Abstract
The nuclear medicine field has seen a rapid expansion of academic and commercial interest in developing artificial intelligence (AI) algorithms. Users and developers can avoid some of the pitfalls of AI by recognizing and following best practices in AI algorithm development. In this article, recommendations on technical best practices for developing AI algorithms in nuclear medicine are provided, beginning with general recommendations and then continuing with descriptions of how one might practice these principles for specific topics within nuclear medicine. This report was produced by the AI Task Force of the Society of Nuclear Medicine and Molecular Imaging.
Keywords: computer/PACS, research methods, statistics, algorithm, artificial intelligence, best practices
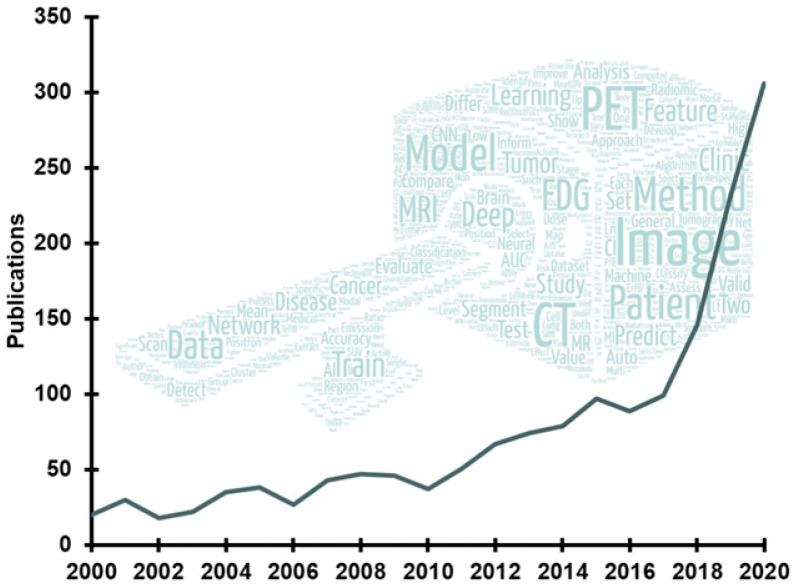
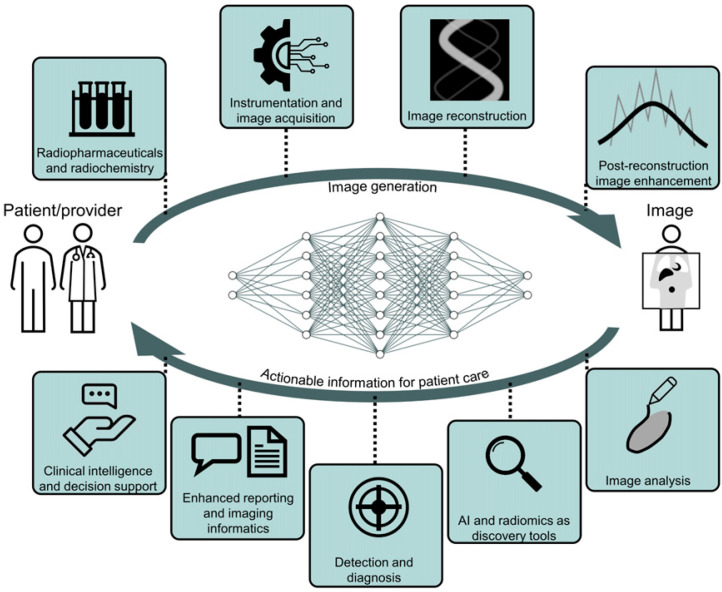
Recent advances in artificial intelligence (AI) algorithms, together with the emergence of highly accessible AI software libraries, have led to an explosion of interest in AI within the nuclear medicine field (Fig. 1). AI, which is the development of computer systems able to perform tasks normally requiring human intelligence, is being explored in nearly every subspecialty in the chain of molecular imaging, from radiochemistry to physician report generation (Fig. 2).
FIGURE 1.
Trend in publications on AI within nuclear medicine according to Scopus (Elsevier). Word cloud contains most commonly used terms in recent abstracts.
FIGURE 2.
AI applications spanning the gamut of nuclear medicine subspecialties.
The hype that propels the development of AI algorithms in nuclear medicine is counterbalanced by concerns about certain pitfalls of AI (1). The enthusiasm for AI is justified given its numerous potential benefits: AI might relieve physicians and staff from repetitive tasks, accelerate time-intensive processes, enhance image quantification, improve diagnostic reproducibility, and deliver clinically actionable information. AI promises to carry nuclear medicine beyond certain human limitations and biases. On the other hand, AI is susceptible to unique biases that are unlike the biases typically associated with human experts. There are also valid concerns about the reproducibility of claims made in many published AI studies (2) and the generalizability of trained algorithms (3). These serious issues must be addressed to ensure that algorithms earn the trust of care providers and care recipients (4).
This report was developed by the AI Task Force of the Society of Nuclear Medicine and Molecular Imaging and lays out good machine learning (ML) practices for algorithm development in nuclear medicine. Standards and recommendations for algorithm development, study design, and scientific reporting can help ensure safe technologies and reproducible gains. The report provides general recommendations for AI algorithm development, followed by recommendations that are specific to the individual subspecialities of nuclear medicine. The report focuses primarily on ML methods, as those are currently the predominant class of AI algorithms being explored in nuclear medicine, although many principles are applicable beyond ML. The target audience of the report is developers, including physicists and clinical scientists, who wish to develop AI algorithms in nuclear medicine, but the report can also benefit users (e.g., physicians) who wish to understand algorithm development. A forthcoming report from the AI Task Force focuses on appropriate methods of evaluating and validating AI algorithms in a clinical setting.
GENERAL RECOMMENDATIONS
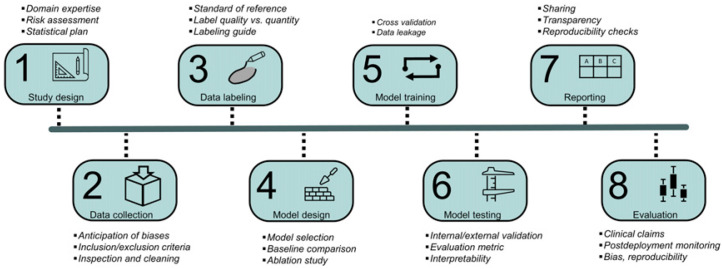
The first part of this report describes the general pipeline of algorithm development (Fig. 3) and provides recommendations that are common to most ML applications in nuclear medicine. The supplemental data (available at http://jnm.snmjournals.org) present a hypothetical tumor segmentation algorithm using a novel architecture (5) trained on a publicly available dataset (6,7) and follows it through all stages of development, from conception through reporting and dissemination, illustrating the recommendations provided here (Supplemental Fig. 1).
