Abstract
Background:
Angiotensin-converting enzyme (ACE) metabolizes a number of important peptides participating in blood pressure regulation and vascular remodeling. Elevated ACE expression in tissues (which is generally reflected by ACE in blood) is associated with increased risk of cardiovascular diseases. Elevated ACE in blood is also a marker for granulomatous diseases.
Methods:
We applied our novel approach—ACE phenotyping—to characterize serum ACE in 300 unrelated patients and to establish normal values for ACE levels. ACE phenotyping includes (a) determination of ACE activity with 2 substrates (Z-Phe-His-Leu [ZPHL] and Hip-His-Leu [HHL]), (b) calculation of a ratio for hydrolysis of ZPHL and HHL, and (c) quantification of ACE immunoreactive protein levels and ACE conformation with a set of monoclonal antibodies (mAbs) to ACE.
Results:
Only a combination of ACE activity determination with 2 substrates and quantification of the amount of ACE immunoreactive protein with mAbs 1G12 and 9B9 allows for the unequivocal detection of the presence of ACE inhibitors in the blood. After excluding such subjects, we were able to establish normal values of ACE in healthy populations: 50%–150% from control pooled serum. This ACE phenotyping approach in screening format with special attention to outliers can also identify patients with various mutations in ACE and may help to identify the as yet unknown ACE secretase or other mechanistic details of precise regulation of ACE expression.
Conclusions:
ACE phenotyping is a promising new approach with potential clinical significance to advance precision medicine screening techniques by establishing different risk groups based on ACE phenotype.
INTRODUCTION
Angiotensin-converting enzyme (ACE, CD143) is a Zn2+ carboxydipeptidase that plays key roles in the regulation of blood pressure and vascular remodeling. ACE is constitutively expressed on the endothelial cell surface, absorptive epithelial and neuroepithelial cells, and cells of the immune system (macrophages, dendritic cells) (1, 2). ACE in blood originates from endothelial cells (3), primarily lung capillary endothelium (4), as a result of proteolytic cleavage by an unidentified but widely postulated ACE secretase (5). In healthy individuals, blood ACE levels are quite stable throughout the lifespan (6). In granulomatous diseases (e.g. sarcoidosis) and Gaucher disease, blood ACE activity is significantly increased (7, 8).
Tissue expression of ACE (and thus blood ACE levels) is under strong genetic influence. The most well-described genetic variant is the ACE I/D polymorphism: the absence (deletion [D]) rather than the presence (insertion [I]) of 287 base markers (Alu repeat) is associated with significantly elevated circulating (9) and tissue (10) ACE levels. Numerous studies have demonstrated associations between the ACE D allele or high plasma ACE levels and increased risk of myocardial infarction in patients (11, 12). ACE I/D polymorphism, or the total level of ACE, is also associated with multiple vascular or renal pathologies, including cardiac hypertrophy and cardiomyopathies, microalbuminuria or nephropathy in patients with type 1 diabetes mellitus, and coronary heart disease in patients with type 2 diabetes mellitus (12, 13). Stimulated by these initial studies, >2000 papers have been published (and several hundreds of thousands of patients were genotyped for the ACE I/D polymorphism) to investigate the role of ACE in tissue in the pathogenesis of various human diseases.
While these studies have demonstrated consistent association of the ACE I/D polymorphism with some diseases, the data remain inconclusive for other pathologic processes. One of the primary limitations is that this particular ACE I/D polymorphism accounts for only 20% of the total variation of serum ACE (and tissue ACE). Therefore, we proposed that a comprehensive assessment of the ACE level (i.e., ACE phenotyping), rather than focusing on ACE genotyping, will be more informative in determining the association of ACE levels with different cardiovascular complications (12).
Diagnostic value may be present not only in detecting increased levels of blood ACE, as in the case of granulomatous diseases, but also in detecting decreased levels of blood ACE in lung cancer, acute respiratory distress syndrome, pneumonia, and thromboembolic pathology (7, 14). These determinations may even prove to be important in COVID-19–associated pneumonia (15) and its thromboembolic complications (16). Consequently, we hypothesize that quantitative determination of blood ACE characteristics will be beneficial. The increased emphasis on personalized and precision medicine approaches (17) also highlights the potential clinical impact of more accurate determination of ACE levels and status (ACE phenotype) (12, 18).
We established a novel approach, blood ACE phenotyping, for the purpose of full characterization of circulating ACE (18). The kinetic and conformational aspects of ACE phenotyping allow for objective identification of patients taking ACE inhibitors and patients with conformationally impaired ACE. The former purpose has potential clinical relevance, as such ACEs can contribute to ACE inhibitor resistance in nonresponders and lead to continuous local increase in angiotensin II formation (19).
This ACE phenotyping approach in screening format with special attention to outliers, in combination with deep genetic analysis (whole exome or whole genome sequencing), also can identify patients with various mutations in ACE and may help to identify the unknown ACE secretase or other mechanistic details of precise regulation of ACE expression. Such an outlier-focused approach also could be applied to analysis of the levels of thousands of other proteins in the blood through aptamer-based technology—SOMAscan (20)—which has already been used for parallel blood proteomic profiling in large population studies (21).
MATERIALS AND METHODS
Chemicals
ACE substrates benzyloxycarbonyl-L-phenylalanyl-L-histidyl-L-leucine (Z-Phe-His-Leu, ZPHL) and hippuryl-L-histidyl-L-leucine (Hip-His-Leu, HHL) were purchased from Bachem Bioscience Inc. and Sigma. Other reagents are from Sigma-Aldrich.
