ABSTRACT
Although antiretroviral therapy (ART) is effective at suppressing HIV replication, a viral reservoir persists that can reseed infection if ART is interrupted. Curing HIV will require elimination or containment of this reservoir, but the size of the HIV reservoir is highly variable between individuals. To evaluate the size of the HIV reservoir, several assays have been developed, including PCR-based assays for viral DNA, the intact proviral DNA assay, and the quantitative viral outgrowth assay (QVOA). QVOA is the gold standard assay for measuring inducible replication-competent proviruses, but this assay is technically challenging and time-consuming. To begin progress toward a more rapid and less laborious tool for quantifying cells infected with replication-competent HIV, we developed the Microwell Outgrowth Assay, in which infected CD4 T cells are co-cultured with an HIV-detecting reporter cell line in a polydimethylsiloxane (PDMS)/polystyrene array of nanoliter-sized wells. Transmission of HIV from infected cells to the reporter cell line induces fluorescent reporter protein expression that is detected by automated scanning across the array. Using this approach, we were able to detect HIV-infected cells from ART-naïve people with HIV (PWH) and from PWH on ART with large reservoirs. Furthermore, we demonstrate that infected cells can be recovered from individual rafts and used to analyze the diversity of viral sequences. Although additional development and optimization will be required for quantifying the reservoir in PWH with small latent reservoirs, this assay may be a useful prototype for microwell assays of infected cells.
IMPORTANCE
Measuring the size of the HIV reservoir in people with HIV (PWH) will be important for determining the impact of HIV cure strategies. However, measuring this reservoir is challenging. We report a new method for quantifying HIV-infected cells that involves culturing cells from PWH in an array of microwells with a cell line that detects HIV infection. We show that this approach can detect rare HIV-infected cells and derive detailed virus sequence information for each infected cell.
KEYWORDS: HIV, reservoir
INTRODUCTION
Despite ongoing efforts, HIV remains an incurable infection leaving people with HIV (PWH) reliant on lifelong regimens of antiretroviral therapy (ART) (1). While ART can prevent infection of CD4 T cells by HIV and subsequent HIV replication within these cells, a subset of cells will enter a state of latent infection (2). In these cells, HIV is reversibly silenced by a combination of factors, including low levels of key HIV-activating transcriptional complexes and covalent modifications to proviral histones that promote heterochromatin formation (3–5). These latently infected cells evade clearance by the host immune system and can seed rebound viremia upon cessation of ART. As such, this reservoir represents the primary barrier to achieving an HIV cure (6, 7). Current HIV cure strategies aim to either induce permanent silencing of HIV or promote induction of viral antigen expression and clearance of infected cells (8–11). A handful of studies have examined the potential of latency reversal in both animal models and PWH and have shown some promise with regard to inducing expression of the reservoir but have thus far not been successful at reducing the size of the reservoir in PWH (12–14). Nevertheless, additional studies will be required to fully assess the potential of this approach. A key complication in terms of quantifying the outcome of a clinical intervention on the HIV reservoir is the complex nature of the reservoir. Of the residual integrated HIV proviruses in people on therapy, only a minor fraction are intact viruses—the remainder containing either large inactivating deletions or hypermutation (15, 16). Furthermore, only a subset of the intact viruses reactivates expression in response to even “potent” latency reversing agents (LRAs) that directly activate T cells through the T cell receptor or protein kinase C agonists such as prostratin (17, 18).
To determine the effectiveness of any HIV cure strategy that targets the reservoir, a simple and scalable assay that measures replication-competent and reactivation-competent viruses would be useful. Thus far, several HIV reservoir assays have been developed. PCR-based assays that measure the latent reservoir by quantifying HIV DNA have been widely used (19). However, PCR-based methods that amplify HIV DNA tend to overestimate the intact reservoir due to the presence of defective proviruses. Droplet digital PCR (ddPCR) methods, such as the intact proviral DNA assay (IPDA), attempt to combat this issue (20). The IPDA amplifies two HIV regions that are preferentially maintained in intact viruses and quantifies proviruses that are ddPCR positive for both. While this allows for a more accurate estimate of the intact reservoir size, some viruses with deletions or mutations in other regions of the HIV genome are still included, likely still leading to an overestimation of the intact reservoir size (20, 21). Additionally, polymorphism at the primer binding site can preclude detection of proviruses by the IPDA. Alternatives to IPDA include the Q4PCR assay which uses a quadruplex PCR reaction and next-generation sequencing to more precisely quantify the reservoir (22). Inducible HIV RNA assays have also been developed (23–25), including the tat/rev-induced limiting dilution assay, which quantifies multiply spliced tat/rev RNA species at limiting dilution (26).
The quantitative viral outgrowth assay (QVOA) has been the gold standard for HIV cure clinical studies thus far (27). The QVOA uses limiting dilution and viral outgrowth within CD4 T cells followed by assessment of p24 production by enzyme-linked immunosorbent assay (ELISA), or by quantitative PCR for viral RNA, and binary scoring of wells. To quantify the size of the reactivation-competent/replication-competent reservoir, maximum likelihood statistics are then applied to the data to calculate the infectious units per million cells (IUPM). While this technique ensures that the estimate only includes replication-competent virus, even with maximum stimulation of CD4 T cells from aviremic participants, in many cases, not all proviruses are reactivated with one round of stimulation (18). This means that the QVOA can underestimate the true size of the viral reservoir. Additionally, the QVOA is technically challenging and requires roughly 14–19 days to complete due the need to culture cells long enough to allow sufficient virus expansion for detection via p24 ELISA. These features limit the scalability of the assay as well as throughput, making its deployment in large clinical trials challenging.
To address this need, we developed a cell-based assay that uses novel CellRaft technology (28) in combination with a highly sensitive Tat-dependent mCherry reporter cell line to detect HIV infection events, enabling quantification of HIV outgrowth from reactivated CD4 T cells from PWH. CellRafts are nanoliter-sized wells (herein referred to as rafts) arranged in polydimethylsiloxane (PDMS)/polystyrene microarrays that allow single-cell resolution microscopy in addition to extraction of individual rafts for sequencing purposes (29, 30). Automated brightfield and fluorescence microscopy scanning of the arrays is enabled by a machine called the AIR system. The CellRaft array provides an environment where reporter cells are segregated from one another by the walls of each individual raft, allowing for analysis of rare events such as HIV viral reactivation while limiting spreading of the virus into neighboring rafts. These events are quantified by automated scanning and image analysis and ultimately used to calculate an IUPM based on the number of donor cells seeded on the array, reducing the physical labor required for the assay.
RESULTS
CellRaft microarrays allow imaging and extraction of individual cells
CellRaft microarray technology enables the analysis of individual cells out of a bulk population via separation by PDMS walls. These arrays enable longitudinal culture of cells within the array and multiple arrays can be incorporated into the volume of a 96-well plate. The compact, biologically compatible design of the materials makes this technology useful for sensitive cell-based assays. In addition, the transparent nature of the devices allows for both brightfield and fluorescence microscopy directly within the array. Individual rafts can then be removed while target cells are still adhered to the raft. This allows extraction of rafts of interest within the array for other biochemical techniques such as PCR analysis of genes of interest (Fig. 1). Critically, these processes are automated using the AIR system, an instrument and software suite that enables image capture and analysis, instrument control, and extraction of individual rafts. Thus, we reasoned that co-culture of HIV-infected cells with a HIV-activated fluorescent reporter cell line in CellRaft arrays might permit a more rapid and sensitive analog of the QVOA. We herein refer to this assay concept as the Microwell Outgrowth Assay (MOA).
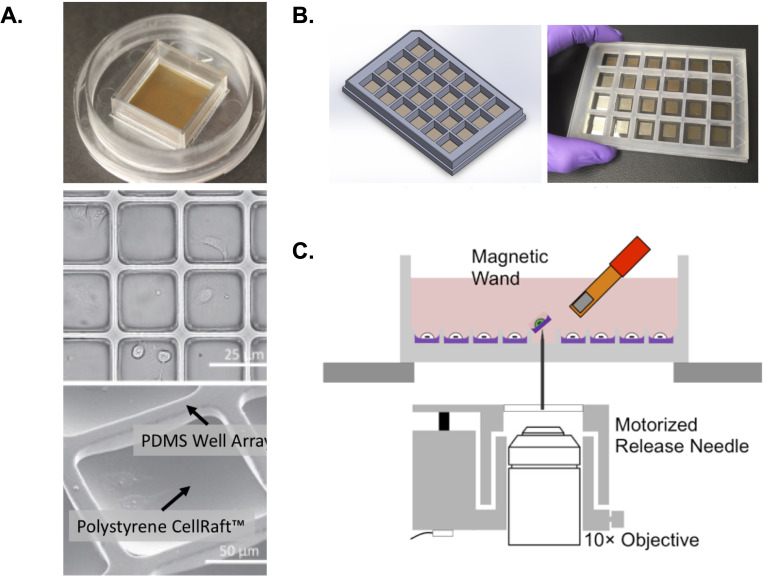
Fig 1.