FIGURE 3.
Pipeline for AI algorithm development together with key considerations of each stage of development.
Study Design
The first step in AI algorithm development is to carefully define the task to be performed by the algorithm (Fig. 3). Investigators should collaborate with relevant stakeholders to understand whether and how the algorithm will be used in practice and then tailor the algorithm to the need. Early and regular feedback from users (e.g., clinicians) throughout the development process is necessary to properly align the algorithm’s functionality with the clinical need. Once the algorithm’s task is defined, studies should then be designed to train and evaluate the algorithm.
It is recommended that nuclear medicine AI studies be classified as either method development studies or evaluation studies, so that each class can be held to unique technical standards (Table 1). Method development studies are defined as studies that introduce a novel method or demonstrate the feasibility of a new application (i.e., proof of concept). Most recently published studies are method development studies. The evidence produced by these studies is insufficient to support a claim about how the trained algorithm is expected to perform clinically, often because of limited datasets and insufficient clinical evaluation techniques. Once an algorithm has shown technical promise in a method development study, the algorithm would then move on to a clinical evaluation phase in which a trained algorithm’s biases and limitations in a clinical task are evaluated to provide evidence substantiating a clinical claim. Evaluation studies must be performed using datasets that are external to the development dataset and should use algorithms that are frozen—that is, are beyond the training stage (e.g., commercial software). Evaluation studies might include reader studies; phantom studies; and, potentially, multicenter masked randomized controlled trials. Both classes of studies play important roles in advancing the field, and well-conducted studies of both classes should have a pathway to publication (potentially even in the same publication, if appropriate). Yet both classes of studies require unique design considerations. By holding both types of study to higher technical standards, it is hoped that the field can better avoid common weaknesses found in AI publications, including poor reproducibility, overly optimistic performance estimation, lack of generalizability, and insufficient transparency. The technical standards for both study types are discussed throughout this report and are summarized in Tables 1 and 2. Requirements for clinical evaluation studies will be further described in a forthcoming companion report from the AI Task Force.
TABLE 1.
Proposed Standards for Development Studies Vs. Evaluation Studies
| Parameter | Development studies | Evaluation studies |
|---|---|---|
| Accessibility of code, models, and executables | Necessary for publication | Encouraged |
| Use of external datasets | Encouraged | Required |
| Subgroup analysis for biases | Encouraged (if applicable) | Required (if applicable) |
| Clinical claims | None | Required |
| Annotation quality | Fair to high | High |
| Ablation studies | Encouraged (if applicable) | Not necessary |
| Comparison of architectures | Encouraged (if applicable) | Not necessary |
| Novelty in technology or application | High (for publication) | Not necessary (for publication) |
| Data splitting | Cross validation | Holdout or external |
TABLE 2.
Summary of Recommendations
| Category | Topic | Recommendation |
|---|---|---|
| Study design | Task definition | Collaborate with domain experts, stakeholders |
| Study types | Identify publications as development studies or evaluation studies | |
| Risk assessment | Assess the degree of risk that algorithm poses to patients and conduct study accordingly | |
| Statistical plan | Preregister statistical analysis plans for prospective studies | |
| Data collection | Bias anticipation | Collect data belonging to classes or groups that are vulnerable to bias |
| Training set size estimation | Estimate size on the basis of trial and error, or prior similar studies | |
| Evaluation of set size estimation* | Use statistical power analysis for guidance | |
| Data decisions | Use justified, objective, and documented inclusion and exclusion criteria | |
| Data labeling | Reference standard | Use labels that are regarded as sufficient standards of reference by the field |
| Label quality | Justify label quality by application, study type, and clinical claim (Fig. 4) | |
| Labeling guide* | Produce detailed guide for labelers in reader studies | |
| Quantity/quality tradeoff | Consider multiple labelers (quality) over greater numbers (quantity) | |
| Model design | Model comparison* | Explore and compare different models for development studies |
| Baseline comparison | Compare complex models with simpler models or standard of care | |
| Model selection | Report model selection and hyperparameter tuning techniques | |
| Model stability | Use repeated training with random initialization when feasible | |
| Ablation study* | Perform ablation studies for development studies focusing on novel architectures | |
| Model training | Cross validation* | Use cross validation for development studies; preserve data distribution across splits |
| Data leakage | Avoid information leaks from test set during model training | |
| Model testing and interpretability | Test set | Use same data and class distribution as for target population; use high-quality labels |
| Target population | Explicitly define target population | |
| External sets | Use external sets for evaluating model sensitivity to dataset shift | |
| Evaluation metric | Use multiple metrics when appropriate; visually inspect model outputs | |
| Model interpretability* | Use interpretability methods for clinical tasks | |
| Reporting and dissemination | Reporting | Follow published reporting guidelines and checklists |
| Sharing* | Make code and models from development studies accessible | |
| Transparency | Be forthcoming about failure modes and population characteristics in training and evaluation sets | |
| Reproducibility checks | Ensure that submitted materials to journals are sufficient for replication | |
| Evaluation† |
*Not all recommendations are applicable to all types of studies.
†Addressed in separate report from AI Task Force.
The pathway that a technology will take to reach clinical adoption should depend on the degree of risk it poses to patients. Risk categories for software have been proposed by the International Medical Device Regulators Forum and adopted by the U.S. Food and Drug Administration (8). Software in the highest risk category will require prospective studies to validate clinical claims. Prospective studies should use preregistered statistical analysis plans (9).
AI algorithms will require postdeployment monitoring to ensure safety and quality. A decline in performance might occur for a variety of reasons, such as new scanners or shifting patient demographics. Developers should plan to seek extensive user feedback and gather performance data after clinical deployment to detect and mitigate algorithm nonconformance and identify opportunities for improvement.
Data Collection
Collecting and labeling data are typically the most time-consuming aspects of algorithm development but also have the greatest dividends. An ML algorithm is ultimately a reflection of its training data, and its performance can be affected by the amount and quality of its training data. In nuclear medicine, collecting large datasets can be challenging because of the lower volumes of examinations compared with other modalities and applications.