Antibodies
Mouse monoclonal antibodies (mAbs) to human ACE were generated to recognize native conformation of the N and C domains of human ACE (22).
Study Participants
The study was approved by the ethics committee of the Medical Center of Moscow University (protocol 9, November 26, 2018). All corresponding procedures were carried out in accordance with the code of ethics of the World Medical Association (Declaration of Helsinki). All patients provided written informed consent to donate serum for ACE characterization.
ACE Activity Assay
Serum was isolated from whole blood by centrifugation after clotting at 4 °C for 30 min. Samples were stored frozen before ACE determination. ACE activity in serum was measured using a fluorometric assay with 2 ACE substrates, 2 mM Z-Phe-His-Leu or 5 mM Hip-His-Leu (23). Briefly, 20 μL aliquots of serum (diluted 1:5 in PBS) were added to 96-well microplates with conical wells, and then 100 μL of ACE substrate was added and incubated for the appropriate time at 37 °C (usually 1 h). Next, 25 μL of 1.4 N NaOH was added to stop the enzymatic reaction and to increase pH. Then, 25 μL of orthophthaldehyde (3.3 mg/mL in methanol or ethanol) was added at 37 °C for complexing with His-Leu (product of the enzymatic reaction). After 10 min at 37 °C, this reaction was stopped by 25 μL of 2.1 N HCl. The protein pellet was formed, which was sedimented by centrifugation of the plate at 2000g for 2 min. The adduct (complex of His–Leu with orthophthaldehyde) was quantified fluorometrically (excitation 365 nm and emission 500 nm). Fluorescence (i.e., production of His–Leu) is linear for at least 1 h with an amount of ACE up to 0.2 mU/assay. The present assay could be run with as little as 0.5 μL of serum by increasing incubation time if necessary. An evaluation of the precision in the normal range (within-run) gave a relative SD ± 6.3%.
ACE activity in individual patients was expressed as percentage from pooled serum (control) collected from sera of healthy donors (purchased from Interstate Blood Bank) or prepared from sera of multiple donors (Medical Center, Moscow). Before pooling, each serum sample was preliminarily tested for the presence of ACE inhibitors or conformationally changed ACEs, as described previously (19). Calculation of the ZPHL/HHL ratio (23) was performed by dividing fluorescence of the sample with ZPHL by that with HHL.
Fluorescence Quenching Assay
Fluorescence quenching was measured as described in the online Supplemental Data.
Immunological Characterization of Blood ACE (Plate Immunoprecipitation Assay)
Microtiter (96-well) plates (high binding; Corning) were coated with anti-ACE mAbs via goat antimouse IgG (IMTEK or Invitrogen) bridge and incubated with serum samples. After washing the unbound ACE with PBS-Tween 20, plate-bound ACE activity was measured by adding a substrate for ACE (Z-Phe-His-Leu) directly into the wells (13). The level of ACE immunoreactive protein, using strong mAb 9B9, was quantified as described previously (13). Conformational fingerprinting of blood ACE with mAbs to ACE was performed and presented, as described previously (18, 22).
RESULTS AND DISCUSSION
Blood ACE Phenotyping: Methodological Aspects
Previously, we developed a new approach to characterize blood ACE in individual patients—blood ACE phenotyping (18, 19). This approach includes not just quantification of ACE activity (with 2 substrates, ZPHL and HHL) but also determination of a novel kinetic parameter—the ratio of the rates of the hydrolysis of these 2 substrates (ZPHL/HHL ratio). This ratio is able to control for the native state of N and C domains of ACE active centers and to reveal the presence of ACE inhibitors (19, 23). The third parameter is the concentration of ACE immunoreactive protein (13). The fourth and most sensitive approach is conformational fingerprinting of ACE using a set of anti-ACE mAbs, which allow for detection of subtle conformational changes in ACE surface topography (19, 22).
In previous fluorimetric ACE activity assays, serum samples were diluted in single glass tubes to a final dilution of serum 1:200 in the reaction mixture during measurement of the fluorescence (24). We modified the method to measure fluorescence of the product of the enzymatic reaction directly in the wells of 96-well plates—analogous to work by Schwager et al. (25) but adapted for measurement of ACE activity in sera samples. Given the high concentration of the protein in the plasma samples (even diluted initially at 1:5), stopping the reaction of generation of the fluorescent adduct (His-Leu with orthophthaldehyde) leads to formation of pellets of precipitated plasma proteins, which required sedimentation by centrifugation of the plate at 2000g for 2 min. For this reason, we used V-shaped 96-well plates for the assay of ACE activity; in this manner, we can measure fluorescence directly in the wells after sedimentation of protein pellets by centrifugation. Therefore, final dilution of the plasma in our plate variant of the assay was only 1:50, and possible quenching of fluorescence by plasma products could be significant and needed to be evaluated (online Supplemental Data, Supplemental Fig. S1).
Identification of Patients with Outlier ACE Phenotype Values
This ACE phenotyping approach was first performed on 100 sera samples from random unrelated patients (women only, group A) (Figs. 1 and 2), and then on 100 sera samples from unrelated patients (both sexes, group B) (Supplemental Figs. S2 and S3), and finally on 100 sera samples taken during annual check-ups of healthy Moscow University personnel (both sexes, group C) (Supplemental Figs. S4 and S5).