CellRaft arrays for the analysis of cell biology in extractable nanowells. (A) Upper panel shows an example of a 100-µm CellRaft array with 40,000 magnetic polystyrene rafts surrounded by a PDMS scaffold. These arrays are loaded into and scanned in an automated AIR scanner. Middle panel shows brightfield scan of a selected region of the array with attached cells visible within each raft. Lower panel shows a close view of a single 100-µm raft with both polystyrene well and PDMS scaffold labeled. (B) Example of a hexaquad array containing 154,000 rafts across 24 reservoirs. (C) Schematic of extraction technology for individual rafts. After selection within the CellRaft AIR software, individual rafts are ejected using a release needle. Due to the intrinsically magnetic nature of the rafts, these ejected rafts can be captured by a magnetic “wand” and moved to a 96-well plate for further analysis.
Generation of a Tat-dependent reporter cell line to detect HIV infection
To achieve a novel, highly sensitive assay for quantification of latently infected cells in patient samples, we first generated a reporter cell line which expresses the fluorescent reporter protein mCherry in response to HIV infection. mCherry was selected due to its superior signal-to-noise ratio in CellRafts compared with green fluorescent protein (GFP). After examining the viability and growth of several adherent cell lines within CellRaft arrays, human osteosarcoma (HOS) cells were selected as the parent cell line for the reporter cell line construction. HOS cells were first transduced with a lentiviral construct containing an mCherry gene with expression controlled by an HIV long terminal repeat (LTR) and the HIV transactivation response element (TAR). An SV40 promoter driving expression of a puromycin resistance gene was included for the selection of the resulting cells (Fig. 2A and B). In this design, the TAR element from the reporter construct serves as a sensor of Tat protein during HIV infection and activates mCherry expression. The bulk puromycin-selected cell population (HOS-LTR-mCherry) was then examined for the presence of HIV-responsive cells by infection with an HIV-GFP reporter virus that had been pseudotyped with the VSV-G envelope protein. Infection with the HIV-GFP virus resulted in a population of cells that was positive for both GFP and mCherry signal and an overall increased population of mCherry+ cells, indicating successful HIV infection coupled with activation of the mCherry reporter. Uninfected control HOS cells showed no mCherry fluorescence, and uninfected HOS-LTR-mCherry cells showed significantly less mCherry activity suggesting the presence of clones with selective activation of the reporter in response to HIV infection (Fig. 2C). Individual Tat-responsive clones within the puromycin-resistant population were then enriched by transient transfection with Tat mRNA after plating on a CellRaft array, allowing us to rapidly screen the clones based on an increase in mCherry expression. Responsive clones were extracted from the array and expanded. We then further screened each clone by infection with VSV-G pseudotyped HIV-GFP, followed by flow cytometry. A clone with a strong mCherry response to HIV infection (A5) was selected for continued engineering (Fig. 2D). To accommodate infection by clinical HIV isolates, the A5 clone was transduced with expression constructs for the CD4, CXCR4, and CCR5 receptors necessary for HIV infection, and stable expression was confirmed by flow cytometry (Fig. 2E). The resulting cell line is herein referred to as MOA cells.
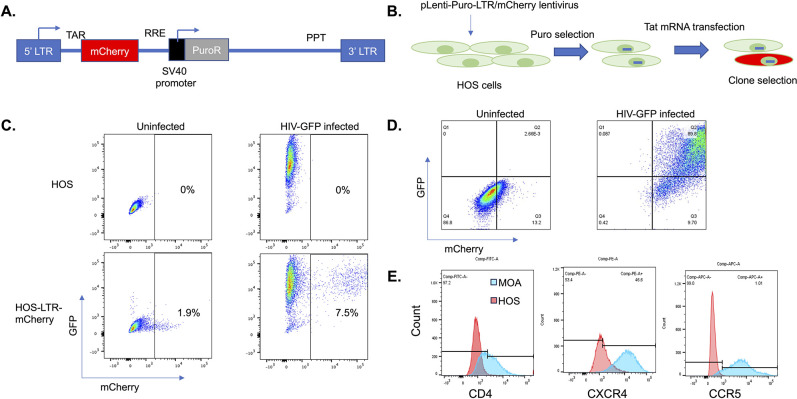
Fig 2.
Generation of an HIV-sensing cell line. (A) Design of HIV reporter construct. An mCherry gene was cloned downstream of an HIV LTR and HIV TAR. Downstream of mCherry, a Rev response element (RRE) and internal SV40 promoter driving a puromycin resistance gene were added. A polypyrimidine tract (PPT) and a 3′ LTR were also included to allow generation of a packageable lentiviral RNA. (B) Overall scheme for generation of an HIV-sensing cell line shown. HOS cells were transduced with lentiviral particles containing the construct from A and selected with puromycin. Cells containing an HIV-responsive insert were enriched by transfection of Tat mRNA and sorting mCherry+ cells using an AIR cell sorter. (C) Flow cytometry of puromycin-resistant HOS cell population in the presence and absence of infection with an HIV-GFP reporter virus. (D) Flow cytometry showing the infection of a highly responsive HOS cell clone (A5) with HIV-GFP. (E) Flow cytometry showing stable expression of HIV receptors CD4, CXCR4, and CCR5 in the HIV-responsive A5 clone.
Quantification of HIV infection in CellRaft arrays
To determine the ability of the MOA cells to detect and quantify replication-competent HIV infection in the context of CellRaft arrays, MOA cells were plated in CellRaft arrays followed by infection of each array with varying volumes of the CXCR4-tropic HIV strain NL4-3. The arrays were then scanned each day for 7 days, and the number of mCherry-positive rafts was quantified for each timepoint (Fig. 3). To select the optimal threshold of mCherry fluorescence to identify infected cells, we compared the number of “above-threshold” rafts on infected and uninfected arrays at different thresholds and selected a threshold (5%) at which the false-positive rate was below one in 10,000 rafts and the majority of true-positive wells were detected (Fig. S1). We then tested different MOA cell plating densities and determined the optimal number of plated cells as four MOA cells per raft to achieve robust detection of infection events (Fig. S2). Also, we compared the signal from infected arrays cultured in different cell media (Dulbecco’s modified Eagle medium (DMEM) vs RPMI] and observed better performance in DMEM (Fig. S3).
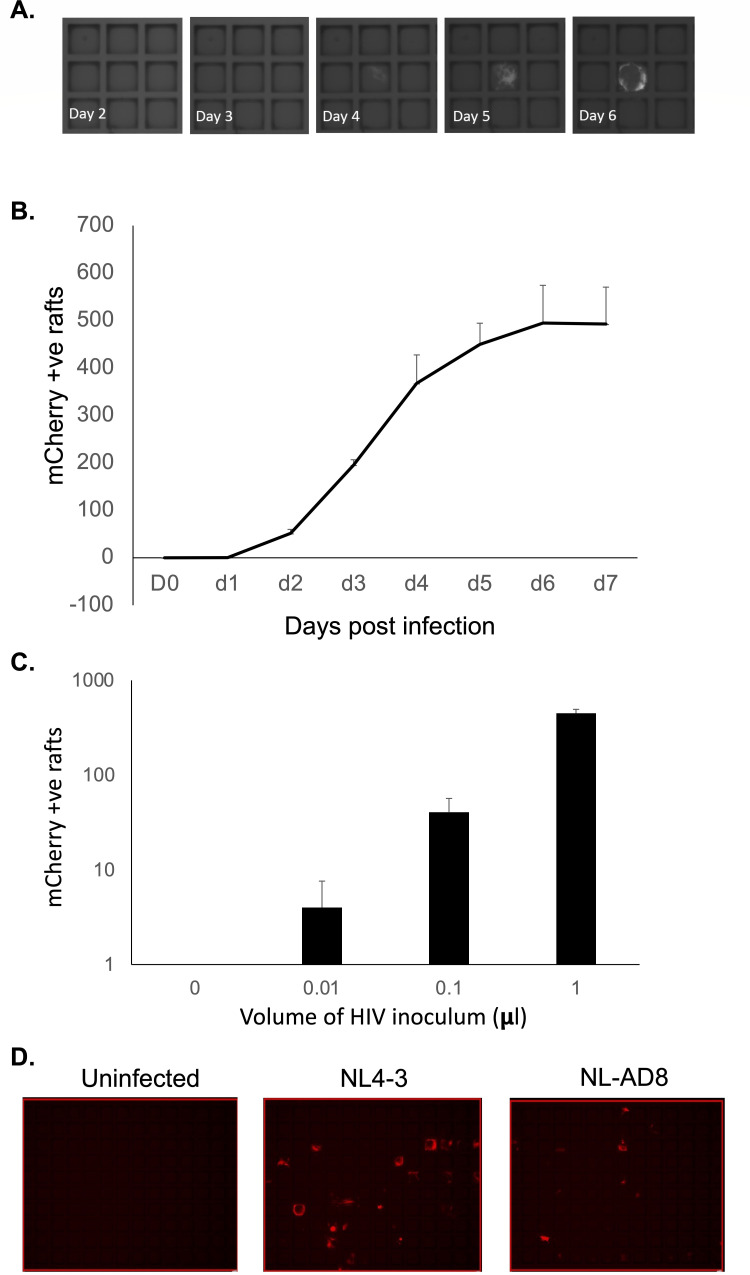
Fig 3.