A data collection strategy should be designed with a goal of avoiding the biases that might result from an insufficiently representative training dataset. Biases can be clinical (how well the training data reflect the clinical condition or pathologic features), technical (scanner models, acquisition protocols, reconstruction settings), demographic (racial and socioeconomic demographics, age, sex, habitus), and selection-based (e.g., tertiary vs. community hospital). For each of these biases, a structural or distribution mismatch between the training and deployment domains can result in unintended model outputs. Datasets should ideally be curated to contain the features and abnormalities that the algorithm is expected to face once deployed. Domain experts (e.g., clinicians) should guide the collection of representative cases.
It is challenging to determine the number of cases needed for algorithm development. For algorithm training, having more data is better, as long as the data are of high quality (i.e., capturing the data distribution of the targeted population). No formal guidelines exist for estimating the size of the training set, although some practical approaches have been described (10), and trial and error are therefore often necessary (11). For evaluation studies, however, sample sizes can be guided by statistical power calculations (12).
Data augmentation can be particularly useful for deep learning applications in nuclear medicine. By synthetically modifying the input data, being careful not to break the association between the input data and their target labels, dataset sizes can be artificially increased (13). Also, using a different dataset to pretrain a model can enhance the model’s capability to learn certain features and associations when labeled data are limited, although there is a risk of model overparameterization (14).
Data Labeling
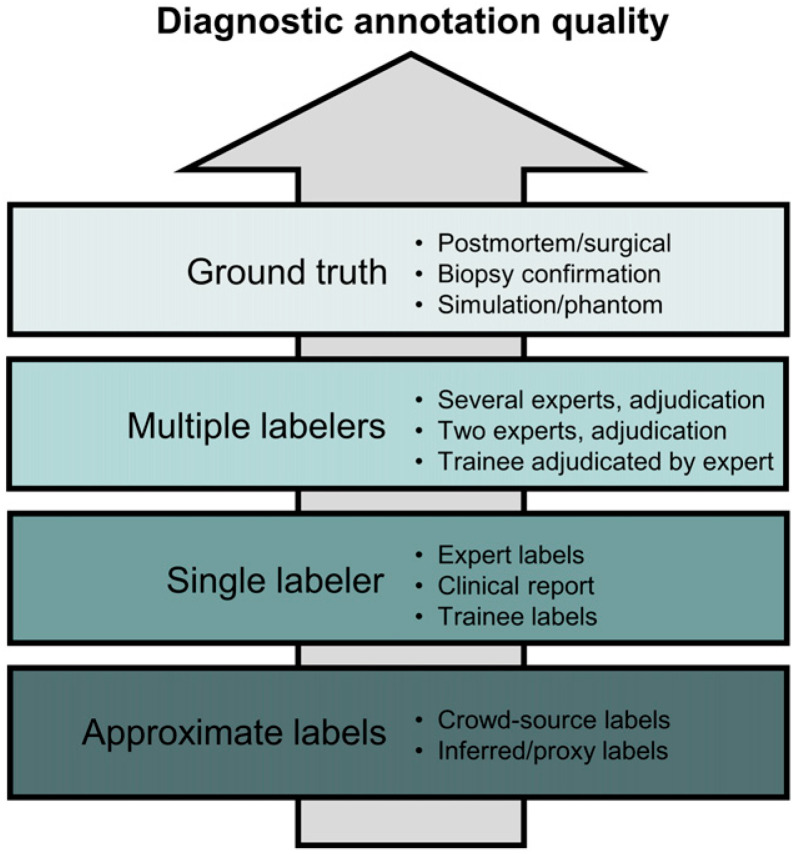
For supervised ML, labels should reflect the desired output of the algorithm in both form and quality. Labels might be generated by expert opinion, computer simulation, or other methods. The labels should be regarded by experts in the field to be sufficient standards of reference. Different labeling techniques are typically possible for a given task, often yielding different degrees of quality as illustrated in Figure 4 for diagnostic applications. When labels are based on expert opinion, it is recommended that a detailed and thorough guide to labeling be developed and discussed among labelers to reduce inter- and intraobserver variability.
FIGURE 4.
Annotation quality as function of different labeling techniques for diagnostic applications. This hierarchy does not imply how useful annotation method is (e.g., expert labels are often more useful than simulations because of limited realism of simulated data).
Because of the high cost of expert labeling, tradeoffs are nearly always made between the number of cases that can be labeled and the quality of those labels. For some tasks, having more labelers per sample can produce greater performance gains than using a larger dataset but with fewer labelers (15,16).
Because of the scarcity of labeled nuclear medicine datasets, methods that minimize labeling efforts and maximize the use of unlabeled data should be considered. Labeling is often a bottleneck in algorithm development, yet troves of unlabeled data sit dormant in clinical databases. Developers should consider data-efficient approaches to algorithm development, including semisupervised learning algorithms (17), active learning, contrastive learning, pretraining with proxy tasks, and self-supervised learning (18).
Model Design
Investigators are often faced with numerous options when selecting or designing a model for a particular task. Options can include supervised or unsupervised learning and use of neural networks or decision trees, among others. Benchmark datasets and data science competitions are useful resources for exploring different options (19).
For development studies, investigators should compare different model types. To avoid unnecessary complexity, investigators using large models are encouraged to also evaluate simpler models as a baseline comparison (e.g., logistic regression (20)). For a fair comparison of models, hyperparameters for all models should be sufficiently tuned. The approach used for hyperparameter optimization, including how many models were trained and compared, should be reported in the publication. For method development studies that introduce a novel architecture, ablation analysis is recommended (21).
When comparing AI models, small performance differences between candidate models have to be carefully interpreted. Random initialization of model weights can result in sizeable performance differences between training sessions even when identical architectures are trained with identical data. If feasible, repeated training with random initialization or with repeated holdout should be performed to provide confidence intervals of a model’s performance, which can be used to more rigorously compare different models.
Model Training
A critical part of model training is the partitioning of labeled datasets into disjoint sets. Each set serves a different purpose: the training set for updating the model’s weights, the validation set for hyperparameter tuning or model selection (if needed), and the testing set for estimating the model’s performance on unseen data. Partitioning a dataset reduces the risk of obtaining overly optimistic performance estimates due to overfitting to its own dataset. For this same reason, careful attention should be paid to preventing information from being leaked from the test set to the model during training. This can happen when, for example, a model is repeatedly retrained after evaluating it on the test set (i.e., tuning to the test set). Investigators should use the validation set to monitor model convergence (i.e., loss curves) to prevent underfitting and overfitting.
Cross validation is recommended for method development studies, whereas holdout or external test sets should be used for evaluation studies. In cross validation, the training, validation, and test datasets are repeatedly sampled from the overall dataset and a different model is trained and evaluated with each sampling. There are several approaches to cross validation (22), some of which are illustrated in Figure 5. Generally, data partitioning should aim to preserve data and class distributions in each of the data splits. A drawback of cross validation is that it creates multiple models and may not be computationally feasible for large models. However, for limited datasets, cross validation produces a less biased estimate of a method’s generalization performance than using 1-time partitions (i.e., holdout testing) (23). The latter should be used in development studies only when cross validation is technically infeasible or for large datasets.
