Abstract
The genomic DNA sequence of herpes simplex virus type 2 (HSV-2) strain HG52 was determined as 154,746 bp with a G+C content of 70.4%. A total of 74 genes encoding distinct proteins was identified; three of these were each present in two copies, within major repeat elements of the genome. The HSV-2 gene set corresponds closely with that of HSV-1, and the HSV-2 sequence prompted several local revisions to the published HSV-1 sequence (D. J. McGeoch, M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor, J. Gen. Virol. 69:1531–1574, 1988). No compelling evidence for the existence of any additional protein-coding genes in HSV-2 was identified.
The complete 152-kbp genomic DNA sequence of herpes simplex virus type 1 (HSV-1) was published in 1988 (56) and since then has been very widely employed in a great range of research on HSV-1. Additionally, results from this most studied member of the family Herpesviridae have fed powerfully into research on other herpesviruses. In contrast, although a substantial number of individual gene sequences have been determined for the other HSV serotype, HSV-2, the complete genome sequence for this virus has not been available hitherto. In this paper we report the sequence of the genome of HSV-2, strain HG52.
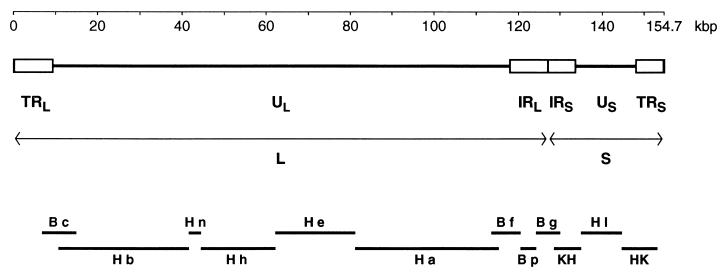
At a gross level the 155-kbp genome of HSV-2 is viewed as consisting of two extended regions of unique sequence (UL and US), each of which is bounded by a pair of inverted repeat elements (TRL-IRL and IRS-TRS) (17, 66) (Fig. 1). There is a directly repeated sequence of some 254 bp at the genome termini (the a sequence), with one or more copies in the opposing orientation (the a′ sequence) at the internal joint between IRL and IRS (21). UL plus its flanking repeats is termed the long (L) region, and US with its flanking repeats is termed the short (S) region. In individual molecules of HSV-2 DNA, the L and S components may be linked with each in either orientation, so that DNA preparations contain four sequence-orientation isomers, one of which is defined as the prototype (66). The sequences of the terminal and internal copies of RL and of RS are considered to be indistinguishable.
FIG. 1.
Overall organization of the genome of HSV-2. The linear double-stranded DNA is represented, with the scale at the top. The unique portions of the genome (UL and US) are shown as heavy solid lines, and the major repeat elements (TRL, IRL, IRS, and TRS) are shown as open boxes. For each pair of repeats the two copies are in opposing orientations. As indicated, TRL, UL, and IRL are regarded as comprising the L region, and IRS, US, and TRS are regarded as comprising the S region. Plasmid-cloned fragments used for sequence determination are indicated at the bottom: BamHI and HindIII fragments are indicated by B and H, respectively, followed by individual fragment designations in lowercase; KH and HK indicate KpnI/HindIII fragments as described in the text.
This paper presents properties of the HSV-2 DNA sequence and our present understanding of its content of protein-coding genes and other elements. We are also interested in comparative analysis of the HSV-1 and HSV-2 genomes to examine processes of molecular evolution which have occurred since the two species diverged, and we intend to pursue this topic in a separate paper.
MATERIALS AND METHODS
Sources of virus and cloned DNA fragments.
HSV-2 strain HG52 was obtained from stocks in the Institute of Virology, Glasgow, United Kingdom (74). HindIII and BamHI fragments of HSV-2 strain HG52 cloned into pAT153 were provided by our colleague A. J. Davison. HindIII/KpnI fragments representing the two ends of US with adjacent parts of RS were cloned into plasmids during the course of the sequence determination project. A plasmid clone of HSV-2 strain 25766 BamHI d was from A. C. Minson.
Sequence determination.
Large plasmid-cloned fragments of HSV-2 DNA were fragmented by sonication and subcloned into M13mp series vectors to give random libraries, and sequences from M13 clones were then determined by standard methods (3, 55). The programs of Staden (67) were used to overlap the shotgun sequence data from sets of M13 clones, to assemble databases for plasmid clones, and to edit the assembled sequences. Sequencing problems associated with G+C-rich DNA were resolved as previously described (55, 56). The complete genome sequence was assembled by utilizing overlaps between sequences represented in neighboring plasmid clones or, where sequences from adjacent plasmid clones abutted but did not overlap, by employing PCR amplification across these junctions with whole virus DNA as the template followed by sequence determination of the PCR product.
Sequence interpretation.
Evaluations of the gene content and other aspects of the HSV-2 sequence were carried out with the Genetics Computer Group program set (30). Database searches used FastA, TfastA, and Blast. The program Diverge (version 9; Genetics Computer Group) was used to compute nonsynonymous (i.e., causing amino acid substitution) and synonymous (silent) substitutional divergences (Ka and Ks, respectively) between pairs of aligned HSV-1 and HSV-2 coding sequences. It should be noted that Ka and Ks are not simply scores of differences: they estimate numbers of mutations that have occurred for each class as a fraction of the total number of sites of the same class and make allowance for multiple hits and differing transition and transversion rates (44).
Nucleotide sequence accession number.
The genome sequence of HSV-2 strain HG52 has been submitted to the EMBL Sequence Library (accession no. Z86099).
RESULTS
Determination of the DNA sequence of HSV-2 strain HG52.
The DNA sequence of HSV-2 strain HG52 was determined by using plasmid-cloned fragments of the genome, as indicated in Fig. 1. The sequences of the central part of US (59), the whole of RL (55), and small parts of UL (7, 55) were reported previously. The US region was completed by using two cloned KpnI/HindIII fragments running from KpnI sites in the flanking RS elements to HindIII sites proximal to each extremity of US; these fragments, together with data from BamHI g (55) for the part of RS adjacent to RL, also provided the RS sequence. The major part of UL was sequenced with clones of HindIII fragments b, n, h, e, and a (17). Locations of genomic termini were obtained from the results of Davison and Wilkie (21).
Because of the accepted equivalence of terminal and internal copies for each pair of major repeats, we did not determine separate sequences for the whole of both copies of each repeat (TRL-IRL and IRS-TRS). Complete sequences were obtained for RL and for RS, and these were supplemented by sequences running from each extremity of UL and US into the adjacent repeat and across the joint between IRL and IRS. For the purpose of assembling the complete genome sequence, internal parts of the determined RL and RS sequences were then used for both the terminal and internal copies. The complete genomic sequence was assembled in the prototypic orientation, with a single copy of the a sequence at each terminus and one in the opposing orientation (a′) at the joint between the L and S regions.
In the genomes of both HSV-1 and HSV-2, there is an origin of DNA replication located near the center of UL (termed OriL) and also a distinct origin (OriS) with copies in both IRS and TRS (see Table 3). Studies on OriL of both HSV-1 and HSV-2 have defined this element as a palindrome of some 136 bp overall, and general experience has been that it is highly prone to deletion from plasmid clones carried in Escherichia coli (47, 64, 80). We therefore expected to obtain a version of the sequence lacking OriL from the HSV-2 HindIII e plasmid used for sequence determination across the OriL locus. In the event, sequences were obtained that represented both a majority population of molecules with OriL deleted and a minority population with OriL intact, as judged by comparison with the analysis by Lockshon and Galloway (47) of OriL in HSV-2 strain 333; i.e., the plasmid preparation contained both intact and deleted versions of the OriL sequence. We incorporated the intact version into our genomic sequence.
TABLE 3.