Detection of HIV infection in CellRaft arrays. MOA cells were plated in a CellRaft array, then infected with HIV (NL4-3), or mock infected. (A) Representative images collected over time showing an individual positive raft. (B) mCherry-positive rafts were counted by scanning each day after infection of the arrays. The average number of positive rafts per array from three arrays is shown. Error bars represent standard deviation of the mean. (C) At day 5 post infection, the number of mCherry-positive rafts was determined for each volume of viral inoculum. The average number of positive rafts per array from three arrays is shown. Error bars represent standard deviation of the mean. (D) MOA cells were plated in CellRaft arrays; then, 1 µL viral inoculum from two different strains of HIV with different co-receptor tropisms (NL4-3, CXCR4 tropic and NL-AD8, CCR5 tropic) corresponding to approximately ~1,000 infectious units was added to the arrays. At 5 days post infection (dpi), the array was scanned for mCherry expression. Representative images are shown.
By examining the mCherry+ wells at each timepoint, we could observe a clear temporal pattern to the emergence of an infected raft signal. While the timing of mCherry expression varied from raft to raft, significant emergence of mCherry+ rafts was apparent by 2 dpi and increased rapidly until 4–6 dpi, at which point the signal plateaued (Fig. 3A and B). Additional mCherry+ rafts were not detected after 7 dpi. When we compared the number of mCherry+ rafts at 5 dpi across different concentrations of viral inoculum, there was a clear linear relationship between the volume of virus added and the number of mCherry+ rafts, suggesting that this approach can accurately and precisely measure small quantities of infectious HIV (Fig. 3C). To confirm that these cells could also mediate infection by a CCR5 tropic strain, we next infected MOA cells in the CellRaft arrays in parallel with 1 µL of viral supernatant for NL4-3 or the CCR5-tropic strain NL-AD8 per array. We observed numerous mCherry+ wells for both strains of HIV, indicating productive HIV infection by both CXCR4 and CCR5-tropic strains (Fig. 3D).
MOA detection of HIV-infected CD4 T cells
We next examined the ability of MOA cells within CellRaft arrays to detect and quantify HIV-infected CD4 T cells. CD4 T cells are the primary target of infection for HIV, and a large fraction of the latent reservoir resides within resting memory CD4 T cells in PWH. Thus, being able to detect infected CD4 T cells would be useful for measuring the abundance of infected cells in samples from PWH. To examine the ability of the MOA assay to detect and quantify HIV-infected cells in CellRaft arrays, we first activated CD4 T cells from healthy donors and then infected them with a replication-competent strain of HIV that encodes Heat Shock Antigen (HSA) in place of the Nef open reading frame. At 48 hours post infection, we measured the abundance of productively infected CD4 T cells (HSA+) by flow cytometry (Fig. 4A). Infected cells were then extensively washed to remove free virus particles, and then, different quantities of the infected cell suspension were co-cultured with MOA cells in a CellRaft array for 5 days. We observed a linear quantitative relationship between the known abundance of infected cells plated in the array and the number of mCherry+ rafts, suggesting the accurate performance of the assay with respect to detecting and quantifying infected cells (Fig. 4B). Furthermore, this assay exhibited a high degree of sensitivity, with as few as 1–2 infected cells per well of a quad array (6,400 rafts per well) being detected above background.
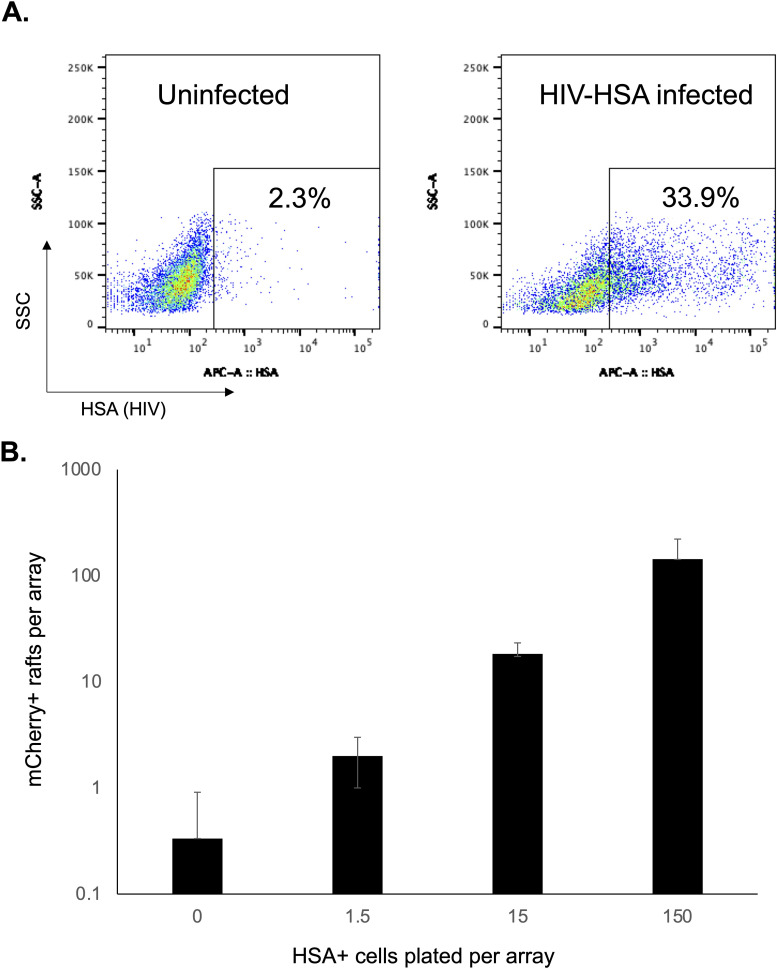
Fig 4.
Detection of HIV-infected CD4 T cells. (A) Primary CD4 T cells from seronegative donors were activated with PHA/IL-2 for 48 hours and then infected with a replication-competent HIV strain that has been modified to encode HSA (HIV-HSA). At 48 hours post infection, the proportion of infected cells was determined by staining for HSA and flow cytometry. (B) MOA cells were plated in CellRaft arrays and overlaid with varying amounts of cells from the infected culture. The arrays were scanned after 5 days of co-culture, and the number of mCherry-positive rafts was determined. Each bar represents the average of three replicate arrays. Error bars represent standard deviation of the mean.
Optimizing MOA/CD4 T co-culture conditions for the microwell outgrowth assay
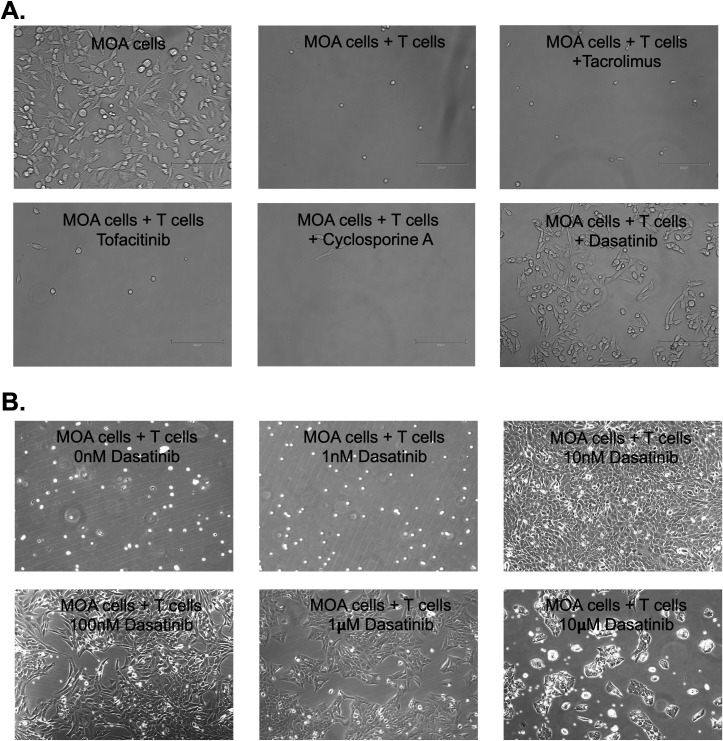
Detection of HIV-infected cells from PWH will require co-culture of high-density CD4 T cells with MOA cells in the CellRaft arrays. To examine the capacity of the rafts to accommodate higher numbers of human CD4 T cells, we plated primary CD4 T cells at different densities per raft and observed that occupancy of the rafts saturated at ~50 T cells per raft (Fig. S4). To induce viral gene expression and replication and transmission of HIV to the MOA cells, the CD4 T cells will also likely need to be activated prior to co-culture. However, when we co-cultured MOA cells with high-density PHA/IL-2-activated CD4 T cells, we observed rapid killing of the MOA cells by the activated T cells (Fig. 5A). This killing did not occur with resting CD4 T cells, demonstrating that activation of the T cells was required and other adherent cell lines such as HEK293T cells and A549 cells were also vulnerable to killing (not shown). Killing became apparent within 3–4 hours of co-culture and resulted in almost complete loss of MOA cells by 24 hours in the presence of activated CD4 T cells. This phenomenon also occurred for CD4 T cells that were activated by CD3/CD28-stimulating beads and by PMA/ionomycin, suggesting that this effect was not specific to PHA/IL-2 activation but was associated with general features of CD4 T cell activation (not shown). Viability of MOA cells was not affected by supernatant from activated CD4 T cells, suggesting that direct contact between CD4 T cells and MOA cells was likely required for killing (not shown). Since the killing likely depended on a signaling pathway that was induced during T cell activation, we tested a panel of inhibitors of T cell signaling pathways to determine whether they could block killing of MOA cells by the activated CD4 T cells. We observed that Dasatinib (Das), a tyrosine kinase inhibitor, was able to rescue MOA cell viability in the presence of activated CD4 T cells, indicating that Dasatinib blocked a pathway that is essential for killing. By examining different Dasatinib concentrations, we further established that 10 nM Dasatinib was sufficient to robustly preserve MOA cell viability during co-culture (Fig. 5B). Higher Dasatinib concentrations (1–10 μM) still protected the MOA cells from CD4 T cells but caused a noticeable reduction in MOA cell growth suggesting possible cytotoxic or cytostatic activity in this concentration range.