FIGURE 5.
Different approaches to cross validation, depending on dataset size and whether model selection is needed. Figure illustrates 5-fold cross validation without model selection/hyperparameter tuning (top), 5-fold cross validation with holdout test set (middle), and nested cross validation (5-fold outer loop, 4-fold inner loop) (bottom).
Federated learning can be considered for multiinstitution studies in which pooling of data across institutions is challenging or prohibited because of privacy concerns. In federated learning, data cohorts reside within their respective institutional boundaries but models and weights are shared across institutions (24).
Model Testing and Interpretability
After model training and selection, the model’s technical performance is determined. Model testing, especially when using the developmental dataset, does not typically result in evidence to substantiate broad clinical claims.
Models are tested using a test dataset, which should be an unseen holdout dataset or—for development studies—may consist of all the data through cross validation (Fig. 5). The test set should have data and class distributions similar to those of the target population. The target population must be explicitly defined (e.g., “Hodgkin lymphoma patients scanned in our department in 2020”). Additional test cohorts that are external to the developmental data are highly desirable, as they provide an estimate of the algorithm’s sensitivity to covariate or dataset shift.
Model performance is quantified using evaluation metrics. Selection of evaluation metrics should be based on how well they reflect the failures and successes of the algorithm for the specific application. However, evaluation metrics are often unable to detect all the ways in which an algorithm fails, and summary statistics can hide meaningful errors (25). Investigators should seek to detect cases of failure and work to understand their causes. This work will often include visual inspection of the model output. It is recommended that challenging cases be included in the test set to probe the model’s limitations. Investigators should also directly compare the AI model’s performance with another acceptable standard, such as the standard of care. It is recommended that subgroup analysis be conducted to identify whether the algorithm is biased against any cohorts.
Investigators should attempt to make their algorithms interpretable to users, especially algorithms that perform clinical tasks (4). Interpretable algorithms attempt to explain their outputs by highlighting the properties of the input data that most impacted the model’s prediction. Interpretability may help identify confounding factors that are unrelated to the task or pathology yet unintentionally guide the model’s predictions (3). Popular approaches include tracking gradients through the network (e.g., gradient-weighted class activation mapping) or iteratively perturbing or occluding parts of the input data (e.g., Shapley additive explanations) (26).
Reporting and Dissemination
The quality of the reporting of AI studies is a key determinant of its subsequent impact on the field. Formal guidelines for reporting of AI studies are emerging (27,28), including some that have been proposed (29–31) and others that are forthcoming (32–34).
For development studies, journals should make publication contingent on the models and either the source codes (preferred) or executables being made accessible. Publications on development studies should contribute to the technical advancement of the field, which is often accomplished only through sharing. Many hosting resources are available for sharing, as listed in Table 3. Investigators should work with institutional review boards to ensure that datasets can be properly anonymized and openly shared. The paucity of large, high-quality multicenter datasets is a major hindrance to the clinical translation of AI tools in nuclear medicine, and open sharing of data would greatly benefit the nuclear medicine community. When data cannot be fully shared for privacy reasons, at least sample data should be made available so that the correct implementation of the model can be tested. Code should come with a modus operandi that does not leave any room for subjective settings, including a data dictionary defining variables and any preprocessing or parameter-tuning instructions.
TABLE 3.
Resources for Hosting and Sharing Code, Models, and Data
| Data type | Resources |
|---|---|
| Code | Git repository hosts (GitHub, GitLab, Bitbucket [Atlassian]), Matlab File Exchange (MathWorks), SourceForge |
| Models, containers, executables | Docker Hub (Docker Inc.), modelhub.ai, Model Zoo, Gradio, TensorFlow Hub, PyTorch Hub, Hugging Face |
| Data | The Cancer Imaging Archive, Kaggle Inc., paperswithcodes.com, LONI Image and Data Archive, Figshare (Figshare) |
In publishing evaluation studies, the scientific contribution is the reporting on the efficacy of a previously reported or commercial algorithm; therefore, referring to the description of the algorithm is deemed sufficient for publication.
Journal editors and reviewers are encouraged to systematically check that all provided materials are sufficient for replicating studies. This step could consist of reproducibility checklists (35) or dedicated data-expert reviewers, similar to statistics reviewers that are solicited for articles involving sophisticated statistical analyses. These demanding but desirable actions have been adopted in other fields and will serve to accelerate development and validation of AI algorithms.
Investigators should be forthcoming about limitations and failures of their algorithm (36). Failure modes should be carefully described, along with positive results. Developers should provide detailed descriptions of the characteristics and limitations of the training and evaluation datasets, such as any missing demographic groups.
Evaluation
Algorithm evaluation refers to the quantification of technical efficacy, clinical utility, biases, and postdeployment monitoring of a trained algorithm. After a successful development study, a trained algorithm should be subjected to a thorough evaluation study. Evaluation studies should involve clinical users of the algorithm and produce evidence to support specific claims about the algorithm. Clinical evaluation of a diagnostic algorithm requires reader studies, in which expert nuclear medicine physicians or radiologists assess how AI algorithms impact image interpretation or clinical decision making, often in comparison to a reference method. There are numerous additional considerations to algorithm evaluation, and a separate forthcoming report from the Society of Nuclear Medicine and Molecular Imaging AI Task Force focuses specifically on these evaluation studies and the claims that result from them.
SPECIFIC APPLICATIONS
The following subsections deal with the application of AI in the various subspecialties of nuclear medicine (Fig. 2). Each section describes how AI might be used in the different domains of nuclear medicine, together with best practices in algorithm development for each type of application and considering the different components of the development pipeline (Fig. 3).
Image Reconstruction
There is great anticipation about the benefits that AI might provide to image reconstruction, including faster reconstruction, improved signal-to-noise ratio, and fewer artifacts. AI might also contribute to different components of image reconstruction, such as direct parametric map estimation, accelerated scatter correction, and attenuation correction for PET/MRI, PET-only, and SPECT-only systems.
In general, 2 classes of approaches are being explored in nuclear medicine reconstruction: those that incorporate neural networks into current physics-based iterative reconstruction methods, and those that directly reconstruct images from projection data (37). Studies on the merits of end-to-end approaches versus penalty-based approaches are needed. Furthermore, for end-to-end algorithms, innovative solutions are needed to handle the large size of 3-dimensional time-of-flight sinograms, as the memory constraints of graphics processing units have limited methods to either single-slice and non–time-of-flight applications or have required sinogram rebinning (38). Solutions might include multi–graphics processing unit parallelization or dimensionality reduction strategies.