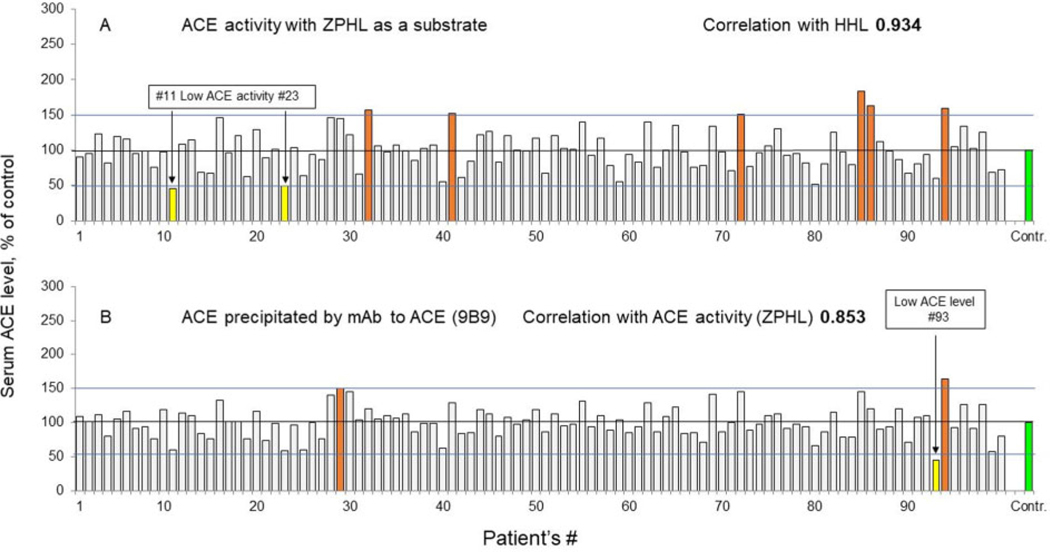
Fig. 1.
ACE activity in group A sera samples. (A), ACE activity in group A (women only) was measured by a spectrofluorometric assay with ZPHL (1 mM) and HHL (2.5 mM, not shown) as substrates. (B), Immunoreactive ACE protein level in group A (women only), ACE was precipitated by mAb 9B9. Data are expressed as percentages of parameters of ACE phenotype from value for control pooled plasma. Bars highlighted with orange, brown, and red indicate samples with values of ACE parameters 120%, 150% and 200%, respectively, from control (shown in green). Bars with significant changes in percentage of ACE precipitation are colored as follows: increase >20%, orange; >50%, brown; >2-fold, red; decrease >50%, yellow. Mean values from 2 to 5 experiments (each made in triplicates); SD (not shown) was <10% in all cases.
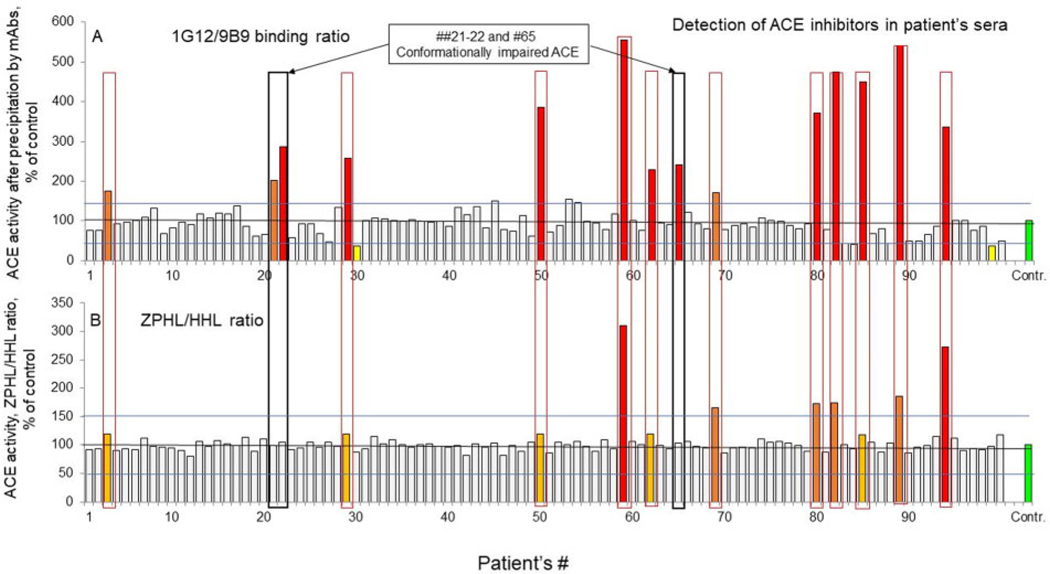
Fig. 2.
Detection of ACE inhibitors in group A sera samples. (A), Ratio of ACE immunoreactive protein levels quantified using mAbs 1G12 and 9B9 (1G12/9B9 binding ratio) in group A (women only). (B), Ratio of the rates of hydrolysis of 2 substrates (ZPHL/HHL ratio) in sera samples from group A (women only). Data are expressed as percentages of parameters of ACE phenotype from value for control pooled plasma. Bars are colored as in Fig.1. Mean values from 2 to 5 experiments (each made in triplicates); SD (not shown) was <10% in all cases.
ACE activity (with ZPHL as a substrate) (Fig. 1, A) varied 3-fold in 100 sera samples from group A (50%–150% of control plasma), which confirmed previously found variations of ACE levels in unrelated (or heathy) populations (6, 13). Two of these plasma samples demonstrated quite low ACE activity, <50% of control values (yellow bars in Fig. 1, A), and one sample with a very low ACE level (Fig. 1, B). This is quite rare because a certain level of ACE is needed for normal embryonic development of kidney proximal tubules (26). Examining such patients may shed light on the mechanism of regulation of ACE expression or even possibly help identify the still unknown ACE secretase. An alternative explanation is that patients with very low ACE levels may have loss of function (LoF) ACE mutations (e.g., stop codon) in one of the alleles of the ACE gene. Six patients (of 100 in group A) had ACE activity >150%, only one of which had a concomitant increase in ACE level, whereas one patient had an opposite situation (elevated ACE level without increased activity) (Fig. 1, brown bars).