Fig 5.
Activated T cells kill MOA reporter cells, but killing is blocked by Dasatinib. (A) MOA cells were plated at 40,000 cells per well in a 12-well plate and then co-cultured with CD4 T cells that been activated with PHA/IL-2 at 500,000 cells per well. At 24 hours post co-culture, the T cells were removed, and the remaining MOA cells were visually examined. In parallel cultures, 10 µM of Tacrolimus, Tofacitinib, Cyclosporine A, or Dasatinib was included. Representative images are shown. (B) Same as A but with varying concentrations of Dasatinib included.
Dasatinib reduces HIV infection of MOA cells but restores detection of infected cells in the presence of high-density activated CD4 T cells
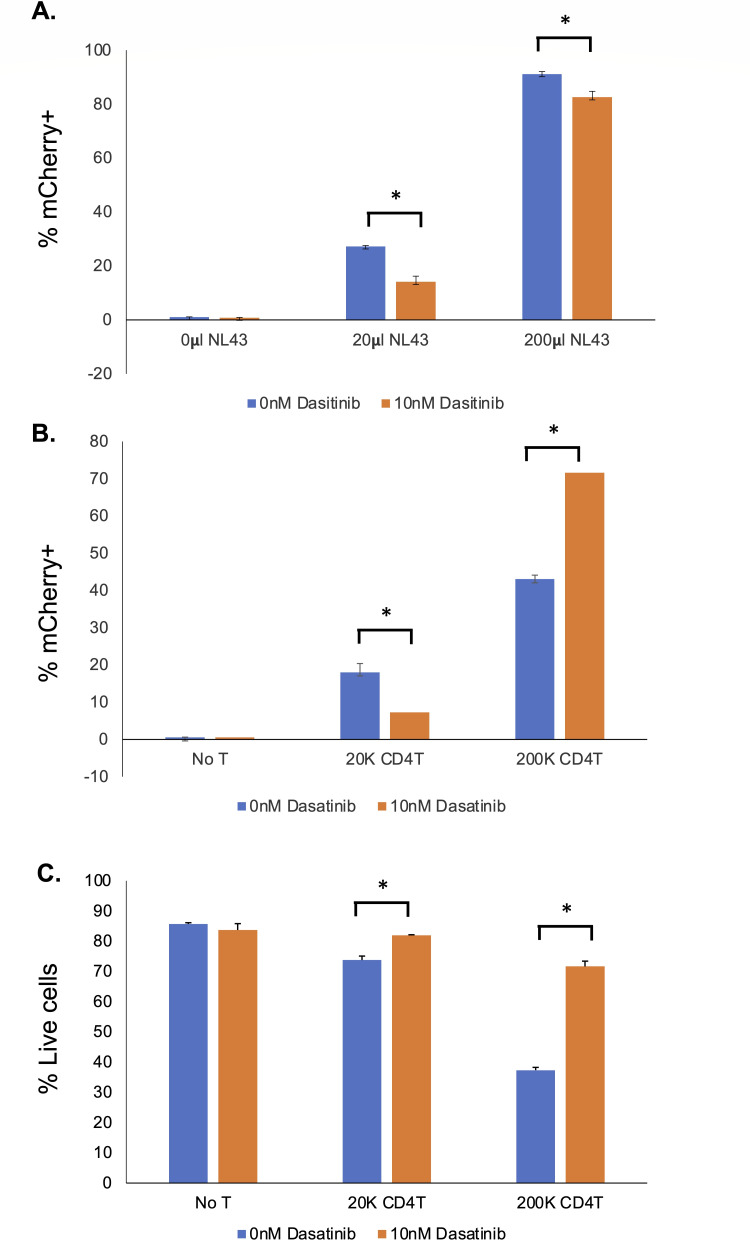
Although Dasatinib potently protects MOA cells from killing by activated CD4 T cells, Dasatinib has also been shown to inhibit HIV infection (31, 32). Therefore, to assess its impact on HIV infection of MOA cells, we infected MOA cells with HIV (NL4-3) in the presence of 10 nM Dasatinib or control vehicle. Notably, treatment with Dasatinib significantly reduced the fraction of mCherry-positive cells by ~50% at low multiplicity of infection (MOI), indicating an antiviral effect at this concentration, although the antiviral effect was less pronounced at higher viral MOI (Fig. 6A). This was also true for MOA cells that were co-cultured with a low density of HIV-infected activated CD4 T cells (Fig. 6B). However, in the presence of high-density activated, ex vivo HIV-infected CD4 T cells, the number of mCherry-positive cells increased significantly in the presence of 10 nM Dasatinib compared with control vehicle. The viability of MOA cells after co-culture with the ex vivo, infected T cells was also measured, and treatment with Dasatinib significantly increased the viability of the MOA cells at high densities of activated CD4 T cells (Fig. 6C). Taken together, these results suggest that 10 nM Dasatinib does have an antiviral effect that could potentially interfere with the sensitivity of the MOA assay, but Dasatinib nevertheless increases transmission of HIV from high-density activated CD4 T cells to MOA cells by preventing T cell-mediated killing of the MOA cells. Dasatinib has been previously reported to inhibit HIV infection by promoting dephosphorylation of the viral restriction factor SAMHD1 (32–34). Interestingly, when we examined the SAMHD1 expression and phosphorylation in MOA cells after 48 hours of 10 nM Dasatinib exposure, we observed that the abundance of both total SAMHD1 and phosphor-SAMHD1 were unchanged, suggesting that a different mechanism contributes to the antiviral effect of Dasatinib on these conditions (Fig. S5).
Fig 6.
Dasatinib reduces HIV infection and transmission but improves MOA cell viability in co-culture and rescues detection of infected cells. (A) MOA cells were plated in 12-well plates at 40,000 cells per well and then infected with different quantities of HIV (NL4-3) in the presence of 10 nM Dasatinib or control vehicle. At 48 hours post infection, the fraction of infected (mCherry-positive) cells was determined by flow cytometry. (B) MOA cells were plated in 12-well plates at 40,000 cells per well and then co-cultured with different amounts of CD4 T cells that had been activated with PHA/IL-2 and then infected with NL4-3 for 48 hours. At 48 hours post co-culture, the fraction of infected (mCherry-positive) MOA cells was determined by flow cytometry. (C) Viability of MOA cells from B was determined by staining with Zombie Violet (ZV) and flow cytometry. ZV-negative cells were determined to be viable cells. Each bar represents an average of replicates. Error bars represent the standard deviation of the mean. Asterisk indicates significant difference between conditions (P < 0.05, Student’s t-test)
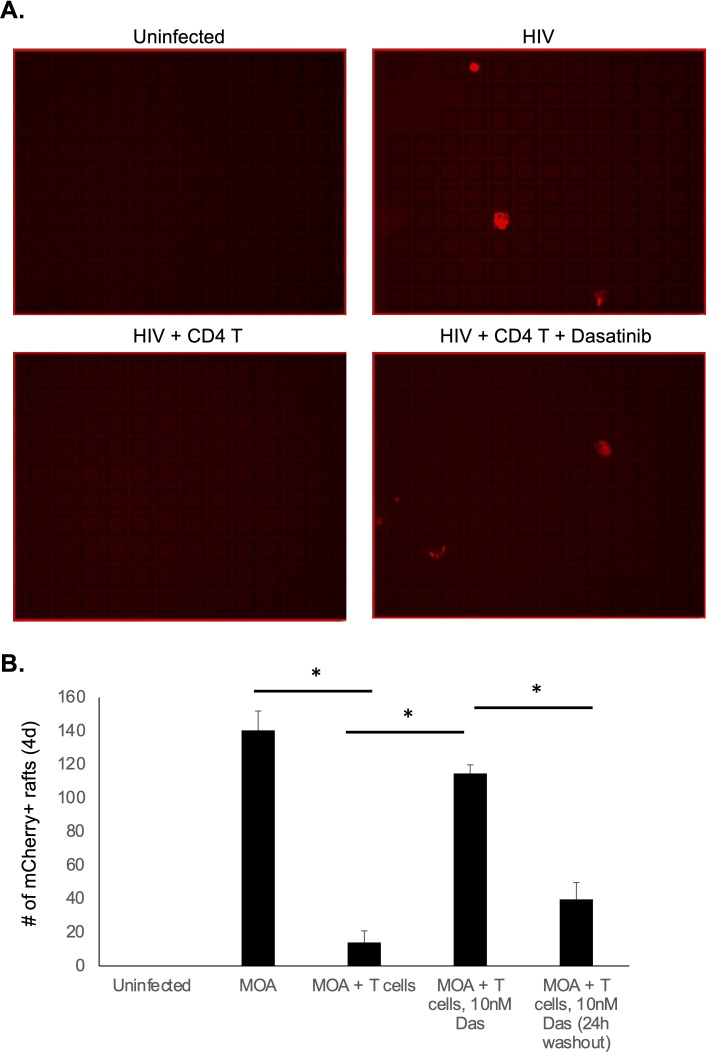
We next tested the ability of Dasatinib to rescue MOA cell viability during high-density CD4 T cell co-culture in the CellRaft arrays. MOA cells were plated at four cells per raft followed by infection with HIV (NL4-3). Immediately after infection, PHA/IL-2-activated T cells were added at high density (50 cells per raft) with or without 10 nM Dasitinib. In one condition, Dasatinib was washed out after 24 hours (Fig. 7). When we quantified the number of mCherry+ rafts at 4 dpi, we observed a potent reduction in the number of infected rafts in the presence of activated CD4 T cells, consistent with killing of the MOA cells by the CD4 T cells. Notably, addition of Dasatinib to the co-culture robustly rescued detection of HIV-infected MOA cells, indicated by the restoration of mCherry+ wells in the presence of Dasatinib. Washout of Dasatinib at 24 hours led to only a partial recovery of mCherry+ rafts, indicating that extended exposure to Dasatinib is required to protect MOA cells from killing by CD4 T cells.