The large impact that AI-based reconstruction methods might have on patient care demands that algorithms be sufficiently validated. Investigators should use figures of merit to evaluate image quality, such as mean-squared error, structural similarity index, or peak signal-to-noise ratio, but should also recognize that these metrics might be misleading, as small, diagnostically important features could potentially be added or removed from images without significantly impacting summary statistics (25). Therefore, evaluation studies will require reader studies with clinically focused tasks (e.g., lesion detection). Models that use anatomic priors (e.g., CT) should be tested for robustness to functional–anatomic misregistration. For development studies, computational model-observer–based studies might prove more economic in identifying promising methods (39).
Overall, comparative studies of different AI-based reconstruction approaches are needed, and evaluation studies should use task-oriented figures of merit and validation methods (i.e., reader studies).
Postreconstruction Image Enhancement
AI methods can enhance reconstructed nuclear medicine images with more favorable qualitative or quantitative properties, with many of the same benefits as AI-based reconstruction, including lower noise, artifact removal, and improved spatial resolution.
Denoising of low-count PET images has been the subject of numerous publications and even commercial software (40). Training data often consist of pairs of images reconstructed from fully sampled and subsampled list-mode data. Subsampling should span the entire length of the examination time so that motion and tracer distribution are consistent between the image pairs. Investigators should compare the performance of denoising networks with other denoising approaches, such as gaussian smoothing and more advanced methods such as nonlocal means. Contrast, feature quantification, and noise levels should be systematically evaluated.
Algorithms might be sensitive to outliers (e.g., implants) or artifacts (e.g., motion) and should always be evaluated on challenging, out-of-distribution cases. For applications that use coregistered CT or MR images as inputs, networks should be evaluated for robustness to misregistration (41).
Traditional figures of merit to evaluate denoising methods may be misleading (42). Metrics such as signal-to-noise ratio, mean squared error, and quantitative bias should be used to evaluate gains in image quality while also ensuring quantitative fidelity. However, these metrics may not reflect the presence or absence of clinically meaningful features. Also, AI can create synthetic-looking or overly smooth images. Thus, evaluation should consist of human observer or model observer studies.
In short, image enhancement algorithms should undergo sensitivity studies and reader evaluation studies, and performance should be compared with existing enhancement methods.
Image Analysis
AI is anticipated to automate several image analysis tasks in nuclear medicine, such as in oncologic imaging (e.g., lesion detection, segmentation, and quantification (43,44)), cardiac imaging (e.g., blood flow analyses), brain imaging (e.g., quantification of neurodegenerative diseases), and dosimetry, among others (44,45). Automation of these tasks has significant potential to save time, reduce interobserver variability, improve accuracy, and fully exploit the quantitative nature of molecular imaging (46,47).
AI-based segmentation algorithms should be task-specific. For instance, segmentation for radiotherapy target volume delineation requires datasets and labeling techniques different from those for segmentation for prediction of overall survival (though they are related). An algorithm might be sufficient for one metric but not another (43). Images from other modalities, such as CT and MRI, that provide complementary high-resolution information can also be considered as inputs to an algorithm if expected to be available clinically.
Segmentation algorithms are typically trained using expert-generated contours. To ensure appropriate and consistent labeling (Fig. 4), clear annotation instructions should be distributed to qualified labelers to guide them on viewing settings, on handling functional-anatomic misregistration, and other conditions that might affect segmentation. Expert contours will inevitably have interobserver variability, which should be measured and used as a point of comparison for automated methods. Various methods exist for creating consensus contours from multiple observers (e.g., simultaneous truth and performance-level estimation algorithm (48)). Investigators should also be aware of the various objective functions and evaluation metrics for segmentation and of the existing guidelines for validation and reporting of autosegmentation methods (49). Because of the sparsity of large, high-quality labeled datasets in nuclear medicine, phantom or realistic simulation data can also be used for model pretraining (47,50).
Overall, the development of AI segmentation algorithms should include meticulous, task-specific labeling practices, and published guidelines for validating and reporting of algorithms should be followed.
AI and Radiomics as a Discovery Tool
AI is expected to play a critical role in assisting physicians and scientists in discovering patterns within large biologic and imaging datasets that are associated with patient outcome. Modern ML methods have shown promise as useful tools to uncover hidden but meaningful relationships within datasets (51). AI is therefore a useful adjunct to radiomics.
First, ML can be used to identify deep radiomic features whose definitions depend completely on the data and on the task, unlike handcrafted radiomic features that are mathematically predefined whatever the data. Second, ML is an effective way to mine large numbers of radiomic features, possibly augmented by other omics or clinical data, to identify associations, reduce redundancy, produce tractable representations in low-dimension spaces, or design prediction models. Unsupervised ML might be used to combine correlated input features into a smaller, more tractable set of factors (52) or to select features relevant to a task. Redundancy in features can arise from technical causes (e.g., mathematic equivalence of radiomics features), from measurement of the same underlying biologic factor, or from a biologic causal relationship (some biologic factor influences multiple feature values). By distinguishing among these 3 situations, investigators can better approach dimensionality reduction (53). For example, mathematic equivalence of radiomics features can be detected by randomly perturbing the image and assessing which correlations persist through the perturbations (54).
The challenge of discovering predictive signatures in high-dimension datasets might necessitate a multistep approach. Investigators might first start with a selection of cases that represent both ends of the label’s range of values, such as short and long survival, to maximize the chances of detecting features associated with outcome but at the cost of low specificity.
After initial discovery, whatever features or relationships have been identified must be rigorously evaluated and scrutinized. Investigators must explore the relationships across the entire dataset using cross validation, aim to understand the underlying cause, and then externally validate these findings, ruling out false-positives or spurious correlations. For example, they can repeat the whole AI-analysis pipeline on sham data (e.g., randomized labels) to determine the baseline false-positive rate for their set of methods and then compare it with the discovery rate found in the real dataset. Investigators should also test different models and architecture to see whether the discovered relationships hold, as it is unlikely that a real association will be identified by only 1 model.
In short, radiomics analysis should include the removal of redundant features, and a multistep approach of discovery (high sensitivity, low specificity) followed by rigorous validation might be considered.