We prefer to express the activity as percentage of (pooled) controls rather than absolute values of ACE activity. There are several commercially available assays for measurement of ACE activity that use different substrates and different detecting compounds, and thus different units (and different ranges) will describe the normal ranges for each type of method. In addition, some methods are based on the detection of immunoreactive ACE proteins by different antibodies (6, 13), and these reference ranges will be expressed in nanograms of ACE per milliliter. Consequently, expressing ACE levels in units (or milliunits) per milliliter is less informative.
A limitation of current clinical testing is that not all laboratories take the fold change in ACE activity into account. As a result, all patients with elevated serum ACE levels are often designated as suspected sarcoidosis. However, if ACE activity or levels are expressed as percentage of the mean, then patients with highly elevated ACE levels (e.g., >4-fold) could be correctly considered as carriers of ACE mutations rather than as patients with sarcoidosis or Gaucher disease. Moreover, if patients have 10-fold increases, identification of the mechanism responsible for such a mutation is possible—for example, elimination of the transmembrane anchor, as we recently reported (18). We prepared the pool of human sera from 40 healthy volunteers, and 2 aliquots from this pool were used in each plate as a control (100%). Therefore, ACE activity in the samples from tested patients were expressed as percentage from this standard (with ACE activity 25.4 mU/mL with ZPHL as a substrate). With this approach, the precision of the assay increased significantly, and the difference between ACE activity or level determination of the same samples in different days (between-run) became <7.5%.
In the second series of 100 sera from unrelated patients of both sexes (group B), we found 2 patients with low ACE levels with concomitant decreases in ACE activity (<50% of control) and 2 patients with elevated ACE activity (>150%) (Supplemental Fig. S2). Finally, in the third series of 100 sera of apparently healthy individuals (group C), we found 3 patients with low blood ACE level, 5 patients with increased ACE activity, and 3 patients with an opposite situation (Supplemental Fig. S4).
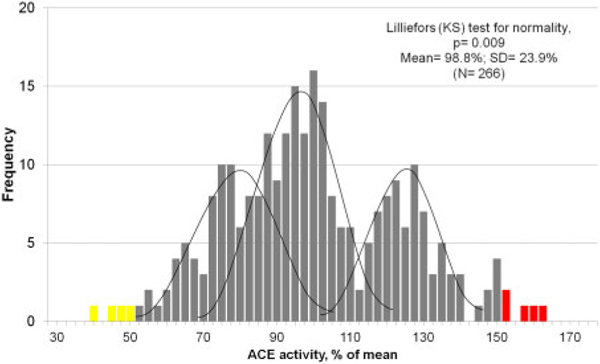
The distribution of ACE activity in 300 individuals is presented in the Fig.3. This activity is rather tri-modal, due to 3 genotypes of ACE gene (II, I/D, and DD). In actuality, the distribution of ACE activity or levels is an integral sum of 3 separate gaussian distributions for each genotype (27).
Fig. 3.
Frequency distribution of ACE activity. Frequency diagram for ACE activity in 266 patients (without ACE inhibitors) with ZPHL as substrate (percentage of mean). Checking the normality of the distribution by the Lilliefors (Kolmogorov-Smirnov) test excludes normality of the frequency diagram due to 3 genotypes of the I/D polymorphism of the ACE gene. The frequency diagram contains 3 gaussian curves corresponding to the expected distributions of ACE activity for the 3 variants of I/D polymorphism. Note: Respondents (n = 34) taking inhibitors ACE are excluded from calculation. Yellow bars indicate that ACE activity is <50% of mean; red bars indicate that ACE level is >150%.
ACE activity determination with another substrate (HHL, not shown) demonstrated excellent correlation (R = 0.934) for women only (group A), with R = 0.952 for both sexes in group B and R = 0.805 for both sexes in group C. However, when we divided fluorescence (i.e., ACE activity) with substrate ZPHL by fluorescence with HHL, their ratio was significantly increased in 11 patients from group A (Fig. 2, B), in 13 patients from group B (Supplemental Fig. S3, B), and in 8 patients from group C (Supplemental Fig. S5, B). Previously, we demonstrated that this increase in the ZPHL/HHL ratio generally reflects the presence of exogenous ACE inhibitors (19, 23). ACE activity determination has been widely used in clinical practice since 1975; however, it has a significant limitation as a screening tool for disease—all patients with ACE inhibitors in their blood have falsely decreased ACE activity, especially with HHL as a substrate (23). Therefore, using 2 substrates for simultaneous determination of ACE activity allows for the objective detection of patients with ACE inhibitors in their blood. This approach dictated to us that the maximal dilution of serum should be 1:50 (despite significant fluorescence quenching; Supplemental Fig. S1) because further dilutions decrease the influence of endo- and exogenous ACE inhibitors on the ZPHL/HHL ratio, thus decreasing the sensitivity of ACE inhibitor detection by this kinetic assay. As one example of the potential clinical utility of the ZPHL/HHL ratio, this assay allows for the detection and quantification of ACE inhibitors in blood samples from hypertensive patients to measure medication compliance and effectiveness; blood pressure control in patients correlates with effective suppression of ACE (28).