Fig 7.
Dasatinib rescues detection of HIV infection with activated T cell co-culture in CellRaft arrays. MOA cells were plated in CellRaft arrays at four cells per raft and then infected with HIV (NL4-3). At 2 hours post infection, media were replaced with fresh media alone or media containing PHA/IL-2-activated T cells at 50 cells per raft with or without 10 nM Das. In one set of arrays, the media with Dasatinib were replaced at 24 hours with media containing control vehicle. At day 4 of co-culture, the arrays were scanned and the quantity of infected rafts (mCherry positive) was quantified for each condition. Representative images from the arrays are shown in A. Quantified results are shown in B Each bar represents the average of three replicates. Error bars represent the standard deviation of the mean. Asterisk indicates statistically significant difference between the conditions (P < 0.05)
Detection and quantification of HIV-infected cells from PWH
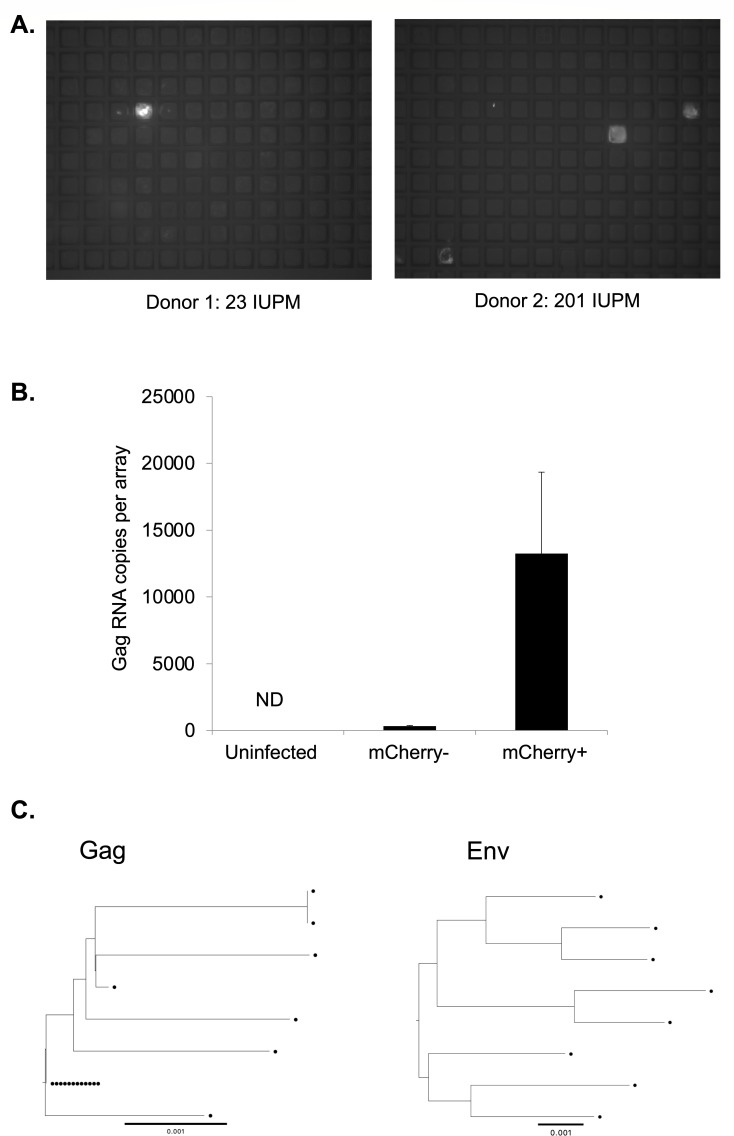
We next examined whether we could use CellRaft arrays to detect and quantify HIV-infected cells from PWH. To test this approach, we co-cultured peripheral blood mononuclear cells (PBMCs) from two chronically infected untreated PWH with MOA cells in CellRaft arrays for 5 days and then scanned the arrays and quantified the number of mCherry-positive rafts. Since these samples contain actively infected cells that can be detected without additional stimulation of the cells, the PBMCs were added without prior activation and without Dasatinib. Significantly, we could detect mCherry+ rafts containing HIV outgrowth events for both donors (Fig. 8A). We then quantified these events for each donor and calculated the number of infected cells per million PBMCs—23 /M for donor 1 and 201 /M for donor 2. To confirm that this approach was accurately detecting HIV-infected cells, we extracted 36 mCherry+ rafts, 12 mCherry− rafts from an infected array, and 12 rafts from an uninfected array and then performed quantitative PCR (qPCR) for HIV Gag RNA. We detected abundant Gag RNA in mCherry+ rafts but not in mCherry− rafts from the same array nor from rafts extracted from an array containing uninfected cells, validating the accuracy of the mCherry signal for identifying infected rafts (Fig. 8B). We then further characterized viral RNA extracted from the infected rafts by sequencing the viral Gag and Envelope regions. From this, we recovered sequences from individual infected cells in the original PBMC sample. Phylogenetic analysis of these sequences revealed a diverse population of infected cells as well as some cells with apparently identical sequences for Gag, while, as expected, Env sequences were more diverse (Fig. 8C). Using this approach to recover and sequence HIV genomes from individual infection events has the potential to be useful for analysis of the genetic diversity of the reservoir.
Fig 8.
Detection of HIV-infected cells from untreated PWH. (A) MOA cells were plated in CellRaft arrays at 4 cells per raft and then overlaid with PBMCs from untreated PWH at a density of 50 cells per raft. After 5 days of co-culture, the arrays were scanned, and mCherry-positive rafts were detected. (B) Sixty rafts were extracted from the arrays including mCherry-positive and mCherry-negative rafts, and RNA was isolated from each raft. The individual rafts were then quantified for HIV Gag RNA by qPCR. (C) Sequencing of partial Gag (HXB2 1195-1726) (left panel) and 3′ Half Genomes (HXB2 4923-9604) (right panel) RNA from virus from extracted mCherry rafts. Each sequence is represented by a circle. The reference bar indicates a phylogenetic distance of 0.001% nucleotide difference.
