reptilian herpesviruses. In: Roizman B, editor. The herpesviruses. Vol. 2. New York, N.Y: Plenum Press; 1983. pp. 319–366. [Google Scholar]
- 20.Wolf K, Darlington R W, Taylor W G, Quimby M C, Nagabayashi T. Herpesvirus salmonis: characterization of a new pathogen of rainbow trout. J Virol. 1978;27:659–666. doi: 10.1128/jvi.27.3.659-666.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]