Locations of protein-coding regions and other features in the HSV-2 genomic sequence
| ORF or feature | Sense | Location
|
Poly(A) signal position | Size (codons) | Divergencesa
|
Comment(s)b | ||
|---|---|---|---|---|---|---|---|---|
| Start | End | Ka | Ks | |||||
| a sequence | 1 | 254 | Terminal direct repeat | |||||
| RL1 | + | 1738 | 261 | 0.272 | 0.660 | Neurovirulence factor | ||
| Exon 1 | + | 440 | 934 | 165 | ||||
| Exon 2 | + | 1089 | 1376 | 96 | ||||
| RL2 | + | 5618 | 825 | 0.316 | 0.464 | Immediate-early protein; modulator of cell state and gene expression | ||
| Exon 1 | + | 2303 | 2377 | 25 | ||||
| Exon 2 | + | 2785 | 3463 | 226 | ||||
| Exon 3 | + | 3645 | 5365 | 574 | ||||
| LAT | − | 7732 | 153827 | LAT initiation site; poly(A) site in circularized genome | ||||
| Start of UL | 9298 | |||||||
| UL1 | + | 9427 | 10098 | 11019 | 224 | 0.247 | 0.554 | Virion surface glycoprotein L |
| UL2 | + | 10211 | 10975 | 11019 | 255 | 0.095 | 0.577 | Uracil-DNA glycosylase |
| UL3 | + | 11033 | 11731 | 11800 | 233 | 0.159 | 0.646 | Nuclear phosphoprotein |
| UL4 | − | 11935 | 12537 | 11846 | 201 | 0.156 | 0.678 | |
| UL5 | − | 12607 | 15249 | 11846 | 881 | 0.065 | 0.480 | Component of DNA helicase-primase |
| UL6 | + | 15248 | 17281 | 18158 | 678 | 0.075 | 0.361 | Minor capsid protein |
| UL7 | + | 17259 | 18146 | 18158 | 296 | 0.125 | 0.379 | |
| UL8 | − | 18410 | 20665 | 18393 | 752 | 0.120 | 0.417 | Component of DNA helicase-primase |
| UL9 | − | 20718 | 23318 | 18393 | 867 | 0.064 | 0.369 | Ori binding protein |
| UL10 | + | 23209 | 24609 | 24629 | 467 | 0.116 | 0.470 | Virion membrane glycoprotein M |
| UL11 | − | 24813 | 25100 | 24809 | 96 | 0.156 | 0.367 | Myristylated tegument protein |
| UL12 | − | 25019 | 26878 | 24809 | 620 | 0.116 | 0.407 | DNase |
| UL13 | − | 26922 | 28475 | 24809 | 518 | 0.079 | 0.369 | Protein kinase; tegument protein |
| UL14 | − | 28232 | 28888 | 24809 | 219 | 0.080 | 0.278 | |
| UL15 | + | 34824 | 734 | 0.030 | 0.411 | Role in DNA packaging | ||
| Exon 1 | + | 28969 | 29997 | 343 | ||||
| Exon 2 | + | 33597 | 34769 | 391 | ||||
| UL16 | − | 30146 | 31261 | 30142 | 372 | 0.147 | 0.328 | Proposed initiator CTG codon |
| UL17 | − | 31366 | 33471 | 30142 | 702 | 0.103 | 0.454 | |
| UL18 | − | 35118 | 36071 | 35049 | 318 | 0.047 | 0.511 | Capsid protein |
| UL19 | − | 36451 | 40572 | 36275 | 1,374 | 0.033 | 0.358 | Major capsid protein (start ATG quoted is second possible) |
| UL20 | − | 40887 | 41552 | 36275 | 222 | 0.092 | 0.448 | Virion membrane protein |
| UL21 | + | 42201 | 43796 | 43808 | 532 | 0.091 | 0.424 | Tegument protein |
| UL22 | − | 44019 | 46532 | 44015 | 838 | 0.129 | 0.521 | Virion membrane glycoprotein H |
| UL23 | − | 46873 | 48000 | 46819, 46806 | 376 | 0.158 | 0.413 | Thymidine kinase (2 possible poly(A) sites) |
| UL24 | + | 47902 | 48744 | 52994 | 281 | 0.132 | 0.511 | |
| UL25 | + | 49037 | 50791 | 52994 | 585 | 0.072 | 0.433 | Virion protein; roles in penetration and virus assembly |
| UL26 | + | 51029 | 52939 | 52994 | 637 | 0.156 | 0.485 | Capsid maturation protease |
| UL26.5 | + | 51953 | 52939 | 52994 | 329 | 0.202 | 0.573 | Capsid assembly protein |
| UL27 | − | 53406 | 56117 | 53367 | 904 | 0.072 | 0.343 | Virion membrane glycoprotein B |
| UL28 | − | 56128 | 58482 | 53367 | 785 | 0.058 | 0.317 | Role in DNA packaging |
| UL29 | − | 58860 | 62447 | 58805 | 1,196 | 0.043 | 0.340 | Single-stranded DNA binding protein |
| OriL | 62862 | 62997 | Origin of DNA replication; location of palindrome given | |||||
| UL30 | + | 63265 | 66984 | 67021 | 1,240 | 0.050 | 0.329 | DNA polymerase catalytic subunit |
| UL31 | − | 66935 | 67849 | 66850 | 305 | 0.065 | 0.330 | |
| UL32 | − | 67845 | 69638 | 66850 | 598 | 0.069 | 0.416 | |
| UL33 | + | 69637 | 70026 | 71464 | 130 | 0.046 | 0.207 | Role in DNA packaging |
| UL34 | + | 70119 | 70946 | 71464 | 276 | 0.154 | 0.576 | Membrane-associated phosphoprotein |
| UL35 | + | 71061 | 71396 | 71464 | 112 | 0.090 | 0.514 | Capsid protein |
| UL36 | − | 71569 | 80934 | 71503 | 3,122 | 0.129 | 0.410 | Very large tegument protein (reiterations omitted for calculation of Ka and Ks) |
| UL37 | − | 81237 | 84578 | 81206 | 1,114 | 0.090 | 0.350 | Tegument protein |
| UL38 | + | 85061 | 86458 | 86578 | 466 | 0.115 | 0.464 | Capsid protein |
| UL39 | + | 87024 | 90449 | 91553 | 1,142 | 0.110 | 0.381 | Ribonucleotide reductase large subunit |
| UL40 | + | 90505 | 91515 | 91553 | 337 | 0.065 | 0.383 | Ribonucleotide reductase small subunit |
| UL41 | − | 91800 | 93275 | 91728 | 492 | 0.082 | 0.498 | Tegument protein; host shutoff factor; defective in HSV-2 (HG52) (see text) |
| UL42 | + | 93769 | 95178 | 95250 | 470 | 0.171 | 0.518 | DNA polymerase subunit |
| UL43 | + | 95433 | 96674 | 96689 | 414 | 0.233 | 0.476 | Probable membrane protein |
| UL44 | + | 96979 | 98418 | 99294 | 480 | 0.209 | 0.447 | Virion membrane glycoprotein C |
| UL45 | + | 98651 | 99166 | 99294 | 172 | 0.138 | 0.644 | Tegument/envelope protein |
| UL46 | − | 99432 | 101597 | 99361 | 722 | 0.139 | 0.404 | Tegument protein |
| UL47 | − | 101685 | 103772 | 99361 | 696 | 0.106 | 0.377 | Tegument protein |
| UL48 | − | 104297 | 105766 | 104201 | 490 | 0.078 | 0.525 | Tegument protein; transactivator of immediate-early genes |
| UL49 | − | 106250 | 107149 | 106213 | 300 | 0.198 | 0.522 | Tegument protein |
| UL49A | − | 107491 | 107751 | 106213 | 87 | 0.330 | 0.394 | Probable virion membrane protein |
| UL50 | + | 107759 | 108865 | 108905 | 369 | 0.151 | 0.529 | Deoxyuridine triphosphatase |
| UL51 | − | 109076 | 109807 | 109048 | 244 | 0.170 | 0.547 | |
| UL52 | + | 109875 | 113072 | 114309 | 1,066 | 0.099 | 0.462 | Component of DNA helicase-primase; ATG initiator codon quoted corresponds to HSV-1 (see text) |
| UL53 | + | 113027 | 114040 | 114309 | 338 | 0.084 | 0.484 | Membrane glycoprotein K |
| UL54 | + | 114589 | 116124 | 116133 | 512 | 0.129 | 0.482 | Immediate-early protein; posttranslational regulator of gene expression |
| UL55 | + | 116342 | 116899 | 116941 | 186 | 0.077 | 0.526 | |
| UL56 | − | 117078 | 117782 | 117044 | 235 | 0.262 | 0.563 | |
| Start of IRL | 117987 | |||||||
| LAT | + | 119518 | 127915 | LAT initiation and poly(A) sites | ||||
| RL2 | − | 121627 | 825 | 0.316 | 0.464 | Immediate-early protein; modulator of cell state and gene expression | ||
| Exon 3 | − | 121885 | 123605 | 574 | ||||
| Exon 2 | − | 123787 | 124465 | 226 | ||||
| Exon 1 | − | 124873 | 124947 | 25 | ||||
| RL1 | − | 125507 | 261 | 0.272 | 0.660 | Neurovirulence factor | ||
| Exon 2 | − | 125874 | 126161 | 96 | ||||
| Exon 1 | − | 126316 | 126810 | 165 | ||||
| a′ sequence | 126996 | 127249 | Opposite-sense copy of sequence directly repeated at genomic termini | |||||
| RS1 | − | 128079 | 132032 | 127956 | 1,318 | 0.146 | 0.296 | Immediate-early protein; transcriptional regulator |
| OriS | 132623 | 132898 | Origin of DNA replication; limits given are for directly repeated 138 nucleotides | |||||
| Start of US | 133707 | |||||||
| US1 | + | 133739 | 134977 | 135019 | 413 | 0.241 | 0.706 | Immediate-early protein; intron in 5′ noncoding region |
| US2 | − | 135167 | 136039 | 135144 | 291 | 0.163 | 0.703 | |
| US3 | + | 136325 | 137767 | 140050 | 481 | 0.150 | 0.474 | Protein kinase |
| US4 | + | 137878 | 139974 | 140050 | 699 | Virion membrane glycoprotein G | ||
| US5 | + | 140287 | 140562 | 143587 | 92 | 0.507 | 0.577 | Putative membrane glycoprotein J |
| US6 | + | 141016 | 142194 | 143587 | 393 | 0.105 | 0.471 | Virion membrane glycoprotein D |
| US7 | + | 142399 | 143514 | 143587 | 372 | 0.225 | 0.637 | Virion membrane glycoprotein I |
| US8 | + | 143843 | 145477 | 146208 | 545 | 0.173 | 0.460 | Virion membrane glycoprotein E |
| US8A | + | 145329 | 145766 | 146208 | 146 | 0.215 | 0.473 | Nucleolar protein |
| US9 | + | 145867 | 146133 | 146208 | 89 | 0.106 | 0.559 | Tegument protein |
| US10 | − | 146628 | 147533 | 146618 | 302 | 0.238 | 0.546 | Virion protein |
| US11 | − | 147247 | 147699 | 146618 | 151 | 0.261 | 0.464 | Nucleolar, RNA binding protein |
| US12 | − | 147778 | 148035 | 146618 | 86 | 0.303 | 0.647 | Immediate-early protein; inhibitor of antigen presentation; intron in 5′ noncoding region |
| Start of TRS | 148036 | |||||||
| OriS | 148844 | 149119 | Origin of DNA replication; limits given are for directly repeated 138 nucleotides | |||||
| RS1 | + | 149710 | 153663 | 153781 | 1,318 | 0.146 | 0.296 | Immediate-early protein; transcriptional regulator |
| a sequence | 154493 | 154746 | Terminal direct repeat | |||||
In a study of HSV-2 gene UL41 (which encodes a protein involved in shutting down host gene expression and is nonessential for growth in tissue culture), Everett and Fenwick (26) showed that in strain HG52 this gene is defective by reason of a frameshift mutation in its coding region (with this conclusion based on sequence data for four independent molecular clones). Our data concurred: relative to HSV-2 strain G, a single nucleotide was missing within a homopolymeric run of G residues at nucleotides 92249 to 92255. This posed for us the question of how best to represent the complete genomic sequence, in particular for lodging in sequence libraries. We decided that it would be most generally useful to have a library entry with a complete reading frame for gene UL41, and we have therefore inserted an extra G at position 92256 in our HSV-2 HG52 sequence.