Another approach that allows for the quantification of ACE levels, even in the presence of ACE inhibitors or in EDTA plasma, is an immunoassay (in which ACE is initially captured from serum or plasma by antibodies). Several immunoassays have been developed, including radioimmunoassay or variations of immunoassays with polyclonal and/or monoclonal antibodies, as reviewed by Danilov et al. (13). However, these radioimmunoassays have limited general utility. They are not ideal for large-scale applications because of the need for large amounts of high-affinity polyclonal antibodies and I125-labeled antigen. In addition, a complex amplification system is necessary for specificity with the sandwich ELISA, and it suffers from the presence of heterophilic antibodies in some samples (13).
To quantify the amount of ACE immunoreactive protein in this study, we applied a new immunoassay variant in which ACE from serum samples is captured by ACE mAbs. After washing away the unbound ACE (and all components of plasma or serum, including possible ACE inhibitors or EDTA), and after precipitation by given mAbs, ACE activity is quantified directly by fluorometry in the wells after adding substrates ZPHL or HHL.
The levels of ACE immunoreactive protein determined with mAb 9B9 (Fig. 1, B; Supplemental Figs. S2, B, and S4, B), which has an epitope on the N domain of ACE, demonstrated excellent correlation with determination of ACE activity with substrate ZPHL for group A (Fig. 1, B; R = 0.853), for group B (Supplemental Fig. S2, B; R = 0.877), and for group C (Supplemental Fig. S4, B; R = 0.556). We also determined the amount of ACE immunoreactive protein with mAb 1G12 (also having an epitope located on the N domain). When we calculated the 1G12/9B9 binding ratio for group A (Fig. 2, A), we observed that 14 samples had highly elevated values of this ratio, as did 13 samples for group B (Supplemental Fig. S3, A) and 8 samples for group C (Supplemental Fig. S5, A). Previously, we demonstrated that an elevated 1G12/9B9 binding ratio also reflects the presence of exogenous ACE inhibitors (19) at a sensitivity of 0.3 nM (whereas the ZPHL/HHL ratio has a sensitivity of ACE inhibitor detection of 3 nM). When we display the 1G2/9B9 binding ratio and ZPHL/HHL ratio on one page (Fig. 2, group A; Supplemental Fig. S3, group B; and Supplemental Fig. S5, group C), it becomes clear that most samples with an elevated 1G12/9B9 ratio also have an increase in the ZPHL/HHL ratio (red boxes). However, a few samples had an increase in the 1G12/9B9 ratio only—not in the ZPHL/HHL ratio (see black boxed samples).
We found previously that there is a small (3%–5%) percentage of individuals (even among healthy army recruits) who have conformationally altered ACE, and the percentage of such conformationally impaired ACE is dramatically increased in uremic patients (19). The screening of unrelated (or healthy) populations for detection of conformationally impaired ACE has potential clinical significance because such patients have a 2- to 4-fold increase in ACE activity with natural substrate angiotensin I (i.e., theoretically resulting in a significant increase in the local concentration of angiotensin II, which is a risk factor for many cardiovascular complications (29)).
Quantification of immunoreactive ACE protein with mAbs other than 9B9 should be done cautiously (Supplemental S2) because precipitation of ACE by some mAbs will be falsely positive because of dissociation of bilirubin from ACE, which impairs binding of these mAbs to ACE (30).
How should elevated ACE levels be interpreted in these patients, as it is hypothesized that ACE levels are increased in those with systemic sarcoidosis (22)? First, it is necessary to mention that 6 patients with elevated activity and/or level of circulating ACE had ACE inhibitors in their blood, which was detected by a simultaneous increase in the 1G12/9B9 and ZPHL/HHL ratios in Fig. 2, Supplemental Figs. S3 and S5, and also corresponds well with declared confirmation of ACE inhibitor intake by these patients (group A: 85 and 94; group B: 18 and 82; group C: 49 and 89). It was demonstrated previously that patients taking ACE inhibitors have elevated levels of ACE protein (40%–50%) in the blood (31, 32). We identified a mechanistic explanation for this observation: after binding to ACE, ACE inhibitors induce local conformational changes, and, as a result, bilirubin dissociates from ACE and flexibility of ACE increases, which increases the rate of ACE shedding (i.e., blood ACE) (30). Therefore, the increase in blood ACE levels in the above-mentioned patients is best interpreted as false positive. It is also necessary to exclude hyperthyroidism (33) and elevated levels of glucocorticoid hormone in the remaining samples because these hormones dramatically increase ACE expression (34), and thus circulating ACE. Interestingly, patient 82 from group B has a history of hyperthyroidism in addition to taking ACE inhibitors. Only after excluding these possible reasons for elevated ACE should we consider these remaining patients as having possible systemic sarcoidosis. Alternatively, these patients may have genetic mutations that raise the ACE level (18).
Why it is clinically necessary to accurately quantify blood ACE levels for situations other than the detection of systemic sarcoidosis? ACE is expressed in many tissues; therefore, it is involved in a wide variety of physiological and pathophysiological processes. Genetic association studies have linked ACE to multiple diseases, including hypertension, myocardial infarction, renal dysfunction, and diabetes (reviewed by Danser et al. (12)), and even Alzheimer disease and schizophrenia (35, 36).