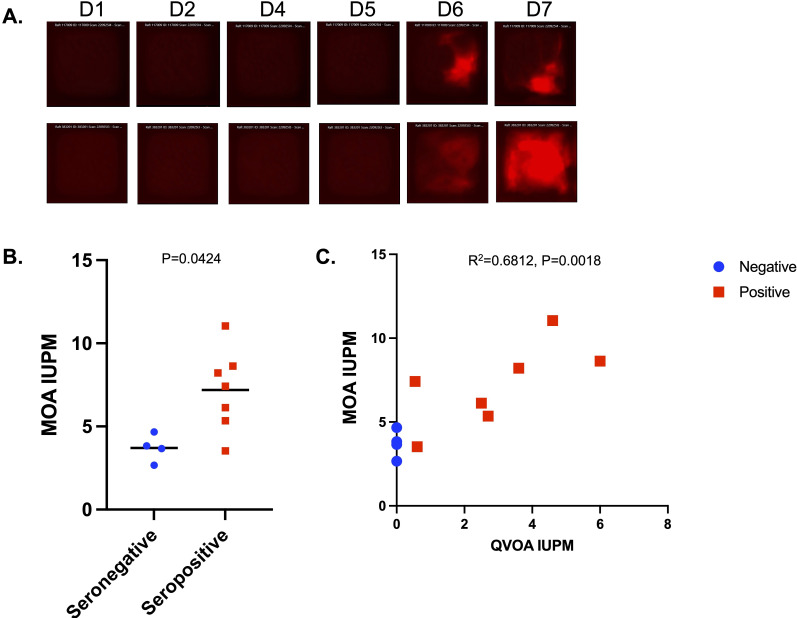
We then examined whether we could detect infected cells in PWH on suppressive antiretroviral therapy. This represents a more challenging goal than for untreated PWH, since infected cells are rare during therapy. To do so, MOA cells were first seeded in hexaquad CellRaft arrays with 154,000 individual rafts. PHA/IL-2 activated CD4 T-cells from PWH on ART were then added at 50 cells per raft (7.5 M cells per array) along with 10 nM Dasatinib to preserve MOA cell viability. Arrays were scanned daily for 7 days of co-culture, and the number of mCherry+ rafts was quantified. The IUPM were then calculated based on the cumulative number of mCherry+ rafts over the time course of the experiment, divided by the total number of CD4 T cells plated. In all, cells from seven PWH on ART were analyzed, along with cells from four seronegative donors that were used as controls. We observed rare outgrowth events in the infected arrays (Fig. 9A), and the number of mCherry wells was significantly greater for the PWH samples than for the seronegative controls, indicating that we were detecting HIV outgrowth (Fig. 9B). Since the samples we selected for analysis had been previously quantified using the QVOA, the IUPM from the MOA assay were compared with the previously measured QVOA IUPM estimates for the same donors (Fig. 9C). This comparison showed a linear relationship between the QVOA and MOA IUPM (r2 = 0.68). A significant background signal in the MOA at this scale due to rare spontaneous mCherry expression in MOA cells led to an overestimation of the IUPM (compared with the QVOA IUPM) and will likely complicate the detection and quantification of the reservoir in PWH with small reservoirs (IUPM < 1). Nevertheless, we were able to detect a clear signal above background from PWH with large reservoirs (IUPM > 2). With further assay optimization, this background fluorescence could potentially be minimized and a more sensitive version of this assay to support clinical trials for HIV cure may be established.
Fig 9.
Detection of HIV-infected cells in PWH on ART. Resting CD4 T cells from seven PWH on ART and four serogenative controls were activated for 48 hours with PHA/IL-2, then washed, and co-plated in hexaquad CellRaft arrays at 50 T cells per raft with MOA cells (4 cells per raft) in the presence of 10 nM Dasatinib. Arrays were scanned daily for 7 days of co-culture. (A) Examples of two rafts with evidence of viral transmission are shown. (B) Comparison of MOA IUPM values for seronegative (blue) and HIV-seropositive donors (red). P value calculated by Mann-Whitney U test. (C) Quantification of total mCherry+ rafts for each donor over the timecourse calculated as IUPM and compared with known QVOA IUPM values for the same donors. Pearson correlation r2 and statistical significance are shown. Serostatus for HIV indicated by color—blue for seronegative and red for seropositive.
DISCUSSION
A key requirement for any HIV cure strategy is an ability to measure changes in the size of the viral reservoir. Several assays have been developed to accomplish this, but each of these assays comes with its own advantages or disadvantages (15, 35). The two most widely used assays to quantitate the HIV reservoir, IPDA and QVOA, have both provided useful insights in studies of PWH. The IPDA is rapid, high-throughput, and scalable but does not completely distinguish between defective and intact viruses and provides no information about the ability of a given provirus to reactivate and reseed infection. While QVOA, on the other hand, more precisely detects intact and replication/reactivation-competent proviruses, it underestimates the true size of the reservoir. Additionally, the assay is time-consuming, technically challenging, and not scalable to large clinical trials. As such, there is still a need for a high-throughput, scalable assay for measuring the reactivation and replication-competent reservoir. The microwell outgrowth assay we have developed aims to achieve this goal. While additional optimization of this approach will be required for clinical deployment, we established an assay that is capable of a comparatively quick (5–7 days) and simple quantification of infected cells from untreated PWH and ART-treated PWH with large reservoirs. Additionally, the extraction capability of the assay design permits recovery of virus sequences from individual cells in clinically relevant samples. However, a significant consideration is the requirement of proprietary technology (AIR machine and CellRaft arrays) for carrying out the MOA assay. Our hope is that this assay may be further optimized and applied to clinical trials to enable HIV cure efforts in the future.
In a direct comparison with the QVOA, we observed a significant correlation between IUPM measured by MOA and IUPM measured by QVOA, indicating measurement of viral outgrowth from the latent reservoir. It is important to note, however, that this analysis depended on the use of samples for PWH with large reservoirs (IUPM > 2), while quantification of the reservoir in PWH with smaller reservoirs was less successful. The MOA nevertheless offers some advantages over the QVOA. The MOA is relatively simple and quick to set up and requires no additional manipulation (e.g., addition of target cells to expand virus) after the initial plating of cells. Scanning and analysis can be carried out in an automated fashion, without the need for additional secondary assays such as p24 ELISA. As such the MOA may be more scalable to a high-throughput format than the QVOA.
It is important to note that the present assay design detects the presence of Tat protein in the MOA cells, which will occur after transmission of a fully replication-competent reactivated virus to the reporter cells but could also potentially occur after release of Tat protein from cells infected with defective viruses and internalization by the reporter cells (36). The contribution of this phenomenon to our results from ART-suppressed PWH is unclear but could conceivably be determined by recovery of viral sequences from mCherry-positive wells and analysis of the intactness of these sequences.
During the course of this study, we also observed that activated human CD4 T cells potently kill the HOS-derived MOA cells. While the precise mechanism behind this killing phenomenon is unknown, it was largely blocked by exposure to 10 nM Dasatinib. Dasatinib is a tyrosine kinase inhibitor suggesting a key role for kinases in MOA cell killing. We also found that 10 nM Dasatinib reduces HIV infection of MOA/HOS cells at this concentration through a mechanism that does not involve changes to the phosphorylation status of the HIV restriction factor SAMHD1. These observations may be useful for other efforts to co-culture activated human T cells with immortalized cell lines. Of note, Dasatinib has been previously shown to inhibit HIV infection and reactivation, and this property may affect the sensitivity of the assay when applied to CD4 T cells from PWH (31, 32). It is possible that an alternative inhibitor that preserves reporter cell viability but lacks Dasatinib’s antiviral properties could be identified.
In its current form, the principal limitation to this assay with respect to reservoir quantification is its sensitivity. While we were able to detect and quantify rare infected cells, the background false-positive rate (three to four IUPM) will make it challenging to use this assay for routine quantification of the reservoir in its current form. As such, additional work will likely be required to reduce this background signal and improve sensitivity. This could potentially be achieved through a number of approaches. Image analysis tools and machine learning could, for example, be used to more accurately distinguish false-positive rafts from true infected rafts. Additional engineering of the MOA cells to eliminate any remaining HIV restriction factors that inhibit outgrowth could enhance the signal-to-background ratio. Further optimization of the reporter transgene or its integration site could also improve the signal-to-background ratio by reducing the number of spontaneous activation events. Finally, further optimization of the CD4 T cell stimulation conditions to reactivate a larger fraction of the reservoir could make this approach practical for quantifying the HIV reservoir.
MATERIALS AND METHODS
CellRaft arrays
CellRaft arrays were provided by Cell Microsystems Inc. (Durham, NC). Three formats of arrays were used—CytoSort arrays, with 40,000 rafts; quad arrays, with four separate arrays of 6,400 rafts; and hexaquad array with 24 separate arrays of 6,400 rafts (154,000 rafts total). To prepare arrays for cell culture, the arrays were washed three times with warm phosphate-buffered saline (PBS) and left in PBS at 37°C overnight or until cells were ready for plating. To plate cells, suspensions of cells were added to each well and allowed to settle into individual rafts. Arrays were scanned on an AIR system (Cell Microsystems) with staged incubation at 37°C and 5% CO2. Scanning was performed in brightfield and in green and red channels (50 ms and 200 ms exposure times, respectively). Arrays were scanned every 24 hours during cell culture beginning at 24 hours post plating. To detect mCherry-positive rafts, Off The Air software (Cell Microsystems) was used in cytometric mode, with the threshold of positivity set at 5%. Rafts that were positive in the mCherry channel at the initial scan were assumed to be debris or false-positive rafts and masked for the duration of the experiment. Rafts that were positive in both the red and green channels were assumed to be autofluorescence debris or dead cells and were also excluded, along with rafts where the mCherry signal was clearly not derived from cells.
Cell lines
HEK293 and HOS cell lines were obtained from ATCC and maintained in DMEM (LifeTech) containing additional 10% FBSs and 10 U/mL Penicillin/Streptomycin (LifeTech).