Detection and Diagnosis
Computer-aided diagnosis and detection have long histories of successes and failures in radiology, but the recent advancements in AI have made widespread use of computer-aided diagnosis and computer-aided detection an approaching reality for nuclear medicine. Automation of diagnostic tasks in nuclear medicine can be challenging, as diagnostic tasks are subjective, have high stakes, and must be incredibly robust to rare cases (e.g., implants or amputations). However, the incentive to develop such tools is strong, with applications including assisted reads (55), tumor detection suggestions, neuro or cardiac diagnosis tools (56), training programs for residents, and many others.
Investigators should select an appropriate labeling technique according to the accuracy that is needed for their computer-aided diagnosis or computer-aided detection application (Fig. 4). Labels from specialists are superior to those from trainees or generalists, and labels resulting from multiple readers (adjudication or consensus) are superior to those from single readers. Labels extracted from clinical reports are considered inferior to those obtained from dedicated research readings (57). Intraobserver and interobserver variability in labels is often an indicator of label quality and should be quantified and reported.
Investigators are encouraged to integrate model interpretability (e.g., Shapley additive explanations) and uncertainty signaling (e.g., Bayesian approximation) into their algorithm. Because diagnostic algorithms will be used under the supervision of a physician, algorithm decisions should ideally be explainable so that clinicians have sufficient information to contest or provide feedback when algorithms fail. Developers also need to be transparent about their algorithm development and evaluation processes, including data sources and training set population characteristics, such as by using reporting checklists such as MI-CLAIM (29). The high visibility and public attention that AI-based diagnostic algorithms receive demands that developers make every effort to be fully transparent.
In short, for computer-aided detection and computer-aided diagnosis algorithms, label quality should be justified by the application (high quality for high-risk applications) and algorithms should be interpretable and fully transparent.
Enhanced Reporting and Imaging Informatics
ML has the potential to transform how the information within diagnostic images is translated into reports and clinical databases. AI can be used to prepopulate radiology reports, assist in real-time report generation, help standardize reporting, and perform structured synoptic reporting (58).
Algorithm development in medical imaging informatics has several unique considerations. A critical challenge is the large heterogeneity in diagnostic reporting standards and practices across institutions, individual physicians, and examination types. Heterogeneity in language can be more challenging for automation than is heterogeneity in medical images. Therefore, training data should be collected from diverse sources and annotators, and studies are expected to require much larger sample sizes than for other applications. Tasks in this domain might be uniquely suitable to unsupervised or semisupervised approaches because of the large volume of unlabeled data available in clinical PACS systems. Various model types will likely be applied in this domain, but language models may need to be adapted to consider the unique nuclear medicine vocabulary that might not be represented in typical medical text corpora (e.g., the term SUV). Because of challenges in deidentification of radiology reports (59), federated learning should be considered to enable privacy-protected multiinstitutional studies. Reporting of model performance should be disaggregated according to data source, originating institution, and annotator.
Clinical Intelligence and Decision Support
Clinical intelligence and decision support are concerned with delivering actionable advice to clinicians after extracting, distilling, and consolidating clinical information across multiple data sources. These systems are expected to pull the most pertinent information generated by a nuclear medicine examination and combine it with other clinical data to best guide patient care. For example, ML can predict future myocardial infarction using PET features combined with other clinical variables (60). The development and validation of clinical decision support systems should be guided by physician needs and clinical experts, involving teams from nearly all sectors of health care.
An algorithm’s ability to explain its decisions is key to safe, ethical, fair, and trustworthy use of AI for decision support, calling for the same recommendations as discussed in the section on detection and diagnosis. An AI model should ideally be able to provide an estimate of uncertainty together with its output, possibly by using Bayesian methods, and be willing to provide a no-decision answer when the model uncertainties are too large to make the output meaningful.
Instrumentation and Image Acquisition
Challenging problems in data acquisition and instrumentation could be well suited to ML-based solutions (61). For example, ML has been used to estimate 2- and 3-dimensional position of interaction for detectors (62). Other promising applications include timing pickoff for detector waveforms, intercrystal scatter estimation, patient motion detection, and the prediction of scanner failure from quality control tracking.
Precise data collection is critical to the success of AI applications within instrumentation. Simulations should be performed using appropriate models that incorporate geometric, physical, and statistical factors underlying image generation. Investigators should consider possible discrepancies between in silico and physical domains and are encouraged to conduct cross-validation studies when possible (61). Physical measurements, such as point source measurements, may require high-precision motion stages and lengthy acquisition studies to collect the full range of training data. Scanner quality control applications will likely require enterprise-level tracking to obtain sufficient data on failure patterns.
Algorithms that process events in real time and need to be implemented on front-end electronics will likely be memory- and operation-limited (63), favoring simpler model architectures. Ablation analysis can help identify more parsimonious models.
Radiopharmaceuticals and Radiochemistry
The potential for AI to challenge the current paradigms in synthesis (64) and administration (65) of radiopharmaceuticals is only beginning to be explored. Potential applications include predicting drug–target interactions (66), predicting and optimizing radiochemical reactions, and performing de novo design of drugs (67), as well as helping optimize radiopharmacy workflows. Proper integration of AI within the radiochemistry and radiopharmacy communities will require collaborations between key stakeholders, including industry, end users, and quality control personnel, as well as experts in information technology, cybersecurity, and regulatory aspects. It is strongly recommended that groups share manufacturing data freely, as this will accelerate innovation by providing large test sets for ML that cannot be sufficiently generated at individual labs (e.g., synthesis module and cyclotron log files).
DISCUSSION AND CONCLUSION
The recommendations listed in this article and summarized in Table 2 are intended to assist developers and users in understanding the requirements and challenges associated with the design and use of AI-based algorithms. They focus on specificities associated with nuclear medicine applications, whereas best practices for software development, data management, security, privacy, ethics, and regulatory considerations are largely covered elsewhere. It is also acknowledged that some standards of today are likely to be superseded by new standards as technologies continue to evolve. These recommendations should serve as a guide to developers and investigators at a time when AI is booming but should not be assumed to be comprehensive or unchanging.
These recommendations were drawn from various sources, including the authors’ collective experiences in academia and industry, as well as other published position papers, and put into the context of nuclear medicine applications. They should be considered an add-on to other guidelines, including forthcoming guidelines from regulatory bodies (68) and relevant working groups (69).
AI is expected to influence and shape the future of nuclear medicine and many other fields. But the potential pitfalls of AI warrant a careful and methodic approach to AI algorithm development and adoption. Standards and guidelines can help nuclear medicine avoid the mismatch between the role that AI is expected to play and what it will actually deliver.