The complete genome sequence of HSV-2 strain HG52 thus obtained comprised 154,746 bp with a G+C content of 70.4%. The sizes and base compositions of the major regions of the HSV-2 genome are shown in Table 1. The elevated G+C compositions of the major repeat elements, in particular of RS, are striking. As with other herpesviruses, the genome of HSV-2 contains families of short reiterated sequences. The copy numbers of these elements are represented in the assembled sequence as found in the plasmid clones sequenced. Details of families of reiterations in HSV-2 RL have been published previously (55). In HSV-2 UL there are two reiterated sets: at nucleotides 72098 to 72266 within the coding region of gene UL36 and at nucleotides 106045 to 106165 between genes UL48 and UL49 (18, 32). There are two regions of reiterated sequences in HSV-2 RS: one lies between the a sequence and the RS1 gene at nucleotides 127672 to 127914 in IRS and 153828 to 154070 in TRS, and the other, a complex assemblage of reiterations previously described by Whitton and Clements (81), is near the RS-US boundaries at nucleotides 133227 to 133645 in IRS and 148097 to 148515 in TRS. In addition, OriS in HSV-2 HG52 is in effect duplicated relative to its HSV-1 strain 17 counterpart and comprises two adjacent similarly oriented copies of a 138-bp sequence (each copy containing an imperfect palindrome) (82). There are no reiterated families in HSV-2 US.
TABLE 1.
Summary of regions in the genomes of HSV-1 and HSV-2
| Region | Virus | Length (bp) | % G+C | No. of genesa |
|---|---|---|---|---|
| RL (as TRL) | HSV-1 | 9,212 | 71.6 | 2 |
| HSV-2 | 9,297 | 75.4 | 2 | |
| UL | HSV-1 | 107,947 | 66.9 | 58 |
| HSV-2 | 108,689 | 68.9 | 58 | |
| US | HSV-1 | 12,980 | 64.3 | 13 |
| HSV-2 | 14,329 | 66.2 | 13 | |
| RS (as TRS) | HSV-1 | 6,677 | 79.5 | 1 |
| HSV-2 | 6,711 | 80.1 | 1 | |
| Whole genome | HSV-1 | 152,261 | 68.3 | 74 |
| HSV-2 | 154,746 | 70.4 | 74 |
Estimates of the number of genes encoding distinct proteins, as discussed in the text.
Evaluation of the gene content of HSV-2.
We have previously published sequences for a number of genes of HSV-2 strain HG52, and other authors have reported sequences for various genes from several HSV-2 strains, as summarized in Table 2.
TABLE 2.
Published sequences of HSV-2 genes
| Gene(s) or elementa | Strain(s) | Length (bp) | Reference |
|---|---|---|---|
| UL1–UL5 (part) | HG52 | 5,829 | 55 |
| UL11–UL13 (part) | HG52 | 2,469 | 24 |
| UL23, UL24 (part) | 333 | 1,601 | 70 |
| UL26, UL26.5 | G | 2,151 | 68 |
| UL27, UL28 (part) | HG52 | 3,325 | 11 |
| UL27, UL28 (part) | 333 | 3,473 | 69 |
| UL28 (part), UL29 | 8620K | 5,105 | 75 |
| OriL | 333 | 326 | 47 |
| UL30, UL31 (part) | 186 | 3,769 | 76 |
| UL38 | G | 2,158 | 84 |
| UL39, UL40 (part) | 333 | 3,998 | 71 |
| UL39 (part)–UL41 (part) | 333 | 1,890 | 28 |
| UL40 (part)–UL42 (part) | HG52, G | 3,653 | 26 |
| UL43 (part)–UL46 (part) | 333 | 3,410 | 72 |
| UL47 (part), UL48 | HG52 | 2,211 | 18 |
| UL48 | 333 | 2,228 | 32 |
| UL49A | HG52 | 2,851 | 7 |
| UL52 (part), UL53 | HG52 | 1,384 | 22 |
| UL53 (part)–RL1 | HG52 | 13,680 | 55 |
| UL54 (part) | HG52 | 773 | 83 |
| LAT, RL2 (part) | 333 | 3,314 | 41 |
| a sequence | HG52 | 348 | 21 |
| OriS | HG52 | 1,100 | 82 |
| US1 (part) | HG52 | 1,560 | 81 |
| US2–US8 (part) | HG52 | 9,629 | 59 |
| US6 | G | 1,635 | 79 |
| US7 | 333 | 1,649 | 36 |
| US8 | 1,880 | 14 | |
| US12 (part) | HG52 | 1,072 | 81 |
Previously published sequences that contributed directly to the complete sequence for HSV-2 (strain HG52) are in boldface.
Protein-coding genes of HSV-2 were identified primarily by comparison with corresponding sequences in the genome of HSV-1, since the DNA sequences for HSV-1 and HSV-2 are closely similar by the standards of the herpesvirus family (see the following section). Corresponding coding regions were in general readily identified by alignment of sections of HSV-1 and HSV-2 DNA sequences and by similarity between encoded amino acid sequences. Translational start and stop signals identified for equivalent HSV-1 and HSV-2 genes mostly correspond closely. In both genomes most genes show a strongly marked pattern of codon usage characterized by a very high proportion of G and C residues in the third positions of the set of codons employed (as expected in a G+C-rich genome, given the properties of the genetic code). As outlined in the following section, the patterns of divergence between the HSV-1 and HSV-2 DNA sequences also support our interpretation of gene content. In addition, we searched the HSV-2 genome sequence for possible additional protein-coding regions by using criteria of codon usage plus similarity and divergence between HSV-1 and HSV-2, and gene US8A was discovered by this route (see below). Finally, most of the proposed HSV proteins have recognizable homologs inferred from DNA sequences for other herpesviruses (for instance, varicella-zoster virus [20] and equine herpesvirus 1 [73]), and this too helps to ensure the validity of coding assignments.
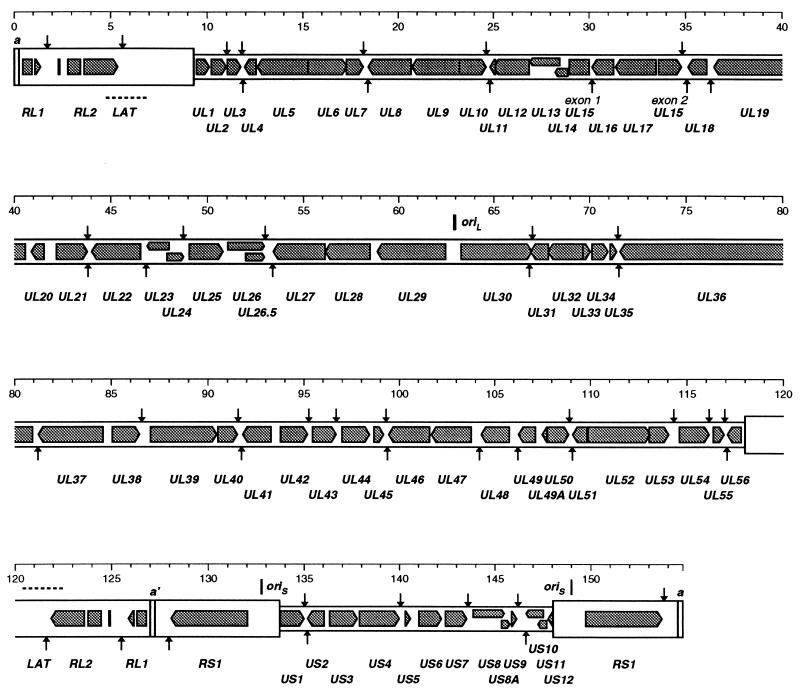
We use for HSV-2 genes the nomenclature previously employed for HSV-1 (56–58, 62) and parts of HSV-2 (7, 55, 59). Table 3 and Fig. 2 present the set of HSV-2 protein-coding genes that we consider, from a critical and conservative viewpoint, can presently be identified with confidence. These number 58 in UL, 2 in each copy of RL, 1 wholly contained within each copy of RS, and 13 in US (of which 2 extend their 5′ noncoding regions into the adjacent RS elements), that is, 74 distinct genes, three of which are present in two copies each. Table 3 also lists the latency-associated transcript (LAT), which we presently regard as probably not protein coding (55). Each HSV-2 gene listed has an HSV-1 homolog, and all occur in locations and orientations corresponding to the HSV-1 versions. On the basis of limited available transcript mapping data for HSV-2 and the occurrence of AATAAA sequences associated with polyadenylation, HSV-1 and HSV-2 appear to have very closely equivalent sets of transcripts, with identical groupings of adjacent, similarly oriented genes into 3′-coterminal transcript families.