Lentivirus generation and transduction
Lentiviruses encoding CD4, CCR5, and CXCR4 along with the selectable resistance markers neomycin, hygromycin, and blasticidin, respectively, were purchased from Genecopeia (Rockville, MD). The LTR-mCherry reporter lentivirus was constructed by modification of the pLenti-Puro plasmid (Addgene) to remove of the RSV/LTR promoter and insertion of an intact HIV LTR-mCherry cassette. An intact HIV 3′ LTR was also inserted downstream of the SV40-puromycin resistance cassette. Lentiviruses for transduction of the LTR-mCherry reporter and expression of HIV receptors (CD4, CXCR4, and CCR5) were generated by transfection of HEK293 cells using the Mirus TransIT transfection kit (Mirus). The resulting supernatant was cleared of debris by low-speed centrifugation (300 g for 5 minutes) and filtration through a 0.45-micron filter. Viral supernatant was added to media on cells in culture followed by selection with the appropriate antibiotic 72 hours post transduction.
HIV particle production and infections
Several different HIV-encoding plasmids were used to generate infectious particles for analysis. For screening transduced HOS cells for a clone that expresses mCherry upon infection, we used a defective HIV clone that encodes a short half-life GFP cassette (37, 38), and packaged the virus using plasmids encoding GagPol (PAX2) and VSV-G (MD2-VSV-G). For replication-competent HIV, we used plasmids for a CXCR4 tropic strain (NL4-3) and a CCR5-tropic strain (NL-AD8), as well as a strain with the gene for heat shock antigen (HSA) within the nef open reading frame (39). Replication-competent HIV plasmids were obtained from Aidsreagent.org. To generate HIV stocks, HEK293T cells were transfected with viral plasmids using Mirus-LT1 reagent (Mirus Bio). At 2 days post transfection, supernatant was harvested and clarified as described above for lentiviruses. To infect MOA cells in the CellRaft arrays, the arrays were first washed three times with warm PBS before plating MOA cells at four cells per raft. After allowing 3–4 hours for the cells to attach, 50% of the media volume was removed and replaced with an equivalent volume of media containing a dilution of the virus stock.
Tat mRNA synthesis
A DNA template for Tat mRNA was created with a 5′ T7 promoter followed by the coding sequence for the Tat protein and an SV40 poly(A) signal (IDT). This sequence was amplified by PCR using the KAPA HiFi HotStart ReadyMix PCR Kit and purified using a GeneJet PCR purification kit. Tat mRNA was then synthesized using the PCR-amplified DNA using the MEGAscript T7 Transcription Kit (Thermo Fisher). The resulting RNA was purified using the RNEasy Plus Mini Kit (Qiagen). Purity and concentration were measured by nanodrop.
Tat mRNA transfection
~20,000 HOS cells transduced with the LTR-mCherry reporter were seeded into a CytoSort CellRaft array. Negative cells were identified by imaging in brightfield and red channels using the AIR system. After 24 hours, cells were transfected with Tat mRNA using the Mirus TransIT-mRNA transfection kit (Mirus). Cells that produced mCherry signal after transfection were isolated from the array and isolated as monoclonal populations into 96-well plates for expansion.
Flow cytometry
To prepare cells for flow cytometry, MOA cells were first removed from plates by washing with PBS then trypsinization with 0.05% trypsin (Gibco). Trypsin was quenched with cell culture media, and the cells were pelleted by low-speed centrifugation (300 g, 5 mins) before fixation with 4% paraformaldehyde. Fixed cells were re-pelleted and re-suspended in PBS with 2% fetal bovine serum and 1 mM ETDA. Cells were then analyzed by flow cytometry using a Fortessa (Becton Dickson) cytometer. For analysis of HIV-HSA-infected cells, the cells were first stained with an anti-HSA antibody (BioLegend) at a 1:100 concentration before fixation and analysis. For analysis of CD4, CXCR4, and CCR5 expression, cells were incubated with antibodies at 1:50 dilution in PBS with 2% FBS and 1 mM EDTA before washing and fixation as described above. Antibody clones used were FITC anti-CD4 (BioLegend, 317408), APC anti-CCR5 (BioLegend, 359121), and PE CXCR4 (BioLegend, 306505).
Primary CD4 T cells
Human blood products from healthy donors were purchased from StemCell Technologies (Vancouver, Canada). PBMCs were first obtained by separation on a Ficoll gradient, followed by platelet removal via low-speed centrifugation, and removal of residual erythrocytes by resuspension in ACK lysis buffer for 5 minutes. Total CD4 T cells were then isolated using a StemCell CD4 isolation kit and purity checked by staining with anti-CD4 and flow cytometry. Aliquots of 10-20 million CD4 T cells were frozen in 10% DMSO and 90% FBS until needed. For activation, CD4 T cells were plated at 2 million cells per mL in the presence of 1.5 mg/mL PHA and 60 U/mL IL-2 for 48 hours, before washing and replating in RPMI media with 60 U/mL IL-2.
Gag RNA qPCR
Cells were sorted into TCL lysis buffer (Qiagen) before extraction of RNA using RNAClean XP beads (Beckman). RNA aliquots were then quantified for HIV Gag RNA using TaqMan Fast Virus 1-step master mix (Thermo) in an Applied Biosystems Real-Time PCR machine.
Primers and probes used were as follows: GAGF: ATCAAGCAGCCATGCAAATGTT; GAGR :CTGAAGGGTACTAGTAGTTCCTGCTATGTC; GAG PROBE: FAM ACCATCAATGAGGAAGCTGCAGAATGGGA BHQ1. A standard curve consisting of a gblock covering 200 nucleotides of Gag was purchased from IDT (Coralville, IA).
PWH samples
PWH stably suppressed on ART (<50 HIV RNA copies/mL) for a minimum of 1 year were recruited through the University of North Carolina (UNC) Global HIV Prevention and Treatment Clinical Trials Unit and the UNC Center for AIDS Research HIV Clinical cohort to donate leukapheresis samples. The study was approved by the UNC Biomedical Institutional Review Board, and all participants provided informed consent. PBMCs were isolated from leukapheresis products by Ficoll gradient. Resting CD4-positive T cells were purified from PBMCs as previously described (40). To estimate the frequency of resting CD4+ T cell replication-competent virus, we performed the QVOA as described elsewhere (27). Maximum likelihood estimation statistics were used to calculate IUPMs (41).
Gag sequencing
Extracted viral RNA was used to produce a Gag amplicon using the Platinum SuperScript III One Step RT PCR System (Thermo Fisher 12-574-026). The 25-µL reaction mix comprised 12.5 µL 2× buffer, 1 µL 20 µM F1195 5′-GTCAGCCAAAATTACCCTATAGTGC, 1 µL 20μM R1726 5′-CAACAAGGTTTCTGTCATCCAATTTTTTAC, 1 µL enzyme mix, and 5 µL template RNA. The PCR reaction conditions were 50°C for 30 min, 94°C for 2 min, 40 cycles of 94°C 15 s, 50°C 30 s, and 68°C for 1 min, followed by 68°C for 5 min. PCR products were visualized on a gel and sequenced with Sanger sequencing. Sequences were trimmed for quality and aligned to produce final sequences using Sequencher 5.4.6, compressed to unique sequences, and aligned using Muscle 5.1. The phylogenetic tree was made using Ninja v1.2.2.
Half genome sequencing
Extracted RNA was converted into cDNA with an Oligo dT primer and Superscript III (Invitrogen). The 3′ half genome was then amplified with two rounds of PCR, the first using 4886F 5′-AATTCAAAATTTTCGGGTTTATTACAG and R3B3R 5′-ACTACTTGAAGCACTCAAGGCAAGCTTTATTG, followed by a nested PCR using 2.VIF1C 5′-GGGTTTATTACAGGGACAGCAGAG and Ofm19 5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA, labeled with unique symmetrical Pacbio barcodes for each amplicon. The amplicons were then pooled, purified with gel extraction, and prepared into a sequencing library using the SMRTbell 3.0 Kit (Pacbio). The library was sequenced on the Pacbio Sequel IIe, and the sequences were processed using SMRTlink 12.0.0 and Sequencher 3.4.1, and phylogenetic trees were constructed using Muscle 5.1 and Ninja v1.2.2.
ACKNOWLEDGMENTS
This work was supported by a Phase 2 SBIR award to Cell Microsystems (Durham NC) and EPB (HHS75N93020C00050).
E.P.B. conceived the study. Wet lab experiments were carried out by A.F., M.M. and E.P.B. Data analyses were carried out by A.F., E.P.B., M.M., and S.J. All authors contributed to writing the manuscript.
Contributor Information
Edward P. Browne, Email: epbrowne@email.unc.edu.
Frank Kirchhoff, Ulm University Medical Center, Ulm, Germany.
DATA AVAILABILITY
Underlying data are available at https://doi.org/10.15139/S3/CIYM4B.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01798-23.