General principles of clinical diagnostics usually categorize values within a “normal range” of parameters as being functionally similar (e.g., in the case of ACE this range includes values between 50% and 150% of control). In reality, patients with 50% of ACE and 150% of ACE are different in important ways. Individuals with ACE levels at the lower end of the normal range (50%–70% of normal value) can climb Mount Everest without oxygen (37) and are associated with improved endurance-sport performance (rowers, bicyclists, marathon runners). In contrast, patients at the upper end of the normal range (130%–150% of ACE) tend to be associated with improved power-sprint performance (38), and they have an elevated probability of dying from myocardial infarction in their sixties (11). However, if they survive, patients at the upper end of the normal range have a higher chance to be centenarians (39). Elevated levels of ACE also are associated with better therapeutic effects of ACE inhibitors (32). Numerous observations suggest that chronic exposure to high tissue and plasma ACE levels can result in an increased risk of vascular diseases, and indeed, the well-proven and powerful therapeutic effect of ACE inhibitors (40) is the best argument for such a conclusion.
Further Characterization of ACE Phenotype Outliers (Whole Exome Sequencing)
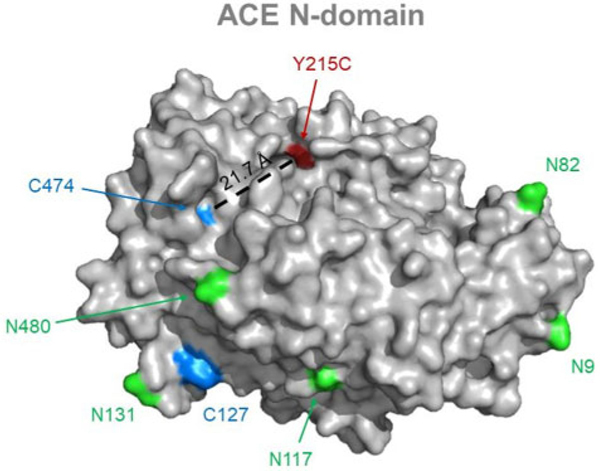
The combination of ACE phenotyping in screening format and whole exome sequencing (in some cases whole genome sequencing) of outliers may help resolve many unclear issues of ACE biology. Therefore, we established an algorithm for further characterization of ACE phenotype outliers (online Supplemental Data, S6) and identified 2 patients with a novel LoF ACE mutation (Y215C, Supplemental Table S1; Fig.4) and putative candidates for the elusive ACE secretase (Supplemental Table S2).
Fig. 4.
Localization of ACE mutation Y215C. ACE N-domain is gray (Protein Data Bank structure 2C6F). Potential glycosylation sites (Asn) are light green, cysteine residues are blue, and ACE mutation Y215C is red; corresponding captions are provided for orientation. Distance between mutation site and the closest cysteine residue is indicated in black.
Detailed analysis of whole exome sequencing data from patients with low blood ACE (and absence of LoF ACE mutations or mutations of the putative ACE secretase[s]) may shed light on the mechanisms that regulate ACE expression via further analysis of LoF mutations in 193 transcription factors with binding sites in the somatic ACE promoter (https://www.genecards.org/cgi-bin/carddisp.pl?gene=ACE).
The novel algorithm that we have established and presented (online Supplemental Data, S6; Table S1 and Table S2) has potential to clarify these unresolved issues of ACE biology, for example, when applied to already established and reported large databases of blood ACE levels (41).
CONCLUSION
We performed complete ACE phenotyping in 300 sera samples of random unrelated patients with a mean age of 44 years (range: 18–65 years). This approach allowed us to objectively detect patients with ACE inhibitors in their blood. We established a normal range of ACE levels for this assay, which varies 3-fold in the tested population: 50%–150% from mean value. We found 17 patients with elevated levels and/or activity of ACE, from which only 11 patients could be considered as possible candidates for further testing for systemic sarcoidosis. Application of our approach potentially would spare 6 of 17 patients with elevated ACE levels from undergoing further unnecessary testing for sarcoidosis.
In addition, 5 patients had decreased ACE levels and/or activity. Further testing of 3 of these subjects (whole exome sequencing) allowed us to suggest that the Y215C substitution in 2 (of 3) patients is another example of a transport-deficient ACE mutation. The combination of ACE phenotyping (with special attention to outliers) together with whole exome or genome sequencing in patients with very low blood ACE levels also may help to identify the still unknown ACE secretase and shed new insights on the overall regulation of ACE expression. Finally, among these 300 unrelated patients, we found 4 individuals with conformationally altered ACE, which, after further investigation, might be important as a risk factor for certain renal and cardiovascular diseases.
In conclusion, the comprehensive method for ACE phenotyping presented in this study combines determination of ACE activity with 2 substrates, and quantification of ACE immunoreactive ACE protein with a set of mAbs to provide valuable information about ACE conformation in each individual. Such an approach holds great promise for significant clinical relevance in our current era of precision medicine.
Supplementary Material
IMPACT STATEMENT.
Elevated expression of angiotensin-converting enzyme (ACE; and blood ACE levels as a reflection of tissue ACE) is a risk factor for cardiovascular diseases; therefore, patients may benefit from detailed description with blood ACE phenotyping. This approach characterizes a patient’s ACE status (ACE concentration, ACE activity, degree of ACE inhibition, and ACE conformation) in a screening format. ACE phenotyping can also identify patients taking ACE inhibitors (i.e., compliance) and those with conformationally altered ACE, providing a potential explanation for nonadherence. This novel approach has clinical potential to advance precision medicine screening techniques.
Acknowledgments:
We are grateful to Dr. Mark Maienschein-Cline, director, Research Informatics Core, Research Resources Center, University of Illinois at Chicago for his analysis of the results of whole exome sequencing of the DNA from 3 patients with low blood ACE levels. We also are grateful to Drs. Laine Francuzevitch and Tatjana Krasnova (Sechenov Moscow Medical University) for providing blood samples from patient 47S and to Dr. Z. Arbieva for the organization of whole exome sequencing.