DISCLOSURE
Tyler Bradshaw receives research support from GE Healthcare. Ronald Boellaard is a scientific advisor (unpaid) for the EARL PET/CT accreditation program. Paul Jacobs is a full-time employee of MIM Software Inc. Sven Zuehlsdorff is a full-time employee of Siemens Medical Solutions USA, Inc. Piotr Slomka has a research grant from Siemens Medical Solutions, is a consultant for IBA, and receives royalties from Cedars-Sinai for nuclear cardiology software. No other potential conflict of interest relevant to this article was reported.
ACKNOWLEDGMENTS
The members of the AI Task Force acknowledge Bonnie Clarke for her excellent support of this project.
REFERENCES
- 1. Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;3:199–217. [Google Scholar]
- 2. Haibe-Kains B, Adam GA, Hosny A, et al. Transparency and reproducibility in artificial intelligence. Nature. 2020;586:E14–E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15:e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buvat I, Orlhac F. The T.R.U.E. checklist for identifying impactful artificial intelligence-based findings in nuclear medicine: is it True? Is it Reproducible? Is it Useful? Is it Explainable? J Nucl Med. 2021;62:752–754. [DOI] [PubMed] [Google Scholar]
- 5. Xue Y, Xu T, Zhang H, Long LR, Huang X. Segan: adversarial network with multi-scale L1 loss for medical image segmentation. Neuroinformatics. 2018;16:383–392. [DOI] [PubMed] [Google Scholar]
- 6. Vallières M, Kay-Rivest E, Perrin LJ, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7:10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andrearczyk V, Oreiller V, Jreige M, et al.Overview of the HECKTOR challenge at MICCAI 2020: automatic head and neck tumor segmentation in PET/CT. In: Lecture Notes in Computer Science. Springer; 2020:1–21.
- 8. Center for Devices and Radiological Health. Software as a medical device (SAMD): clinical evaluation—guidance for industry and Food and Drug Administration staff. Food and Drug Administration website. https://www.fda.gov/media/100714/download. Published December 8, 2017. Accessed December 13, 2021.
- 9. Nosek BA, Ebersole CR, DeHaven AC, Mellor DT. The preregistration revolution. Proc Natl Acad Sci USA. 2018;115:2600–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dirand A-S, Frouin F, Buvat I. A downsampling strategy to assess the predictive value of radiomic features. Sci Rep. 2019;9:17869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riley RD, Ensor J, Snell KIE, et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. [DOI] [PubMed] [Google Scholar]
- 12. Smith SM, Nichols TE. Statistical challenges in “big data” human neuroimaging. Neuron. 2018;97:263–268. [DOI] [PubMed] [Google Scholar]
- 13. Hwang D, Kim KY, Kang SK, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–1629. [DOI] [PubMed] [Google Scholar]
- 14. Raghu M, Zhang C, Kleinberg J, Bengio S. Transfusion: understanding transfer learning for medical imaging. arXiv.org website. https://arxiv.org/abs/1902.07208. Published February 14, 2019. Revised October 29, 2019. Accessed December 13, 2021.
- 15. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 16. Krause J, Gulshan V, Rahimy E, et al. Grader variability and the importance of reference standards for evaluating machine learning models for diabetic retinopathy. Ophthalmology. 2018;125:1264–1272. [DOI] [PubMed] [Google Scholar]
- 17. Cheplygina V, de Bruijne M, Pluim JPW. Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal. 2019;54:280–296. [DOI] [PubMed] [Google Scholar]
- 18. Zhu J, Li Y, Hu Y, Ma K, Zhou SK, Zheng Y. Rubik’s Cube+: a self-supervised feature learning framework for 3D medical image analysis. Med Image Anal. 2020;64:101746. [DOI] [PubMed] [Google Scholar]
- 19. Home page. Papers with Code website. https://paperswithcode.com/datasets. Accessed December 13, 2021.
- 20. Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. [DOI] [PubMed] [Google Scholar]
- 21. Zhao K, Zhou L, Gao S, et al. Study of low-dose PET image recovery using supervised learning with CycleGAN. PLoS One. 2020;15:e0238455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raschka S. Model evaluation, model selection, and algorithm selection in machine learning. arXiv.org website. https://arxiv.org/abs/1811.12808. Published November 13, 2018. Revised November 11, 2020. Accessed December 13, 2021.
- 23. Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010;4:40–79. [Google Scholar]
- 24. Li T, Sahu AK, Talwalkar A, Smith V. Federated learning: challenges, methods, and future directions. IEEE Signal Process Mag. 2020;37:50–60. [Google Scholar]
- 25. Yang J, Sohn JH, Behr SC, Gullberg GT, Seo Y. CT-less direct correction of attenuation and scatter in the image space using deep learning for whole-body FDG PET: potential benefits and pitfalls. Radiol Artif Intell. 2020;3:e200137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huff DT, Weisman AJ, Jeraj R. Interpretation and visualization techniques for deep learning models in medical imaging. Phys Med Biol. 2021;66:04TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cruz Rivera S, Liu X, Chan A-W, Denniston AK, Calvert MJ; SPIRIT-AI and CONSORT-AI Working Group. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Lancet Digit Health. 2020;2:e549–e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK, SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26:1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Norgeot B, Quer G, Beaulieu-Jones BK, et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med. 2020;26:1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sengupta PP, Shrestha S, Berthon B, et al. Proposed requirements for cardiovascular imaging-related machine learning evaluation (PRIME): a checklist. JACC Cardiovasc Imaging. 2020;13:2017–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hernandez-Boussard T, Bozkurt S, Ioannidis JPA, Shah NH. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc. 2020;27:2011–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sounderajah V, Ashrafian H, Aggarwal R, et al. Developing specific reporting guidelines for diagnostic accuracy studies assessing AI interventions: the STARD-AI Steering Group. Nat Med. 2020;26:807–808. [DOI] [PubMed] [Google Scholar]
- 33. Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–1579. [DOI] [PubMed] [Google Scholar]
- 34. DECIDE-AI Steering Group. DECIDE-AI: new reporting guidelines to bridge the development-to-implementation gap in clinical artificial intelligence. Nat Med. 2021;27:186–187. [DOI] [PubMed] [Google Scholar]
- 35. Pineau J, Vincent-Lamarre P, Sinha K, et al.Improving reproducibility in machine learning research (a report from the NeurIPS 2019 Reproducibility Program). arXiv.org website. https://arxiv.org/abs/2003.12206. Published March 27, 2020. Revised December 30, 2020. Accessed December 13, 2021.