Fig. S1 to S5.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Margolis DM. 2014. How might we cure HIV? Curr Infect Dis Rep 16:392. doi: 10.1007/s11908-014-0392-2 [DOI] [PubMed] [Google Scholar]
- 2. Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. doi: 10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Lint C, Bouchat S, Marcello A. 2013. HIV-1 transcription and latency: an update. Retrovirology 10:67. doi: 10.1186/1742-4690-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mbonye U, Karn J. 2017. The molecular basis for human immunodeficiency virus latency. Annu Rev Virol 4:261–285. doi: 10.1146/annurev-virology-101416-041646 [DOI] [PubMed] [Google Scholar]
- 5. Turner A-M, Margolis DM. 2017. Chromatin regulation and the histone code in HIV latency. Yale J Biol Med 90:229–243. [PMC free article] [PubMed] [Google Scholar]
- 6. Chun T-W, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 8. Margolis DM, Archin NM, Cohen MS, Eron JJ, Ferrari G, Garcia JV, Gay CL, Goonetilleke N, Joseph SB, Swanstrom R, Turner A-M, Wahl A. 2020. Curing HIV: seeking to target and clear persistent infection. Cell 181:189–206. doi: 10.1016/j.cell.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Margolis DM. 2022. Latency reversal and clearance of persistent HIV infection. Methods Mol Biol 2407:375–389. doi: 10.1007/978-1-0716-1871-4_25 [DOI] [PubMed] [Google Scholar]
- 10. Rasmussen TA, Tolstrup M, Søgaard OS. 2016. Reversal of latency as part of a cure for HIV-1. Trends Microbiol 24:90–97. doi: 10.1016/j.tim.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 11. Moranguinho I, Valente ST. 2020. Block-and-lock: new horizons for a cure for HIV-1. Viruses 12:1443. doi: 10.3390/v12121443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uldrick TS, Adams SV, Fromentin R, Roche M, Fling SP, Gonçalves PH, Lurain K, Ramaswami R, Wang C-CJ, Gorelick RJ, Welker JL, O’Donoghue L, Choudhary H, Lifson JD, Rasmussen TA, Rhodes A, Tumpach C, Yarchoan R, Maldarelli F, Cheever MA, Sékaly R, Chomont N, Deeks SG, Lewin SR. 2022. Pembrolizumab induces HIV latency reversal in people living with HIV and cancer on antiretroviral therapy. Sci Transl Med 14:eabl3836. doi: 10.1126/scitranslmed.abl3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, Mattingly C, Ho PT, Schoof N, Cammon CG, et al. 2020. Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 578:160–165. doi: 10.1038/s41586-020-1951-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Z, Simonetti FR, Siliciano RF, Laird GM. 2018. Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 15:21. doi: 10.1186/s12977-018-0404-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho Y-C, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siliciano JD, Siliciano RF. 2021. Low inducibility of latent human immunodeficiency virus type 1 proviruses as a major barrier to cure. J Infect Dis 223:13–21. doi: 10.1093/infdis/jiaa649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambrechts L, Cole B, Rutsaert S, Trypsteen W, Vandekerckhove L. 2020. Emerging PCR-based techniques to study HIV-1 reservoir persistence. Viruses 12:149. doi: 10.3390/v12020149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, et al. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. doi: 10.1038/s41586-019-0898-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kinloch NN, Ren Y, Conce Alberto WD, Dong W, Khadka P, Huang SH, Mota TM, Wilson A, Shahid A, Kirkby D, Harris M, Kovacs C, Benko E, Ostrowski MA, Del Rio Estrada PM, Wimpelberg A, Cannon C, Hardy WD, MacLaren L, Goldstein H, Brumme CJ, Lee GQ, Lynch RM, Brumme ZL, Jones RB. 2021. HIV-1 diversity considerations in the application of the intact proviral DNA assay (IPDA). Nat Commun 12:165. doi: 10.1038/s41467-020-20442-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaebler C, Lorenzi JCC, Oliveira TY, Nogueira L, Ramos V, Lu C-L, Pai JA, Mendoza P, Jankovic M, Caskey M, Nussenzweig MC. 2019. Combination of quadruplex qPCR and next-generation sequencing for qualitative and quantitative analysis of the HIV-1 latent reservoir. J Exp Med 216:2253–2264. doi: 10.1084/jem.20190896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M, Coffin JM, Mellors JW. 2014. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 111:7078–7083. doi: 10.1073/pnas.1402873111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yucha RW, Hobbs KS, Hanhauser E, Hogan LE, Nieves W, Ozen MO, Inci F, York V, Gibson EA, Thanh C, Shafiee H, El Assal R, Kiselinova M, Robles YP, Bae H, Leadabrand KS, Wang S, Deeks SG, Kuritzkes DR, Demirci U, Henrich TJ. 2017. High-throughput characterization of HIV-1 reservoir reactivation using a single-cell-in-droplet PCR assay. EBioMedicine 20:217–229. doi: 10.1016/j.ebiom.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massanella M, Yek C, Lada SM, Nakazawa M, Shefa N, Huang K, Richman DD. 2018. Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine 36:113–121. doi: 10.1016/j.ebiom.2018.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin A-G, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJO, Miller MD, Hazuda DJ, Deeks SG, Sékaly R-P, Chomont N. 2015. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2:874–883. doi: 10.1016/j.ebiom.2015.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. 2014. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. J Virol 88:14070–14077. doi: 10.1128/JVI.01900-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cortés-Llanos B, Jain V, Cooper-Volkheimer A, Browne EP, Murdoch DM, Allbritton NL. 2023. Automated microarray platform for single-cell sorting and collection of lymphocytes following HIV reactivation. Bioeng Transl Med 8:e10551. doi: 10.1002/btm2.10551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, Phillips C, Xu W, Pai J-H, Dhopeshwarkar R, Sims CE, Allbritton N. 2010. Micromolded arrays for separation of adherent cells. Lab Chip 10:2917. doi: 10.1039/c0lc00186d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cortés-Llanos B, Wang Y, Sims CE, Allbritton NL. 2021. A technology of a different sort: microraft arrays. Lab Chip 21:3204–3218. doi: 10.1039/d1lc00506e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vigón L, Martínez-Román P, Rodríguez-Mora S, Torres M, Puertas MC, Mateos E, Salgado M, Navarro A, Sánchez-Conde M, Ambrosioni J, et al. 2021. Provirus reactivation is impaired in HIV-1 infected individuals on treatment with dasatinib and antiretroviral therapy. Biochem Pharmacol 192:114666. doi: 10.1016/j.bcp.2021.114666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bermejo M, López-Huertas MR, García-Pérez J, Climent N, Descours B, Ambrosioni J, Mateos E, Rodríguez-Mora S, Rus-Bercial L, Benkirane M, Miró JM, Plana M, Alcamí J, Coiras M. 2016. Dasatinib inhibits HIV-1 replication through the interference of SAMHD1 phosphorylation in CD4+ T cells. Biochem Pharmacol 106:30–45. doi: 10.1016/j.bcp.2016.02.002 [DOI] [PubMed] [Google Scholar]
- 33. Williams E, Szaniawski MA, Martins LJ, Innis EA, Alcamí J, Hanley TM, Spivak AM, Coiras M, Planelles V. 2022. Dasatinib: effects on the macrophage phospho proteome with a focus on SAMHD1 and HIV-1 infection. Clin Res HIV AIDS 8:1053. [PMC free article] [PubMed] [Google Scholar]
- 34. Szaniawski MA, Spivak AM, Cox JE, Catrow JL, Hanley T, Williams E, Tremblay MJ, Bosque A, Planelles V. 2018. SAMHD1 phosphorylation coordinates the anti-HIV-1 response by diverse interferons and tyrosine kinase inhibition. mBio 9:e00819-18. doi: 10.1128/mBio.00819-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, Chomont N, Bar KJ, Yu XG, Lichterfeld M, et al. 2020. Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat Med 26:1339–1350. doi: 10.1038/s41591-020-1022-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mele AR, Marino J, Chen K, Pirrone V, Janetopoulos C, Wigdahl B, Klase Z, Nonnemacher MR. 2018. Defining the molecular mechanisms of HIV-1 Tat secretion: PtdIns(4,5)P2 at the epicenter. Traffic 19:655–665. doi: 10.1111/tra.12578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang H-C, Xing S, Shan L, O’Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 119:3473–3486. doi: 10.1172/JCI39199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peterson JJ, Lewis CA, Burgos SD, Manickam A, Xu Y, Rowley AA, Clutton G, Richardson B, Zou F, Simon JM, Margolis DM, Goonetilleke N, Browne EP. 2023. A histone deacetylase network regulates epigenetic reprogramming and viral silencing in HIV-infected cells. Cell Chem Biol 30:1617–1633. doi: 10.1016/j.chembiol.2023.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marodon G, Landau NR, Posnett DN. 1999. Altered expression of CD4, CD54, CD62L, and CCR5 in primary lymphocytes productively infected with the human immunodeficiency virus. AIDS Res Hum Retroviruses 15:161–171. doi: 10.1089/088922299311583 [DOI] [PubMed] [Google Scholar]
- 40. Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. 2009. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol 83:4749–4756. doi: 10.1128/JVI.02585-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Macken C. 1999. Design and analysis of serial limiting dilution assays with small sample sizes. J Immunol Methods 222:13–29. doi: 10.1016/s0022-1759(98)00133-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5.
Data Availability Statement
Underlying data are available at https://doi.org/10.15139/S3/CIYM4B.