Role of Sponsor:
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Nonstandard Abbreviations:
- ACE
angiotensin-converting enzyme
- D
deletion
- I
insertion
- ZPHL
benzyloxycarbonyl-L-phenylalanyl-L-histidyl-L-leucine (Z-Phe-His-Leu)
- HHL
dhippuryl-L-histidyl-L-leucine (Hip-His-Leu)
- mAb
monoclonal antibody
- LoF
loss of function
Footnotes
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest: Employment or Leadership: None declared. Consultant or Advisory Role: A.P. Bobkov, Lomonosov Moscow State University. Stock Ownership: None declared. Honoraria: None declared. Research Funding: This research has been funded in part by the University of Illinois at Chicago Center for Clinical and Translational Science (CCTS) award UL1TR002003. Expert Testimony: None declared. Patents: None declared.
SUPPLEMENTAL MATERIAL
Supplemental material is available at The Journal of Applied Laboratory Medicine online.
REFERENCES
- 1.Sturrock ED, Anthony CS, Danilov SM. Peptidyl-dipeptidase A/angiotensin I-converting enzyme. In: Rawlings ND, Salvesen GS, editors. Handbook of proteolytic enzymes. Vol. 1. Amsterdam (Netherlands):Academic Press; 2013. p. 480–94. [Google Scholar]
- 2.Bernstein KE, Ong FS, Blackwell WL, Shah KH, Giani JF, Gonzalez-Villalobos RA, et al. A modern understanding of the traditional and nontraditional biological functions of angiotensin-converting enzyme. Pharmacol Rev 2012;65: 1–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ching SF, Hayes LW, Slakey LL. Angiotensin-converting enzyme in cultured endothelial cells. Synthesis, degradation, and transfer to culture medium. Arteriosclerosis 1983;3:581–8. [DOI] [PubMed] [Google Scholar]
- 4.Metzger R, Franke FF, Bohle RM, Alhenc-Gelas F, Danilov SM. Heterogeneous distribution of angiotensin I-converting enzyme (CD143) in the human and rat vascular systems: vessel, organ and species specificity. Microvasc Res 2011;81:206–15. [DOI] [PubMed] [Google Scholar]
- 5.Parkin ET, Turner AJ, Hooper NM. Secretase-mediated cell surface shedding of the angiotensin-converting enzyme. Protein Pept Lett 2004;11:423–32. [DOI] [PubMed] [Google Scholar]
- 6.Alhenc-Gelas F, Richard J, Courbon D, Warnet JM, Corvol P. Distribution of plasma angiotensin I-converting enzyme levels in healthy men: relationship to environmental and hormonal parameters. J Lab Clin Med 1991;117:33–9. [PubMed] [Google Scholar]
- 7.Rømer FK. Clinical and biochemical aspects of sarcoidosis. With special reference to angiotensin-converting enzyme (ACE). Acta Med Scand 1984;690 Suppl:3–96. [PubMed] [Google Scholar]
- 8.Lieberman J Enzymes in sarcoidosis. Angiotensin-converting enzyme (ACE). Clin Lab Med 1989;9:745–55. [PubMed] [Google Scholar]
- 9.Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, Soubrier F. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 1990; 86:1343–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danser AH, Schalekamp MA, Bax WA, van den Brink AM, Saxena PR, et al. Angiotensin-converting enzyme in the human heart. Effect of the deletion/insertion polymorphism. Circulation 1995;92:1387–8. [DOI] [PubMed] [Google Scholar]
- 11.Cambien F, Poirier O, Lecerf L, Evans A, Cambou JP, Arveiler D, et al. Deletion polymorphism in the gene for angiotensin-converting enzyme is a potent risk factor for myocardial infarction. Nature 1992;359:641–4. [DOI] [PubMed] [Google Scholar]
- 12.Danser AH, Batenburg WW, van den Meiraker AH, Danilov SM. ACE phenotyping as a first step toward personalized medicine for ACE inhibitors. Why does ACE genotyping not predict the therapeutic efficacy of ACE inhibition? Pharmacol Ther 2007;113:607–18. [DOI] [PubMed] [Google Scholar]
- 13.Danilov S, Savoie F, Lenoir B, Jeunemaitre X, Azizi M, Tarnow L, et al. Development of enzyme-linked immunoassays for human angiotensin I converting enzyme suitable for large-scale studies. J Hypertens 1996;14:719–27. [DOI] [PubMed] [Google Scholar]
- 14.Drouet L, Baudin B, Baumann FC, Caen JP. Serum angiotensin-converting enzyme: an endothelial cell marker. Application to thromboembolic pathology. J Lab Clin Med 1988;112:450–7. [PubMed] [Google Scholar]
- 15.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020;180: 934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lodigiani C, Iapichino G, Carenzo L, Cecconi M, Ferrazzi P, Sebastian T, et al. Venous and arterialthromboembolic complications in COVID-19 patients admitted to an academic hospital in Milan, Italy. Thromb Res 2020;191: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.König IR, Fuchs O, Hansen G, von Mutius E, Kopp MV. What is precision medicine? Eur Respir J 2017;50: 1700391. [DOI] [PubMed] [Google Scholar]
- 18.Danilov SM, Jain MS, Petukhov PA, Goldman C, DiSantoRose M, Vancavage R, et al. Novel ACE mutations mimicking sarcoidosis by increasing blood ACE levels. Transl Res 2021;230:5–20. [DOI] [PubMed] [Google Scholar]