FIG. 2.
Layout of genes and other elements in the genome of HSV-2. The genome is shown expanded from the representation in Fig. 1, with unique regions as narrow open boxes and major repeats as wider boxes, on four successive lines; sequence numbering (in kilobases) is indicated above each section. Positions of the terminal repeat (a) sequences and the internal inverted copy (a′) are marked. Protein-coding regions and orientations for recognized genes as listed in Table 3 are shown as grey-filled arrow shapes (non-3′ exons are shown as box shapes). Locations of proposed transcript polyadenylation signals (AATAAA and variants thereof) are marked by small arrows (for rightward transcripts these are above the genome, and for leftward transcripts they are below). The positions of origins of replication are marked.
When we published the complete genome sequence of HSV-1 in 1988, we identified 70 genes encoding distinct proteins (56). As of the middle of 1997, four further HSV-1 protein-coding genes that we consider should be definitely added to the list have been identified, namely, UL26.5 (45, 63), UL49A (5, 7), RL1 (15, 23), and US8A; all of these have HSV-2 homologs. Given the conservation of gene content between HSV-1 and HSV-2 and the various facets of evidence supporting assignments, we have confidence in the reality of all the gene assignments listed in Table 3. The status of possible additional protein-coding genes is discussed below.
Comparisons of the HSV-1 and HSV-2 genomes.
As noted above, the genomic sequences of HSV-1 and HSV-2 are closely related. Comparison of HSV-2 sequence data with the previously determined sequence of HSV-1 strain 17 was therefore included as a late-stage check during the sequencing process. This revealed occasional problems in the HSV-2 data that required further attention and also led to the uncovering of several errors in the published HSV-1 sequence. Changes to UL56 and RL1 of HSV-1 were reported previously (23, 55), and we now add corrections in the UL14, UL46, and US8A coding sequences; these are all described in Table 4 and in the next section. To the best of our understanding, these changes do not affect interpretation of HSV-1 genome organization outside the reading frames named. The revisions to the HSV-1 sequence have been communicated to the EMBL Sequence Library (accession no. X14112).
TABLE 4.
Corrections to the genome sequence of HSV-1 strain 17
| Gene | Locationa | Alteration | Reference |
|---|---|---|---|
| RL1 (TRL copy) | 780 | Change T to C | 23 |
| 818 | Change C to G | ||
| 823 | Delete CG | ||
| UL14 | 28866 | Change C to GCG | This paper |
| UL46 | 100407 | Insert G | This paper |
| 100446 | Delete G | ||
| UL56 | 116344 | Insert CG | 55 |
| RL1 (IRL copy) | 125547 | Delete CG | 23 |
| 125553 | Change G to C | ||
| 125591 | Change A to G | ||
| US8A | 143173 | Insert G | This paper |
Numbers refer to the DNA sequence of HSV-1 (strain 17) as listed in reference 56.
HSV-1 and HSV-2 comprise the most closely related pair of herpesviruses for which complete genome sequences are presently known, but their genomes are nonetheless substantially diverged; in an analysis of herpesvirus phylogeny, we estimated the divergence date of the two HSV lineages as 8 million years ago (53, 54). Comparisons of aspects of their genome structures should help in interpreting the contents of the genomes and in assessing differences in functional capabilities of the viruses and will also be of interest in illuminating a relatively early stage of evolutionary divergence. In this paper our attention is primarily on aspects of gene content and on a comparative description of the two genomes.
The determined lengths of the HSV-1 and HSV-2 sequences are 152,261 and 154,746 bp, respectively. Neither of these values is unique, since high-frequency variation in lengths of homopolymer runs and in copy numbers of families of short reiterated sequences will act to distribute lengths of genome molecules over a range of perhaps several hundred base pairs. As shown in Table 1, the major difference in length is located in the US region, which in HSV-2 is 1,349 bp longer. This is almost all attributable to the US4 gene, which in HSV-1 appears to have suffered a large internal deletion (50, 59). UL is 742 bp longer in HSV-2, the net result of many small differences in lengths (in both directions) between corresponding coding and noncoding constituents of the two UL sequences. The sizes determined for each of the major repeats are very close for the two genomes. As was previously well known from buoyant density analyses of the virus DNAs, HSV-2 has a slightly more G+C-rich genome than HSV-1 (2.1% higher overall from the sequences). The difference is distributed across the genome, being greatest in RL and least in RS (Table 1). Within coding regions, the effect is most pronounced in the sets of nucleotides constituting the third positions of codons (data not shown).
Characteristics of the aligned pairs of protein-coding regions were examined for all genes (except US4, which in HSV-1 is grossly truncated). This set of alignments required introduction of gapping characters representing 2% of the total alignment length. Excluding such gapped regions, the overall incidence of identical aligned nucleotides was 83%. Noncoding regions are typically much less conserved; this topic was previously discussed in some detail for the major part of US (59) and for RL, which contains the most diverged regions of the genomes (55). The families of short reiterations found in both genomes are conserved only partially in their locations and are distinct in their sequences.
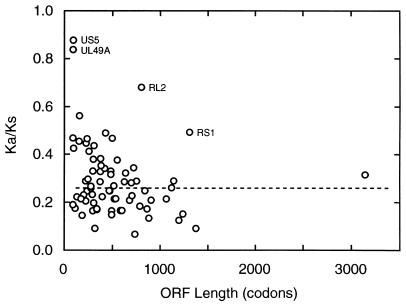
Nucleotide substitutional differences between aligned pairs of coding regions were examined for all genes, except US4, in terms of whether they correspond to a substitution at the amino acid level. The general expectation is that across the gene set the synonymous divergence, Ks, should be approximately constant and that Ks should be greater than the nonsynonymous divergence, Ka. Table 3 lists Ka and Ks for pairs of aligned HSV-1 and HSV-2 gene sequences. In every case Ks is higher than Ka. There is substantial variation within each of the Ks and Ka datasets; in both sets differences from the mean diminish sharply with increasing size of open reading frames (ORFs), suggesting pronounced stochastic effects, especially with the values for the smaller ORFs. Number averages taken over the whole set are 0.47 for Ks and 0.14 for Ka. Plotting the Ka/Ks ratio against ORF length gives a good view of the relative magnitudes of the two measures of divergence for single genes and the spread of values, as shown in Fig. 3. There are four outliers in this plot that have notably high Ka/Ks ratios for their ORF lengths, namely, UL49A, US5, RL2, and RS1. We consider that these can all be adequately rationalized in terms of specific peculiarities and do not have any wider significance for our present purposes. The first two are very small genes considered to encode membrane proteins (7, 58), and the high relative incidence of nonsilent mutations probably reflects both stochastic effects and nonstringent requirements for amino acid sequence conservation. For RL2 and RS1 the atypical substitution ratios may be associated with the genes’ locations in G+C-rich major repeat elements, where recombinatory processes are thought to be particularly active (57), and also with the sensitivity of procedures estimating Ka and Ks toward inaccuracy with sequences of highly biased base composition (39). It is clear from this analysis that there are no genes showing strong positive selection effects overall (i.e., with Ka greater than Ks). For the great majority of identified genes in HSV-1 and HSV-2, the pattern of nucleotide substitutions accumulated during the divergent evolution of the two genomes is thus thoroughly consistent with coding assignment.
FIG. 3.
Plot of Ka/Ks ratio versus length of coding region for each aligned pair of HSV-1 and HSV-2 genes. The ratio of nonsynonymous to synonymous divergences (Ka/Ks; see Table 3) for pairs of HSV-1 and HSV-2 coding sequences (excluding UL26.5 and US4) is plotted against the corresponding ORF length (average of HSV-1 and HSV-2 lengths). The median value for Ka/Ks is indicated with a dashed line. Plotted points for four outliers with high Ka/Ks values are annotated with gene names.
Notes on individual genes and proteins of HSV-1 and HSV-2.
In this section we present background, evaluation, and comments on certain genes of HSV-1 and HSV-2 where sequence determination and interpretation raised particular points of difficulty or interest not previously discussed. Table 3 includes a summary of gene functions, mostly as understood from studies on HSV-1 (19, 52, 60).
(i) Gene UL14.