- 36. Reuzé S, Orlhac F, Chargari C, et al. Prediction of cervical cancer recurrence using textural features extracted from 18F-FDG PET images acquired with different scanners. Oncotarget. 2017;8:43169–43179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reader AJ, Corda G, Mehranian A. D, Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci. 2021;5:1–25. [Google Scholar]
- 38. Whiteley W, Luk WK, Gregor J. DirectPET: full-size neural network PET reconstruction from sinogram data. J Med Imaging (Bellingham). 2020;7:032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu Z, Rahman MA, Schindler T, Laforest R, Jha AK. A physics and learning-based transmission-less attenuation compensation method for SPECT. In: Proceedings of SPIE, Medical Imaging 2021: Physics of Medical Imaging. SPIE; 2021:1159512. [DOI] [PMC free article] [PubMed]
- 40. Katsari K, Penna D, Arena V, et al. Artificial intelligence for reduced dose 18F-FDG PET examinations: a real-world deployment through a standardized framework and business case assessment. EJNMMI Phys. 2021;8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu W, Onofrey JA, Lu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64:165019. [DOI] [PubMed] [Google Scholar]
- 42. Yu Z, Rahman MA, Schindler T, et al. AI-based methods for nuclear-medicine imaging: need for objective task-specific evaluation [abstract]. J Nucl Med. 2020;61(suppl 1):575. [Google Scholar]
- 43. Weisman AJ, Kim J, Lee I, et al. Automated quantification of baseline imaging PET metrics on FDG PET/CT images of pediatric Hodgkin lymphoma patients. EJNMMI Phys. 2020;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Capobianco N, Meignan M, Cottereau A-S, et al. Deep-learning 18F-FDG uptake classification enables total metabolic tumor volume estimation in diffuse large B-cell lymphoma. J Nucl Med. 2021;62:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weisman AJ, Kieler MW, Perlman SB, et al. Convolutional neural networks for automated PET/CT detection of diseased lymph node burden in patients with lymphoma. Radiol Artif Intell. 2020;2:e200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weisman AJ, Kieler MW, Perlman S, et al. Comparison of 11 automated PET segmentation methods in lymphoma. Phys Med Biol. 2020;65:235019. [DOI] [PubMed] [Google Scholar]
- 47. Leung KH, Marashdeh W, Wray R, et al. A physics-guided modular deep-learning based automated framework for tumor segmentation in PET. Phys Med Biol. 2020;65:245032. [DOI] [PubMed] [Google Scholar]
- 48. Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hatt M, Lee JA, Schmidtlein CR, et al. Classification and evaluation strategies of auto-segmentation approaches for PET: report of AAPM task group no. 211. Med Phys. 2017;44:e1–e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Z, Laforest R, Mhlanga J, et al.Observer study-based evaluation of a stochastic and physics-based method to generate oncological PET images. Proceedings of SPIE, Medical Imaging 2021: Image Perception, Observer Performance, and Technology Assessment. SPIE; 2021:1159905.
- 51. Pierson E, Cutler DM, Leskovec J, Mullainathan S, Obermeyer Z. An algorithmic approach to reducing unexplained pain disparities in underserved populations. Nat Med. 2021;27:136–140. [DOI] [PubMed] [Google Scholar]
- 52. Peeters CFW, Übelhör C, Mes SW, et al.Stable prediction with radiomics data. arXiv.org website. https://arxiv.org/abs/1903.11696. Published March 27, 2019. Accessed December 13, 2021.
- 53. Pfaehler E, Mesotten L, Zhovannik I, et al. Plausibility and redundancy analysis to select FDG-PET textural features in non-small cell lung cancer. Med Phys. 2021;48:1226–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2019;130:2–9. [DOI] [PubMed] [Google Scholar]
- 55. Li Z, Kitajima K, Hirata K, et al. Preliminary study of AI-assisted diagnosis using FDG-PET/CT for axillary lymph node metastasis in patients with breast cancer. EJNMMI Res. 2021;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Betancur J, Hu L-H, Commandeur F, et al. Deep learning analysis of upright-supine high-efficiency SPECT myocardial perfusion imaging for prediction of obstructive coronary artery disease: a multicenter study. J Nucl Med. 2019;60:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bluemke DA, Moy L, Bredella MA, et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the Radiology editorial board. Radiology. 2020;294:487–489. [DOI] [PubMed] [Google Scholar]
- 58. Panayides AS, Amini A, Filipovic ND, et al. AI in medical imaging informatics: current challenges and future directions. IEEE J Biomed Health Inform. 2020;24:1837–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steinkamp JM, Pomeranz T, Adleberg J, Kahn CE, Cook TS. Evaluation of automated public de-identification tools on a corpus of radiology reports. Radiol Artif Intell. 2020;2:e190137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwiecinski J, Tzolos E, Meah M, et al.Machine-learning with 18F-sodium fluoride PET and quantitative plaque analysis on CT angiography for the future risk of myocardial infarction. J Nucl Med. April 23, 2021. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 61. Arabi H, Zaidi H. Applications of artificial intelligence and deep learning in molecular imaging and radiotherapy. Eur J Hybrid Imaging. 2020;4:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gong K, Berg E, Cherry SR, Qi J. Machine learning in PET: from photon detection to quantitative image reconstruction. Proc IEEE. 2020;108:51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Müller F, Schug D, Hallen P, Grahe J, Schulz V. A novel DOI positioning algorithm for monolithic scintillator crystals in PET based on gradient tree boosting. IEEE Trans Radiat Plasma Med Sci. 2019;3:465–474. [Google Scholar]
- 64. de Almeida AF, Moreira R, Rodrigues T. Synthetic organic chemistry driven by artificial intelligence. Nat Rev Chem. 2019;3:589–604. [Google Scholar]
- 65. Nelson SD, Walsh CG, Olsen CA, et al. Demystifying artificial intelligence in pharmacy. Am J Health Syst Pharm. 2020;77:1556–1570. [DOI] [PubMed] [Google Scholar]
- 66. Wen M, Zhang Z, Niu S, et al. Deep-learning-based drug-target interaction prediction. J Proteome Res. 2017;16:1401–1409. [DOI] [PubMed] [Google Scholar]
- 67. Ståhl N, Falkman G, Karlsson A, Mathiason G, Boström J. Deep reinforcement learning for multiparameter optimization in de novo drug design. J Chem Inf Model. 2019;59:3166–3176. [DOI] [PubMed] [Google Scholar]
- 68.Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD): discussion paper and request for feedback. U.S. Food and Drug Administration website. https://beta.regulations.gov/document/FDA-2019-N-1185-0001. Published April 2, 2019. Accessed December 13, 2021.
- 69. IEEE artificial intelligence medical device working group. IEEE Standards Association website. https://sagroups.ieee.org/aimdwg/. Accessed December 13, 2021.