- 19.Petrov MN, Shilo VY, Tarasov AV, Schwartz DE, Garcia JGN, Kost OA, et al. Confor-mational changes of blood ACE in chronic uremia. PLoS One 2012;7:e49290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 2010;5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361: 769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danilov SM, Balyasnikova IV, Danilova AS, Naperova IA, Arablinskaya E, Borisov SE, et al. Conformational fingerprinting of the angiotensin I-converting enzyme (ACE). 1. Application in sarcoidosis. J Proteome Res 2010; 9:5782–93. [DOI] [PubMed] [Google Scholar]
- 23.Danilov SM, Balyasnikova IV, Albrecht RFII, Kost OA. Simultaneous determination of ACE activity with 2 substrates provides information on the status of somatic ACE and allows detection of inhibitors in human blood. J Cardiovasc Pharmacol 2008;52:90–103. [DOI] [PubMed] [Google Scholar]
- 24.Friedland J, Silverstein E. A sensitive fluorimetric assay for serum angiotensin converting enzyme. Am J Clin Pathol 1976;66:416–24. [DOI] [PubMed] [Google Scholar]
- 25.Schwager SL, Carmona AK, Sturrock ED. A high-throughput fluorimetric assay for angiotensin I-concerting enzyme. Nat Protocols 2006;1:1961–4. [DOI] [PubMed] [Google Scholar]
- 26.Gribouval O, Morinière V, Pawtowski A, Arrondel C, Sallinen SL, Saloranta C, et al. Spectrum of mutations in the renin-angiotensin system genes in autosomal recessive renal tubular dysgenesis. Hum Mutat 2012;33: 316–26. [DOI] [PubMed] [Google Scholar]
- 27.Cambien F, Costerousse O, Tiret L, Poirier O, Lecerf L, Gonzales MF, et al. Plasma level and gene polymorphism of angiotensin-converting enzyme in relation to myocardial infarction. Circulation 1994;90:669–76. [DOI] [PubMed] [Google Scholar]
- 28.Jones ES, Lesosky M, Blockman M, Castel S, Decloedt EH, Schwager SLU, et al. Thera-peutic drug monitoring of amlodipine and the Z-FHL/HHL ratio: adherence tools in patients referred for apparent treatment-resistant hypertension. S Afr Med J 2017;107:887–91. [DOI] [PubMed] [Google Scholar]
- 29.Te RL, van Esch JH, Roks AJ, van den Meiracker AH, Danser AH. Hypertension: renin-angiotensin-aldosterone system alterations. Circ Res 2015;116:960–75. [DOI] [PubMed] [Google Scholar]
- 30.Danilov SM, Lunsdorf H, Akinbi HT, Nesterovitch AB, Epshtein Y, Letsiou E, et al. Lysozyme and bilirubin bind to ACE and regulate its conformation and shedding. Sci Rep 2016;6:34913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boomsma F, de Bruyn JH, Derkx FH, Schalekamp MA. Opposite effects of captopril on angiotensin I-converting enzyme “activity” and “concentration”; relation between enzyme inhibition and long-term blood pressure response. Clin Sci (Lond) 1981;60:491–8. [DOI] [PubMed] [Google Scholar]
- 32.Danilov SM, Tovsky SI, Schwartz DE, Dull RO. ACE phenotyping as a guide toward personalized therapy with ACE inhibitors. J Cardiovasc Pharmacol Ther 2017; 22:374–86. [DOI] [PubMed] [Google Scholar]
- 33.Grönhagen-Riska C, Fyhrquist F, Välimäki M, Lamberg BA. Thyroid hormones affect serum angiotensin I converting enzyme levels. Acta Med Scand 1985;17:59–64. [DOI] [PubMed] [Google Scholar]
- 34.Barreto-Chaves MLM, Anèas I, Krieger JE. Glucocorticoid regulation of angiotensin-converting enzyme in primary culture of adult cardiac fibroblasts. Am J Physiol (Regul Integr Comp Physiol) 2001;280:R25–R32. [DOI] [PubMed] [Google Scholar]
- 35.Kehoe PG. The renin-angiotensin-aldosterone system and Alzheimer s disease? J Renin Angiotensin Aldosterone Syst 2003;4:80–93. [DOI] [PubMed] [Google Scholar]
- 36.Gadelha A, Vendramini AM, Yonamine CM, Nering M, Berberian A, Suiama MA, et al. Convergent evidences from human and animal studies implicate angiotensin I-converting enzyme activity in cognitive performance in schizophrenia. Transl Psychiatry 2015;5:e691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montgomery HE, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, et al. Human gene for physical performance. Nature 1998;393:221–2. [DOI] [PubMed] [Google Scholar]
- 38.Puthucheary Z, Skipworth, Rawal J, Loosemore M, Van Someren K, Montgomery HE. The ACE gene and human performance: 12 years on. Sports Med 2011;41: 433–48. [DOI] [PubMed] [Google Scholar]
- 39.Schächter F, Faure-Delanef L, Guénot F, Rouger H, Froguel P, Lesueur-Ginot L, et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet 1994;6:29–32. [DOI] [PubMed] [Google Scholar]
- 40.Menard J, Patchett AA. Angiotensin-converting enzyme inhibitors. Adv Prot Chem 2001;56:13–75. [DOI] [PubMed] [Google Scholar]
- 41.Emilsson V, Gudmundsson EF, Aspelund T, Jonsson BG, Gudjonsson A, Launer LJ, et al. Antihypertensive medication uses and serum ACE2 levels: ACEIs/ARBs treatment does not raise serum levels of ACE2. Preprint at https://www.medrxiv.org/content/10.1101/2020.05.21.20108738v1 (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.