The sequences of HSV-1 and HSV-2 showed a relative frameshift near the 5′ end of the UL14 ORF, which was resolved by the finding of an error in the HSV-1 sequence on reexamination of the region (Table 4). The UL14 ORF of HSV-1 now starts at an ATG upstream of the previously proposed start, so that 14 codons at the 5′ end of the old version are replaced by 18 new codons.
(ii) Gene UL16.
The HSV-2 UL16 ORF does not possess any candidate ATG for translational initiation, and we have assigned translational initiation to the codon CTG (Leu) aligned with the ATG of HSV-1 UL16. Sequence analysis of this region for the distinct HSV-2 strain 25766 gave a sequence identical to that of HSV-2 HG52. We note that CTG (CUG) is a known start codon for eukaryotic genes and indeed is the most frequently described among non-AUG start codons (8).
(iii) Gene UL19.
The position of the start codon for the HSV-2 UL19 ORF as listed in Table 3 is equivalent to that proposed for HSV-1. There is another possible ATG start codon in the HSV-2 sequence, 10 codons upstream and in frame, which lacks an HSV-1 counterpart.
(iv) Gene UL36.
In both HSV-1 and HSV-2 the very large UL36 ORF (encoding a tegument protein) contains a set of reiterated sequences, which are presumed to be translated. The HSV-1 set encodes the amino acid sequence (PQ)35, while the distinct set in HSV-2 encodes (PQPPL)11. This region in the protein thus has the appearance of a flexible linker, whose requirement for conservation of sequence and length is not stringent.
(v) Genes UL46 and UL47.
There are published sequences for genes UL46 and UL47 for both HSV-1 strain 17 and HSV-1 strain F, and for both genes the pairs of aligned sequences show relative frameshifting differences (56, 61). We have corrected a local frameshifted region affecting codons 170 to 182 in UL46 of HSV-1 strain 17 after redetermining the sequence for the region (Table 4). For both genes, comparisons with the HSV-2 versions suggest that differences remaining between the sequences of the two HSV-1 strains probably represent frameshifts in the HSV-1 strain F data.
(vi) Gene UL52.
The HSV-2 UL52 ORF of 1,066 codons as listed in Table 3 is the distal portion of an ATG-initiated, 1,240-codon ORF that extends over most of the adjacent, oppositely oriented UL51 ORF. The upstream part was discounted because it does not have an equivalent in HSV-1 or other herpesviruses, and the start codon given in Table 3 corresponds closely to that in HSV-1.
(vii) Gene US8A.
US8A is an additional reading frame proposed for both HSV-2 and HSV-1, overlapping the 3′ portion of the US8 ORF and extending into the region between US8 and US9. The presence of a HSV-1 gene (US8.5) in this location and detection of its product in infected cells were described by Georgopoulou et al. (31). Our assignment differs in that the sequence correction shown in Table 4 changes the reading frame from codon 144 of the 159-codon HSV-1 US8A ORF. Among members of the Simplexvirus genus, DNA sequences corresponding to this genomic region are also available for herpesvirus B and for simian agent 8 (25, 40). We have found counterparts of US8A (not detected by the original investigators) in both these sequences. In both cases frameshift corrections have to be proposed, but the conservation of encoded protein sequences is compelling for the C-terminal region, as shown in Fig. 4. In the Varicellovirus genus (the other genus in the Alphaherpesvirinae subfamily), no convincing homologs were found, although there is a possible counterpart in equine herpesvirus 1 (gene 75) (73) with no sequence similarity but in a corresponding genomic location. Our current evaluation is that the US8A gene family is probably specific to the Simplexvirus genus. Its function remains obscure.
FIG. 4.
US8A amino acid sequences for HSV-1, HSV-2, and two simian alphaherpesviruses. Proposed amino acid sequences for US8A in HSV-1 and HSV-2 are aligned together with sequences of counterparts for herpesvirus B (BV) and simian agent 8 (SA8) as interpreted from the DNA sequence data of references 40 and 25, respectively. In the consensus (Cons) line, completely conserved residues are indicated in uppercase and residues conserved in three of the sequences are indicated in lowercase. The position corresponding to the end of the US8 ORF in the DNA sequences (−1 frame relative to US8A) is marked (– – – |). In BV and SA8 a region of uncertainty representing the limits for a proposed shift of reading frame in the reported DNA sequence is shown as bounded by //// and \\\\, with inserted gapping characters indicated by dots. The position corresponding to a presumptive frameshift in SA8 only is marked by •. The location corresponding to the identically placed end of the ORF in all four sequences is marked ∗.
(viii) Gene US12.
The products of US12 (ICP47 or Vmw12) have been shown for both HSV-1 and HSV-2 to interdict antigen presentation in infected cells by binding to the TAP transporter (27, 29, 34, 35, 85). Optimization of alignment of the two coding sequences (by using Bestfit with default and also other values for gapping parameters) indicated a relative deletion of 13 nucleotides in the HSV-1 sequence around codon 59, which was judged to be convincing in terms of the incidence of identical aligned residues that it gave (62% in the 23 following codons, versus 72% in the 58 upstream codons, in heteropolymeric sequences and without any other gapping). This implies that the two sequences would be read in different frames for the distal 24 and 23 codons of the HSV-1 and HSV-2 ORFs, respectively (29). No error was apparent in either DNA sequence, and both US12 proteins have been sufficiently characterized to give good confidence in the interpretation of the coding arrangements. Thus, the shift of reading frame is taken as genuine, being the result of a mutation that has occurred in one of the HSV lineages, and this is consistent with experimental data showing that the C-terminal parts of the proteins are not required for TAP binding (27, 29, 35, 85). This is the only unambiguous instance of a frameshift within still-functional coding regions that emerged from comparisons of the two HSV genomic sequences.
US12 gene homologs are known only for HSV-1 and HSV-2 and appear to be specific to the Simplexvirus genus. It thus seems possible that this small gene evolved de novo within this virus group. Nuclear magnetic resonance and circular dichroism analyses of the HSV-2 US12 protein indicate an absence of stable secondary structure (6), so the protein may be better regarded as an oligopeptide with a TAP binding epitope than as a typical globular protein. This view is also thoroughly compatible with the small size of the peptide chain and the lack of conservation in the C-terminal part and is particularly attractive in considering evolution of the gene de novo.
Evaluation of possible additional genes of HSV-1 and HSV-2.
In addition to the 74 distinct genes identified in this paper, other HSV-1 genes have been proposed in recent years. For some of the latter there are readily visible equivalents in the HSV-2 sequence, and for others there are not. The possible extra genes of HSV-1 and HSV-2 fall into three classes with respect to their relationship to genes in the canonical list of Table 3. In the first class are those with coding regions based on a translational start internal to and in frame with an established coding ORF, and in the second class are genes that occupy the same DNA sequence as a recognized coding ORF but in the opposite sense. Finally, some proposed ORFs are in regions distinct from established ORFs.
The use of an internal start in an ORF is now well established for the UL26.5 gene in HSV-1 (46, 65) and HSV-2 (68) and for homologs in other herpesviruses (37). In summary, the 5′ portion of the UL26 ORF encodes a proteinase activity, and the 3′ portion (UL26.5) encodes a scaffolding component for capsid assembly. Two transcripts are produced, which initiate translation, respectively, at the 5′ end of the ORF (UL26) and internally (UL26.5). Processing of the resulting translation products is carried out by the proteinase. Evidently the proteinase and scaffolding functions are each active both in the product representing the whole UL26 ORF and in the relevant separate domain.
The next best characterized case in HSV-1 concerns gene UL12, encoding DNase. It has long been known, for both HSV-1 and HSV-2, that there is a transcript initiated within the UL12 ORF with an ATG codon appropriately located in the UL12 reading frame to act as a translational initiator in this RNA (24). It has recently been shown for HSV-1 that this 3′ portion of the UL12 ORF (termed UL12.5) is translated as expected from the transcript described and that the protein product is present in virions (9, 49). However, no distinct phenotype has yet been shown to depend on expression from the UL12.5 start, and we have therefore omitted UL12.5 from our gene list, at least for now. A less well characterized but formally similar case concerns the so-called UL8.5 ORF of HSV-1 (actually the 3′ portion of UL9) (4), for which a late transcript, an initiator ATG in the UL9 ORF, and a protein product have been identified. We note that an equivalent ATG exists in the HSV-2 sequence (nucleotides 22178 to 22176). The same phenomenon has been described for HSV-1 US1 (12), but in this case there is not an equivalently placed ATG in HSV-2. The final example concerns HSV-1 gene UL15, which has two exons, both protein coding. A protein species smaller than the product of the complete gene that apparently is translated from all or part of the downstream exon only and is probably the product of a separate translational initiation event and separate mRNA has been observed; no distinct phenotype associated with this UL15.5 protein has been detected (1, 2, 86). There are several ATG codons in the HSV-1 exon 2 ORF that might act as translational initiators for the protein, and all are conserved in HSV-2.
Turning to the class of proposed genes with ORFs antisense to already recognized functional ORFs, we address two instances where the HSV-1 loci have been investigated experimentally, namely, UL43.5 and ORF P. We note first that ORFs other than authenticated protein-coding ORFs are rather common in the HSV genomes, as a consequence of the high G+C contents and the resulting low incidence of adventitious stop codons. This topic is developed further in Discussion.
Ward and colleagues (78) have described expression in HSV-1-infected cells of a protein from a 311-codon ORF antisense to UL43, termed UL43.5. The protein was produced late in infection and was associated with nuclear structures. The UL43 region is not essential for growth of virus in tissue culture (48). There is no counterpart in the HSV-2 sequence to the ATG that in HSV-1 opens the UL43.5 ORF, and the equivalent HSV-2 reading frame has stop codons at positions corresponding to codons 16 and 68 in the HSV-1 ORF. A resemblance to a motif of aminoacyl-tRNA synthetase noted in the HSV-1 UL43.5 amino acid sequence (78) is absent from the HSV-2 counterpart, and its locus lies between the two stop codons mentioned. After the second stop codon, the HSV-2 reading frame is open until a location equivalent to five codons past the end of the HSV-1 ORF, but the first potential initiating ATG is well downstream in this HSV-2 ORF at a position equivalent to codon 163 of HSV-1 UL43.5. The HSV-2 sequence thus does not appear to be a likely candidate for a protein-coding region.
ORF P lies in the RL element of HSV-1 within the region traversed by LATs and in the LAT orientation. It is antisense to and largely overlaps the ORF of gene RL1 (43). Mutation of an ICP4 binding motif in the proposed promoter for ORF P allowed expression of the ORF in infected cells, giving a protein of 248 residues in HSV-1 strain F and 233 residues in HSV-1 strain 17 (with the difference resulting from distinct copy numbers of a reiterated element). The ORF P protein was observed to associate with splicing factors, and it was proposed that it could have a gene-regulatory function, perhaps in latently infected cells (10, 42). In HSV-2 the 5′ portion of the ORF P region is preserved, with a putative ATG initiator codon corresponding to that in HSV-1. This 5′ part of ORF P corresponds on the opposite strand to a part of the RL1 coding sequence specifying a 63-amino-acid sequence that is highly conserved among HSV-1, HSV-2, and cellular homologs (16, 51). Thus, by the criteria of sequence interpretation, the similarity of the 5′ parts of the ORF P frames in HSV-1 and HSV-2 appears to be primarily as a consequence of coding requirements of the RL1 gene. The HSV-2 RL1 gene contains an intron (33, 55), unlike its HSV-1 equivalent, and in the ORF P phase the intron sequence causes divergence from ORF P of HSV-1 after a position corresponding to codon 48 of HSV-1 ORF P. After a 50-codon stretch traversing the intron sequence (which consists mostly of reiterations of a 19-bp sequence), the HSV-2 ORF reenters sequence with an HSV-1 counterpart but is now out of phase with HSV-1 ORF P, and it then terminates after 130 codons in all. These pronounced differences render it rather unlikely that HSV-1 ORF P and the HSV-2 ORF encode equivalent functional proteins.
In the third class of potential additional genes, a number of coding regions in HSV-1 that are unrelated to other ORFs have been proposed. These include ORFs in the LAT region of RL (77) and an ORF across OriS (38). These are not conserved convincingly in HSV-2, as previously discussed for LAT (55).
This section has outlined the background for certain HSV-1 candidate genes and a comparative interpretation of corresponding loci in the HSV-2 genome. We found that this analysis fell short of yielding firm conclusions on protein coding and functionality, in that there was no strong support for extra HSV-2 genes but negative evaluations were not definitive; discussion of this topic is continued below.
DISCUSSION
As presented in this paper, HSV-1 and HSV-2 possess corresponding sets of 74 genes that encode distinct proteins. From the characteristics of the genomic sequences, from the comparative analyses described above, and from published experimental work on many genes, particularly of HSV-1, we have good confidence that these gene assignments are valid. The next question that arises about the gene sets of the viruses is whether the present listings are complete. Several extra genes have been proposed for HSV-1, and in some cases apparently substantive experimental evidence is published. We have found evaluation of the limits of the protein-coding capacities to be a complex and vexed matter, as outlined below.
Dealing first with proposed genes in which the coding sequence comprises a distal, in-frame subsection of the coding sequence of another gene, we noted above that most of the HSV-1 cases have equivalents in the HSV-2 sequence (“UL8.5,” UL12.5, and UL15.5, as well as the thoroughly characterized UL26.5), while US1 does not. However, all that is required for an equivalent downstream ORF to be registered for HSV-2 is conservation of a Met codon at the appropriate location. We are concerned that designating such sections of ORFs as distinct genes should be a matter of some caution. The UL26.5 gene is presently exceptional in this class in that an explicit functional basis is known, which clearly justifies the separate name. It appears that in the other examples, the novel transcript and/or protein species are typically observed late in the infectious cycle, that is, in a situation where control of virus gene expression may be collapsing, so it could be argued that functional significance is dubious. It may be that experimental observation of distinct, explicable phenotypes associated with a complete ORF (or, in the case of UL15, set of exons) and a sub-ORF will provide the only definitive evidence for these cases, and we consider that this state has not been attained for any of the proposed genes in this class, in either HSV-1 or HSV-2.
Turning to additional HSV-2 genes based on distinct ORFs (including antisense ORFs), the primary examples from HSV-1 in this class are UL43.5 and ORF P. As outlined above, neither of these has a compellingly conserved counterpart in HSV-2, so our evaluation is that the existence of functional HSV-2 homologs is unlikely in these cases. The topic of possible extra coding sequences in both genomes can be broadened: as already noted, both HSV-1 and HSV-2 sequences possess many ORFs in addition to those in the canonical gene set. To illustrate this point, Table 5 lists by length class the numbers of complete ORFs and of ATG-initiated ORFs for the HSV-2 sequence and compares these with the numbers of identified genes: the incidence of unassigned ORFs, up to quite large sizes, is striking. These data can be viewed in two complementary ways. First, they provide context, demonstrating that there is nothing out of the ordinary about any given moderately sized ORF that lacks a gene assignment. Second, they suggest, at least superficially, that here is a potential resource which evolution might turn to a protein-coding role in at least some cases.
TABLE 5.
ORFs in the HSV-2 sequence
| Length range (codons) | No. of:
|
|||
|---|---|---|---|---|
| ORFsa
|
Genes as M-ORF equivalentsb | Unassigned M-ORFs | ||
| C-ORFs | M-ORFs | |||
| 51–100 | 752 | 200 | 6 | 194 |
| 101–200 | 416 | 143 | 7 | 136 |
| 201–400 | 212 | 79 | 25 | 54 |
| 401–600 | 52 | 24 | 16 | 8 |
| 601–800 | 23 | 12 | 10 | 2 |
| 801–1,000 | 7 | 7 | 4 | 3 |
| 1,001–1,200 | 5 | 3 | 3 | 0 |
| 1,201–1,400 | 7 | 4 | 4 | 0 |
| >1,400 | 1 | 1 | 1 | 0 |
ORFs lying wholly within RL or RS were counted once only. C-ORF denotes a complete ORF, and M-ORF denotes an ATG-initiated ORF.
Data for genes are based on Table 3, but with reading frames scored for length as their M-ORF equivalents.
Our evaluation of the presence of protein-coding regions primarily involved criteria based on characteristics of DNA and protein sequences, as distinct from experimental observation of transcripts and proteins or from functional studies. We were thus concerned with evolutionary conservation and divergence of coding DNA sequences, with similarities in predicted protein sequences, and with patterns of codon usage and amino acid composition (these last two are not detailed in this paper). This approach clearly distinguished properties of the standard protein-coding ORFs from those of other frames and regions. However, a potential weakness is that it might fail to discern functional coding regions that are far from the norm for these criteria. For instance, there might exist genuinely novel protein-coding sequences that have arisen only recently on an evolutionary time scale and are therefore present in only one of the two HSV genomes and are as yet unmarked by evolutionary change, and these could present as ORFs with atypical characteristics. We thus consider that analysis of sequence characteristics gives a clear positive evaluation of coding potential, but a negative indication must in principle be less than definitive. We have to conclude that if candidate genes of atypical structure are to be validated, then discriminating experimental analysis has to represent the final criterion. Again, we consider it appropriate to adopt a conservative outlook on adding candidate genes to the canonical list. While our main concern in this paper is with HSV-2’s genetic content, we regard the most-studied novel genes of HSV-1, UL43.5 and ORF P, as also still being of tentative status. For these and other antisense transcripts, the possibility of a regulatory role through annealing to complementary, sense transcripts remains a possibility, separate from any direct protein-coding function (13).
Concerning the possible correlation of genome contents with biological differences between HSV-1 and HSV-2, we consider it quite likely that, notwithstanding these unresolved aspects concerning the standing of extra genes, it might well turn out to be the case that there are no HSV-1 or HSV-2 genes that lack a homolog in the other virus. Nor is there any substantive indication in nucleotide substitution patterns (i.e., Ka high relative to Ks) of genes under strong differential selection between the two lineages, which might reflect evolutionary divergence into particular niches in the host organism. There is one major difference between genes that does command attention in this context: relative to HSV-2, there is a large deletion in the HSV-1 US4 gene, encoding virion glycoprotein G (59), and it remains plausible that this could correspond with some key difference (for instance, in cell tropisms). Nonetheless, our judgment is that differences in the behaviors of the viruses are likely for the most part to arise from multiple local differences in sequences of many proteins.
The HSV-2 genome sequence should provide a powerful aid to work on HSV-2 per se, integrating the previous quite extensive but incomplete and scattered information on sequences of HSV-2 genes. The analyses described in this paper were in essence comparative, with interpretation of the HSV-2 sequence depending heavily on the wider knowledge of HSV-1 gene content, and with availability of the two genomic sequences, significantly diverged but still quite close in evolutionary terms, allowing some interesting insights. Finally, the sequence for HSV-2 completes (belatedly) the set of determined genomic sequences for all of the eight known human herpesviruses.
ACKNOWLEDGMENTS
We thank A. J. Davison for supplying most of the HSV-2 plasmids used and for critical discussions.
B.C.B. was supported by SmithKline Beecham during part of the project.
REFERENCES
- 1.Baines J D, Cunningham C, Nalwanga D, Davison A. The UL15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the UL15 gene product. J Virol. 1997;71:2666–2673. doi: 10.1128/jvi.71.4.2666-2673.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Poon A P W, Rovnak J, Roizman B. The herpes simplex virus UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J Virol. 1994;68:8118–8124. doi: 10.1128/jvi.68.12.8118-8124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bankier A T, Barrell B G. Sequencing single-stranded DNA using the chain-termination method. In: Howe C J, Ward E S, editors. Nucleic acids sequencing: a practical approach. Oxford, United Kingdom: IRL Press; 1989. pp. 37–78. [Google Scholar]
- 4.Baradaran K, Dabrowski C E, Schaffer P A. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9 and UL10 genes and identification of a novel delayed-early gene product, OBPC. J Virol. 1994;68:4251–4261. doi: 10.1128/jvi.68.7.4251-4261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker D E, Roizman B. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992;66:562–566. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barlow, P. N., H. W. M. Moss, and D. J. McGeoch. Unpublished data.
- 7.Barnett B C, Dolan A, Telford E A R, Davison A J, McGeoch D J. A novel herpes simplex virus gene (UL49A) encodes a putative membrane protein with counterparts in other herpesviruses. J Gen Virol. 1992;73:2167–2171. doi: 10.1099/0022-1317-73-8-2167. [DOI] [PubMed] [Google Scholar]
- 8.Boeck R, Kolakofsky D. Positions +5 and +6 can be major determinants of the efficiency of non-AUG initiation codons for protein synthesis. EMBO J. 1994;13:3608–3617. doi: 10.1002/j.1460-2075.1994.tb06668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronstein J C, Weller S K, Weber P C. The product of the UL12.5 gene of herpes simplex virus type 1 is a capsid-associated nuclease. J Virol. 1997;71:3039–3047. doi: 10.1128/jvi.71.4.3039-3047.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruni R, Roizman B. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc Natl Acad Sci USA. 1996;93:10423–10427. doi: 10.1073/pnas.93.19.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bzik D J, Debroy C, Fox B A, Pederson N E, Person S. The nucleotide sequence of the gB glycoprotein gene of HSV-2 and comparison with the corresponding gene of HSV-1. Virology. 1986;155:322–333. doi: 10.1016/0042-6822(86)90196-0. [DOI] [PubMed] [Google Scholar]
- 12.Carter K L, Roizman B. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J Virol. 1996;70:172–178. doi: 10.1128/jvi.70.1.172-178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter K L, Ward P L, Roizman B. Characterization of the products of the UL43 gene of herpes simplex virus 1: potential implications for regulation of gene expression by antisense transcription. J Virol. 1996;70:7663–7668. doi: 10.1128/jvi.70.11.7663-7668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi S Y, Seong Y R, Lee E K, Chon S K, Yoo W D, Lee C K, Im D S. The nucleotide sequence of the glycoprotein E gene of herpes simplex virus type 2 and its structural characteristics in comparison with the gE of herpes simplex virus type 1. Mol Cells. 1996;6:145–152. [Google Scholar]
- 15.Chou J, Roizman B. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+ J Virol. 1990;64:1014–1020. doi: 10.1128/jvi.64.3.1014-1020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou J, Roizman B. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc Natl Acad Sci USA. 1994;91:5247–5251. doi: 10.1073/pnas.91.12.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortini R, Wilkie N M. Physical maps for HSV type 2 DNA with five restriction endonucleases. J Gen Virol. 1978;39:259–280. doi: 10.1099/0022-1317-39-2-259. [DOI] [PubMed] [Google Scholar]
- 18.Cress A, Triezenberg S J. Nucleotide and deduced amino acid sequences of the gene encoding virion protein 16 of herpes simplex virus type 2. Gene. 1991;103:235–238. doi: 10.1016/0378-1119(91)90278-j. [DOI] [PubMed] [Google Scholar]
- 19.Davison A J. Herpesvirus genes. Rev Med Virol. 1993;3:237–244. [Google Scholar]
- 20.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 21.Davison A J, Wilkie N M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981;55:315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- 22.DebRoy C. Nucleotide sequence of the herpes simplex virus type 2 syn gene that causes cell fusion. Gene. 1990;88:275–277. doi: 10.1016/0378-1119(90)90043-q. [DOI] [PubMed] [Google Scholar]
- 23.Dolan A, McKie E, MacLean A R, McGeoch D J. Status of the ICP34.5 gene in herpes simplex virus type 1 strain 17. J Gen Virol. 1992;73:971–973. doi: 10.1099/0022-1317-73-4-971. [DOI] [PubMed] [Google Scholar]
- 24.Draper K G, Devi-Rao G, Costa R H, Blair E D, Thompson R L, Wagner E K. Characterization of the genes encoding herpes simplex virus type 1 and 2 alkaline exonucleases and overlapping proteins. J Virol. 1986;57:1023–1036. doi: 10.1128/jvi.57.3.1023-1036.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberle R, Zhang M, Black D H. Gene mapping and sequence analysis of the unique short region of the simian herpesvirus SA8 genome. Arch Virol. 1993;130:391–411. doi: 10.1007/BF01309669. [DOI] [PubMed] [Google Scholar]
- 26.Everett R D, Fenwick M L. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J Gen Virol. 1990;71:1387–1390. doi: 10.1099/0022-1317-71-6-1387. [DOI] [PubMed] [Google Scholar]
- 27.Früh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampé R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature (London) 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 28.Galloway D A, Swain M A. Organization of the left-hand end of the herpes simplex virus type 2 BglII N fragment. J Virol. 1984;49:724–730. doi: 10.1128/jvi.49.3.724-730.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galocha B, Hill A, Barnett B C, Dolan A, Raimondi A, Cook R F, Brunner J, McGeoch D J, Ploegh H L. The active site of ICP47, a herpes simplex virus-encoded inhibitor of the major histocompatibility complex (MHC)-encoded peptide transporter associated with antigen processing (TAP), maps to the NH2-terminal 35 residues. J Exp Med. 1997;185:1565–1572. doi: 10.1084/jem.185.9.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genetics Computer Group. Program manual for the GCG package, version 7. Madison, Wis: Genetics Computer Group; 1991. [Google Scholar]
- 31.Georgopoulou U A, Michaelidou B, Roizman B, Mavromara-Nazos P. Identification of a new transcriptional unit that yields a gene product within the unique sequences of the short component of the herpes simplex virus 1 genome. J Virol. 1993;67:3961–3968. doi: 10.1128/jvi.67.7.3961-3968.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaves R F, O’Hare P. Sequence, function and regulation of the Vmw65 gene of herpes simplex virus type 2. J Virol. 1991;65:6705–6713. doi: 10.1128/jvi.65.12.6705-6713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harland J E, Bdour S, Brown S M, MacLean A R. The herpes simplex virus type 2 strain HG52 RL1 gene contains a 154 bp intron as predicted from sequence analysis. J Gen Virol. 1996;77:481–484. doi: 10.1099/0022-1317-77-3-481. [DOI] [PubMed] [Google Scholar]
- 34.Hill A B, Barnett B C, McMichael A J, McGeoch D J. HLA class I molecules are not transported to the cell surface in cells infected with herpes simplex virus types 1 and 2. J Immunol. 1994;152:2736–2741. [PubMed] [Google Scholar]
- 35.Hill A, Jugovic P, York I, Ruus G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature (London) 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 36.Hodgman T C, Minson A C. The herpes simplex virus type 2 equivalent of the herpes simplex virus type 1 US7 gene and its flanking sequences. Virology. 1986;153:1–11. doi: 10.1016/0042-6822(86)90002-4. [DOI] [PubMed] [Google Scholar]
- 37.Holwerda B C. Herpesvirus proteases: targets for novel antiviral drugs. Antiviral Res. 1997;35:1–21. doi: 10.1016/s0166-3542(97)00012-0. [DOI] [PubMed] [Google Scholar]
- 38.Hubenthal-Voss J, Starr L, Roizman B. The herpes simplex virus origins of DNA synthesis in the S component are each contained in a transcribed open reading frame. J Virol. 1987;61:3349–3355. doi: 10.1128/jvi.61.11.3349-3355.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ina Y. Pattern of synonymous and non-synonymous substitutions: an indicator of mechanisms of molecular evolution. J Genet. 1996;75:91–115. [Google Scholar]
- 40.Killeen A M, Harrington L, Wall L V M, Kelly D C. Nucleotide sequence analysis of a homologue of herpes simplex virus type 1 gene US9 found in the genome of simian herpes B virus. J Gen Virol. 1992;73:195–199. doi: 10.1099/0022-1317-73-1-195. [DOI] [PubMed] [Google Scholar]
- 41.Krause P R, Ostrove J M, Straus S E. The nucleotide sequence, 5′ end, promoter domain, and kinetics of expression of the gene encoding the herpes simplex virus type 2 latency-associated transcript. J Virol. 1991;65:5619–5623. doi: 10.1128/jvi.65.10.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lagunoff M, Randall G, Roizman B. Phenotypic properties of herpes simplex virus 1 containing a derepressed open reading frame P gene. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagunoff M, Roizman B. Expression of a herpes simplex virus 1 open reading frame antisense to the γ134.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J Virol. 1994;68:6021–6028. doi: 10.1128/jvi.68.9.6021-6028.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li W H. Unbiased estimation of the rates of synonymous and non-synonymous substitution. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Roizman B. The herpes simplex virus 1 gene encoding a protease also contains within its coding domain the gene encoding the more abundant substrate. J Virol. 1991;65:5149–5156. doi: 10.1128/jvi.65.10.5149-5156.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F, Roizman B. Characterization of the protease and other products of amino-terminus-proximal cleavage of the herpes simplex virus UL26 protein. J Virol. 1993;67:1300–1309. doi: 10.1128/jvi.67.3.1300-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lockshon D, Galloway D A. Cloning and characterization of oriL2, a large palindromic DNA replication origin of herpes simplex virus type 2. J Virol. 1986;58:513–521. doi: 10.1128/jvi.58.2.513-521.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacLean C A, Efstathiou S, Elliott M L, Jamieson F E, McGeoch D J. Investigation of herpes simplex virus type 1 genes encoding multiply-inserted membrane proteins. J Gen Virol. 1991;72:897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- 49.Martinez R, Shao L, Bronstein J C, Weber P C, Weller S K. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology. 1996;215:152–164. doi: 10.1006/viro.1996.0018. [DOI] [PubMed] [Google Scholar]
- 50.McGeoch D J. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J Gen Virol. 1990;71:2361–2367. doi: 10.1099/0022-1317-71-10-2361. [DOI] [PubMed] [Google Scholar]
- 51.McGeoch D J, Barnett B C. Neurovirulence factor. Nature (London) 1991;353:609. doi: 10.1038/353609b0. [DOI] [PubMed] [Google Scholar]
- 52.McGeoch D J, Barnett B C, MacLean C A. Emerging functions of alphaherpesvirus genes. Semin Virol. 1993;4:125–134. [Google Scholar]
- 53.McGeoch D J, Cook S. Molecular phylogeny of the Alphaherpesvirinae subfamily and a proposed evolutionary timescale. J Mol Biol. 1994;238:9–22. doi: 10.1006/jmbi.1994.1264. [DOI] [PubMed] [Google Scholar]
- 54.McGeoch D J, Cook S, Dolan A, Jamieson F E, Telford E A R. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J Mol Biol. 1995;247:443–458. doi: 10.1006/jmbi.1995.0152. [DOI] [PubMed] [Google Scholar]
- 55.McGeoch D J, Cunningham C, McIntyre G, Dolan A. Comparative sequence analysis of the long repeat regions and adjoining parts of the long unique regions in the genomes of herpes simplex viruses types 1 and 2. J Gen Virol. 1991;72:3057–3075. doi: 10.1099/0022-1317-72-12-3057. [DOI] [PubMed] [Google Scholar]
- 56.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 57.McGeoch D J, Dolan A, Donald S, Brauer D H K. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGeoch D J, Dolan A, Donald S, Rixon F J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181:1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- 59.McGeoch D J, Moss H W M, McNab D, Frame M C. DNA sequence and genetic content of the HindIII l region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68:19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- 60.McGeoch D J, Schaffer P A. Herpes simplex virus. In: O’Brien S J, editor. Genetic maps—locus maps of complex genomes. 6th ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 1.147–1.154. [Google Scholar]
- 61.McKnight J L C, Pellett P E, Jenkins F J, Roizman B. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate alpha-trans-inducing factor-dependent activation of alpha genes. J Virol. 1987;61:992–1001. doi: 10.1128/jvi.61.4.992-1001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry L J, McGeoch D J. The DNA sequence of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:2831–2846. doi: 10.1099/0022-1317-69-11-2831. [DOI] [PubMed] [Google Scholar]
- 63.Preston V G, Rixon F J, McDougall I M, McGregor M, Al-Kobaisi M. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology. 1992;186:87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- 64.Quinn J P, McGeoch D J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985;13:8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson B J, McCann III P J, Matusick-Kumar L, Preston V G, Gao M. Na, an autoproteolytic product of the herpes simplex virus type 1 protease, can functionally substitute for the assembly protein ICP35. J Virol. 1997;71:1683–1687. doi: 10.1128/jvi.71.2.1683-1687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 67.Staden R. Computer handling of DNA sequencing projects. In: Bishop M J, Rawlings C J, editors. Nucleic acid and protein sequence analysis: a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 173–217. [Google Scholar]
- 68.Steffy K R, Schoen S, Chen C-M. Nucleotide sequence of the herpes simplex virus type 2 gene encoding the protease and capsid protein ICP35. J Gen Virol. 1995;76:1069–1072. doi: 10.1099/0022-1317-76-4-1069. [DOI] [PubMed] [Google Scholar]
- 69.Stuve L L, Brown-Shimer S, Pachl C, Najarian R, Dina D, Burke R L. Structure and expression of the herpes simplex virus type 2 glycoprotein gB gene. J Virol. 1987;61:326–335. doi: 10.1128/jvi.61.2.326-335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swain M A, Galloway D A. Nucleotide sequence of the herpes simplex virus type 1 thymidine kinase gene. J Virol. 1983;46:1045–1050. doi: 10.1128/jvi.46.3.1045-1050.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Swain M A, Galloway D A. Herpes simplex virus specifies two subunits of ribonucleotide reductase encoded by 3′-coterminal transcripts. J Virol. 1986;57:802–808. doi: 10.1128/jvi.57.3.802-808.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swain M A, Peet R W, Galloway D A. Characterization of the gene encoding herpes simplex virus type 2 glycoprotein C and comparison with type 1 counterpart. J Virol. 1985;53:561–569. doi: 10.1128/jvi.53.2.561-569.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 74.Timbury M C. Temperature-sensitive mutants of herpes simplex virus type 2. J Gen Virol. 1971;13:373–376. doi: 10.1099/0022-1317-13-2-373. [DOI] [PubMed] [Google Scholar]
- 75.Toh Y, Lui Y, Tanaka S, Mori R. Nucleotide sequence of the major DNA-binding protein gene of herpes simplex virus type 2 and a comparison with the type 1 counterpart. Arch Virol. 1993;129:183–196. doi: 10.1007/BF01316894. [DOI] [PubMed] [Google Scholar]
- 76.Tsurumi T, Maeno K, Nishiyama Y. Nucleotide sequence of the DNA polymerase gene of herpes simplex virus type 2 and comparison with the type 1 counterpart. Gene. 1987;52:129–137. doi: 10.1016/0378-1119(87)90039-4. [DOI] [PubMed] [Google Scholar]
- 77.Wagner E K, Devi-Rao G, Feldman L T, Dobson A T, Zhang Y-F, Flanagan W M, Stevens J G. Physical characterization of the herpes simplex virus latency-associated transcript in neurons. J Virol. 1988;62:1194–1202. doi: 10.1128/jvi.62.4.1194-1202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ward P L, Barker D E, Roizman B. A novel herpes simplex virus type 1 gene, UL43.5, maps antisense to the UL43 gene and encodes a protein which colocalizes in nuclear structures with capsid proteins. J Virol. 1996;70:2684–2690. doi: 10.1128/jvi.70.5.2684-2690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watson R J. DNA sequence of the herpes simplex virus type 2 glycoprotein D gene. Gene. 1983;26:307–312. doi: 10.1016/0378-1119(83)90203-2. [DOI] [PubMed] [Google Scholar]
- 80.Weller S K, Spadaro A, Schaffer J E, Murray A W, Maxam A M, Schaffer P A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985;5:930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whitton J L, Clements J B. The junctions between the repetitive and the short unique sequences of the herpes simplex virus genome are determined by the polypeptide-coding regions of two spliced immediate-early mRNAs. J Gen Virol. 1984;65:451–466. doi: 10.1099/0022-1317-65-3-451. [DOI] [PubMed] [Google Scholar]
- 82.Whitton J L, Clements J B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984;12:2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whitton J L, Rixon F J, Easton A J, Clements J B. Immediate-early mRNA-2 of herpes simplex viruses types 1 and 2 is unspliced: conserved sequences around the 5′ and 3′ termini correspond to transcription regulatory signals. Nucleic Acids Res. 1983;11:6271–6287. doi: 10.1093/nar/11.18.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yei S, Chowdhury S I, Bhat B M, Conley A J, Wold W S M, Batterson W. Identification and characterization of the herpes simplex virus type 2 gene encoding the essential capsid protein ICP32/VP19c. J Virol. 1990;64:1124–1134. doi: 10.1128/jvi.64.3.1124-1134.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 86.Yu D, Sheaffer A K, Tenney D J, Weller S K. Characterization of ICP6:lacZ insertion mutants of the UL15 gene of herpes simplex virus type 1 reveals the translation of two proteins. J Virol. 1997;71:2656–2665. doi: 10.1128/jvi.71.4.2656-2665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]